Abstract
Many biologically active proteins, which are usually called intrinsically disordered or natively unfolded proteins, lack stable tertiary and/or secondary structure under physiological conditions in vitro. Their functions complement the functional repertoire of ordered proteins, with intrinsically disordered proteins (IDPs) often being involved in regulation, signaling and control. Their amino acid sequences and compositions are very different from those of ordered proteins, making reliable identification of IDPs possible at the proteome level. IDPs are highly abundant in various human diseases, including neurodegeneration and other protein dysfunction maladies and, therefore, represent attractive novel drug targets. Some of the aspects of IDPs, as well as their roles in neurodegeneration and protein dysfunction diseases, are discussed in this article, together with the peculiarities of IDPs as potential drug targets.
Keywords: disorder-based drug discovery, disorder prediction, intrinsically disordered protein, molten globule, premolten globule, random coil
Intrinsically disordered proteins
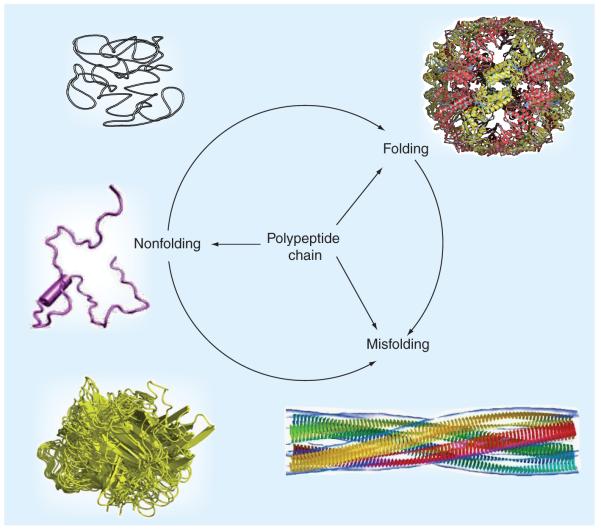
Proteins play a number of crucial roles in the maintenance of life, and protein dysfunctions may cause the development of various pathological conditions. For a very long time it has been believed that the specific functionality of a given protein is predetermined by its unique 3D structure [1,2]. However, Figure 1 shows that, although many proteins are indeed predisposed to fold into unique structures, which evolved to possess unique biological functions, some proteins can misfold either spontaneously or owing to a number of factors, such as mutations and other genetic alterations, problematic processing or post-translational modifications, or exposure to harmful environmental conditions. Such misfolding is now considered a crucial early step in the development of various protein conformation diseases [3]. Finally, many proteins (now widely known as intrinsically disordered proteins [IDPs]) possess no definite ordered 3D structure, but still play important biological roles. Figure 1 shows that protein disorder is a multifaced phenomenon. IDPs can be disordered as a whole or can contain intrinsically disordered regions (IDRs) of different length. IDPs, being mobile, flexible and dynamic, might have very different structural features, ranging from the collapsed molten globule-like conformation to the extended premolten globule-like, and even coil-like, conformations [4-7]. The discovery and characterization of these proteins is becoming one of the fastest growing areas of protein science and they are now recognized as having important biological functions [6,8-19]. Structural flexibility and plasticity originating from the lack of a definite ordered 3D structure are believed to represent the major functional advantages for these proteins, enabling them to interact with a broad range of binding partners, including other proteins, membranes, nucleic acids and various small molecules [20-22].
Figure 1. Fate of a polypeptide chain.
The three structures on the left representing typical intrinsically disordered proteins with different disordered levels (from top to bottom): native coil, native premolten globule and native molten globule. Top right structure illustrates a well-folded protein, whereas the bottom right structure represents one of the products of protein misfolding – a molecular model of the compact, 4-protofilament insulin fibril.
Modified from [221].
Some illustrative biological activities of IDPs include regulation of cell division, transcription and translation, signal transduction, storage of small molecules, chaperone action and regulation of the self-assembly of large multiprotein complexes, such as the ribosome [6,8-13,16-19,23-30]. The functional diversity provided by IDPs is believed to complement functions of ordered proteins.
Unbound IDPs are disordered in solution. However, they often perform their biological functions by binding to their specific partners. This binding involves a disorder-to-order transition, in which either an entire IDP or its region adopts a highly structured conformation [31-38]. In this way, IDPs play diverse roles in regulating the function of their binding partners and in promoting the assembly of supramolecular complexes. Furthermore, because sites within their polypeptide chains are highly accessible, IDPs can undergo extensive post-translational modifications, such as phosphorylation, acetylation and/or ubiquitination (sumoylation), allowing for modulation of their biological activity or function. IDPs were also shown to be highly associated with numerous human diseases (disorders), giving rise to the ‘disorder in disorders’ (D2) concept [39]. According to this concept, IDPs are abundantly involved in the development of various diseases, owing to their unique structural and functional properties. Such diseases, therefore, may originate from misidentification, misregulation and mis-signaling owing to the misfolding of causative IDPs [39].
Computer-aided search for IDPs
The disorderedness of a given protein is linked to the peculiarities of its amino acid sequence. In fact, IDPs exhibit low sequence complexity, are generally enriched in polar and charged residues, and are depleted of hydrophobic residues (other than proline). These features are consistent with their inability to fold into globular structures and form the basis of computational tools for disorder prediction [6,13,40-43]. These same computational tools can also be utilized for the large-scale discovery of IDPs in various proteomes.
Similar to the ‘normal’ foldable proteins, whose correct folding into the rigid biologically active conformation is determined by amino acid sequence, the absence of rigid structure in the ‘non-traditional’ nonfoldable IDPs is encoded in the specific features of their amino acid sequences. In fact, some of the IDPs have been discovered because of their unusual amino acid sequence compositions, and the absence of regular structure in these proteins has been explained by the specific features of their amino acid sequences, including the presence of numerous uncompensated charged groups (i.e., a large net charge at neutral pH arising from the extreme pI values in such proteins [44-46]), and a low content of hydrophobic amino acid residues [44,46]. From the physical viewpoint, a combination of low hydrophobicity with high net charge represents an obvious prerequisite for intrinsic unfoldedness: high net charge leads to charge–charge repulsion, and low hydrophobicity means less driving force for protein compaction. In other words, these features are characteristic for highly extended IDPs with the coil-like (or close to coil-like) structures. Obviously, such highly disordered proteins represent only a small subset of the IDP realm. One of the first disorder predictors, charge–hydropathy (CH) plot, was built based on these features [40].
More detailed comparisons of amino acid compositions of ordered and IDPs and regions revealed that IDPs are significantly depleted in balky hydrophobic (Ile, Leu and Val) and aromatic amino acid residues (Trp, Tyr and Phe), which would normally form the hydrophobic core of a folded globular protein, and also possess low content of Cys and Asn residues [6]. The depletion of IDPs in Cys is also crucial, as this amino acid residue is known to have a significant contribution to the protein conformation stability via the disulfide bond formation or being involved in coordination of different prosthetic groups. These depleted residues – Trp, Tyr, Phe, Ile, Leu, Val, Cys and Asn – were proposed to be called order-promoting amino acids. On the other hand, IDPs were shown to be substantially enriched in polar, disorder-promoting amino acids (Ala, Arg, Gly, Gln, Ser, Glu and Lys) and also in the hydrophobic but structure-breaking Pro [6,47,48]. Note that these biases in the amino acid compositions of IDPs are also consistent with the low overall hydrophobicity and high net charge, characteristic of the extended IDPs, also known as natively unfolded proteins [40]. Since the amino acid sequences of the IDPs and IDRs differ dramatically from those of the ordered proteins and regions, these amino acid sequence differences were used to develop various predictors of intrinsic disorder, utilizing various prediction ideas and different computing techniques. The current list of intrinsic disorder predictors includes more than 50 specialized computational tools [49].
The proteome-wide application of disorder predictors revealed that IDPs are highly abundant in nature. At the proteome level, many proteins were not only predicted to have long disordered regions (~55% of eukaryotic proteins are predicted to contain at least one disordered region that is at least 30 amino acids in length [6,50]), but were shown to be disordered along their entire lengths (~25% of eukaryotic proteins are predicted to be fully disordered) [7,35].
Experimental search for IDPs
Being characterized by specific (and somewhat unique) amino acid sequences, IDPs possess a number of very distinctive structural properties that can be implemented for their discovery. The specific structural features include, but are not limited to, sensitivity to proteolysis [51], aberrant migration during SDS-PAGE [52], insensitivity to denaturing conditions [53], as well as definitive disorder characteristics visualized by CD spectropolarimetry, NMR spectroscopy, small-angle x-ray scattering, hydrodynamic measurement, fluorescence, Raman and infrared spectroscopies [5,54]. Structurally, IDPs range from completely unstructured polypeptides, to extended partially structured forms, to compact disordered ensembles containing substantial secondary structure (Figure 1) [6,11,12,25,55]. Many proteins contain mixtures of ordered and disordered regions. Extended IDPs are known to possess the atypical conformational behavior (e.g., ‘turn-out’ response to acidic pH and high temperature, and insensitivity to high concentrations of strong denaturants), which is determined by the peculiarities of their amino acid sequences and the lack of ordered 3D structure [56]. These unique structural features of extended IDPs and their specific conformational behavior were shown to be useful in elaborating the experimental techniques for the large-scale identification of these important members of the protein kingdom. Three related methods were introduced: a method based on the finding that many proteins that fail to precipitate during perchloric acid or trichloroacetic acid treatment were IDPs [57], a method utilizing the fact that IDPs possessed high resistance toward the aggregation induced by the heat treatment [57-59], and a method based on heat treatment coupled with a novel 2D gel methodology to identify IDPs in cell extracts [58]. It is anticipated that these methodologies, combined with the highly sensitive mass spectrometry (MS)-based techniques, can be used for the detection and functional characterization of IDPs in various proteomes.
Bioinformatics approaches for the large-scale discovery of the disease-related IDPs
Misfolding (the failure of a specific peptide or protein to adopt its functional conformational state) and related dysfunction of many proteins were considered a major cause for the development of different pathological conditions. Such misfolding and dysfunction can originate from point mutation(s) or result from an exposure to internal or external toxins, impaired post-translational modifications (e.g., phosphorylation, advanced glycation, deamidation and racemization), an increased probability of degradation, impaired trafficking, lost binding partners or oxidative damage. All these factors can act independently or in association with one another.
Although the formation of various aggregates represents the most visible consequence of protein misfolding, and although these aggregates form the basis for the development of various protein deposition diseases, the pathogenesis of many more human diseases does not depend on aggregation, but rather on protein dysfunction. To support this hypothesis, three computational approaches were elaborated for estimating the abundance of IDPs in various pathological conditions. The first approach is based on the assembly of specific datasets of proteins associated with a given disease, and the computational analysis of these datasets using a number of disorder predictors [27,39,60,61]. In essence, this is an analysis of individual proteins, extended to a set of independent proteins. The second approach utilized a network of genetic diseases, where the related proteins are interlinked within one disease and between different diseases [62]. The third approach is based on the evaluation of the association between a particular protein function (including the disease-specific functional keywords) with the level of intrinsic disorder in a set of proteins known to carry out this function [16-18]. These three approaches are briefly described later.
The easiest way to evaluate the abundance of intrinsic disorder in a given disease is based on a simple two-stage protocol, where a set of disease-related proteins is first assembled by searching various databases, and then the collected group of proteins is analyzed for intrinsic disorder. The depth of this analysis is based on the breadth of the search for the disease-related proteins and on the number of different computational tools utilized to find disordered proteins/regions [27,39,60,61,63,64]. Using this approach, it has been shown that many proteins associated with cancer, neurodegenerative diseases and cardiovascular disease are highly disordered, being depleted in major order-promoting residues (Trp, Phe, Tyr, Ile and Val) and enriched in some disorder-promoting residues (Arg, Gln, Ser, Pro and Glu). A high level of intrinsic disorder, and a substantial number of potential interaction sites, were also found using a set of computational tools, and many proteins were predicted to be wholly disordered. Overall, these studies clearly showed that intrinsic disorder is highly prevalent in proteins associated with human diseases, comparable to that of signaling proteins and significantly exceeding the levels of intrinsic disorder in eukaryotic proteins and in nonhomologous structured proteins.
A set of IDPs associated with human genetic diseases was assembled via the analysis of a specific network, which was built to estimate whether human genetic diseases and corresponding disease genes are related to each other at a higher level of cellular and organism organization. This network represented a bipartite graph with a network of genetic diseases, the ‘human disease network’ (HDN), where two diseases were directly linked if there was a gene that was related to both of them, and a network of disease genes, the ‘disease gene network’ (DGN), where two genes were directly linked if there was a disease to which they were both related to [65]. This framework, called the human diseasome, systematically linked the human disease phenome (which includes all the human genetic diseases) with the human disease genome (which contains all the disease-related genes) [65]. The analysis of HDN revealed that of 1284 genetic diseases, 867 had at least one link to other diseases and 516 diseases formed a giant component, suggesting that the genetic origins of most diseases were, to some extent, shared with other diseases. In the DGN, 1377 of 1777 disease genes were shown to be connected to other disease genes, and 903 genes belonged to a giant cluster of the HDN. The vast majority of genes associated with genetic diseases were nonessential, and showed no tendency to encode hub proteins [65]. The large-scale analysis of the abundance of intrinsic disorder in transcripts of the various disease-related genes was performed using a set of computational tools, which uncovers several important features [62,63]:
Intrinsic disorder is common in proteins associated with many human genetic diseases
Different disease classes vary in the IDP contents of their associated proteins
Molecular recognition features, which are relatively short loosely structured protein regions within mostly disordered sequences, and which gain structure upon binding to partners, are common in the diseasome, and their abundance correlates with the intrinsic disorder level
Some disease classes have a significant fraction of genes affected by alternative splicing, and the alternatively spliced regions in the corresponding proteins are predicted to be highly disordered and, in some diseases, contain a significant number of molecular recognition features (MoRFs), which are structure-prone segments located within long disordered regions
Correlations were found among the various diseasome graph-related properties and intrinsic disorder. In agreement with earlier studies, hub proteins were shown to be more disordered.
Another approach is a computational tool elaborated for the evaluation of a correlation between the functional annotations in the Swiss-Prot database and the predicted intrinsic disorder [16-18]. The approach is based on the hypothesis that if a function described by a given keyword relies on intrinsic disorder, then the keyword-associated protein would be expected to have a greater level of predicted disorder compared with the protein randomly chosen from the Swiss-Prot database. To test this hypothesis, functional keywords associated with 20 or more proteins in Swiss-Prot were found, and corresponding keyword-associated datasets of proteins were assembled. Next, for each keyword-associated set, a length-matching set of proteins was randomly drawn from the Swiss-Prot, and order–disorder predictions were carried out for the keyword-associated sets, as well as for the random sets [16-18]. The application of this tool revealed that, out of 710 Swiss-Prot keywords, 310 functional keywords were associated with ordered proteins, 238 functional keywords were attributed to disordered proteins, and the remaining 162 keywords yielded ambiguity in the likely function–structure associations [16-18]. It has also been shown that keywords describing various diseases were strongly correlated with proteins predicted to be disordered. Contrary to this, no disease-associated proteins were found to be strongly correlated to the absence of disorder [17].
Illustrative examples of IDPs in neurodegeneration
The large class of human neurodegenerative maladies includes many acquired neurological diseases, with distinct phenotypic and pathologic expressions, all characterized by the pathological conditions in which cells of the brain and spinal cord are lost. As neurons are not readily regenerated, over time their deterioration leads to dysfunctions and disabilities. Neurodegenerative diseases can be divided into two groups according to their phenotypic effects: conditions causing problems with movements, and conditions affecting memory and leading to dementia. Neurodegeneration is a slow process, which begins long before the patient experiences any symptoms. It can take months or even years before visible outcomes of this degeneration are felt and diagnosed. Symptoms are usually noticed when many cells die or fail to function and a part of the brain begins to cease functioning properly. For example, the symptoms of Parkinson’s disease (PD) become apparent after more than 70% dopaminergic neurons die in the substantia nigra (a small area of cells in the mid-brain affected by PD).
Many of the well-known neurodegenerative diseases represent a set of proteinopathies, which can be classified and grouped based on the causative proteins. In fact, neurodegenerative maladies constitute a subset of a broader class of human diseases, known as protein conformational or protein-misfolding diseases. These disorders arise from the failure of a specific peptide or protein to adopt its native functional state. The obvious consequences of misfolding are protein aggregation (and/or fibril formation), loss of function and gain-of-toxic function. Some proteins have an intrinsic propensity to assume a pathologic conformation, which becomes evident with aging or at persistently high concentrations.
Often, neurodegenerative diseases are protein-deposition maladies, characterized by the formation and accumulation of extracellular amyloid fibrils or intracellular inclusions with amyloid-like characteristics. Protein deposition diseases can be sporadic (85%), hereditary (10%) or even transmissible, as in the case of prion diseases (5%) [66]. Although these diseases are very different clinically, they share similar molecular mechanisms, where a specific protein or protein fragment changes from its natural soluble form into insoluble fibrils. It has been pointed out that, prior to fibrillation, amyloidogenic polypeptides may be rich in β-sheet, α-helix, β-helix, or contain both α-helices and β-sheets. They may be globular proteins with rigid 3D structure or that belong to the class of natively unfolded (or intrinsically unstructured) proteins [67]. The molecular mechanisms of fibrillation of IDPs and ordered proteins are different [67]. In ordered proteins, the first critical step in fibrillogenesis is the partial unfolding [67-76], whereas the earliest stage of fibrillation of IDPs is their partial folding [67].
An incomplete list of human neurodegenerative diseases associated with IDPs includes Alzheimer’s disease (AD; deposition of amyloid-β, tau-protein and α-synuclein fragment [NAC] [77,78]); taupathies, including progressive supranuclear palsy, corticobasal degeneration, Niemann–Pick disease type C, subacute sclerosing panencephalitis, argyrophilic grain disease, myotonic dystrophy and motor neuron disease with neurofibrillary tangles (NFTs; accumulation of tau-protein in form of NFTs [79]); Down’s syndrome (nonfilamentous amyloid-β deposits [80]); synucleinopathies, such as PD, dementia with Lewy body (LB; also known as diffuse LB disease, the LB variant of AD, senile dementia of the LB type, and PD in AD), multiple system atrophy (MSA) and Hallervorden–Spatz disease (deposition of α-synuclein in a form of LB, or Lewy neurites [LNs] [81]); prion diseases (deposition of PrPSC [82]); and a family of polyQ diseases, a group of neurodegenerative disorders caused by expansion of GAC trinucleotide repeats coding for polyQ in the gene products [83], and amyotrophic lateral sclerosis (ALS), familial or sporadic forms that are characterized by the presence of intracellular inclusions composed of superoxide dismutase (SOD1) [84] and transactive response (TAR) DNA-binding protein 43 (TDP-43) [85].
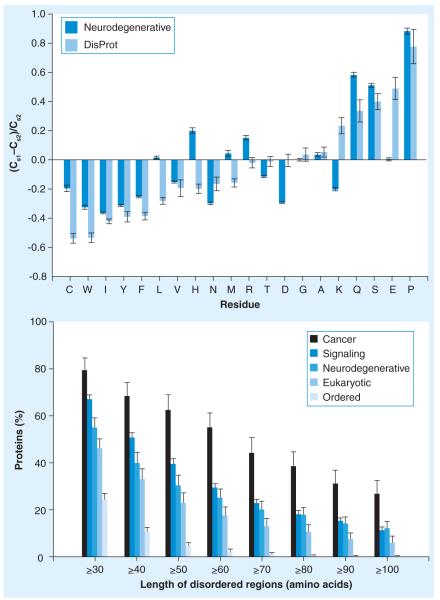
The fact that the causative agents of various neurodegenerative diseases are IDPs is illustrated by Figure 2A, which compares the amino acid compositions of several neurodegeneration-related proteins with the compositions of ordered proteins from a protein data bank (PDB) [64]. The corresponding data for the typical IDPs from the DisProt database [86] are shown for comparison. The calculations were performed using a normalization procedure, elaborated for analysis of IDPs [87,88]. In brief, compositional profiling is based on the evaluation of the (Cs1−Cs2)/Cs2 values, where Cs1 is a content of a given residue in a set of interest (proteins associated with neurodegenerative diseases or typical IDPs from DisProt), and Cs2 is the corresponding value for the set of ordered proteins. In this presentation, negative values correspond to residues that are depleted in a given dataset, in comparison to a set of ordered proteins, whereas the positive values correspond to the residues that are over-represented in the set. Figure 2A clearly shows that neurodegeneration-related proteins are very different from typical ordered proteins, and generally follow the trends expected for IDPs (with some exceptions). In general, they are depleted in major order-promoting residues (C, W, I, Y, F, V and N) but are highly enriched in the major disorder-promoting residues (Q, S, R and P). This suggests that the neurodegeneration-related proteins are characterized by a high level of intrinsic disorder. This conclusion is further illustrated by Figure 2B, which represents the results of evaluation of intrinsic disorder in a set of 689 proteins related to neurodegenerative diseases as percentages of proteins with at least 30 consecutive residues predicted to be disordered. Figure 2B confirms that intrinsic disorder is highly prevalent in neurodegenerative disease-related proteins, comparable to that of signaling and cancer-related proteins [89], and significantly exceeds the level of intrinsic disorder in eukaryotic proteins from Swiss-Prot, and in nonhomologous structured proteins from the PDB [64].
Figure 2. Peculiarities of amino acid sequences of intrinsically disordered proteins associated with neurodegeneration.
(A) Compositional profiling of proteins involved in neurodegenerative disease. Compositional profiling is based on the evaluation of the (Cs1−Cs2)/Cs2 values, where Cs1 is a content of a given residue in a set of interest (proteins associated with neurodegenerative diseases or typical intrinsically disordered proteins [IDPs] from DisProt), and Cs2 is the corresponding value for the set of ordered proteins. In this presentation, enrichment or depletion in each amino acid type appears as a positive or negative bar, respectively. Amino acids are indicated by the single-letter code and ordered according to their disorder promoting strength. Corresponding data for well-characterized intrinsically disordered regions from DisProt are also shown. (B) Abundance of intrinsic disorder in proteins associated with neurodegenerative diseases. Percentages of disease-associated proteins with at least 30–100 consecutive residues predicted to be disordered. The error bars represent 95% CIs and were calculated using 1000 bootstrap re-sampling. Corresponding data for signaling and ordered proteins are shown for the comparison. Analyzed protein sets included 1786 proteins associated with cancer, 2329 proteins involved in cellular signaling, 689 proteins involved in the neurodegenerative diseases, 53,630 nonredundant eukaryotic proteins from Swiss-Prot, and 1138 non-homologous ordered proteins from PDB Select 25 (this dataset contained only the ordered parts of the proteins). Redrawn from [64].
α-synuclein & synucleinopathies
α-synuclein, being one of the most studied disease-related IDPs, is a protein that links various synucleinopathies [90,91]. It possesses little-to-no ordered structure under the ‘physiological’ conditions in vitro [92]. At neutral pH, α-synuclein is essentially disordered, but is slightly more compact than a random coil [92]. This conclusion confirms the results of pulsed-field gradient NMR, which also shows that α-synuclein is slightly collapsed [93]. A high-resolution NMR analysis of the protein revealed that α-synuclein is largely unfolded in a solution, but exhibits a region between residues 6 and 37 with a preference for helical conformation [94]. Conformational analysis of α-synuclein, under a variety of environments, showed that the structure of this protein is extremely sensitive to the environment. It adopts a variety of structurally unrelated conformations, including the substantially unfolded state, an amyloidogenic partially folded conformation, and different α-helical or β-structural species, folded to a different degree, both monomeric and oligomeric [90-92,95]. Furthermore, it may also form several morphologically different types of aggregates, including oligomers (spheres or doughnuts), amorphous aggregates and amyloid-like fibrils [92]. Based on this astonishing conformational behavior, the concept of a protein chameleon was proposed, according to which the structure of α-synuclein, to a dramatic degree, depends on the environment, and the choice between different conformations is determined by the peculiarities of protein surroundings [92].
Amyloid β-protein, tau & AD
Alzheimer’s disease is characterized biochemically by the accumulation of two types of proteinaceous inclusions: extracellular amyloid deposits – senile plaques in the cerebral cortex and vasculature – and intracellular NFTs (which comprise the paired helical filaments [PHFs]) [96]. Amyloid deposits in AD contain the amyloid β-protein (Aβ), which is a 40–42 residue peptide produced by endoproteolytic cleavage of the amyloid precursor protein (APP). PHFs are assembled from a hyperphosphorylated form of the microtubular protein tau. Structurally, both of these proteins are intrinsically disordered. In fact, Aβ appears to be unfolded at the beginning of fibrillation under physiological conditions. NMR studies have shown that monomers of Aβ1–40, or Aβ1–42 possess no α-helical or β-sheet structure [97] (i.e., they exist predominantly as random coil-like, highly extended chains). It is worth noting that, although APP is predicted to be highly disordered, both Aβ1–40 and Aβ1–42 peptides are rather hydrophobic and, therefore, are not completely consistent with the definition of extended IDPs, which are characterized by the low content of hydrophobic amino acid residues. Partial folding to the premolten globule-like conformation has been detected at the earliest stages of Aβ fibrillation [97]. Similarly, prior to aggregation, tau protein was shown to be in a mostly random coil-like state [98]. Analysis of the primary structure revealed a very low content of hydrophobic amino acids and a high content of charged residues, which was sufficient to explain the lack of folding [98]. Based on the detailed analysis of tau fibrillation in the presence of anionic inducers using a set of spectroscopic techniques, it has been concluded that the fibrillation inducer (anionic surfactants) stabilized a monomeric partially folded species with the structural characteristics of a premolten globule state [99]. The stabilization of this intermediate was sufficient to trigger the fibrilliation of the protein [99].
Prion protein & prion diseases
Prion diseases are a group of incurable and fatal neurodegenerative maladies in mammals. These diseases, collectively referred to as the transmissible spongiform encephalopathies (TSEs; including Creutzfeldt–Jakob disease [CJD], Gerstmann–Sträussler–Scheinker [GSS] disease, fatal familial insomnia [FFI] and kuru in humans; scrapie in sheep; bovine spongiform encephalopathy [BSE] in cattle; and chronic wasting disease [CWD] in mule deer and elk [100]), are caused by the pathological deposition of the prion protein (PrP) in its aggregated form. The N-terminal region of approximately 100 amino acids in PrPC (from amino acid 23 to 126) is largely unstructured [101]. Investigations of the steps required for prion propagation and neurodegeneration indicated that the last 50 residues in the disordered N-terminal region play a particularly important role in the interaction of PrPC with PrPSc, which leads to the conversion of the former to the latter [102,103]. Those residues are largely unordered or weakly helical in the full-length PrPC [104,105], but are predicted to be β-structure in PrPSc [106].
Polyglutamine repeat diseases & their causative proteins
There are at least nine hereditary diseases in which the expansion of a CAG repeat in the gene (which correspond to the expansion of the polyglutamine repeat in the corresponding protein) leads to neurodegeneration [107,108]. These polyglutamine repeat diseases include Huntington’s disease (HD), Kennedy disease (also known as spinal and bulbar muscular atrophy), spinocerebellar ataxia type 1 (SCA1), dentatorubral–pallidoluysian atrophy (DRPLA), spinocerebellar ataxia type 2 (SCA2), Machado–Joseph disease (MJD/SCA3), SCA6, SCA7 and SCA17. In HD, the CAG repeat that encodes the polyQ region is the part of exon 1 in the 3140-residue huntingtin protein [109]. The polyQ repeat varies between 16 and 37 residues in healthy individuals, and individuals who are afflicted by disease have repeats of more than 38 residues. The mechanistic hypothesis linking CAG repeat expansion to toxicity involves the tendency of longer polyQ sequences, regardless of protein context, to form insoluble aggregates [83,110-117]. Biophysical characterization of polyglutamine peptides with lengths of 5, 15, 28 and 44 residues, revealed their highly disordered nature despite the length of the polyQ stretch, suggesting that length dependence of the disease is not related to a conformational change in the monomeric states of expanded polyQ sequences [115]. By contrast, there was a dramatic acceleration in the spontaneous formation of amyloid-like aggregates for polyQ peptides longer than 37 residues. In addition to the obvious intrinsically disordered nature of polyglutamine stretches, several proteins responsible for the pathogenesis of polyQ repeat diseases were shown or predicted to be either completely disordered or to contain long disordered regions [64]. The examples include androgen receptor in SBMA [118], atrophin-1 in DRPLA [119], ataxin-2 in SCA2 [120], ataxin-3 in SCA3/MJD [121], P/Q-type calcium channel α1A subunit in SCA6 [64], ataxin-7 in SCA7 [64,122] and TATA-box-binding protein in SCA17 [64,123].
Abri, Adan & familial dementias
Familial British dementia (FBD) and familial Danish dementia (FDD) are autosomal dominant diseases linked to a genetic defect in the BRI2 gene. FBD is a neurodegenerative malady with the onset at approximately the fifth decade of life and full penetrance by the age of 60 years characterized by the presence of amyloid deposits comprised of the protein subunit (termed ABri) in cerebral blood vessels and brain parenchyma, which coexist with NFTs in limbic areas [124]. In FDD, the deposited amyloid protein, ADan, is the C-terminal proteolytic fragment of a genetically altered BRI2 precursor molecule [125]. Structural analysis revealed that, similar to Aβ, ABri [126] and ADan [127] are typical natively unfolded proteins.
SOD1, TDP-43, progranulin, angiogenin, FUS/TLS, senataxin & SMN in amyotrophic lateral sclerosis
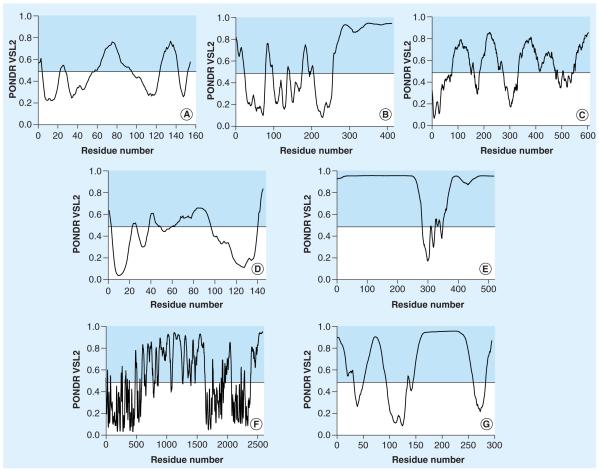
Amyotrophic lateral sclerosis is a neurodegenerative disorder affecting upper and lower motor neurons and results in fatal paralysis; it is one of the most common motor neuron diseases in the USA. Although the occurrence of ALS is predominantly sporadic, approximately 10% of cases are familial, with approximately 20% of these familial cases being caused by mutations in the gene that encodes the antioxidant enzyme copper-zinc superoxide dismutase (SOD1) [84,128]. SOD1 is an abundant protein found in the cytosol, the nucleus, peroxisomes and the mitochondrial intermembrane space of human cells. The primary function of SOD1 is to serve as an antioxidant enzyme, lowering the steady-state concentration of superoxide via the catalysis of the dismutation of superoxide to oxygen and hydrogen peroxide. Each subunit of the SOD1 homodimer can coordinate one Cu2+ and one Zn2+ ion, and several ALS-related mutations were shown to diminish the affinity of this protein for Cu2+ or Zn2+ [84]. Structural analysis of the human wild-type SOD1 and three pathogenic variants of SOD1 (A4V, G93R and H48Q), which cause familial ALS, revealed that these proteins are partially unfolded at physiological temperature (e.g., it is locally unfolded at residues 21–53 and 117–144) and unfold noncooperatively [129]. In agreement with these experimental data, computational analysis revealed that the regions 22–30, 55–95 and 123–142 of human SOD1 are likely to be intrinsically disordered (Figure 3A).
Figure 3. Predicted intrinsic disorder in proteins associated with amyotrophic lateral sclerosis.
Abundance of intrinsic disorder in human proteins associated with amyotrophic lateral sclerosis was evaluated using the PONDR® VSL2 algorithm [214], which is one of the most accurate predictors of intrinsic disorder for proteins containing both ordered and disorder regions. Each plot represents a distribution of the predicted intrinsic disorder propensity (PONDR VSL2 score) within the amino acid sequence of a given protein. Analyzed proteins are: (A) SOD1, (B) TDP-43, (C) progranulin, (D) angiogenin, (E) fused in sarcoma/translated in liposarcoma, (F) senataxin and (G) survival motor neuron protein. Shaded areas in each plot correspond to the scores associated with predicted intrinsic disorder.
In addition to mutations in SOD1, mutations in TARDBP, the gene encoding TDP-43, have been discovered in sporadic and familial ALS [130]. It is now recognized that TDP-43 is a nuclear protein that is involved in several major neurodegenerative diseases, such as frontotemporal lobar degeneration with ubiquitin (FTLD-U), a neurodegenerative condition that predominantly affects behavior, social awareness and language, and ALS. In fact, TDP-43 was recognized as the primary protein component of intracellular ubiquitinated inclusions in most cases of these two disorders [131,132]. As discussed in a review by Buratti and Baralle, TDP-43 is a DNA-, RNA- and protein-binding nucleoprotein involved in the regulation of numerous processes, such as transcription, splicing, cell cycle regulation, apoptosis, miRNA biogenesis, mRNA transport to, and local translation at, the synapse, and scaffolding for nuclear bodies [133]. TDP-43 has multiple alternatively spliced isoforms, and contains two RNA-recognition motifs, and a C-terminal glycinerich region that is involved in protein binding [134], and is crucial for protein solubility and cellular localization [135]. Figure 3B shows that human TDP-43 is a highly disordered protein, containing approximately 50% predicted disordered residues, mostly localized to its C-terminal half.
Progranulin (PGRN) is a secreted precursor protein that is ubiquitously expressed and contains tandem repeats of a unique (ten- or) 12-Cys motif, which are proteolytically cleaved to form seven granulin (Grn) peptides (Grn A to G). Both full-length PGRN and the Grn peptides are involved in a wide variety of biological functions, starting at embryonic development and including cell cycle regulation, wound repair, tumor growth and inflammation, sometimes with opposite effects [136]. High-resolution NMR analysis revealed that only the three human Grns, hGrnA, hGrnC and hGrnF, contained relatively well-defined 3D structures in solution, whereas four other granulins represented mixtures of poorly structured disulfide isomers. Structurally, folded granulins hGrnA, hGrnC and hGrnF were characterized by a stable stack of two β-hairpins in their N-terminal subdomains, whereas their C-terminal subdomains were highly dynamic [137]. Figure 3C support these experimental data, and shows that, according to the PONDR VSL2 analysis, human PGRN is predicted to contain a number of long disordered regions (e.g., regions 76–169, 180–270, 328–426 and 530–593).
Another protein with mutations implemented in ALS is angiogenin (ANG), which is a 14.1-kDa protein that belongs to the pancreatic ribonuclease superfamily [138]. NMR analysis revealed that human ANG is a relatively well-folded protein, with increased dynamics in some regions (e.g., backbone atoms of ~five residues at both N- and C-termini, and regions 59–70 and 82–95 were shown to possess relatively high root-mean square deviations from their position in the mean structure) [139]. Figure 3D shows that these regions are predicted to be intrinsically disordered.
Some familial ALS patients were reported to have mutations in fused in sarcoma/translated in liposarcoma (FUS/TLS) protein [140,141]. FUS/TLS is a 526-amino acid-long nuclear protein, characterized by an N-terminal serine, tyrosine, glycine and glutamine-rich region, an RNA-recognition motif (RRM), a C2/C2 zinc finger motif, multiple RGG-repeat regions and a nuclear localization sequence (NLS) at the extreme C-terminus [138]. FUS/TLS is involved in multiple biological processes, and is proposed to serve as an RNA chaperone [138], since it is engaged in rapid nucleo–cytoplasmic shuttling [142]. Unfortunately, as of yet, FUS/TLS has not been structurally characterized. However, Figure 3E clearly shows that this protein is predicted to be mostly disordered.
Mutations in the senataxin (SETX) are known to result in the development of an autosomal dominant form of juvenile ALS, with slow progressive distal muscle weakness and an onset prior to 25 years of age [143]. SETX is a ubiquitously expressed 302.8-kDa protein consisting of 2677 amino acids, which, at the C-terminus, contains a classical seven-motif domain characteristic for RNA/DNA helicases. Although not much is known about structural properties of this large protein, Figure 3F shows that it has a large number of long disordered regions, especially in the central and C-terminal regions.
Finally, the high risk of developing ALS is associated with the abnormal copy number of the survival motor neurons (SMN1) gene [144]. There are two copies of the SMN gene in humans, SMN1 and SMN2. SMN2 differs from SMN1 by a C to T transition in exon 7, resulting in a predominant skipping of exon 7 in the SMN2 transcript [145]. The SMN1 is a 294-amino acid-long protein found in the cytoplasm and nucleus, especially in discrete Gemini of Cajal bodies (Gems) [146]. It is an important part of the assemblyosome, called the SMN complex, involved in the creation of the ring-shaped core domain of the small nuclear (sn)RNPs [138], which combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex where pre-mRNA splicing occurs. Structurally, human SMN1 protein contains a SMN protein-interacting protein 1 binding site (residues 13–44), an ordered Tudor domain (residues 91–151), a smith antigen B binding site (residues 240–267), a region required for interaction with heterogeneous nuclear ribonucleoprotein Q encoded by SYNCRIP (residues 279–294), and several polyproline regions (residues 195–203, 217–226 and 244–248) in the C-terminal domain. Data shown in Figure 3G illustrate that human SMN1 protein is predicted to have several disordered regions. Some of the identified binding sites of SMN1 are located within, or in the close proximity to, the long disordered regions.
IDPs in other neurodegenerative diseases
The major causative agents of many neurodegenerative diseases are frequently disordered [64]. In addition to the relatively well-documented examples outlined previously, intrinsic disorder was predicted and, in part, confirmed experimentally (mostly based on the aberrant mobility on SDS-gel electrophoresis) for mitochondrial DNA polymerase γ in Alpers disease [147], DNA excision repair protein ERCC-6 in Cockayne syndrome [148], survival of motor neurons protein and spinal muscular atrophy [149], various septins [150] and members of the neurotrophin family [151].
Illustrative examples of IDPs in protein-dysfunction diseases
p53 & cancer
p53 (also known as tumor protein 53 or tumor antigen p53) is a tumor-suppressor protein encoded in humans by the TP53 gene [152]. p53 is located at the center of a large signaling network, regulating expression of genes involved in such cellular processes as cell cycle progression, apoptosis induction, DNA repair and response to cellular stress [153]. p53 has been demonstrated to induce or inhibit over 150 genes, including p21, GADD45, MDM2, IGFBP3 and BAX [154]. Since p53 plays a crucial role in conserving stability by preventing genome mutation, it has been described as ‘the guardian of the genome’, ‘the guardian-angel gene’ and the ‘master watchman’ [155]. Therefore, when p53 function is lost, either directly through mutation or indirectly through several other mechanisms, the cell often undergoes cancerous transformation [156]. Mutations in the p53 gene are associated with 50–55% of human cancers, and cancers showing mutations in p53 are found in colon, lung, esophagus, breast, liver, brain, reticuloendothelial and hemopoietic tissues [156]. These mutations frequently occur in the DNA-binding domain of p53, destabilizing it to a great degree [157-159] and, therefore, clearly representing a case of the protein-misfolding disease.
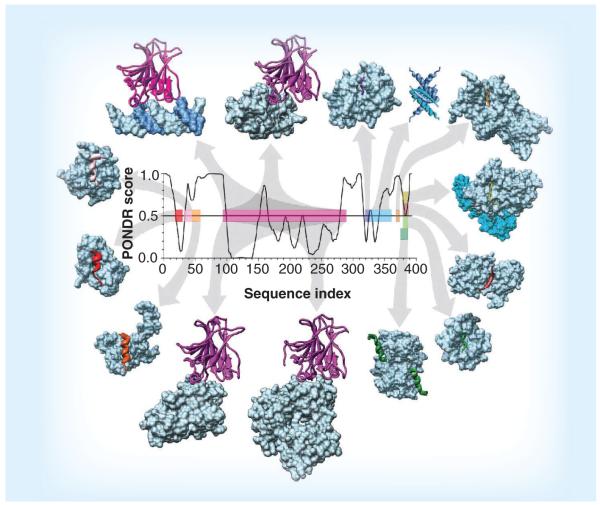
As p53 is one of the most studied proteins, it represents an excellent illustrative example of the D2 concept. This is summarized in Figure 4, which shows the currently available information on p53 interactions and structure [21]. There are four domains in p53: the unstructured N-terminal transactivation domain, the structured central DNA-binding domain, and the unstructured C-terminal tetramerization and regulatory domain [21]. At the transactivation region, p53 interacts with TFIID, TFIIH, Mdm2, RPA, CBP/p300 and CSN5/Jab1, among many other proteins [153]. At the C-terminal domain, it interacts with GSK3β, PARP-1, TAF1, TRRAP, hGcn5, TAF, 14–3–3, S100B(ββ) and several other proteins [153]. In Figure 4, some of these interactions are mapped to regions of the p53 sequence, together with the order/disorder tendencies of p53 as revealed by PONDR® VLXT. Figure 4 shows that many complexes are formed that involve the disordered regions of p53. An interesting aspect of these many partnerships is that, for each interaction, only a short region of p53 typically becomes structured upon binding. Examples include the mdm2-binding motif of the N-terminal domain, the tetramerization domain, and the short segment near the p53 C-terminus, which binds to four different partners. Because this segment is unstructured to begin with, it can adopt different conformations when binding to the different partners. For this particular example, the disordered segment adopts a short α-helix, a β-strand and two different coils upon binding with its four different partners [21].
Figure 4. Intrinsic disorder in p53 and interactions with different binding partners.
A structure versus disorder prediction on the p53 amino acid sequence is shown in the center of the figure, along with the structures of various regions of p53 bound to 14 different partners. Disorder predictions were done using the PONDR VLXT algorithm. The plot represents the distribution of the disorder propensity (PONDR score, up = disorder, down = order) within the amino acid sequence of p53. The results of the disorder predictions (the central region was predicted to be ordered, whereas the N- and C-termini were predicted to be disordered) have been confirmed experimentally for p53. The various regions of p53 are color coded to show their structures in the complex, and to map the binding segments to the amino acid sequence. Starting with the p53–DNA complex (top-left: magenta = protein; blue = DNA), and moving in a clockwise direction, the Protein Data Bank IDs and partner names are given as follows for the 14 complexes: 1tsr–DNA, 1gzh–53BP1, 1q2d–gcn5, 3sak–p53 (tetrametization domain), 1xqh–set9, 1h26–cyclinA, 1ma3–sirtuin, 1jsp–CBP bromo domain, 1dt7–s100ββ, 2h1l–sv40 large T antigen, 1ycs–53BP2, 2gs0–PH, 1ycr–Mdm2 and 2b3g–rpa70.
Reproduced from [7].
Disorder comes in waves: intrinsic disorder in p53 upstream regulators & downstream effectors
p53 is an important hub protein located at the middle of the regulatory network that includes a complex array of upstream regulators and downstream effectors [160]. This central position helps p53 in integrating a wide range of signals to promote adaptive responses to stress. As has been pointed out in the review by Lu et al. [161], there are three major regulatory mechanisms in the upstream control of the p53 regulatory network: stabilization, antirepression and promoter-specific activation [162]. The first two mechanisms, p53 stabilization and antirepression, are based on involvement of the Mdm proteins as key regulators. The activity of Mdm proteins is, in turn, regulated by their interaction with p19ARF, also known as cyclin-dependent kinase inhibitor 2A [161]. Post-translational modifications (e.g., phosphorylation by checkpoint kinase 2 [CHK2] and other kinases) are also crucial for p53 activation, and for apoptosis induction in response to ionizing radiation [163]. Furthermore, the p53 interaction with some other upstream regulators, such as ataxia–telangiectasia mutated protein (ATM), and ataxia–telangiectasia and Rad3-related protein (ATR), are important for the induction of the UV radiation-induced cell death [164].
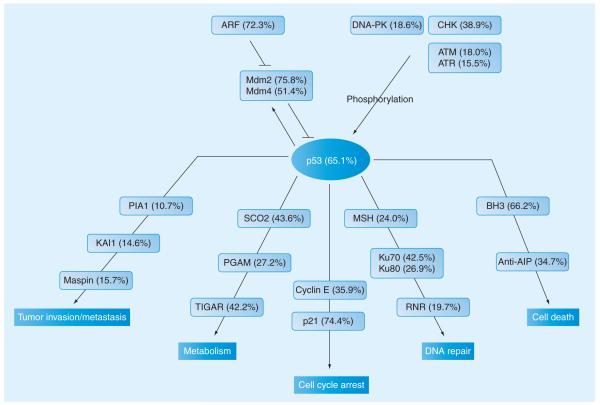
Activated p53 is crucial for widely diverse stress responses, the outcomes of which depend on the involvement of p53 transcriptional targets (downstream effectors) [161]. Depending on a particular set of these downstream effectors, activated p53 is known to initiate apoptosis by targeting BH3-only Bcl-2 subfamily, and antagonists of the inhibitor of apoptosis (anti-IAP) to promote DNA repair by targeting DNA mismatch-repair protein MSH2, ribonucleotide reductase (RNR) and ssDNA-dependent ATP-dependent helicase Ku70/Ku80, and to mediate cell cycle arrest by targeting cyclin E and cyclin-dependent kinase inhibitor 1 (or protein p21). It also controls crucial metabolic pathways by regulating the expression of several genes, such as phosphoglycerate mutase (PGAM), TP53-induced glycolysis and apoptosis regulator (TIGAR), and synthesis of cytochrome c oxidase 2 (SCO2), each linked to the processes of glycolysis and oxidative phosphorylation [165]. It also regulates tumor invasion and/or metastasis by modulating expression of the tumor metastasis suppressor gene KAI1, plasminogen activator inhibitor 1 (PAI1), and maspin [166].
Many of the aforementioned upstream regulators of p53 activity and downstream effectors were predicted to be noticeably disordered. This is illustrated by Figure 5, which is an oversimplified representation of the p53 regulatory network. The figure shows that both regulators and effectors of p53 are predicted to contain noticeable amount of disorder. However, not all members of the p53 regulatory network are disordered to a similar degree. There is a wide range of predicted disorder in this network (from as low as 10.7% in PIA1 to as high as 75.8% in Mdm2). The abundance of disorder seems to be correlated with pathways; for example, in the upstream regulators, the members of the ARF–Mdm pathway are more greatly disordered than the CHK2/DNA–PK/ATM/ATR group. Similarly, p53 effectors responsible for the cell death and the cell cycle arrest contain, on average, more predicted intrinsic disorder than proteins responsible for the suppression of tumor invasion. These data clearly show that intrinsic disorder plays a crucial role in all the stages of p53 functioning, such as p53 activation, actual p53 function and functions of various p53 effectors.
Figure 5. Abundance of intrinsic disorder within the p53 regulatory network.
Selected upstream regulators, downstream effectors and output terms are shown. A simplified evolutionary schematic of the p53 regulatory network, which was presented in [162], was used as a basis for this figure. Color-gradient coding related to the range of conservation of various members of the p53 regulatory network in vertebrates and in invertebrates is removed. The new color coding corresponds to the various pathways within the p53 network. Several p53 effectors related to the regulation of metabolism were added to the original plot together with the new output term ‘tumor invasion/metastasis’. Percentages in brackets reflect the information on abundance of predicted disorder in various members of this network.
c-Myc & cancer
Deregulation of the c-Myc transcription factor is involved in many types of cancer, making this oncoprotein an attractive target for drug discovery. In order to bind DNA, regulate target gene expression and function in most biological contexts, c-Myc must dimerize with Max, which lacks a transactivation segment. The interaction regions of both Max and c-Myc are disordered as monomers. They undergo mutual coupled binding and folding when their zipper domains interact to form a helical coiled coil [167].
PDE4D & stroke
Cyclic AMP is a second messenger that mediates physiological responses to host hormones, neurotransmitters and autacoids, and broadly suppresses the activity of immune and inflammatory cells. cAMP-specific 3′,5′-cyclic phosphodiesterase 4D (CN4D, also known as PDE4D) is a member of a large protein family that regulates the level of cAMP in the cell [168]. It has been indicated that the gene encoding PDE4D is strongly associated with carotid and cardiogenic stroke, forms of stroke related to atherosclerosis [169]. A substantial disregulation of multiple PDE4D isoforms in individuals affected by stroke was observed. Based on these findings it has been proposed that PDE4D is involved in the pathogenesis of stroke, possibly through atherosclerosis, which is the primary pathological process underlying ischemic stroke [169]. Furthermore, the selective inhibitors of the PDE4 family provide potent anti-inflammatory agents [168]. PDE4D contains three functional domains: a conserved catalytic core (>50% sequence identity through family), a regulatory N-terminus and the C-terminus. The catalytic domain was predicted to be ordered and was perfectly aligned with the determined structure [61]. Both termini contain long regions of predicted disorder. However, important functions, such as phosphorylation, membrane targeting and intramolecular inhibitory function, are associated with the N-terminal fragment, whereas the C-terminal region is thought to be involved in dimerization [170]. The N-terminal domain of PDE4D (residues 1–136) was shown to be able to bind to SH3 domains of certain proteins, including src family tyrosyl kinase lyn, fyn and src, as well as abl tyrosyl kinase and the cytoskeletal protein fodrin [171].
IRS1 & diabetes
IRS1 is one of several proteins activated by the binding of insulin to insulin receptor (IR) on a cell’s surface. IR is a tyrosine kinase that phosphorylates several proteins, including IRS1 [172], Gab1, isoforms of Shc, p60dok, APS and Cbl [173]. Certain proteins with Src homology 2 (SH2) domains then recognize the phophorylated tyrosines and interact with the aforementioned proteins. These SH2-containing proteins include both adaptor molecules and enzymes. The interaction of IRS1 and Shc with Grb2 leads to activation of the MAP kinase pathway via Ras, allowing cell growth and differentiation [174].
IRS1 also interacts with the p85 regulatory portion of PI3K, through pYMXM and pYXXM motifs, to activate it [175]. PI3K then phosphorylates phosphoinositides on the 3-position, generating PtdIns(3,4,5)P3, which binds to the pleckstrin homology domain on the AGC serine/threonine kinase family member, phosphoinositide-dependent kinase 1 (PDK1), which activates the serine/threonine kinase Akt/PKB [176]. Activation of Akt leads to translocation of Glut4 vesicles for glucose transport, through a currently unknown mechanism [177].
The activity of PI3K is controlled by two different phosphatases, PTEN [178] and the SH2 domain-containing inositol-5-phosphatase (SHIP2) [179]. Disruption of these two proteins, either through gene disruption or by reducing mRNA expression, yielded mice with increased insulin sensitivity [180]. Finally, IR and its substrates are dephosphorylated by phosphatases, such as protein tyrosine phosphatase 1B (PTP1B). PTP1B has received much attention because, when knocked out, both IR and IRS proteins assume higher phosphorylated states, which contribute to increased insulin sensitivity and reduced occurrence of dietrelated obesity [181]. Although IRS1 is predicted to be a highly disordered protein (834 out of 1242 residues are predicted to be disordered), its N-terminal domain adopts a folded crystallizable structure, and its short fragment located near residue 900, folds upon binding to IGF-1 receptor kinase [182].
IDPs as novel drug targets
The possibility of interrupting the action of disease-associated proteins is an extremely attractive objective for the development of new drugs. The rational design of enzyme inhibitors depends on the classical view of protein function, which states that 3D structure is an obligatory prerequisite for function. Computational evaluations can be used to search a protein’s interaction surface for sites that will potentially bind small molecules [183], provided that a reliable 3D structure is available. While this approach has led to many successful drug molecules that target enzymatic domains, it has influenced thinking with regard to all types of protein functions, including functions that depend on intrinsic disorder. Although structure-aided design techniques have been modified and extended for the discovery of molecules that function as inhibitors of protein–protein interactions, the search for such inhibitors has been mostly unsuccessful, in large part because many of the approaches for discovery of such inhibitors were derived directly from the techniques used for the discovery of enzyme inhibitors [184]. However, there is a fundamental difference between the action mechanisms of these two types of effectors. To be an effective blocker of the enzymatic activity, the small molecules have to bind a contiguous epitope of five or more residues [185], whereas many protein–protein interfaces are much more complex and consist of discontinuous, or a combination of multiple contiguous, epitopes [186].
Since disordered proteins often bind their partners via relatively short molecular-recognition motifs, which become ordered upon binding [35,36,187], and since IDPs are involved in two types of protein–protein interactions, where either both partners are disordered or an IDP interacts with the ordered partner, there are two general concepts for targeting interactions involving IDPs. One approach is similar to traditional drug-discovery approaches, where a small molecule targets the binding site on the structured partner. In another approach, small molecules target possible binding sites on the IDP. An interesting twist of this second approach is that targeting disordered regions can be described as induction of non-native structure to prevent function.
An illustrative example of the first approach is a set of small molecules that bind to the structured protein Mdm2, an E3 ubiquitin ligase and, thereby, prevents the binding of this protein to a disordered region of the tumor-suppressor p53. Mdm2 associates with a short stretch of p53, residues 13–29 (Figure 3), which is located within the transactivation domain. As a result, p53 cannot activate or inhibit other genes when Mdm2 is bound. Furthermore, Mdm2 binding leads to p53 ubiquitination and, thus, targets it for destruction. Mdm2 also contains a nuclear export signal, which causes p53 to be transported out of the nucleus. Because of the importance of the p53–MDM2 interaction, this system has been investigated as a possible drug target and several successful peptide inhibitors of this interaction have been created [188-191]. In addition to the peptide inhibitors, several small, drug-like molecules have been found to block the p53–MDM2 interaction [192-195]. More importantly, computational analysis of p53 using a set of disorder predictors suggested that the disordered sites of druggable protein–protein interactions are predictable [21]. This druggable p53–MDM2 interaction involves a disorder-to-order transition, principles of which are generally understood and, therefore, can be used to find similar drug targets [196].
In the second approach, the protein–protein interaction is prevented via small molecules interacting directly with the disordered binding regions of proteins. These small molecules induce misfolding of disordered regions to non-native structures incompatible with complexation. This approach is illustrated by a set of small molecules that inhibit the interaction between c-Myc and its obligate heterodimerization partner, Max [197-207]. Recently, the effective c-Myc inhibition was achieved via the disruption of the formation of the c-Myc–Max complex, induced by the c-Myc interaction with small-molecule inhibitors [199,204]. These molecules were shown to bind to one of three discrete sites within the 85-residue bHLHZip domain of c-Myc, a region that is disordered before it binds to Max. Inhibitor binding induced only local conformational changes, which preserved the overall disorder of c-Myc and inhibited the interaction with Max. Therefore, rational approaches to the inhibition of protein–protein interactions involving IDPs may be possible through the targeting of disordered sequence [167,206] or structured partner [196].
Summary & conclusion
Intrinsically disordered proteins are highly abundant in nature and are frequently associated with various human diseases, including protein-dysfunction diseases and neurodegeneration. A high degree of association between protein intrinsic disorder and human diseases is owing to structural and functional peculiarities of IDPs, which are typically involved in cellular regulation, recognition and signal transduction. Structurally, IDPs represent a diverse group of proteins without a unique 3D structure, and disorder might be collapsed (as in native molten globules) or extended (as in native random coils or in native premolten globules). Amino acid sequences of IDPs are characterized by several unique features, which provide a solid background for the reliable identification of these proteins, and are utilized in the development of various computational tools for the prediction and characterization of a disorder at the proteome level.
The major functional advantages of IDPs are based on their structural flexibility and plasticity, originating from the lack of a definite ordered 3D structure. This plasticity is believed to represent the major functional advantage for IDPs, providing them with a unique capability to interact with a broad range of binding partners, including other proteins, membranes, nucleic acids and various small molecules.
Many IDPs and IDRs undergo disorder-to-order transition as a result of interaction with their specific binding partners. The amino acid sequences of foldable IDRs (known as MoRFs, among other terms) are characterized by specific features. These features were utilized for the reliable identification of MoRFs from the amino acid sequence alone.
So far, three computational approaches were elaborated for estimating the abundance of IDPs in various pathological conditions. In the first approach, specific datasets of proteins associated with a given disease were assembled and analyzed using various disorder predictors. In the second approach, the network of genetic diseases linking protein mutations with diseases was analyzed. In the third approach, the association between a particular protein function (including the disease-specific functional keywords), and the level of intrinsic disorder in a set of proteins known to carry out this function, was analyzed.
Intrinsically disordered proteins are unique novel drug targets. Although this field is still at its early stages, two methodologically different approaches for the IDP-based drug discovery have already been developed. In the first approach, a small molecule targets a binding site at the ordered binding partner, and prevents this partner from interaction with IDP. In the second approach, a small molecule directly interacts with IDP and induces conformation that is incompatible with binding.
Expert commentary
The past decade has witnessed a dramatic increase in the appreciation of the protein intrinsic disorder phenomenon. The discovery and characterization of IDPs is becoming one of the fastest-growing areas of protein science. This field continues to develop at an accelerating pace, since many such proteins with no unique structures have important biological functions and are involved in various diseases. Novel computational and experimental tools are being constantly elaborated for more accurate predictions of IDPs and IDRs, and for more precise characterizations of their structural and functional properties.
Biological functions of IDPs & IDRs
It is now well accepted that the century-old lock-and-key paradigm of protein functionality, where the function of a protein is closely linked to its rigid 3D structure, needs to be re-examined to include IDPs and their numerous functions. In fact, the conformational plasticity of IDPs and their intrinsic lack of rigid structure results in a number of exceptional functional advantages, providing them with unique capabilities to act in functional modes not achievable by ordered proteins [6-10,12,15,19,21,23,26,28,34]. Since these advantages were systemized in several recent reviews, only several illustrative examples are provided. Many IDPs contain multiple functional elements, which are often relatively short. Given the existence of multiple functions in a single disordered region, and given that each element is localized over a few amino acids, alternative splicing could readily generate a set of protein isoforms, having a highly diverse collection of regulatory elements [208]. The ability of many IDPs/IDRs to fold at binding to natural partners decouples the specificity and strength of binding, giving rise to the high-specificity–low-affinity interactions. Furthermore, a continuum of binding strength is expected to exist, since binding regions containing predominant preformed structure contribute more free energy to binding, and those regions with little or no preformed structure contribute little free energy to binding [7].
One IDR can bind to multiple partners and, consequently, gain very different structures [21]. The overall repertoire of binding modes utilized by IDPs is very broad as well, ranging from binding-induced gaining of local structure on the surface of a binding partner, to folding of a whole molecule, and from wrapping around the binding partner, to penetrating deep inside the binding partner. IDPs can form highly stable complexes, or be involved in signaling interactions, where they undergo constant ‘bound–unbound’ transitions, thus acting as dynamic and sensitive ‘on–off’ switches. In general, the action of IDPs illustrates a new meaning of the ‘reversibility’ concept. The ability of these proteins to return to the highly flexible conformations after the completion of a particular function, and their predisposition to gain different conformations depending on the environmental peculiarities, are unique physiological properties of IDPs, which allow them to exert different functions in different cellular contests according to a specific conformational state.
Analyzing structural properties of IDPs
Structurally, IDPs and IDRs are considered as natural analogues of various partially folded conformations of globular proteins [4-7]. However, this is just an approximation that should not be regarded as the final truth. In fact, IDPs, with their exceptional variability in amino acid compositions and sequences, are expected to wander over much greater areas of the polypeptide conformational space than ordered proteins. The characterization of the conformational properties and functions of IDPs continues to represent a major challenge. The set of experimentally characterized IDPs is rapidly growing and several experimental approaches sensitive to the intrinsic disorder of a given protein, or its part, have been developed. Ironically, the vast majority of current knowledge about IDPs was accumulated using experimental tools elaborated for the analysis of ordered proteins. The choice of suitable techniques for the characterization of IDPs is based on the experience retrieved from the studies on structure and folding of ordered proteins. In fact, the conclusion about the disordered nature of a given protein is often based on the lack of a specific signal expected to be seen by a given technique if the protein were ordered. This inevitably results in rather frequent misidentifications and misclassifications.
One illustrative example of how dangerous this approach could be is now provided. Size-exclusion chromatography is frequently used for the estimation of protein molecular mass. However, this technique physically separates proteins by their hydrodynamic volumes, rather than by masses. The hydrodynamic volume of a polypeptide chain depends on its folding degree. Therefore, nonfolded IDPs, with their increased hydrodynamic volumes relative to volumes of ordered proteins of similar molecular masses, are characterized by increased apparent molecular masses, as evaluated by gel filtration. For example, for a 100-residue-long polypeptide with a real molecular mass of 11 kDa, the apparent molecular mass of a molten globular conformation estimated from the hydrodynamic data (e.g., gel filtration) is 16 kDa; for the native premolten globule, it is 29 kDa, whereas, for the native coil, the gel-filtration column will be approximately 40 kDa. For a 500 residue-long IDP with a real molecular mass of 55 kDa these numbers are approximately 90, 180 and 375 kDa if the protein is a native molten globule, a native premolten globule or a native coil, respectively [7]. Obviously, the use of the gel filtration as a tool for the evaluation of protein molecular masses will produce erroneous conclusions regarding the existence of the specific oligomers for monomeric proteins, with different degrees of disorderedness. Therefore, extreme caution should be taken when interpreting the experimental data for IDPs.
Computational analysis of IDPs
The development of computational tools for disorder prediction is based on the hypothesis that the absence of rigid structure in the IDPs is encoded in the peculiarities of their amino acid sequences. First predictors have already achieved accuracies slightly over 70%. The fact that the predictors’ accuracy was significantly higher than expected by chance suggested that the information for failure to fold into a 3D structure is, indeed, somehow encoded within the amino acid sequence. Over the years, the prediction accuracy has continued to increase, and recently several machine-learning algorithms for predicting structure and disorder have achieved accuracies exceeding 80% [209]. Taking into account the numerous efforts made over the past decade to develop predictors of intrinsic disorder (currently, there are more than 50 specialized computational tools for disorder prediction [49]), the gain of a mere 10% in the accuracy of disorder prediction seems to be a moderate success.
However, it is very likely that the accuracy of ‘general’ disorder prediction has natural limits. These limits are only partly determined by the obvious ‘noise’ in the datasets used for training of disorder predictors; another reason is that intrinsic disorder is a highly heterogeneous phenomenon and, structurally IDPs/IDRs can range from relatively ordered collapsed molten globule-like species to highly disordered, coil-like or premolten globule-like conformations [4-7]. Furthermore, research shows that IDRs and IDPs are very sensitive to changes in amino acid sequences and to the peculiarities of their environment [210]. On the other hand, the environmental conditions for living organisms vary naturally, from highly acidic to highly alkaline, and from cold to hot. They also can be characterized by extremely high salinity or by extremely high hydrostatic pressure. All this could have profound effects on protein structure, dynamics and function and, therefore, can influence the accuracy of disorder prediction. Finally, some IDPs/IDRs never fold, and their disordered ensembles are the biologically active states, while other IDPs/IDRs undergo function-related structural transitions. Obviously, all these structural-, functional- and environment-specific peculiarities are also encoded in the amino acid sequences. For example, a recent analysis revealed that the disordered regions of the α-helical bundle integral membrane proteins, those of the β-barrel integral membrane proteins and those of the water-soluble proteins, all exhibit statistically distinct amino acid compositional biases [211]. Another example is foldable IDRs (or MoRFs), which are mostly disordered while free in solution, but are able to gain unique structure when bound to specific partners. These regions where shown to be characterized by specific and identifiable sequence features [35,36,38,212]. However, none of these structural-, functional- or environment-specific sequence features are explicitly considered by current ‘generalized’ disorder predictors, thus affecting their overall accuracy. For example, short disordered regions have distinct sequence biases, and are known to be less accurately predictable than the long regions by current predictors [213,214]. Therefore, the possible future gain in disorder prediction accuracy is potentially associated with the development of specialized predictors utilizing unique environment-, structure- and function-specific features of IDP sequences.
The large number of currently available disorder predictors, with their great variability of prediction ideas, computing techniques and training attributes, makes the choice of a suitable tool a difficult task. The problem is further aggravated by the fact that different predictors often give different (at times, dramatically different) answers for a given protein. One of the possible solutions for this problem is the development of so-called meta-predictors, where the predicted results from a collection of individual predictors are used as the inputs for a meta-predictor. This approach often results in improvement of the prediction accuracy, since the different predictors are orthogonal (i.e., they utilize complementary information arising from different sequence features, different prediction models and different training sets) [215-218].
IDPs in diseases
Application of various bioinformatics tools revealed that IDPs are commonly associated with various diseases. On the other hand, the results of this analysis (Figure 2B) show that there is no significant difference in the abundance of long disordered regions between the normal proteins involved in cell signaling and the pathological proteins responsible for the development of various diseases (e.g., cancer, neurodegenerative diseases, cardiovascular disease and diabetes). This clearly shows that high levels of intrinsic disorder in proteins are not sufficient to cause the development of disease.
The maladies considered in this review have been frequently termed conformational diseases, since they are characterized by the conformational changes, misfolding, and often aggregation, of an underlying protein. One should keep in mind that there is a clear distinction between misfolded and unfolded proteins, which can be illustrated in terms of their energy landscapes. The energy landscape of an unfolded protein (including extended coil-like and pore-molten globule-like IDPs) is characterized by numerous local energy minima, leading to a highly frustrated system without any stable well-folded conformation [39]. This is in great contrast to the funnelled energy landscape of a globular protein, which is characterized by a deep energy minimum and, therefore, resembles a funnel [219,220]. Misfolded protein forms wrong intramolecular interactions and gets kinetically trapped in a low energy minimum. Therefore, misfolding is related to the perturbed protein homeostasis and occurs when a protein fails to keep its intrinsically disordered state (for IDPs), or, for ordered proteins, fails to fold normally (i.e., to achieve its native folded 3D structure). In order to regain the ability to fold normally or to regain native intrinsically disordered state, the misfolded protein has to climb out this low-energy minimum. In other words, it has to unfold (at least partially) to eliminate wrong intramolecular interactions formed during the misfolding process.
To better understand conformational diseases, perturbed protein functionality must also be considered, as the aforementioned high level of intrinsic disorder in pathogenic proteins is a reflection of the involvement of these proteins in some crucial signaling functions. Therefore, the misregulation and misfolding of these proteins can result in their dysfunction, ultimately leading to the development of life-threatening pathological conditions. For example, functions ascribed to α-synuclein, a protein involved in several neurodegenerative diseases, including the binding of fatty acids and metal ions, regulation of certain enzymes, transporters and neurotransmitter vesicles, and regulation of neuronal survival (reviewed in [81,90,91,95]). Furthermore, α-synuclein possesses an amazing structural plasticity, adopting a series of different monomeric and oligomeric conformations, as well as assembling in various insoluble aggregates (reviewed in [90-92,95]). The choice between these conformations is determined by the peculiarities of the protein environment, suggesting that α-synuclein has an exceptional ability to fold in a template-dependent manner [90-92,95]. Based on these and similar observations, it was hypothesized that the development of the conformational diseases may originate from protein dysfunctions associated with misidentification, misregulation and mis-signaling events, and accompanied by the protein misfolding. In other words, mutations and/or changes in the environment may result in protein confusion, reducing its capability to recognize proper binding partners, and leading to the formation of nonfunctional complexes and deadly aggregates [28].
IDPs as drug targets
The modulation of protein–protein interactions of pathogenic proteins is an appealing opportunity for the development of new drugs. However, the vast majority of currently available drugs target the active site of enzymes, presumably since these are the only proteins for which the lock-and-key function paradigm is generally applicable and, therefore, the structure-based rational drug design could be used. The limited success of drug development efforts that are targeted to disrupt specific protein–protein interactions [184] is determined (at least in part) by the failure to recognize the important role of intrinsic disorder in protein function. This means that novel, nonstandard solutions are needed to resolve this problem and to find effective inhibitors of protein–protein interactions. Obviously, structure-based approaches do not work well for objects without fixed structure, but might work for the disordered subjects, which are able to gain folded structure on binding to specific sites located on surface of the ordered interaction partners. Therefore, the successful approach for these cases can utilize the traditional drug discovery modus operandi, where a small molecule acts as a competitive inhibitor that targets the binding site on the structured partner [192-195]. An alternative approach is the ‘medicated dysfunction’, where, to prevent function, a small molecule targets a possible binding site on the IDP, and induces misfolding of this region into the non-native dysfunctional conformation [167,206]. Obviously, these two directions are just the first steps, and some radically new ways of thinking about approaches for the nontraditional inhibition of protein–protein interactions are needed.
The aforementioned advances in understanding of intrinsic disorder phenomenon are necessary prerequisites for the development of new computational and experimental tools, aiming for the structural and functional characterization of IDPs and IDRs, for evaluating the abundance of IDPs/IDRs in various human diseases, and for better understanding of the roles of IDP dysfunctions in the pathogenesis of these diseases. This will create a venue to design novel drugs successfully targeting disorder-based protein–protein interactions.
Five-year view
In the upcoming years, more-sensitive and accurate computational and experimental tools will be developed for finding and characterizing IDPs individually and at the proteome level. New IDP functions will be found, and a better understanding of the interconnection between IDP structure, function, dysfunction and various human diseases will be achieved.
A dramatic increase in the number of experimentally characterized IDPs is expected. These new examples will facilitate the development of novel experimental and computational tools, and their application for subsequent discovery of new members of the IDP family. New tools will be created to analyze the structure and function of IDPs in their natural environments (i.e., inside the living cell).
A better understanding of the relationship between the intracellular environment (e.g., reactive oxygen species levels, cellular confinement and macromolecular crowding) and protein structure and function, will also be achieved. The role of post-transcriptional modifications in the regulation and modulation of structure and function of various IDPs will be analyzed in detail.
Intrinsically disordered proteins will become important and accepted targets for the development of novel drugs aiming at the modulation of protein functions. New small molecules will be discovered, which will not only inhibit the action of IDPs, but will be able to produce a wide range of functional modulation (e.g., enhancing IDP function via stabilizing the functional conformations). Intrinsic disorder will be utilized for the development of drugs that target given protein–protein interactions in a tissue-specific manner, opening a unique opportunity for the highly targeted modulation of the IDP function in a given tissue and not affecting the functionality of this same protein in other tissues.
Key issues.
Intrinsic disorder is highly abundant in nature, especially among the proteins involved in signaling and regulation.
Intrinsically disordered proteins (IDPs) possess numerous unique features at the levels of amino acid composition, sequence and overall structure, which are used for their experimental and computational identification.
Various computational and experimental tools are developed for the identification and characterization of IDPs.
Many disease-related proteins are IDPs.
IDPs represent a set of novel and very promising targets for the development of drugs aiming at the inhibition or modulation of protein-based interactions.
Acknowledgements
This review would not be possible without many colleagues who, over the years, contributed to the introduction and development of the intrinsically disordered protein field. Numerous helpful and thought-promoting discussions with my colleagues and friends are deeply acknowledged. I express my deepest gratitude to Alexey Uversky for careful reading and editing the manuscript.
Footnotes
Financial & competing interests disclosure
This work was supported in part by the Program of the Russian Academy of Sciences for the ‘Molecular and cellular biology’, by grants R01 LM007688-01A1 and GM071714-01A2 from the National Institutes of Health, and by the grant EF 0849803 from the National Science Foundation. I gratefully acknowledge the support of the IUPUI Signature Centers Initiative. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber. Dt. Chem. Ges. 1894;27:2985–2993. [Google Scholar]
- 2.Lemieux UR, Spohr U. How Emil Fischer was led to the lock and key concept for enzyme specificity. Adv. Carbohydrate Chem. Biochem. 1994;50:1–20. [PubMed] [Google Scholar]
- 3.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999;24(9):329–332. doi: 10.1016/s0968-0004(99)01445-0. • Provides a comprehensive overview of the protein-misfolding phenomenon and links misfolding with protein evolution and pathogenesis of conformational diseases.
- 4.Uversky VN. Protein folding revisited. A polypeptide chain at the folding–misfolding–nonfolding cross-roads: which way to go? Cell Mol. Life Sci. 2003;60(9):1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of Protein Folding. Wiley-VCH, Verlag GmbH & Co.; Weinheim, Germany: 2005. pp. 271–353. [Google Scholar]
- 6.Dunker AK, Lawson JD, Brown CJ, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. • Provides a comprehensive overview of intrinsically disordered proteins (IDPs) and their functions.
- 7.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. • Represents a first systematic classification of biological functions of IDPs.
- 9.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 10.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. • Provides the first systematic analysis of the abundance and role of intrinsic disorder in hub proteins.
- 11.Dunker AK, Obradovic Z. The protein trinity – linking function and disorder. Nat. Biotechnol. 2001;19(9):805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 12.Dunker AK, Oldfield CJ, Meng J, et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9(Suppl. 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys. J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008;4(4):229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 16.Vucetic S, Xie H, Iakoucheva LM, et al. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J. Proteome Res. 2007;6(5):1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie H, Vucetic S, Iakoucheva LM, et al. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007;6(5):1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie H, Vucetic S, Iakoucheva LM, et al. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007;6(5):1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog. Biophys. Mol. Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell RB, Gibson TJ. A careful disorderliness in the proteome: sites for interaction and targets for future therapies. FEBS Lett. 2008;582(8):1271–1275. doi: 10.1016/j.febslet.2008.02.027. • Provides analysis of IDPs as potential drug targets.
- 21.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl. 1):S1. doi: 10.1186/1471-2164-9-S1-S1. • Represents a comprehensive analysis of the role of intrinsic disorder in one-to-many and many-to-one binding scenarios.
- 22.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 23.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. • Represents the first comprehensive review of IDPs and suggests re-evaluation of the protein structure–function paradigm.
- 24.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27(10):527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 25.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 27.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 28.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 29.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 31.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl Acad. Sci. USA. 1996;93(21):11504–11509. doi: 10.1073/pnas.93.21.11504. • One of the first systematic studies of structural properties of an IDP.
- 32.Lacy ER, Filippov I, Lewis WS, et al. p27 binds cyclin–CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 2004;11(4):358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 33.Lacy ER, Wang Y, Post J, et al. Molecular basis for the specificity of p27 toward cyclin-dependent kinases that regulate cell division. J. Mol. Biol. 2005;349(4):764–773. doi: 10.1016/j.jmb.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12(1):54–60. doi: 10.1016/s0959-440x(02)00289-0. • Represents a systematic overview of the coupled folding and binding of IDPs.
- 35.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry. 2005;44(37):12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining α-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46(47):13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohan A. The School of Informatics. Indiana University; IN, USA: 2006. MoRFs: a dataset of molecular recognition features; p. 59. [Google Scholar]
- 38.Vacic V, Oldfield CJ, Mohan A, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. • Introduces the D2 concept to emphasize the abundance and role of intrinsic disorder in human diseases.
- 40.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 41.Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65(1):1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 42.Dosztanyi Z, Sandor M, Tompa P, Simon I. Prediction of protein disorder at the domain level. Curr. Protein Pept. Sci. 2007;8(2):161–171. doi: 10.2174/138920307780363406. [DOI] [PubMed] [Google Scholar]
- 43.Dosztanyi Z, Tompa P. Prediction of protein disorder. Methods Mol. Biol. 2008;426:103–115. doi: 10.1007/978-1-60327-058-8_6. [DOI] [PubMed] [Google Scholar]
- 44.Gast K, Damaschun H, Eckert K, et al. Prothymosin α: a biologically active protein with random coil conformation. Biochemistry. 1995;34(40):13211–13218. doi: 10.1021/bi00040a037. [DOI] [PubMed] [Google Scholar]
- 45.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35(43):13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 46.Hemmings HC, Jr, Nairn AC, Aswad DW, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Purification and characterization of the phosphoprotein from bovine caudate nucleus. J. Neurosci. 1984;4(1):99–110. doi: 10.1523/JNEUROSCI.04-01-00099.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams RM, Obradovi Z, Mathura V, et al. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac. Symp. Biocomput. 2001:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- 48.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 49.He B, Wang K, Liu Y-L, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19(8):929–949. doi: 10.1038/cr.2009.87. • Provides a comprehensive overview of computational tools for prediction of intrinsic disorder in proteins.
- 50.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 51.Hubbard SJ, Beynon RJ, Thornton JM. Assessment of conformational parameters as predictors of limited proteolytic sites in native protein structures. Protein Eng. 1998;11(5):349–359. doi: 10.1093/protein/11.5.349. [DOI] [PubMed] [Google Scholar]
- 52.Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10(7):1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reeves R, Nissen MS. Purification and assays for high mobility group HMG-I(Y) protein function. Methods Enzymol. 1999;304:155–188. doi: 10.1016/s0076-6879(99)04011-2. [DOI] [PubMed] [Google Scholar]
- 54.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62(1):24–45. doi: 10.1002/prot.20750. • Represents a comprehensive overview of experimental techniques used for structural analysis of intrinsically disordered proteins and conformational transitions related to their functioning.
- 55.Uversky VN. What does it mean to be natively unfolded? Eur. J. Biochem. 2002;269(1):2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 56.Uversky VN. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28(7-8):305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 57.Cortese MS, Baird JP, Uversky VN, Dunker AK. Uncovering the unfoldome: enriching cell extracts for unstructured proteins by acid treatment. J. Proteome Res. 2005;4(5):1610–1618. doi: 10.1021/pr050119c. [DOI] [PubMed] [Google Scholar]
- 58.Csizmok V, Szollosi E, Friedrich P, Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol. Cell Proteomics. 2006;5(2):265–273. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Galea CA, Pagala VR, Obenauer JC, Park CG, Slaughter CA, Kriwacki RW. Proteomic studies of the intrinsically unstructured mammalian proteome. J. Proteome Res. 2006;5(10):2839–2848. doi: 10.1021/pr060328c. [DOI] [PubMed] [Google Scholar]
- 60.Uversky VN, Roman A, Oldfield CJ, Dunker AK. Protein intrinsic disorder and human papillomaviruses: increased amount of disorder in E6 and E7 oncoproteins from high risk HPVs. J. Proteome Res. 2006;5(8):1829–1842. doi: 10.1021/pr0602388. [DOI] [PubMed] [Google Scholar]
- 61.Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45(35):10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- 62.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics. 2009;10(Suppl. 1):S12. doi: 10.1186/1471-2164-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uversky VN, Oldfield CJ, Midic U, et al. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;10(Suppl. 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 65.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc. Natl Acad. Sci. USA. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 67.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698(2):131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 1998;8(1):101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 69.Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des. 1998;3(1):R9–23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 70.Bellotti V, Mangione P, Stoppini M. Biological activity and pathological implications of misfolded proteins. Cell Mol. Life Sci. 1999;55(6–7):977–991. doi: 10.1007/s000180050348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. I. Systemic amyloidoses. Med. Sci. Monitor. 1999;5:1001–1012. [Google Scholar]
- 72.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. II. Localized amyloidosis and neurodegenerative disordres. Med. Sci. Monitor. 1999;5:1238–1254. [Google Scholar]
- 73.Lansbury PT., Jr. Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc. Natl Acad. Sci. USA. 1999;96(7):3342–3344. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rochet JC, Lansbury PT., Jr. Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol. 2000;10(1):60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 75.Dobson CM. The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356(1406):133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zerovnik E. Amyloid–fibril formation. Proposed mechanisms and relevance to conformational disease. Eur. J. Biochem. 2002;269(14):3362–3371. doi: 10.1046/j.1432-1033.2002.03024.x. [DOI] [PubMed] [Google Scholar]
- 77.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120(3):885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 78.Ueda K, Fukushima H, Masliah E, et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl Acad. Sci. USA. 1993;90(23):11282–11286. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. A68: a major subunit of paired helical filaments and derivatized forms of normal tau. Science. 1991;251(4994):675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 80.Wisniewski KE, Dalton AJ, McLachlan C, Wen GY, Wisniewski HM. Alzheimer’s disease in Down’s syndrome: clinicopathologic studies. Neurology. 1985;35(7):957–961. doi: 10.1212/wnl.35.7.957. [DOI] [PubMed] [Google Scholar]
- 81.Dev KK, Hofele K, Barbieri S, Buchman VL, van der Putten H. Part II: α-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacology. 2003;45(1):14–44. doi: 10.1016/s0028-3908(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 82.Prusiner SB. Shattuck lecture – neurodegenerative diseases and prions. N. Engl. J. Med. 2001;344(20):1516–1526. doi: 10.1056/NEJM200105173442006. [DOI] [PubMed] [Google Scholar]
- 83.Zoghbi HY, Orr HT. Polyglutamine diseases: protein cleavage and aggregation. Curr. Opin. Neurobiol. 1999;9(5):566–570. doi: 10.1016/S0959-4388(99)00013-6. [DOI] [PubMed] [Google Scholar]
- 84.Valentine JS, Doucette PA, Zittin Potter S. Copper–zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 85.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol. 2010;6(4):211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sickmeier M, Hamilton JA, LeGall T, et al. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Williams RM, Obradovic Z, Mathura V, et al. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac. Symp. Biocomput. 2001:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- 88.Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 90.Uversky VN. α-synuclein misfolding and neurodegenerative diseases. Curr. Protein Pept. Sci. 2008;9(5):507–540. doi: 10.2174/138920308785915218. [DOI] [PubMed] [Google Scholar]
- 91.Uversky VN. Neuropathology, biochemistry and biophysics of α-synuclein aggregation. J. Neurochem. 2007;103(1):17–37. doi: 10.1111/j.1471-4159.2007.04764.x. [DOI] [PubMed] [Google Scholar]
- 92.Uversky VN. A protein-chameleon: conformational plasticity of α-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn. 2003;21(2):211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 93.Morar AS, Olteanu A, Young GB, Pielak GJ. Solvent-induced collapse of α-synuclein and acid-denatured cytochrome c. Protein Sci. 2001;10(11):2195–2199. doi: 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307(4):1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 95.Uversky VN, Eliezer D. Biophysics of Parkinson’s disease: structure and aggregation of α-synuclein. Curr. Protein Pept. Sci. 2009;10(5):483–499. doi: 10.2174/138920309789351921. • Represents a systematic analysis of structural properties of α-synuclein.
- 96.Clark CM, Ewbank D, Lee VM-Y, Trojanowski JQ. Molecular pathology of Alzheimer’s disease: neuronal cytoskeletal abnormalities. In: Growdon JH, Rossor MN, editors. The Dementias. Vol. 19 of Blue Books of Practical Neurology. Butterworth–Heinemann; MA, USA: 1998. pp. 285–304. [Google Scholar]
- 97.Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J. Mol. Biol. 2001;312(5):1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 98.von Bergen M, Barghorn S, Jeganathan S, Mandelkow EM, Mandelkow E. Spectroscopic approaches to the conformation of tau protein in solution and in paired helical filaments. Neurodegener. Dis. 2006;3(4–5):197–206. doi: 10.1159/000095257. [DOI] [PubMed] [Google Scholar]
- 99.Chirita CN, Congdon EE, Yin H, Kuret J. Triggers of full-length tau aggregation: a role for partially folded intermediates. Biochemistry. 2005;44(15):5862–5872. doi: 10.1021/bi0500123. [DOI] [PubMed] [Google Scholar]
- 100.Aronoff-Spencer E, Burns CS, Avdievich NI, et al. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39(45):13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413(2):282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 102.Scott M, Groth D, Foster D, et al. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73(5):979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 103.Telling GC, Scott M, Mastrianni J, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83(1):79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 104.Donne DG, Viles JH, Groth D, et al. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl Acad. Sci. USA. 1997;94(25):13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.James TL, Liu H, Ulyanov NB, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl Acad. Sci. USA. 1997;94(19):10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93(3):337–348. doi: 10.1016/s0092-8674(00)81163-0. • Provides a nice overview of prion protein biology.
- 107.Cummings CJ, Zoghbi HY. Trinucleotide repeats: mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. • Provides a comprehensive review of the role of glutamine repeats in the development of inherited neurodegenerative diseases.
- 108.Cummings CJ, Zoghbi HY. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 2000;9(6):909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 109.Perutz MF. Glutamine repeats and inherited neurodegenerative diseases: molecular aspects. Curr. Opin. Struct. Biol. 1996;6(6):848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 110.Ross CA, Wood JD, Schilling G, et al. Polyglutamine pathogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999;354(1386):1005–1011. doi: 10.1098/rstb.1999.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Preisinger E, Jordan BM, Kazantsev A, Housman D. Evidence for a recruitment and sequestration mechanism in Huntington’s disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999;354(1386):1029–1034. doi: 10.1098/rstb.1999.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wanker EE. Protein aggregation and pathogenesis of Huntington’s disease: mechanisms and correlations. Biol. Chem. 2000;381(9–10):937–942. doi: 10.1515/BC.2000.114. [DOI] [PubMed] [Google Scholar]
- 113.McCampbell A, Taylor JP, Taye AA, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 2000;9(14):2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 114.McCampbell A, Fischbeck KH. Polyglutamine and CBP: fatal attraction? Nat. Med. 2001;7(5):528–530. doi: 10.1038/87842. [DOI] [PubMed] [Google Scholar]
- 115.Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 2001;311(1):173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 116.Chen S, Berthelier V, Hamilton JB, O’Nuallain B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry. 2002;41(23):7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- 117.Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid β-peptide of amyloid plaques. Proc. Natl Acad. Sci. USA. 2002;99(8):5596–5600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lavery DN, McEwan IJ. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry. 2008;47(11):3360–3369. doi: 10.1021/bi702221e. • Represents a comprehensive analysis of structural properties of human androgen receptor.
- 119.Yazawa I, Nukina N, Hashida H, Goto J, Yamada M, Kanazawa I. Abnormal gene product identified in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) brain. Nat. Genet. 1995;10(1):99–103. doi: 10.1038/ng0595-99. [DOI] [PubMed] [Google Scholar]
- 120.Albrecht M, Golatta M, Wullner U, Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur. J. Biochem. 2004;271(15):3155–3170. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- 121.Masino L, Musi V, Menon RP, et al. Domain architecture of the polyglutamine protein ataxin-3: a globular domain followed by a flexible tail. FEBS Lett. 2003;549(1–3):21–25. doi: 10.1016/s0014-5793(03)00748-8. [DOI] [PubMed] [Google Scholar]
- 122.Palhan VB, Chen S, Peng GH, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl Acad. Sci. USA. 2005;102(24):8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Friedman MJ, Wang CE, Li XJ, Li S. polyglutamine expansion reduces the association of TATA-binding protein with DNA and induces DNA binding-independent neurotoxicity. J Biol. Chem. 2008;283(13):8283–8290. doi: 10.1074/jbc.M709674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Plant GT, Revesz T, Barnard RO, Harding AE, Gautier-Smith PC. Familial cerebral amyloid angiopathy with nonneuritic amyloid plaque formation. Brain. 1990;113(Pt 3):721–747. doi: 10.1093/brain/113.3.721. [DOI] [PubMed] [Google Scholar]
- 125.Vidal R, Revesz T, Rostagno A, et al. A decamer duplication in the 3′ region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred. Proc. Natl Acad. Sci. USA. 2000;97(9):4920–4925. doi: 10.1073/pnas.080076097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Srinivasan R, Jones EM, Liu K, Ghiso J, Marchant RE, Zagorski MG. pH-dependent amyloid and protofibril formation by the ABri peptide of familial British dementia. J. Mol. Biol. 2003;333(5):1003–1023. doi: 10.1016/j.jmb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 127.Surolia I, Reddy GB, Sinha S. Hierarchy and the mechanism of fibril formation in ADan peptides. J. Neurochem. 2006;99(2):537–548. doi: 10.1111/j.1471-4159.2006.04072.x. [DOI] [PubMed] [Google Scholar]
- 128.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 129.Durazo A, Shaw BF, Chattopadhyay M, et al. Metal-free superoxide dismutase-1 and three different amyotrophic lateral sclerosis variants share a similar partially unfolded beta-barrel at physiological temperature. J. Biol. Chem. 2009;284(49):34382–34389. doi: 10.1074/jbc.M109.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 132.Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351(3):602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 133.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 134.Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83(1):130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 135.Ayala YM, Zago P, D’Ambrogio A, et al. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 2008;121(Pt 22):3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 136.Sleegers K, Cruts M, Van Broeckhoven C. Molecular pathways of frontotemporal lobar degeneration. Annu. Rev. Neurosci. 2010 doi: 10.1146/annurev-neuro-060909-153144. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 137.Tolkatchev D, Malik S, Vinogradova A, et al. Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci. 2008;17(4):711–724. doi: 10.1110/ps.073295308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van Blitterswijk M, Landers JE. RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics. 2010 doi: 10.1007/s10048-010-0239-4. DOI: 10.1007/s10048-010-0239-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 139.Lequin O, Thuring H, Robin M, Lallemand JY. Three-dimensional solution structure of human angiogenin determined by 1H,15N-NMR spectroscopy – characterization of histidine protonation states and pKa values. Eur. J. Biochem. 1997;250(3):712–726. doi: 10.1111/j.1432-1033.1997.00712.x. [DOI] [PubMed] [Google Scholar]
- 140.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 141.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo–cytoplasmic shuttling. J. Cell Sci. 1997;110(Pt 15):1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 143.Chen YZ, Bennett CL, Huynh HM, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74(6):1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Corcia P, Camu W, Halimi JM, et al. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology. 2006;67(7):1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- 145.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 146.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15(14):3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 147.Hayashi M, Imanaka-Yoshida K, Yoshida T, et al. A crucial role of mitochondrial Hsp40 in preventing dilated cardiomyopathy. Nat. Med. 2006;12(1):128–132. doi: 10.1038/nm1327. [DOI] [PubMed] [Google Scholar]
- 148.Selzer RR, Nyaga S, Tuo J, et al. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res. 2002;30(3):782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kerr DA, Nery JP, Traystman RJ, Chau BN, Hardwick JM. Survival motor neuron protein modulates neuron-specific apoptosis. Proc. Natl Acad. Sci. USA. 2000;97(24):13312–13317. doi: 10.1073/pnas.230364197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Garcia W, de Araujo AP, Neto Mde O, et al. Dissection of a human septin: definition and characterization of distinct domains within human SEPT4. Biochemistry. 2006;45(46):13918–13931. doi: 10.1021/bi061549z. [DOI] [PubMed] [Google Scholar]
- 151.Paoletti F, Konarev PV, Covaceuszach S, et al. Structural and functional properties of mouse proNGF. Biochem. Soc. Trans. 2006;34(Pt 4):605–606. doi: 10.1042/BST0340605. [DOI] [PubMed] [Google Scholar]
- 152.Isobe M, Emanuel BS, Givol D, Oren M, Croce CM. Localization of gene for human p53 tumour antigen to band 17p13. Nature. 1986;320(6057):84–85. doi: 10.1038/320084a0. [DOI] [PubMed] [Google Scholar]
- 153.Anderson CW, Appella E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and nongenotoxic stress. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. Academic Press; NY, USA: 2004. pp. 237–247. • Represents a comprehensive overview of the p53 tumor-suppressor functions.
- 154.Zhao R, Gish K, Murphy M, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14(8):981–993. [PMC free article] [PubMed] [Google Scholar]
- 155.Read AP, Strachan T. Human Molecular Genetics 2. Wiley; NY, USA: 1999. [Google Scholar]
- 156.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 157.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. • Nice review of the structural biology of the tumor suppressor p53.
- 158.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53 and cancer-associated mutants. Adv. Cancer Res. 2007;97:1–23. doi: 10.1016/S0065-230X(06)97001-8. [DOI] [PubMed] [Google Scholar]
- 159.Joerger AC, Fersht AR. Structure–function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26(15):2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 160.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 161.Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat. Rev. Cancer. 2009;9(10):758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 162.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Peters M, DeLuca C, Hirao A, et al. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc. Natl Acad. Sci. USA. 2002;99(17):11305–11310. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14(6):1129–1138. doi: 10.1038/sj.cdd.4402115. [DOI] [PubMed] [Google Scholar]
- 165.Corcoran CA, Huang Y, Sheikh MS. The regulation of energy generating metabolic pathways by p53. Cancer Biol. Ther. 2006;5(12):1610–1613. doi: 10.4161/cbt.5.12.3617. [DOI] [PubMed] [Google Scholar]
- 166.Zou Z, Gao C, Nagaich AK, et al. p53 regulates the expression of the tumor suppressor gene maspin. J. Biol. Chem. 2000;275(9):6051–6054. doi: 10.1074/jbc.275.9.6051. [DOI] [PubMed] [Google Scholar]
- 167.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009;131(21):7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 168.Torphy TJ. Phosphodiesterase isozymes: molecular targets for novel antiasthma agents. Am. J. Respir. Crit. Care Med. 1998;157(2):351–370. doi: 10.1164/ajrccm.157.2.9708012. [DOI] [PubMed] [Google Scholar]
- 169.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35(2):131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 170.Kovala T, Sanwal BD, Ball EH. Recombinant expression of a type IV, cAMP-specific phosphodiesterase: characterization and structure-function studies of deletion mutants. Biochemistry. 1997;36(10):2968–2976. doi: 10.1021/bi9613483. [DOI] [PubMed] [Google Scholar]
- 171.Beard MB, O’Connell JC, Bolger GB, Houslay MD. The unique N-terminal domain of the cAMP phosphodiesterase PDE4D4 allows for interaction with specific SH3 domains. FEBS Lett. 1999;460(1):173–177. doi: 10.1016/s0014-5793(99)01335-6. [DOI] [PubMed] [Google Scholar]
- 172.White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell. Biochem. 1998;182(1–2):3–11. [PubMed] [Google Scholar]
- 173.Pessin JE, Saltiel AR. Signaling pathways in insulin action: molecular targets of insulin resistance. J. Clin. Invest. 2000;106(2):165–169. doi: 10.1172/JCI10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 175.Myers MG, Jr, Backer JM, Sun XJ, et al. IRS-1 activates phosphatidylinositol 3′-kinase by associating with src homology 2 domains of p85. Proc. Natl Acad. Sci. USA. 1992;89(21):10350–10354. doi: 10.1073/pnas.89.21.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alessi DR, James SR, Downes CP, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 1997;7(4):261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 177.Saltiel AR. New perspectives into the molecular pathogenesis and treatment of Type 2 diabetes. Cell. 2001;104(4):517–529. doi: 10.1016/s0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 178.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell. 1998;2(6):887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 179.Wada T, Sasaoka T, Ishiki M, et al. Role of the Src homology 2 (SH2) domain and C-terminus tyrosine phosphorylation sites of SH2-containing inositol phosphatase (SHIP) in the regulation of insulin-induced mitogenesis. Endocrinology. 1999;140(10):4585–4594. doi: 10.1210/endo.140.10.7028. [DOI] [PubMed] [Google Scholar]
- 180.Clement S, Krause U, Desmedt F, et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409(6816):92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 181.Elchebly M, Payette P, Michaliszyn E, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283(5407):1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 182.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat. Struct. Biol. 2001;8(12):1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 183.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7(9):1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cochran AG. Antagonists of protein–protein interactions. Chem. Biol. 2000;7(4):R85–R94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 185.Rodi DJ, Agoston GE, Manon R, Lapcevich R, Green SJ, Makowski L. Identification of small molecule binding sites within proteins using phage display technology. Comb. Chem. High Throughput Screen. 2001;4(7):553–572. doi: 10.2174/1386207013330779. [DOI] [PubMed] [Google Scholar]
- 186.Blundell TL, Burke DF, Chirgadze D, et al. Protein–protein interactions in receptor activation and intracellular signalling. Biol. Chem. 2000;381(9–10):955–959. doi: 10.1515/BC.2000.117. [DOI] [PubMed] [Google Scholar]
- 187.Garner E, Romero P, Dunker AK, Brown C, Obradovic Z. Predicting binding regions within disordered proteins. Genome Inform. Ser. Workshop Genome Inform. 1999;10:41–50. [PubMed] [Google Scholar]
- 188.Bottger A, Bottger V, Sparks A, Liu WL, Howard SF, Lane DP. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol. 1997;7(11):860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 189.Wasylyk C, Salvi R, Argentini M, et al. p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene. 1999;18(11):1921–1934. doi: 10.1038/sj.onc.1202528. [DOI] [PubMed] [Google Scholar]
- 190.Chene P, Fuchs J, Bohn J, Garcia-Echeverria C, Furet P, Fabbro D. A small synthetic peptide, which inhibits the p53–hdm2 interaction, stimulates the p53 pathway in tumour cell lines. J. Mol. Biol. 2000;299(1):245–253. doi: 10.1006/jmbi.2000.3738. [DOI] [PubMed] [Google Scholar]
- 191.Garcia-Echeverria C, Chene P, Blommers MJ, Furet P. Discovery of potent antagonists of the interaction between human double minute 2 and tumor suppressor p53. J. Med. Chem. 2000;43(17):3205–3208. doi: 10.1021/jm990966p. [DOI] [PubMed] [Google Scholar]
- 192.Chene P. Inhibition of the p53–MDM2 interaction: targeting a protein–protein interface. Mol. Cancer Res. 2004;2(1):20–28. [PubMed] [Google Scholar]
- 193.Klein C, Vassilev LT. Targeting the p53–MDM2 interaction to treat cancer. Br. J. Cancer. 2004;91(8):1415–1419. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vassilev LT. Small-molecule antagonists of p53–MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3(4):419–421. • Represents an overview of small-molecule antagonists of p53-MDM2 binding as potential drugs.
- 195.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 196.Cheng Y, LeGall T, Oldfield CJ, et al. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24(10):435–442. doi: 10.1016/j.tibtech.2006.07.005. • Describes a new intrinsic disorder-based approach in drug disovery – rational drug design via IDP.
- 197.Pescarolo MP, Bagnasco L, Malacarne D, et al. A retro-inverso peptide homologous to helix 1 of c-Myc is a potent and specific inhibitor of proliferation in different cellular systems. FASEB J. 2001;15(1):31–33. doi: 10.1096/fj.00-0422fje. [DOI] [PubMed] [Google Scholar]
- 198.Berg T, Cohen SB, Desharnais J, et al. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl Acad. Sci. USA. 2002;99(6):3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc–Max interaction and function. Oncogene. 2003;22(40):6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 200.Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem. Biol. 2006;13(7):745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 201.Mo H, Henriksson M. Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc. Natl Acad. Sci. USA. 2006;103(16):6344–6349. doi: 10.1073/pnas.0601418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. A credit-card library approach for disrupting protein–protein interactions. Bioorg. Med. Chem. 2006;14(8):2660–2673. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 203.Bagnasco L, Tortolina L, Biasotti B, et al. Inhibition of a protein–protein interaction between INI1 and c-Myc by small peptidomimetic molecules inspired by helix-1 of c-Myc: identification of a new target of potential antineoplastic interest. FASEB J. 2007;21(4):1256–1263. doi: 10.1096/fj.06-7082com. [DOI] [PubMed] [Google Scholar]
- 204.Wang H, Hammoudeh DI, Follis AV, et al. Improved low molecular weight Myc–Max inhibitors. Mol Cancer Ther. 2007;6(9):2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]
- 205.Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg. Med. Chem. Lett. 2009;19(3):807–810. doi: 10.1016/j.bmcl.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 206.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 2008;15(11):1149–1155. doi: 10.1016/j.chembiol.2008.09.011. • Describes molecular mechanisms of the small molecules c-Myc–Max inhibitors.
- 207.Mustata G, Follis AV, Hammoudeh DI, et al. Discovery of novel Myc–Max heterodimer disruptors with a three-dimensional pharmacophore model. J. Med. Chem. 2009;52(5):1247–1250. doi: 10.1021/jm801278g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Romero PR, Zaidi S, Fang YY, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Noivirt-Brik O, Prilusky J, Sussman JL. Assessment of disorder predictions in CASP8. Proteins. 2009;77(Suppl. 9):210–216. doi: 10.1002/prot.22586. [DOI] [PubMed] [Google Scholar]
- 210.Mohan A, Uversky VN, Radivojac P. Influence of sequence changes and environment on intrinsically disordered proteins. PLoS Comput. Biol. 2009;5(9):e1000497. doi: 10.1371/journal.pcbi.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xue B, Li L, Meroueh SO, Uversky VN, Dunker AK. Analysis of structured and intrinsically disordered regions of transmembrane proteins. Mol. Biosyst. 2009;5(12):1688–1702. doi: 10.1039/B905913J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mohan A, Oldfield CJ, Radivojac P, et al. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362(5):1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 213.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Radivojac P, Obradovic Z, Smith DK, et al. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13(1):71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24(11):1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 216.Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved disorder prediction by combination of orthogonal approaches. PLoS ONE. 2009;4(2):e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all: consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009;583(9):1469–1474. doi: 10.1016/j.febslet.2009.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence–structure relationship. Proc. Natl Acad. Sci. USA. 1992;89(18):8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dill KA, Chan HS. From levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4(1):10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 221.Uversky VN. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010;2010:568068. doi: 10.1155/2010/568068. [DOI] [PMC free article] [PubMed] [Google Scholar]