Summary
Nucleotide binding-leucine rich repeat (NB-LRR) proteins function as intracellular receptors for the detection of pathogens in both plants and animals. Despite their central role in innate immunity, the molecular mechanisms that govern NB-LRR activation are poorly understood. The Arabidopsis NB-LRR protein RPS5 detects the presence of the Pseudomonas syringae effector protein AvrPphB by monitoring the status of the Arabidopsis protein kinase PBS1. AvrPphB is a cysteine protease that targets PBS1 for cleavage at a single site within the activation loop of PBS1. Using a transient expression system in the plant Nicotiana benthamiana and stable transgenic Arabidopsis plants we found that both PBS1 cleavage products are required to activate RPS5 and can do so in the absence of AvrPphB. We also found, however, that the requirement for cleavage of PBS1 could be bypassed simply by inserting five amino acids at the PBS1 cleavage site, which is located at the apex of the activation loop of PBS1. Activation of RPS5 did not require PBS1kinase function, thus RPS5 appears to sense a subtle conformational change in PBS1, rather than cleavage. This finding suggests that NB-LRR proteins may function as fine-tuned sensors of alterations in the structures of effector targets.
Introduction
Plants employ a two-tiered immune system to detect microbial pathogens and mount appropriate responses (Dodds et al., 2010). The first tier is comprised of transmembrane pattern recognition receptors (PRRs) that detect conserved pathogen- (or microbe-) associated molecular patterns (PAMPs/MAMPs). Upon PAMP perception, the PRRs activate defense responses including an increase in extracellular pH, release of reactive oxygen species (ROS), callose deposition, changes in hormone levels, and activation of signal transduction pathways. In plants this is known as basal resistance, or PAMP-triggered immunity (PTI) (Jones et al., 2006). To be successful, pathogens must suppress or evade the PTI system. Suppression of PTI by gram-negative bacterial pathogens requires a suite of effector proteins delivered by the Type III secretion system into the cytoplasm of the plant cell (Cunnac et al., 2009). These effectors target a diverse array of host proteins and signal transduction pathways, often interfering with PTI by modulating key regulators of PTI resistance pathways. Plants, in turn, employ Resistance (R) proteins to detect pathogen effectors, a phenomenon that is termed effector-triggered immunity (ETI) and represents the second tier of the plant immune system (Chisholm et al., 2006, Jones et al., 2006). Upon effector detection, R proteins initiate several responses including ROS production and activation of signal transduction pathways, but the hallmark of R protein-triggered immunity is the hypersensitive response (HR), a type of programmed cell death localized to the site of pathogen detection (Greenberg, 1997).
The majority of plant R proteins are variations on a common architecture consisting of carboxy-terminal leucine-rich repeats (LRRs), a central nucleotide binding site (NB) domain, and a variable amino-terminal domain. NB-LRR proteins can be subdivided into two groups, the TIR-NB-LRR proteins, which contain sequences shared among Toll, interleukin receptor, and R-proteins (TIR) at their N-terminus, or the non-TIR-NB-LRR proteins, which most commonly contain coiled-coil motifs at their N-terminus. NB-LRR proteins detect pathogen effector molecules either directly, by interacting with the effector molecules, or indirectly, by monitoring the functional activity of pathogen effectors (DeYoung et al., 2006).
For direct interactors, the LRR domain appears to play a key role in recognizing the pathogen effector as this domain has been shown to interact with effector proteins in yeast two-hybrid experiments and the LRR domain of several NB-LRR proteins have been shown to be under diversifying selection (DeYoung et al., 2006, Dodds et al., 2006, Noel et al., 1999, McDowell et al., 1998, Meyers et al., 1998). NB-LRR proteins that utilize indirect detection mechanisms monitor host proteins targeted by the pathogen effector. For example, the Arabidopsis proteins RPM1 and RPS2 detect the presence of the Pseudomonas syringae effector proteins AvrRpm1, AvrB, and AvrRpt2 by monitoring RIN4, the presumed virulence target of these effectors. AvrRpm1 and AvrB interact with RIN4 promoting its phosphorylation, which results in activation of RPM1-mediated defense responses (Mackey et al., 2002, Chung et al., 2011, Liu et al., 2011). AvrRpt2 uses cysteine protease activity to cleave RIN4 and RIN4 disappearance results in RPS2 activation, initiating defense responses (Axtell et al., 2003a, Axtell et al., 2003b, Mackey et al., 2003). Likewise, the Arabidopsis NB-LRR protein RPS5 detects the presence of the P. syringae effector AvrPphB by associating with and monitoring the status of the protein kinase PBS1, a target of AvrPphB (Ade et al., 2007, Shao et al., 2003, Shao et al., 2002). AvrPphB is a cysteine protease and upon entering the plant cell, cleaves PBS1 at a single position within the activation loop of PBS1. PBS1 cleavage results in activation of RPS5, triggering defense responses including the HR.
PBS1 belongs to a subfamily of receptor-like cytoplasmic kinases (RLCK family VII), of which there are 45 in Arabidopsis. Nine of these PBS1-like proteins have recently been shown to be substrates of AvrPphB, including BIK1 and PBL1 (Zhang et al., 2010). Both BIK1 and PBL1 can physically associate with the flagellin receptor, FLS2, and AvrPphB inhibits defense responses activated by FLS2, as well as defenses induced by the PAMPs EfTu and chitin (Zhang et al., 2010). These data indicate that this family of kinases plays a general role in PAMP signaling via interaction with PAMP receptors. However, pbs1 null mutants are only slightly reduced in PAMP responses, suggesting that there may be some redundancy in function between PBS1, BIK1, and/or PBL1 in terms of PAMP signaling (Zhang et al., 2010).
For NB-LRR proteins that detect pathogen effectors indirectly, the N-terminal domain appears to be important for interacting with the effector target, while the LRR domain may play a role in negatively regulating R-protein activity (Ade et al., 2007, Mackey et al., 2002, Mucyn et al., 2006, Rairdan et al., 2006, Moffett et al., 2002, Tao et al., 2000, Weaver et al., 2006, Bendahmane et al., 2002). The amino-terminal coiled-coil domains of both RPM1 and RPS5 are known to interact with the effector targets that they monitor, RIN4 and PBS1, respectively (Ade et al., 2007, Mackey et al., 2002). Furthermore, Pto, a tomato protein that binds to two pathogen effectors, AvrPto and AvrPtoB, interacts with the amino-terminus of the NB-LRR protein Prf, activating defense signaling upon effector detection (Mucyn et al., 2006). Similarly, the amino-terminal TIR domain of the tobacco N protein binds to the N receptor-interacting protein (NRIP1), which in turn interacts with the tobacco mosaic virus 50 kDa helicase protein that triggers N-mediated resistance (Caplan et al., 2008).
Our previous studies have described the cleavage of PBS1 by AvrPphB and the complex intramolecular interactions that regulate RPS5 function (Ade et al., 2007, Shao et al., 2003). Cleavage of PBS1 by AvrPphB is required for activation of RPS5, but how these cleavage products induce RPS5 activity is not understood. Although the site of PBS1 cleavage by AvrPphB is known and the cleavage products can be detected in planta, the requirement for the individual cleavage products themselves has not been determined. Furthermore, the mechanism by which NB-LRR proteins are activated is not well characterized. Here we show that both PBS1 cleavage products are required for activation of RPS5. Furthermore, we characterize the domains of PBS1 that are required for cleavage by AvrPphB and activation of RPS5. Finally, we show that the requirement for cleavage of PBS1 can be bypassed by insertion of 5 alanine residues at the cleavage site. Collectively, these data indicate that RPS5 is activated by a conformational change in the kinase domain of PBS1.
Results
Both amino- and carboxy-terminal PBS1 cleavage products are required for RPS5 activation
To assess the role of the individual PBS1 cleavage products in the activation of RPS5, we designed constructs to express the two halves of PBS1 corresponding to the AvrPphB-induced cleavage products (amino acids 1-243 and amino acids 244-456). Each half was fused to a three-copy human influenza hemagglutinin (3xHA) epitope tag to facilitate detection by immunoblotting. These tags were placed on the N-terminal and C-terminal ends of PBS1 to avoid altering the amino acid sequence at the AvrPphB cleavage site. To avoid disturbing predicted N-terminal acylation motifs, the N-terminal 3xHA tag was inserted after the predicted acylation target sequence of PBS1 (amino acids 1-7) in the N-terminal clone. We refer to the proteins expressed from these constructs as ‘engineered cleavage products’ to distinguish them from the cleavage products produced by AvrPphB proteolytic activity.
Previous experiments have indicated that adding a C-terminal 3xHA tag to full-length PBS1 (PBS1:HA) does not adversely affect the ability of PBS1 to activate RPS5 (Ade et al., 2007). Therefore, the 3xHA tag should not affect the function of the C-terminal engineered cleavage product. However, the effect of the placement of the HA tag with respect to the N-terminal engineered cleavage product was unknown. We therefore created a construct to express full-length PBS1 containing the 3xHA epitope tag inserted between amino acids 7 and 8 (referred to as HA:PBS1, henceforth). This construct was transiently co-expressed with RPS5 fused to five copies of the human cMyc epitope tag (RPS5:Myc) and AvrPphB in Nicotiana benthamiana under the control of a dexamethasone-inducible promoter and their ability to activate HR-like programmed cell death was assessed. We have previously shown that AvrPphB-mediated activation of RPS5 can be reconstituted in N. benthamiana using a similar transient expression system (Ade et al., 2007). HA:PBS1 retained full function when compared to the PBS1:HA control, indicating that placement of the HA tag after the acylation target sequence was not deleterious to PBS1 activation of RPS5 (Figure S1).
We next tested whether the PBS1 N- and C-terminal engineered cleavage products could activate RPS5-mediated HR. When either N- or C-terminal engineered cleavage product was co-expressed with RPS5:Myc in the absence of AvrPphB, no HR was detected (Figure 1). However, co-expression of the N- and C-terminal engineered cleavage products with RPS5 elicited the HR in the absence of AvrPphB and this HR was dependent on RPS5. Therefore we conclude that both cleavage products are required to activate RPS5 and that activation is not dependent on AvrPphB.
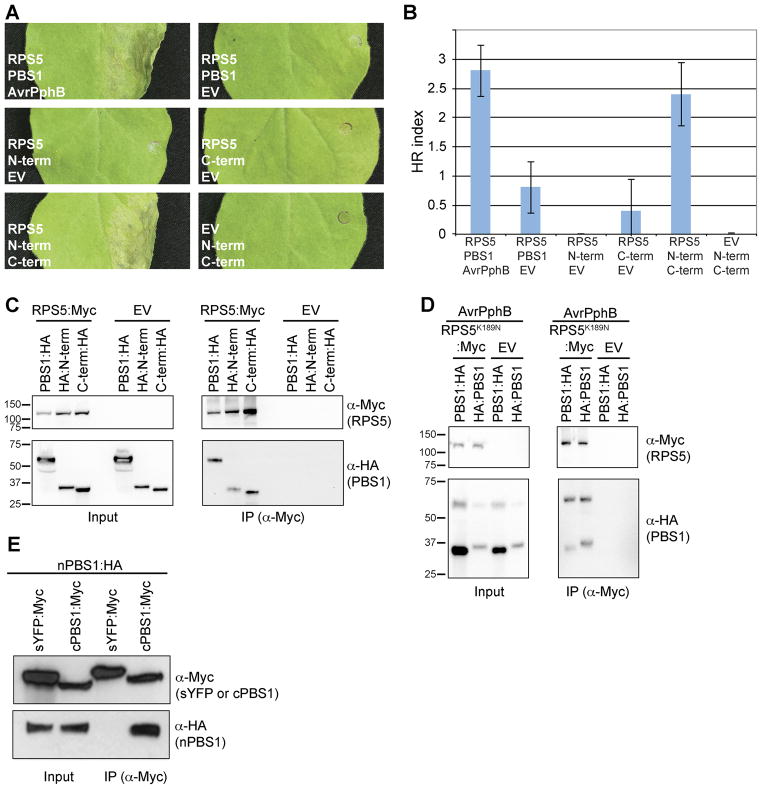
Fig. 1. Both PBS1 cleavage products are necessary and sufficient for activating RPS5 and associate with RPS5.
A. Activation of RPS5 by co-expression with both PBS1 cleavage products. The indicated proteins were transiently co-expressed in N. benthamiana. Leaves were photographed 24 h after transgene induction. N-term, PBS1 N-terminal engineered cleavage product; C-term, PBS1 C-terminal engineered cleavage product; EV, empty vector.
B. Quantification of HRs induced by co-expression of RPS5 and PBS1 cleavage products. The HR was quantified as described in Experimental Procedures, with sample sizes of 5 injections. Error bars indicate standard error.
C. Co-immunoprecipitation of PBS1 cleavage products with RPS5. The indicated PBS1 constructs were transiently co-expressed with RPS5:Myc or empty vector.
D. AvrPphB and RPS5K189N:Myc in N. benthamiana. Proteins were immunoprecipitated with anti-cMyc (IP). As a control for expression, a portion of each sample was taken prior to immunoprecipitation (input). Immunoblots were performed with the antibodies indicated on the right. The slower migrating band in panel C detected in the anti-HA blot corresponds to full-length PBS1 due to incomplete cleavage by AvrPphB.
E. Co-IP of engineered PBS1 cleavage products with each other. The indicated constructs were transiently co-expressed in N. benthamiana and immunoprecipitated with anti-cMyc. sYFP is used as a negative control and is targeted to the plasma membrane by an N-terminal fusion with the first 20 amino acids of RPS5 (Qi et al., 2012).
To confirm that the PBS1 cleavage products are sufficient to activate RPS5, we generated transgenic Arabidopsis plants in which the two engineered cleavage products were expressed under dexamethasone-inducible promoters in a single construct. This construct was transformed into both wild-type and rps5 mutant Arabidopsis plants using Agrobacterium-mediated transformation. Transgenic lines of both genotypes grew normally in the absence of dexamethasone application. When sprayed with a dexamethasone solution (50 μM), however, leaves of the wild-type transgenic plants became yellow and necrotic within three days, while the rps5 lines remained green and healthy (Figure S2). Thus, the PBS1 cleavage products can activate RPS5 in Arabidopsis even when RPS5 is expressed at native levels.
Both amino- and carboxy-terminal PBS1 cleavage products interact with RPS5
Because both the N- and C-terminal PBS1 engineered cleavage products were required for activation of RPS5, we hypothesized that both cleavage products would physically interact with RPS5. To test this, we transiently co-expressed RPS5:Myc with either the N-terminal or C-terminal engineered cleavage product in N. benthamiana and assayed for physical association using co-immunoprecipitation (co-IP) analysis on protein extracts from whole leaves. Both N-terminal and C-terminal PBS1 engineered cleavage products immunoprecipitated with RPS5:Myc, but were not precipitated in the absence of RPS5 (Figure 1C). Furthermore, these experiments were performed in the absence of AvrPphB suggesting that PBS1 cleavage products do not require AvrPphB for interaction with RPS5.
Considering that the engineered PBS1 cleavage products associated with RPS5, we wanted to determine if the AvrPphB-induced PBS1 cleavage products behaved in a similar manner. We co-expressed a P-loop mutant form of RPS5 (RPS5K189N:Myc) and AvrPphB with either HA:PBS1 or PBS1:HA in N. benthamiana leaves and assessed protein interaction using co-IP analysis. The P-loop mutant form was used to avoid the rapid and specific degradation of RPS5 that occurs during RPS5-mediated HR (Ade et al., 2007). Again, both N- and C-terminal cleavage products were immunoprecipitated with RPS5K189N:Myc, but not in the absence of RPS5K189N:Myc (Figure 1D). Furthermore, as PBS1 cleavage by AvrPphB was incomplete, immunoprecipitation of the full-length HA:PBS1 or PBS1:HA could be used as an internal control. Both uncleaved HA:PBS1 and uncleaved PBS1:HA immunoprecipitated with RPS5K189N:Myc, but not in the absence of RPS5K189N:Myc.
The ability of the engineered PBS1 cleavage products to activate RPS5 suggested that the two cleavage products may re-associate to form an intact kinase structure when co-expressed. We therefore performed a co-IP analysis using an N-terminal engineered cleavage product tagged with HA and a C-terminal engineered cleavage product tagged with Myc. As a negative control for this experiment, we used a derivative of super Yellow Fluorescent Protein (sYFP) in which YFP was fused to the first 20 amino acids of RPS5 at its N-terminus and Myc at its C-terminus. We have shown previously that this protein is targeted to the plasma membrane (Qi et al., 2012). The N-terminal engineered cleavage product co-immunoprecipitated with the C-terminal engineered cleavage product, but not with sYFP (Figure 1E), indicating that the two halves of PBS1 can re-associate inside the plant cell.
Defining PBS1 domains required for HR activation
PBS1 contains a central kinase domain and previous studies have indicated that kinase activity is important for RPS5-mediated HR (Shao et al., 2003, Swiderski et al., 2001). Flanking the PBS1 kinase domain are N- and C-terminal domains whose functions are unknown (Figures 2A and S3; amino acids 1-88 and 361-456, respectively). Because no additional protein motifs were found in the PBS1 N- and C-terminal domains, we sought to determine if these regions contained previously undescribed motifs. If the N-and C-terminal non-kinase domains are important for PBS1 function, we hypothesized that they would be conserved among PBS1 orthologs from divergent plant species. To identify PBS1 orthologous sequences, we used TBLASTN to search the NCBI nr database with qualifiers, Viridiplantae not Arabidopsis, using PBS1 protein sequence as a query. Additional sequences were identified using the Eukaryotic Gene Orthologues (EGO) database (Computational Biology and Functional Genomics Laboratory, Dana-Farber Cancer institute and Harvard School of Public Health). Apparent full-length sequences were selected from taxa representing several plant families and aligned (Figure S3). The alignment revealed several blocks of sequence homology in both N- and C-terminal domains that were conserved across taxa from mosses to flowering plants, suggesting that these motifs have been maintained for more than 400 million years (Steemans et al., 2009). Additional or extended conserved sequence blocks common to dicot, but not monocot taxa were also identified.
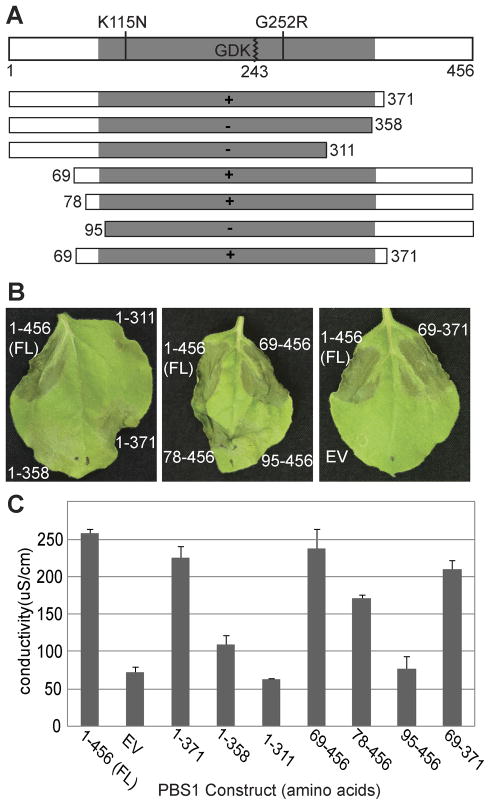
Fig. 2. The amino- and carboxy-terminal non-kinase domain portions of PBS1 are dispensable for RPS5 activation.
A. Schematic representation of the PBS1 protein showing relevant mutations and truncation constructs. The kinase domain is indicated by gray shading and the full-length protein is 456 amino acids long. The AvrPphB recognition sequence (GDK) is indicated in the kinase domain. Cleavage occurs after lysine 243 and is marked with a zigzag line. Missense substitutions are indicated above the protein diagram and are marked by a straight line. C-terminal and N-terminal truncation constructs are indicated by rectangles with numbers indicating the position of the terminal amino acid residue. Ability to induce visible HR when co-expressed with AvrPphB and RPS5 in N. benthamiana (panel B) is indicated by ‘+’ and ‘−’ symbols inside the rectangles.
B. The indicated PBS1 constructs were transiently expressed with RPS5 and AvrPphB in N. benthamiana and assessed for HR elicitation. PBS1 constructs were named according to the position of the amino acids present (e.g 69-371 contains amino acids 69 through 371). For amino-terminal truncations, a methionine was added for translation initiation. For carboxy-terminal truncations, a stop codon was added for translation termination.
C. Quantification of the HR using electrolyte leakage. Error bars indicate standard deviation. Conductivity was measured after floating leaf disks in deionized water for 14 hours following transgene induction.
To determine the functional significance of the conserved motifs in the PBS1 N- and C-terminal domains, we created a truncation series, deleting portions of the N- and/or C-termini of PBS1 (Figure 2A). These constructs were transiently co-expressed with RPS5 and AvrPphB in N. benthamiana and assayed for their ability to activate the HR (Figure 2B). To quantify the HR-inducing capacity of each derivative, we performed electrolyte leakage analysis (Figure 2C). N-terminal truncations lacking amino acids 1-68 (PBS169-456) or 1-77 (PBS178-456) showed only a slight reduction in HR activity as measured by electrolyte leakage, while the truncation lacking amino acids 1-94 (PBS195-456) exhibited a near complete loss of HR (Figure 2). This loss of HR-inducing activity was not due to protein destabilization as all truncations accumulated to similar levels, as will be discussed further below and shown in Figure 4. The C-terminal truncation lacking amino acids 372-456 (PBS11-371) exhibited no noticeable reduction in HR activity, while the truncations lacking amino acids 359-456 (PBS11-358) or 312-456 (PBS11-311) exhibited a marked reduction in HR activity. Furthermore, a truncation lacking both amino acids 1-68 and 372-456 (PBS169-371) exhibited no noticeable reduction in HR activity. The N-terminus of PBS1 contains a predicted palymitoylation motif that is conserved among its orthologues (Figure S3); thus any requirement for this motif in the localization of PBS1 has likely been obviated by overexpression in this assay.
Fig. 4. Loss of PBS1 function by truncation corresponds to loss of cleavage by AvrPphB.
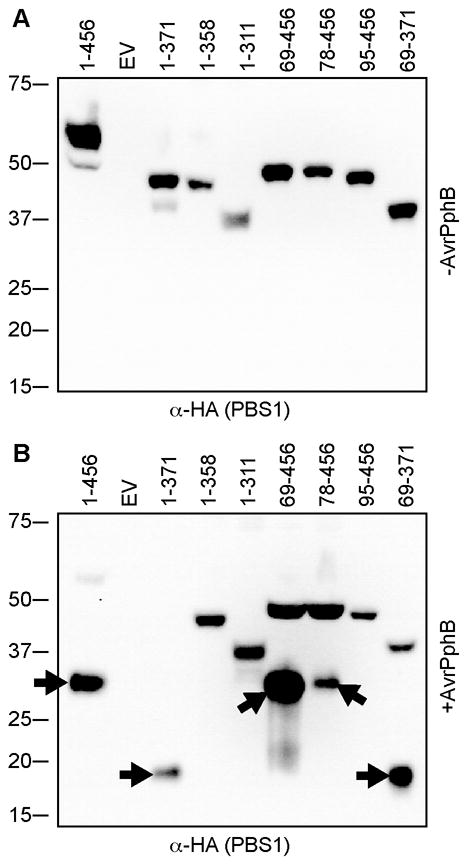
PBS1 constructs were transiently expressed in N. benthamiana in the absence or presence of AvrPphB.
A. Immunoblot of PBS1 truncation products expressed in the absence of AvrPphB showing that all expressed and migrated at the expected sizes.
B. Immunoblot of PBS1 truncation products expressed in the presence of AvrPphB. Only a subset of PBS1 truncations were cleaved by AvrPphB as evidenced by appearance of a faster migrating band (indicated with arrows). EV, empty vector.
Upon comparing the location of the truncation with the HR activity level, two things were clear. First, the N- and C-terminal non-kinase domains of PBS1 were dispensable for RPS5-mediated HR. Second, truncation of either end of the kinase domain, as defined by the conserved kinase motifs, resulted in severe reduction in HR activity. PBS195-456 and PBS11-358 lack only 6 and 3 residues in the conserved Kinase I and Kinase XI motifs (Figure S3), respectively, yet their function was significantly diminished.
PBS1 kinase activity is only partially required for RPS5 activation in N. benthamiana
The above results suggested that PBS1 kinase activity may be required to activate RPS5-mediated HR in N. benthamiana. Consistent with this hypothesis, we have previously shown that kinase inactive alleles of PBS1, PBS1K115N and PBS1G252R are unable to complement pbs1-1 in transiently or stably transformed Arabidopsis lines (Shao et al., 2003, Swiderski et al., 2001). To address whether PBS1 kinase activity is required to activate RPS5 in our N. benthamiana assay, we transiently co-expressed RPS5, AvrPphB, and PBS1K115N or PBS1G252R and assessed their ability to activate RPS5-mediated HR. Both PBS1K115N and PBS1G252R were capable of eliciting HR at levels above background (Figure 3A and B). Although their activation of the HR appears somewhat reduced when compared to wild-type PBS1, it is clear that they retain at least partial function. These results indicate that PBS1 kinase activity is not absolutely required for activation of RPS5-mediated HR, at least when PBS1 is over-expressed.
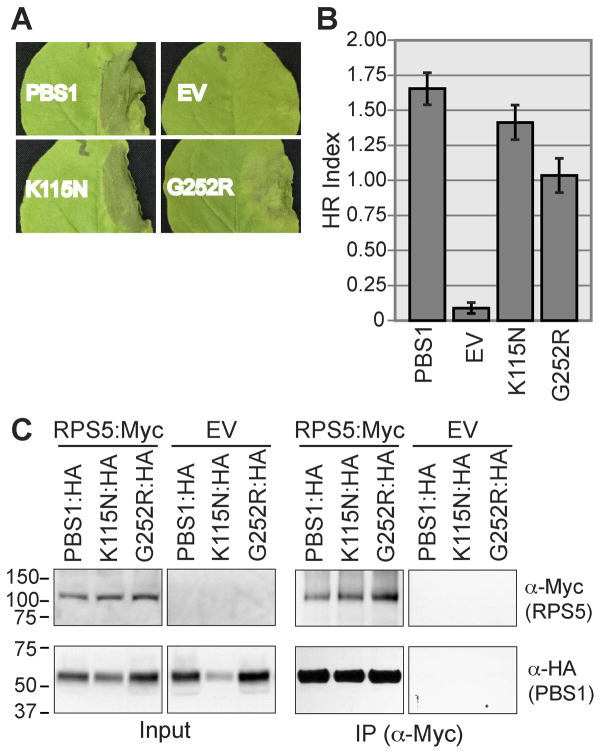
Fig. 3. Kinase inactive variants of PBS1 activate the HR and interact with RPS5.
A. The indicated PBS1 constructs were transiently expressed with RPS5 and AvrPphB in N. benthamiana and assessed for HR elicitation. A representative leaf for each construct was photographed 24 h after transgene induction. EV, empty vector.
B. The HR was quantified as described in Experimental Procedures, with sample sizes of 56–58 injections. Error bars indicate standard error.
C. The indicated constructs were transiently co-expressed in N. benthamiana. Proteins were immunoprecipitated with anti-cMyc (IP). Immunoblots were performed with the antibodies indicated on the right. EV, Empty Vector; K115N, PBS1K115N; G252R, PBS1G252R.
We have previously reported that kinase inactive forms of PBS1 have reduced ability to co-immunoprecipitate with RPS5 (Ade et al., 2007). This observation could account for the reduced ability of kinase inactive PBS1 to activate RPS5 in N. benthamiana. We therefore assessed the interaction of the kinase-inactive PBS1 variants with RPS5 using different vectors and epitope tags than used in the Ade et al. study in which the kinase-inactive forms of PBS1 accumulated to lower levels than wild-type PBS1. Using the new PBS1 constructs, we obtained more robust expression of the mutant forms of PBS1 (PBS1K115N and PBS1G252R), and both kinase inactive variants co-immunoprecipitated with RPS5 at levels similar to wild-type PBS1 (Figure 3C). Thus, PBS1 kinase activity is not required for interaction with RPS5 when overexpressed in N. benthamiana. It remains possible, however, that autophosphorylation of PBS1 could affect the strength of this interaction, which would become apparent at native protein levels.
Truncation of PBS1 kinase domain residues results in loss of cleavage by AvrPphB and loss of RPS5 activation
Our finding that PBS1 kinase activity is not required for full RPS5 activation lead us to question why truncations into the kinase domain of PBS1 eliminate its ability to activate RPS5. Two plausible hypotheses are that these truncations are unstable, or they are poor substrates for AvrPphB cleavage. To test these hypotheses, we transiently expressed the PBS1 truncation series with AvrPphB in N. benthamiana. Total protein extracts were prepared four hours after induction of protein expression. Although there was some variability in protein accumulation, all of the truncations were detectable by immunoblot analysis in the presence and absence of AvrPphB (Figure 4). However, only a subset of the truncated proteins was cleaved by AvrPphB. Significantly, constructs that lacked the ability to induce the HR corresponded exactly to the constructs that were unable to be cleaved by AvrPphB. Hence, for the PBS1 truncations, loss of activity could be completely explained by loss of cleavage by AvrPphB.
PBS1 containing an insertion of 5 alanine residues at the PBS1 cleavage site activates RPS5 in the absence of cleavage
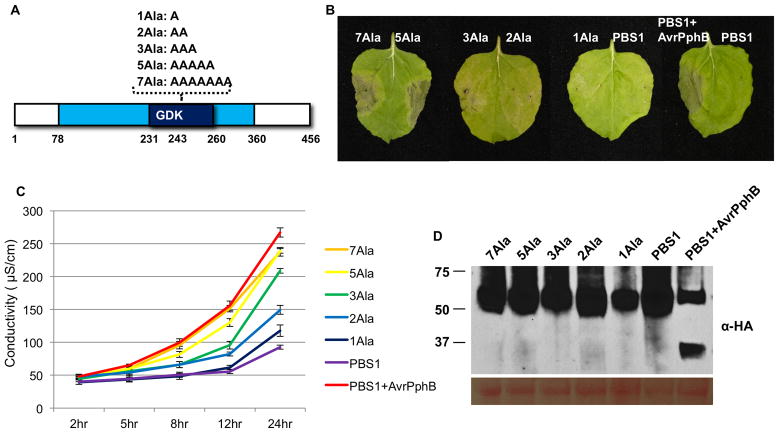
The correlation between PBS1 cleavage and RPS5 activation further supported our conclusion that PBS1 cleavage is a key step in activating RPS5. We developed two alternative hypotheses for how cleavage activates RPS5. The first was that cleavage creates two free ends with a negative and positive charge and these free ends then associate with the LRR domain of RPS5 to activate it. Our second hypothesis was that cleavage results in an overall conformational change in PBS1 that enables PBS1 to specifically associate with the LRR domain, independent of the free ends. If the latter hypothesis were correct, we speculated that insertion of amino acids at the site of cleavage might mimic cleavage in regards to RPS5 activation. We therefore constructed a series of PBS1 derivatives with insertions of one, two, three, five, or seven alanine residues at the cleavage site (lysine 243; Figure 5). We then co-expressed these PBS1 derivatives with RPS5 in N. benthamiana in the absence of AvrPphB. Significantly, PBS1 derivatives with either seven or five alanine insertions strongly induced RPS5-mediated HR (Figure 5B). Derivatives with three or two alanines induced a weaker HR, while PBS1 with a single alanine insertion appeared unable to activate RPS5. Consistent with the visible symptoms, PBS1 derivatives with seven and five alanine insertions induced almost as much electrolyte leakage as wild-type PBS1 cleaved by AvrPphB (Figure 5C). The derivatives with three or two alanine insertions induced electrolyte leakage intermediate between the positive and negative controls, while the single alanine insertion derivative behaved like wild-type PBS1 through 12 hours post-induction, and showed slightly elevated electrolyte leakage at 24 hours post-induction. Immunoblot analysis showed that all PBS1 derivatives expressed well (Figure 5D), thus the difference in HR induction was not due to differences in protein accumulation. We thus conclude that RPS5 is activated by a conformational change in PBS1 that results from cleavage of the activation loop, rather than the generation of free ends. This likely explains the requirement for both cleavage products when activating RPS5.
Fig. 5. Insertion of alanine residues at the PBS1 cleavage site activates RPS5.
A. Schematic representation of synthetic PBS1 constructs. The kinase domain (amino acids 78-360) and activation segment (amino acids 231-260) of PBS1 are represented by a light blue box and dark blue box, respectively. Synthetic PBS1 constructs containing the indicated alanine insertions after lysine 243 are shown above the diagram.
B. HR phenotypes of Agrobacterium-infiltrated N. benthamiana leaves. The indicated synthetic and wild type PBS1:HA constructs were transiently co-expressed with RPS5-Myc or with RPS5-Myc and AvrPphB. 50 μM dexamethasone was sprayed 40 hours after injection and photographs were taken 24 hr later. This experiment was repeated three times with similar results.
C. Quantification of the HR triggered by the PBS1 constructs described in (B) using electrolyte leakage. Error bars indicate standard deviation. Conductivity was measured at the indicated time points after dexamethasone treatment. This experiment was repeated once with a similar result.
D. Expression of PBS1 constructs described in (B) was confirmed by immunoblot analysis with anti-HA antibody. The same blot was stained with Ponceau S solution to show equal loading of protein samples. Samples were taken 4 hr after dexamethasone treatment.
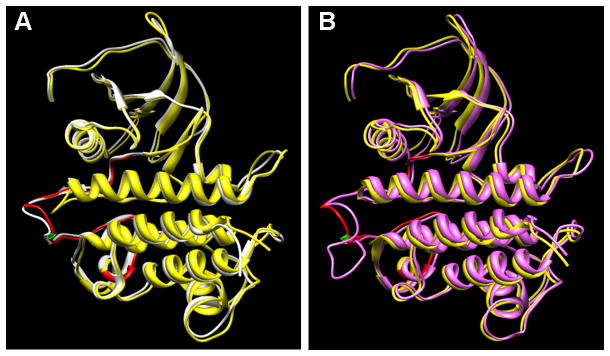
To gain insight into how insertion of alanine residues after lysine 243 might affect the structure of PBS1 we generated a model for PBS1 using the Pto kinase from tomato as a template, whose structure has recently been determined (Dong et al., 2009, Xing et al., 2007). These two kinases aligned well, allowing us to predict the structure of the wild-type PBS1 kinase domain with confidence (Figure 6A). Notably, lysine 243 is positioned at the apex of the A-loop region of the PBS1 activation segment (the activation segment is defined as the region between and including the conserved DFG and APE tripeptide motifs; (Nolen et al., 2004)). Insertion of five amino acids in this location (Figure 6B) would likely affect the position of the entire activation segment, and possibly cause more general conformational changes as it could disrupt critical bridges between residues in the P+1 loop and the catalytic loop (e.g. threonine 253 in the P+1 loop is predicted to form hydrogen bonds with aspartate 213 and lysine 215 in the catalytic loop when PBS1 is in an active conformation (Nolen et al., 2004)). It is worth noting that the PBS1 and Pto activation segments are identical in length (30 amino acids), and this length is well-conserved among plant cytoplasmic receptor-like protein kinases, with the majority being either 29 or 30 amino acids (data not shown), suggesting that longer activation segments disrupt function.
Fig. 6. Predicted structure of the PBS1 kinase domain aligned with the known structure of Pto.
A. Ribbon diagrams are shown for both PBS1 (yellow) and Pto (white). The activation segment of PBS1 is colored in red, with the AvrPphB cleavage site (lysine 243) indicated in green.
B. Wild-type PBS1 is shown in yellow and PBS1 with an insertion of five alanines at the AvrPphB cleavage site in magenta. Note that the computer model attempts to maximize the alignment between the two forms of PBS1, but it is likely that the five alanine insertion would cause greater conformational changes than indicated in the model.
Discussion
Despite detailed mutant and domain interaction analysis, relatively little is known about how NB-LRR proteins are activated. Work by Tameling et al. (2002) has suggested that the identity of nucleotide bound to the NB domain may dictate the activity state of the protein. For the I-2 and Mi proteins, ATP appears to be bound in the active protein, while ADP is bound in the inactive protein. Mutations in I-2 that reduce nucleotide binding correlate with a loss of function while mutations that reduced hydrolysis correlate with autoactivation (Tameling et al., 2002, Tameling et al., 2006). Equivalent mutations in several plant NB-LRR proteins, including RPS5, result in loss of function and autoactivation, respectively (Ade et al., 2007, DeYoung et al., 2006, van Ooijen et al., 2007). Further supporting this model, wild-type M protein of flax purified from yeast was found to preferentially bind ADP, whereas an autoactive mutant form bound ATP (Williams et al., 2011). It has thus been hypothesized that complex inter- and intramolecular interactions may alter the conformation of the NB-LRR protein, promoting ATP binding, hydrolysis, or exchange of ADP for ATP (Collier et al., 2009, Lukasik et al., 2009). For RPS5, we have shown that deletion of the LRR domain results in autoactivity (Ade et al., 2007). Similarly, a mutation of aspartic acid to glutamic acid at position 266, which is predicted to reduce ATP hydrolysis, also confers autoactivity to RPS5. Taking these points into account, we propose a model in which the RPS5 LRR domain influences the NB structure such that exchange of ADP for ATP is prevented in the absence of AvrPphB. Because PBS1 cleavage is required for activation of wild-type RPS5, it would follow that PBS1 cleavage promotes a conformational change in RPS5, perhaps through relieving the negative regulation of the RPS5 LRR domain resulting in an exchange of ADP for ATP and activation of RPS5.
Previously it was unknown whether the physical act of PBS1 cleavage or merely the presence of the cleavage products was sufficient for RPS5 activation. Furthermore, it was unclear if AvrPphB was required in some capacity in addition to cleavage of PBS1. Here we have shown that PBS1 cleavage products and not AvrPphB are required for activation of RPS5. Furthermore, we have established that insertion of just five alanine residues at the cleavage site of PBS1 will also activate RPS5. These results are consistent with a model in which cleavage of PBS1 causes a subtle conformational change that then allows PBS1 to interact with RPS5 in such a way that it promotes nucleotide exchange. More generally, this represents an attractive model for NB-LRR protein regulation whereby interaction with modified host protein (or effector protein) causes a conformational change in the NB-LRR protein leading to activation.
It is interesting to note that AvrPphB appears to require an intact, but not necessarily active, kinase domain for recognition of its substrate. Deletion of even three amino acids from the last conserved kinase domain of PBS1 or six amino acids from the first conserved kinase domain of PBS1 results in loss of cleavage by AvrPphB, despite the fact that the truncated PBS1 proteins are expressed to significant levels (Figure 4 and Figure S3). Truncations containing only 14 or 17 additional amino acids at the C- or N-termini, respectively, were recognized and cleaved by AvrPphB, suggesting that a complete kinase domain is critical for recognition. However, kinase-inactive missense mutations of PBS1 are efficiently cleaved by AvrPphB, thus kinase activity is not required for AvrPphB cleavage (Shao et al., 2003). A three amino acid motif (GDK) adjacent to the cleavage site is necessary for cleavage of PBS1 by AvrPphB (Shao et al., 2003). The data presented here indicate that residues distant from the GDK motif are also crucial for PBS1 recognition by AvrPphB. It is possible that AvrPphB requires specific contact points throughout the PBS1 kinase domain for proper docking or orientation before cleavage is possible. Alternatively, the kinase domain truncations may alter the conformation of the PBS1 protein, making the GDK motif less accessible to AvrPphB. However, it is unlikely that the PBS1 truncations are grossly misfolded as this typically results in targeted degradation, lending support to the hypothesis that an intact kinase domain is necessary for AvrPphB recognition. Interestingly, this differs from AvrRpt2, which only requires a 12 amino acid sequence for cleavage (Chisholm et al., 2005).
Previous experiments have shown that PBS1 kinase activity is required for activation of RPS5 in response to AvrPphB in Arabidopsis (Shao et al., 2003, Swiderski et al., 2001). Here we show that the requirement for PBS1 kinase activity can be partially overcome by overexpression of PBS1, at least in N. benthamiana. What then is the role of PBS1 kinase activity relative to RPS5 activation? Perhaps the simplest explanation is that PBS1 kinase activity stabilizes interaction with RPS5. This effect may not be evident when PBS1 and RPS5 are transiently overexpressed, as increased protein levels may drive sufficient interaction. Indeed, we have detected interaction of kinase-inactive forms of PBS1 with RPS5 in our transient expression system (Figure 3). In endogenous tissues, NB-LRR proteins are expressed at relatively low levels and RPS5 is no exception (Tornero et al., 2002, Holt et al., 2005). Thus, at endogenous expression levels, PBS1 kinase activity may be critical to drive steady state interaction with RPS5. Another possibility, however, is that PBS1 is trans-phosphorylated by an endogenous N. benthamiana kinase overcoming the requirement for PBS1 autophosphorylation.
A second example of an NB-LRR protein requiring a protein kinase for recognition of pathogen effectors is the tomato Prf protein, which requires the Pto kinase for recognition of the P. syringae effectors AvrPto and AvrPtoB (Salmeron et al., 1996, Mucyn et al., 2006). Unlike PBS1, however, kinase inactive Pto is unable to activate Prf when co-expressed with avrPto or avrPtoB in N. benthamiana (Mucyn et al., 2006, Mucyn et al., 2009). Similar to PBS1, though, subtle changes in Pto structure can activate Prf, as over expression of Pto mutants containing aspartate substitutions at several different positions within the P+1 loop of Pto can activate Prf (Rathjen et al., 1999, Wu et al., 2004). These substitutions would be predicted to disrupt key interactions between the P+1 loop and catalytic loop, and thus likely cause general conformational changes in the Pto kinase structure. Activation of Prf by these conformational changes in Pto does not require the kinase activity of Pto (Rathjen et al., 1999, Wu et al., 2004); thus, RPS5 and Prf both appear to be activated by conformational changes in the kinase that they ‘guard’. That relatively subtle changes in the conformation of effector targets can activate NB-LRR proteins, and hence programmed cell death, highlights the sensitivity of the plant innate immune system.
Experimental procedures
Plant material and growth conditions
N. benthamiana seed was obtained from D. Baulcombe. The rps5-2 and pbs1-1 Arabidopsis mutants have been described previously (Warren et al. 1998, 1999). The transgenic A. thaliana line expressing PBS1:HA was provided by Robert Dowen (Dowen et al., 2009). N. benthamiana and A. thaliana plants were grown under a 9-h photoperiod at 24°C in Metro-Mix 360 potting mixture (Sun Gro Horticulture, www.sungro.com).
Plasmid construction
The RPS5:Myc and AvrPphB constructs have been described previously (Ade et al., 2007). The construct encoding PBS1 with an N-terminal HA tag (HA:PBS1) was created as follows. The PBS1 sequence from bases 22-1370 was PCR-amplified from a cDNA template using primers BD162 and BD164 (primer sequences are provided in Table S1), which exclude the putative acylation target sequence and introduce ClaI and SpeI restriction sites at the 5′ and 3′ ends, respectively. The resulting product was cloned into the ClaI/SpeI sites of the pBluescriptII KS+ plasmid (Stratagene, La Jolla, CA). A three-copy HA tag was PCR-amplified from a previously described construct (Ade et al., 2007) using primers BD165 and BD166, which introduce a 5′ SpeI restriction site followed by PBS1 bases 1-21 (putative acylation sites) and a 3′ ClaI restriction site, respectively. This product was cloned into the SalI/ClaI sites of the pBluescriptII KS+ clone containing PBS1 bases 21-1370. The resulting construct was cut with SalI/SpeI and cloned into the XhoI/SpeI sites of the dexamethasone-inducible binary vector pTA7002 (Aoyama et al., 1997).
Constructs encoding full-length PBS1 or PBS1 truncations were created in a three-step process. First, PBS1 sequences were PCR-amplified from a cDNA template using primers that introduce 5′ SalI and 3′ NotI restriction sites and remove the stop codon (and, in the case of N-terminal truncations, introduce a 5′ ATG codon), then cut and cloned into the SalI/NotI sites of pBluescriptII SK+. Next, a three-copy HA tag flanked by NotI and NheI/NotI restriction sites was excised from a previously described construct (Ade et al., 2007) using NotI and inserted into the PBS1 pBluescriptII KS+ full-length and truncation clones. Finally, the resulting clones were cut with SalI/NheI to release the tagged PBS1 fragment and inserted into the XhoI/SpeI sites of pTA7002. The bases included in each clone (corresponding amino acids, and primers used) were as follows: 1-1368 (1-456, BD93-BD94); 1-1113 (1-371, BD93-BD98); 1-1074 (1-358, BD93-BD185); 1-933 (1-311, BD93-BD186); 205-1368 (69-456, BD97-BD94); 232-1369 (78-456, BD178-BD94); 283-1369 (95-456 BD179-94); and 205-1113 (69-371, BD97-BD98).
Constructs encoding PBS1 engineered cleavage products were created in a two-step process. The sequences corresponding to the N-terminal engineered cleavage product were amplified from the HA:PBS1 pBluescriptII KS+ construct described above using the standard M13 reverse primer and primer BD184, which introduces a 3′ stop codon followed by a NheI restriction site. This amplified fragment also included the 5′ SalI restriction site followed by the PBS1 myristoylation target sequence and three-copy HA tag present in the HA:PBS1 pBluescriptII KS+ clone. The sequences corresponding to the C-terminal engineered cleavage product were amplified from the PBS1:HA pBluescriptII KS+ construct described above using BD183, which introduces a SalI restriction site followed by an ATG codon, and the standard M13 forward primer including the 3′ three-copy HA tag followed by a NheI restriction site present in the PBS1:HA pBluescriptII KS+ clone. The resulting clones were cut with SalI/NheI to release the tagged PBS1 fragments and inserted into the XhoI/SpeI sites of pTA7002. The bases included in each clone (and corresponding amino acids) were as follows: 1-21[3xHA]22-729 (1-7[3xHA]8-243); 730-1369 (244-456).
Constructs encoding PBS missense mutations were generated by excising a fragment of PBS1 encoding either K115N or G252R mutations from previously described constructs (Shao et al., 2003) using flanking restriction sites and ligating the resulting fragment into the same sites of PBS1:HA pBluescriptII KS+. The resulting clones were cut with SalI/NheI to release the tagged mutant PBS1 fragments and inserted into the XhoI/SpeI sites of pTA7002.
Engineered PBS1 cleavage products with a Myc tag (as opposed to the HA tagged forms described above) were generated using a modified multi-site Gateway (Invitrogen) cloning system. First the target regions (N-term PBS1: nucleotides 1-729, aa 1-243, primers DQ111 and DQ132 ; and C-term PBS1: nucleotides 730-1368, aa 244-456, primers DQ114 and DQ133) were PCR-amplified and cloned into the Gateway vectors pBSDONR P1-P4 (an ampicillin resistant derivative of pDONR221 P1-P4) and the 5xMyc and 3xHA tags were cloned into the entry vector pBSDONR P4r-P2 (an ampicillin resistant derivative of pDONR221 P4r-P3r in which the attP3r sequence was replaced with attP2) using Gateway BP Clonase II (Invitrogen). The expression constructs were then generated by mixing the desired pBSDONR-P1-P4 constructs, pBSDONR-P4r-P2 constructs, and destination vector in a molar ratio of 2:2:1 followed by LR-clonase catalyzed in vitro recombination. The N-term PBS1:Myc, and C-term PBS1:Myc fusions were recombined into the dexamethasone-inducible expression vector pTA7002. All constructs were verified by sequencing.
To generate PBS1 entry clones harboring 7 alanine, 5 alanine, 3 alanine, 2 alanine and 1 alanine insertions at the site of AvrPphB cleavage, PCR was performed with primers overlapping the cleavage site and containing the desired number of alanine codons (Table S1 primers SHP26-SHP41) using the template pBSDONR PBS1. Multi-site Gateway LR Clonase (Invitrogen) reactions were performed in a molar ratio of 2:2:1 to recombine the entry clones and 3xHA into the pTA7002 destination vector.
To generate a clone expressing both engineered PBS1 cleavage products simultaneously, we used the modified multi-site Gateway cloning system described above, but with three donor plasmids and pBAV154 as destination vector (pBAV154 is a derivative of pTA7001, carries a dexamethasone inducible promoter upstream of the recombination site, and confers basta resistance in plants; Vinatzer et al. 2006). The three donor plasmids contained N-term PBS1:HA in pBSDONR P1-P4, a fragment amplified from pBAV154 containing a nopaline synthase 3′ terminator sequence and the dexamethasone inducible promoter sequence in pBSDONR P4r-P3r, and C-term PBS1:Myc in pBSDONR P3-P2. These four plasmids were mixed and recombined using LR clonase (Invitrogen), to generate the final construct in which the engineered cleavage products were expressed from identical, but separate, dexamethasone inducible promoters. All junctions were verified by sequencing, and protein expression verified by transient expression in N. benthamiana. The construct was then transformed into wild-type Col-0 and rps5 mutant (T-DNA insertion line SALK_015294C) Arabidopsis using the floral dip method (Clough and Bent, 1998).
Transient expression assays in N. benthamiana
Agrobacterium tumefaciens GV3101 (pMP90) strains transformed with the dexamethasone-inducible constructs described above were grown and prepared for transient expression as previously described (Wroblewski et al., 2005). Agrobacterium cultures were resuspended in water at an OD600 of 1.0 and, for co-expression of multiple constructs, suspensions were mixed in equal ratios.
Bacterial suspension mixtures were infiltrated by needle-less syringe into expanding leaves of 4-week-old N. benthamiana. Protein expression was induced by spraying the leaves with 50 μM dexamethasone 40 h after injection. Samples were collected for protein extractions between 4 and 6 h after dexamethasone application and were flash-frozen in liquid nitrogen. For evaluation of hypersensitive response, leaves were scored 24 h after dexamethasone application. For semiquantitative assessment of HR strength, phenotypes were scored as follows: no discernable difference from uninjected tissue = 0, small patches of necrosis (<25% injected area) = 1, moderate patches of tissue collapse (25–75% of injected area) = 2, large patches to complete collapse (>75% of injected area) = 3.
Electrolyte leakage assays
The relevant combinations of Agrobacterium strains were infiltrated into N. benthamiana leaves as described above. At 2 hr post DEX induction, leaf discs (6 mm in diameter) were collected from the infiltrated area using a cork borer. A total of 9 discs from 6 individual leaves of two different plants were included for each replication and three independent replications were included for each treatment. The discs were then washed briefly in distilled water and gently dried on paper towel. The leaf discs were floated in 5 ml of distilled water containing 0.01% of Silwet-L77. Conductivity was measured using a VWR Traceable Pen Conductivity Meter at the indicated times post dexamethasone induction.
Immunoprecipitations and immunoblots
Immunoprecipitations were performed as previously described (Shao et al., 2003) using c-Myc Monoclonal Antibody-Agarose Beads (Clontech, www.clontech.com). The immunocomplexes were resuspended in 50 μL of 1×SDS loading buffer and boiled for 5 min while total proteins were mixed with 4×SDS loading buffer at a ratio of 3:1 and boiled for 5 min. Immunocomplexes and/or total proteins were separated by electrophoresis in a 4–20% gradient Tris-HEPES-SDS polyacrylamide gel (Thermo Fisher Scientific, www.thermo.com). Proteins from duplicate gels and filters were transferred to a nitrocellulose membrane and probed with Anti-c-Myc-peroxidase (Roche, www.roche.com) or Anti-HA peroxidase (Sigma-Aldrich, www.sigmaaldrich.com).
Sequence analysis and alignment
PBS1 and putative orthologous sequences were aligned with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using default parameters (Chenna et al., 2003), and the display output was generated with Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html) using default parameters.
PBS1 structure modeling
To create a model for the structure of the PBS1 kinase domain, the PBS1 amino acid sequence was aligned with the Pto amino acid sequence using ClustalX (Larkin et al., 2007) and the alignment trimmed to positions 41-302 in Pto. Secondary structure was predicted for both proteins using the PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/) (Jones, 1999, Bryson et al., 2005) to help validate the alignment. The previously described structure for Pto (PDB code 2QKW) was obtained from the RCSB PDB database (www.pdb.org) (Berman et al., 2000). The tertiary structure for PBS1 was then modeled using the MODELLER software (http://www.salilab.org/modeller/) (Eswar et al., 2006) as implemented through the UCSF Chimera interface (http://www.cgl.ucsf.edu/chimera/) (Pettersen et al., 2004) and using Pto as the known structure for comparison. Structures were visualized and manipulated using UCSF Chimera.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers NM_121319 (Arabidopsis PBS1, At5g13160); FJ014791 (Glycine max PBS1); XM_002514818 (Ricinus communis PBS1); XM_002304628.1 (Populus trichocarpa PBS1); AK326615 (Solanum lycopersicum PBS1); NM_001053478 (Oryza sativa PBS1, Os02g0513000); BT063797 (Zea mays PBS1); TC82077 (Pinus taeda PBS1, tentative consensus from Dana-Farber Cancer Institute Pinus Gene Index); XM_001769533 (Physcomitrella patens PBS1); NC_003070 (RPS5); AJ870974 (AvrPphB); AAB47421 (S. lycopersicum Pto).
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the United States National Institutes of Health (grant GM046451 to RWI and GM080912-01A1 to BJD). We thank Dr. Ullrich Dubiella for helpful discussions and comments on the manuscript, and Robert Dowen for providing seed of PBS1:HA transgenic plants. We also thank the Arabidopsis Biological Resource Center at Ohio State for providing seed of the rps5 T-DNA insertion mutant.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Addition of an N-terminal or C-terminal HA Tag Does Not Affect PBS1 Activation of RPS5.
A. Schematic of HA epitope tagged constructs. To create an N-terminally tagged version of PBS1, without disrupting potential acylation target sequences, we inserted three copies of the HA epitope tag sequence between PBS1 codons 7 and 8. The C-terminally tagged version of PBS1 was created by addition of the 3x HA epitope to the C-terminus of PBS1.
B. PBS1:HA expressed under the native PBS1 promoter complements the pbs1-1 mutation in Arabidopsis. Bars represent bacterial population levels at three days post inoculation with Pseudomonas syringae strains DC3000(pVSP61) (=empty vector) and DC3000(avrPphB) in pbs1-1 mutant Arabidopsis transformed with an empty vector (EV) construct (pJH212B) or with PBS1:HA (Dowen et al., 2009). Error bars represent standard deviation.
C. Representative images of HRs induced by the indicated constructs transiently co-expressed in N. benthamiana with RPS5 and AvrPphB. EV, empty vector (pTA7002).
D. Quantification of the HRs in N. benthamiana induced by the indicated constructs co-expressed with RPS5 and AvrPphB. Bars represent mean phenotype score for 31 leaves. Error bars indicate standard error. Phenotype scoring was as follows: no discernable difference from uninjected tissue = 0, small patches of necrosis (<25% injected area) = 1, moderate patches of tissue collapse (25–75% of injected area) = 2, large patches to complete collapse (>75% of injected area) = 3.
Fig. S2. Engineered PBS1 cleavage products activate RPS5 in Arabidopsis.
A. Wild-type Col-0 (left) and rps5 mutant Arabidopsis (right) were transformed with a construct expressing both N- and C-terminal PBS1 cleavage products under control of dexamethasone-inducible promoters. Three-week old seedlings were sprayed with 50 μM dexamethasone and photographed four days later. Each pot contains T2 plants (second generation) derived from a single T1 parent and is representative of at least three independent T1 lines. All plants were selected for presence of the transgene prior to dexamethasone treatment.
B. Immunoblot showing that engineered cleavage products are expressed at approximately the same levels in wild-type and rps5 mutant Arabidopsis genotypes shown in panel A.
Fig. S3. Alignment of PBS1 orthologues.
Amino acid sequences were aligned using ClustalW2, with default parameters selected. The alignment output was converted to a visual representation using Boxshade 3.21 using the default parameters. Conserved protein kinase domains of PBS1 are indicated with brackets above the alignment. Potential palmitoylation (P) sites are indicated above the alignment. The position of the N- and C-terminal truncations of PBS1 are indicated above the alignment with a vertical line and the position of the terminal amino acid. The activation segment is indicated by the red bar and the site of cleavage by AvrPphB is indicated by a vertical arrow within the red bar.
Table S1. Primers Used for Plasmid Construction.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003a;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-Specified Disease Resistance in Arabidopsis Is Coupled to the AvrRpt2-Directed Elimination of RIN4. Cell. 2003b;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci U S A. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, et al. Specific Threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Moffett P. NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 2009;14:521–529. doi: 10.1016/j.tplants.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Opin Microbiol. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dong J, Xiao F, Fan F, Gu L, Cang H, Martin GB, Chai J. Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell. 2009;21:1846–1859. doi: 10.1105/tpc.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Engel JL, Shao F, Ecker JR, Dixon JE. A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J Biol Chem. 2009;284:15867–15879. doi: 10.1074/jbc.M900519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT. Programmed cell death in plant-pathogen interactions. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:525–545. doi: 10.1146/annurev.arplant.48.1.525. [DOI] [PubMed] [Google Scholar]
- Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E, Takken FL. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12:427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW. Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell. 1998;10:1833–1846. doi: 10.1105/tpc.10.11.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. Embo J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn TS, Wu AJ, Balmuth AL, Arasteh JM, Rathjen JP. Regulation of tomato Prf by Pto-like protein kinases. Mol Plant Microbe Interact. 2009;22:391–401. doi: 10.1094/MPMI-22-4-0391. [DOI] [PubMed] [Google Scholar]
- Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell. 1999;11:2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qi D, Deyoung BJ, Innes RW. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 2012 doi: 10.1104/pp.112.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell. 2006;18:2082–2093. doi: 10.1105/tpc.106.042747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW. Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. Embo J. 1999;18:3232–3240. doi: 10.1093/emboj/18.12.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- Steemans P, Herisse AL, Melvin J, Miller MA, Paris F, Verniers J, Wellman CH. Origin and radiation of the earliest vascular land plants. Science. 2009;324:353. doi: 10.1126/science.1169659. [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001;26:101–112. doi: 10.1046/j.1365-313x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, et al. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006;140:1233–1245. doi: 10.1104/pp.105.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl J. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, van den Burg HA, Cornelissen BJC, Takken FLW. Structure and function of resistance proteins in solanaceous plants. Annual Review of Phytopathology. 2007;45:43–72. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Teitzel GM, Lee MW, Jelenska J, Hotton S, Fairfax K, et al. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol. 2006;62:26–44. doi: 10.1111/j.1365-2958.2006.05350.x. [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RF, Merritt PM, Holub E, Innes RW. Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics. 1999;152:401–412. doi: 10.1093/genetics/152.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Swiderski MR, Li Y, Jones JDG. The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J. 2006;47:829–840. doi: 10.1111/j.1365-313X.2006.02834.x. [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sornaraj P, Decourcy-Ireland E, Menz RI, Kobe B, Ellis J, et al. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, while wild-type M protein has a preference for binding ADP. Mol Plant Microbe Interact. 2011 doi: 10.1094/MPMI-03-11-0052. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore RW. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnology Journal. 2005;3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- Wu AJ, Andriotis VM, Durrant MC, Rathjen JP. A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell. 2004;16:2809–2821. doi: 10.1105/tpc.104.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Zou Y, Liu Q, Liu J, Luo X, Huang Q, et al. The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature. 2007;449:243–247. doi: 10.1038/nature06109. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.