Abstract
The neutron structure of wild type human carbonic anhydrase II at pH 7.8 has been determined to 2.0 Å resolution. Detailed analysis and comparison to the previously determined structure at pH 10.0 shows important differences in protonation of key catalytic residues in the active site as well as a rearrangement of the hydrogen bonded water network. For the first time, a completed hydrogen bonded network stretching from the Zn-bound solvent to the proton shuttling residue His64 has been directly observed.
Keywords: neutron diffraction, hydrogen bond, carbonic anhydrase, deuterium, proton transfer
Human carbonic anhydrase II (HCA II) is a 29 kDa monomeric Zn-metalloenzyme that catalyzes the reversible hydration of CO2 into HCO3− and a proton, and is found prominently in red blood cells and secretory tissues. HCA II is one of the fastest enzymes known with a kcat for CO2 hydration of 106/sec and its activity is close to diffusion limited (1). HCA II serves as a relatively simple model enzyme for the study of long-range proton transfer mechanisms in more complex systems, such as ATP synthase, bacteriorhodopsin, and cytochrome c oxidase. There is also increasing industrial interest in using engineered carbonic anhydrase for carbon dioxide extraction from flue gas in coal-fired power plants (2). Analyses of HCA II neutron structures provide unique data and insights into proton transfer mechanisms and could have broad applicability to many other systems.
All CAs identified to date use a metal-hydroxide mechanism to mediate the two-step ping-pong catalytic mechanism (3). The first step of catalysis by HCA II is a nucleophilic attack on incoming CO2 by a Zn-bound OH− to produce HCO3−. The binding of HCO3− at the metal is weak and accordingly is easily displaced by a water molecule. The second step activates the metal-bound water to OH− through a series of proton transfers (1, 4). This is thought to occur through a network of well-ordered H-bonded waters (4, 5). This network stretches from the metal to the proton shuttling residue, His64, and is H-bonded to several hydrophilic residues that line the active site (Figure S1; 6, 7). In structural studies residue His64 has been observed to occupy two distinct positions, termed the inward and outward conformations, pointing towards and away from the Zn active site, respectively (8, 9). Exactly how protons move in the active site of proteins is not clear but it has been proposed that a Grotthus proton hopping mechanism or the formation of a series of Zundel (H5O2+) or Eigen cations (H9O4+) may be factors (10). It has been postulated that the inward conformation of His64 might be poised to accept the excess proton from the water network, while the outward conformation is ready for proton shuttling to the bulk solvent. This observed flexibility of His64 is most likely related to its protonation state (7, 11). The enzyme displays very strong pH dependence for both kcat and kcat/KM that are defined by a single ionization corresponding to a pKa of ~7 (1).
The details of CO2 binding and subsequent hydration are clearer since the structure determination of CO2 at the active site revealed a side-on binding coordination (12). Currently neutron crystallography studies on HCA II have focused on the structural details of the active site hydrophilic residues and water molecules involved in proton transfer. As such the analysis of the neutron structure at high pH (~pH 10) has been previously reported (13).
The approach taken in the current study was to change the pH of the crystal as a way to modulate the changes in protonation that occur during catalysis. Discussed here is the pH 7.8 crystal structure that is compared to the previously determined pH 10.0 form. The advantage of neutron crystallography is that H and its isotope D diffract neutrons very strongly and to about the same extent as the heavier atoms found in proteins such as N, C, or O (scattering lengths in fm: H = −3.74, D = 6.67, N = 9.4, C = 6.6, O = 5.8). Accordingly, H and D atoms appear as negative or positive peaks, respectively, in nuclear scattering density maps. In this study the crystal was subject to H/D vapor exchange, to replace labile H with D, thus promoting strong positive scattering from D atoms. The benefit of using vapor exchange instead of soaking is that there is less chance of damage to the large crystals required for neutron studies. Using neutron diffraction on H/D exchanged samples it is possible, even at medium (2.0 – 2.2 Å) resolution, to directly observe the protonation states of amino acid residues (eg. neutral vs. charged histidine residues) and to distinguish between D3O+, D2O, and OD− (14,15). The room temperature neutron structures were complemented with X-ray data from a similar crystal prepared and H/D exchanged in the same manner. Using the joint neutron and X-ray methodology for model refinement leads to more accurate structures in that X-rays provide highly accurate information for the heavier atoms in proteins while neutrons are used to refine the positions of H/D atoms, effectively increasing the data-to-parameter ratio for refinement. In this way the data are highly complementary and allow a detailed understanding of hydration and H-bonding in proteins (16).
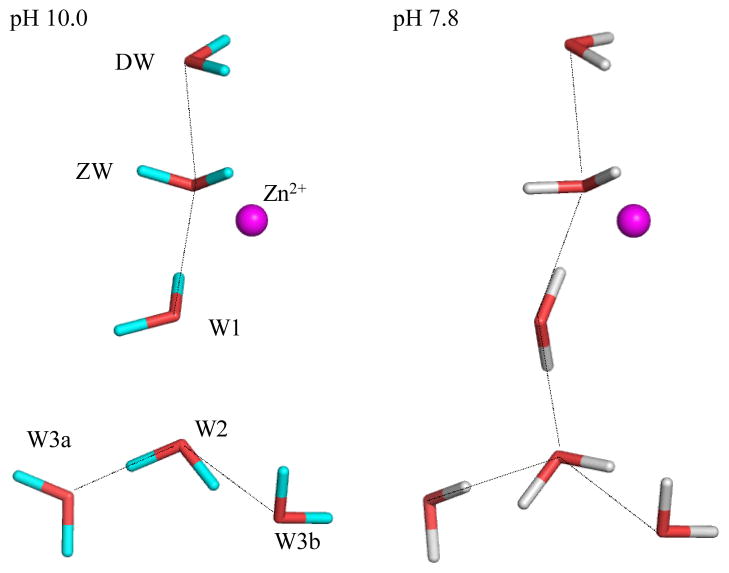
Careful analyses of omit and difference nuclear density maps revealed several interesting features in the pH 7.8 structure that are different from those at high pH. The most interesting one - from a proton transfer point of view - is that the water network has rearranged in response to the reduced pH and forms a completed hydrogen bonded network between the Zn-bound solvent (ZW) and W2 not observed before (Figure 1).
Figure 1.
Comparison stick diagram of the water networks in the active site of HCA II determined by neutron crystallography at pH 10 (left) and 7.8 (right). Zn is included for reference and waters are as labeled. Observed hydrogen bonds are shown as black lines, active site amino acid residues are omitted for clarity.
At high pH, W1 was acting as an H-bond donor to the Zn-bound water (ZW) and Thr200 (left panel in Figure 1). In this configuration there is a break in the H-bonded water network between W1 and W2. This can, in part, explain the lower observed rate constant for proton transfer between His64 and the zinc-bound solvent molecule. At pH 7.8 of the current structure, the positions of the water termed “Deep Water” (DW) because of its location in the active site and ZW are unchanged. However, W1 has flipped to engage in an H-bond to W2 and now acts as an H-bond acceptor to Thr200 (right panel in Figure 1). Accordingly, W2 and W3a have changed orientation in a compensatory manner to remain H-bonded to each other and W1. This rearrangement positions one of the W2 D atoms to act as an H-bond donor to His64 (right panel Figure 1). This represents an unbranched proton transfer pathway between ZW and His64, something that is thought to be essential for efficient proton transfer (17). Previous computational studies have implied that proton transfer occurs through W3a, but this observation supports the notion that proton transfer most likely occurs through W2 (11). It also provides a clue to the specific participation of an explicit water molecule in proton transfer. The cluster of H-bonds between W3b, Asn62, and Asn67 are completely unchanged from the high pH structure. Table 1 shows the O…O distances and O-D…O H-bond donor acceptor angles between waters in the pH 10 and 7.8 structures. Within the structural coordinate errors at 2.0 Å resolution, the O distances are identical while the D atoms rearrange.
TABLE 1.
Table shows O…O distances and angles between H-bond donor-acceptors for the water networks determined at pH 10 and pH 7.8.
| pH 10.0 | O…O in Å (O-D…O in °) | pH 7.8 | O…O in Å (O-D…O in °) |
|---|---|---|---|
| W1-ZW | 2.5 (170) | W1-ZW | 2.7 (170) |
| W1-W2 | 2.8 (n/a) | W1-W2 | 2.7 (170) |
| W2-W3a | 2.7 (150) | W3a-W2 | 2.9 (150) |
| W2-W3b | 2.8 (160) | W2-W3b | 2.7 (170) |
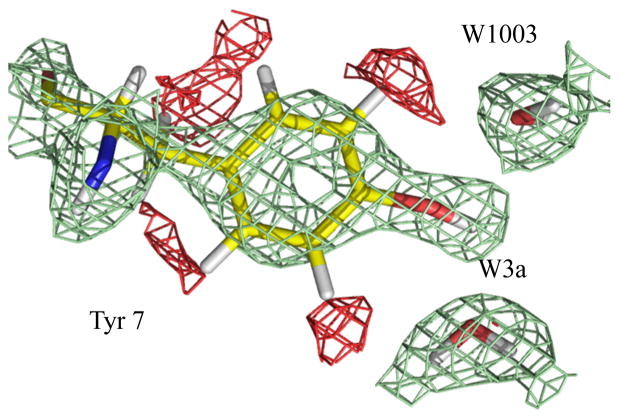
The fact that the solvent O positions are unchanged while only the D atoms are re-organizing is consistent with the significant role of the hydrophilic residues of the active site, such as Asn62 and Asn67, in stabilizing the positions of water molecules in the structure. The change in H-bonding of the water structure is most likely due to the significant changes in the charged states of two key residues, Tyr7 and His64, demonstrating the effect of changing electrostatics in the active-site cavity. An unanticipated result observed in the high pH structure was that Tyr7 was deprotonated, implying that this residue has a lower pKa than that for free Tyr in solution (Figure 2) (13).
Figure 2.
Tyr7 protonated in the pH 7.8 structure, and can act as a bifurcated H-bond donor to W3a and W1003. Positive 2Fo−Fc nuclear maps are shown in green (contoured at 1.5s) with the negative 2Fo−Fc nuclear map shown in red (contoured at 2.0s), indicating the unexchanged/non-labile H density.
As anticipated Tyr7 is indeed protonated at pH 7.8 (Figure 2) but it was thought that it would still act as H-bond acceptor from W3a and that the phenolic D atom would point towards the buried water W1003 (13). Instead, at pH 7.8 Tyr7 OD makes a bifurcated H-bond to buried water W1003 and W3a (Figure 2). The O…O distances between the phenolic O of Tyr7 to W1003 and W3a are 2.6 Å each. This significant electrostatic change from the negatively charged deprotonated oxygen to neutral protonated OD group may be causing W3a to flip and establish an H-bond with W2 (Figures 1–3). This, along with the changes observed in His64, is causing a cascade of flipping and re-establishing of H-bonds along the water network. This raises interesting implications for Tyr affecting proton transfer at high pH in vitro, such as the possibility that Tyr7 may act as a proton trap at high pH.
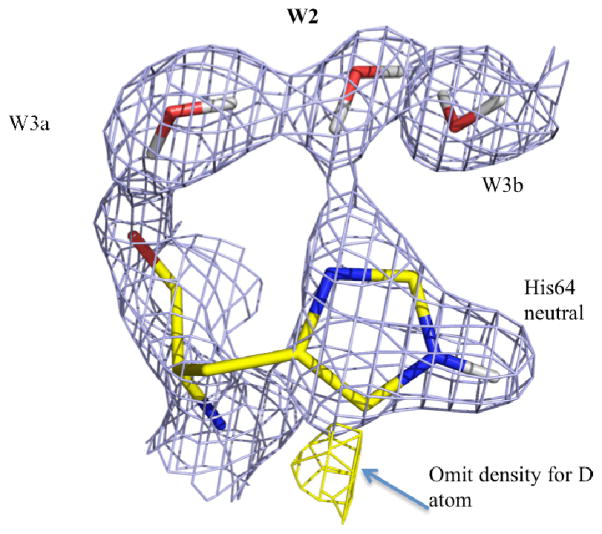
Figure 3.
The predominant conformation of the His64 side chain is neutral and in the “inward” position. The 2Fo−Fc nuclear map (contoured at 1.5σ) is shown in blue with the positive omit Fo−Fc nuclear density map (contoured at 3.0σ) in yellow, indicating the D atom density for the flipped neutral histidine conformation. Waters are as labeled, H atoms on the His imidazole ring are omitted for clarity.
As expected, at high pH His64 was observed to be neutral and in a single inward conformation. At pH 7.8 His64 occupies three superimposed forms: the predominant one is neutral (~60% occupancy), another one is flipped and neutral (~30% occupancy, Figure S3), and finally we observed a minor flipped charged species (~10% occupancy, not shown). The predominant form directs the Nδ1 electron lone pair toward W2’s D atom, indicating formation of an H-bond between W2 and His64 (Figure 3). The orientations of the 30% neutral and the minor protonated form, with their rings flipped ~180°, are similar to that observed for the neutral imidazole ring at high pH. The observation of the minor protonated species is consistent with the pKa of His64 as ~ 7 considering the pH of the crystal (pH 7.8).
These observations reveal important contributions to the understanding of the molecular details of proton transfer in HCA II. A change in pH in the crystals has permitted the observation of what might happen during catalysis. This has allowed the observation, for the first time, of a completed H-bonded network connecting the ZW to His64. In addition, His64 was observed in an intermediate state where a minor form is protonated, possibly poised to rotate to the outward conformation to deliver the excess proton to the bulk solvent.
To further probe the role of Tyr7 during catalysis 13C-labeled Tyr HCA II is being prepared for pH titrations using NMR to look at the pKa of all 7 Tyr and the active pursuit of a low pH crytal form to shed more structural light on proton transfer.
Supplementary Material
Acknowledgments
Funding Sources
The PCS is funded by the Office of Biological Environmental of the U.S. Dept. of Energy. SZF is partially funded by LANL LDRD Early Career grant # 20110535ER. PL and MM are partially funded by NIH-NIGMS GM071939. RM and DNS are partially funded through NIH-NIGMS GM25154.
ABBREVIATIONS
- HCA II
human carbonic anhydrase II
- H-bond
hydrogen bond
- PCS
Protein Crystallography Station
Footnotes
Details of crystallization, data collection, and structure refinement; crystallographic data table for both room temperature neutron and X-ray diffraction data. Additional figures of waters, His64, and active site. This material is available free of charge via the Internet at http://pubs.acs.org. Structural coordinates and experimental data have been deposited in the Protein Data Bank as PDB ID 3TMJ.
References
- 1.Silverman DN, Lindskog S. Acc Chem Res. 1988;21:30–36. [Google Scholar]
- 2.Favre N, Christ ML, Pierre AC. J Mol Cat B: Enzymatic. 2009;60:163–170. [Google Scholar]
- 3.Christianson DW, Fierke CA. Acc Chem Res. 1996;29:331–339. [Google Scholar]
- 4.Silverman DN, McKenna R. Acc Chem Res. 2007;40:669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 5.Venkatasubban KS, Silverman DN. Biochemistry. 1980;19:4984–4989. doi: 10.1021/bi00563a008. [DOI] [PubMed] [Google Scholar]
- 6.Tu CK, Silverman DN, Forsman C, Jonsson B-H, Lindskog S. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 7.Fisher SZ, Tu CK, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Biochemistry. 2007;46:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 8.Nair SK, Christianson DW. J Amer Chem Soc. 1991;113:9455–9458. [Google Scholar]
- 9.Fisher SZ, Hernandez-Prada J, Tu CK, Duda D, Yoshioka C, An H, Govindasamy L, Silverman DN, McKenna R. Biochemistry. 2005;44:1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 10.Davidson VL. Nature Chem. 2011;3:662–663. doi: 10.1038/nchem.1122. [DOI] [PubMed] [Google Scholar]
- 11.Maupin CM, McKenna R, Silverman DN, Voth GA. J Amer Chem Soc. 2009;131:7598–7608. doi: 10.1021/ja8091938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domsic JF, Avvaru BS, Kim CU, Gruner SM, Agbandje-McKenna M, Silverman DN, McKenna R. J Biol Chem. 2008;382:30766–30771. doi: 10.1074/jbc.M805353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher SZ, Kovalevsky AY, Domsic JF, Mustyakimov M, McKenna R, Silverman DN, Langan PA. Biochemistry. 2010;49:415–421. doi: 10.1021/bi901995n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovalevsky AY, Hanson BL, Mason SA, Yoshida T, Fisher SZ, Mustyakimov M, Forsyth VT, Blakeley MP, Keen DA, Langan PA. Angew Chem Int Ed. 2011;50:7520–7523. doi: 10.1002/anie.201101753. [DOI] [PubMed] [Google Scholar]
- 15.Kovalevsky AY, Katz AK, Carrell HL, Hanson L, Mustyakimov M, Fisher SZ, Coates L, Schoenborn BP, Bunick GJ, Glusker J, Langan P. Biochemistry. 2008;47:7595–7597. doi: 10.1021/bi8005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams PD, Mustyakimov M, Afonine PV, Langan PA. Acta Cryst D. 2009;65:567–573. doi: 10.1107/S0907444909011548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Q, Karplus M. J Phys Chem B. 2003;107:1071–1078. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.