Abstract
With the advent of high-throughput DNA sequencing, more than 4,000 bacterial genomes have been sequenced and are publicly available. We report a user-friendly web platform, ssGeneFinder Webserver (http://147.8.74.24/ssGeneFinder/), which is updated weekly for the automated pangenomic selection of specific targets for direct PCR detection and the identification of clinically important bacteria without the need of gene sequencing. To apply the ssGeneFinder Webserver for identifying specific targets for Salmonella enterica serovar Typhi, we analyzed 11 S. Typhi genomes, generated two specific targets, and validated them using 40 S. Typhi, 110 non-Typhi Salmonella serovars (serovar Paratyphi A, n = 4; Paratyphi B, n = 1; Typhimurium, n = 5; Enteritidis, n = 12; non-Paratyphi group A, n = 6; non-Paratyphi group B, n = 29; non-Paratyphi group C, n = 12; non-Typhi group D, n = 35; group E and others, n = 6), 115 Enterobacteriaceae isolates (Escherichia, n = 78; Shigella, n = 2; Klebsiella, n = 13; Enterobacter, n = 9; others, n = 13), and 66 human stool samples that were culture negative for S. Typhi. Both targets successfully detected all typical and atypical S. Typhi isolates, including an H1-j flagellin gene mutant, an aflagellated mutant which reacted with 2O Salmonella antiserum, and the Vi-negative attenuated vaccine strain Ty21a. No false positive was detected from any of the bacterial isolates and stool samples. DNA sequencing confirmed the identity of all positive amplicons. The PCR assays have detection limits as low as 100 CFU per reaction and were tested using spiked stool samples. Using a pangenomic approach, ssGeneFinder Webserver generated targets specific to S. Typhi. These and other validated targets should be applicable to the identification and direct PCR detection of bacterial pathogens from uncultured, mixed, and environmental samples.
INTRODUCTION
The detection and identification of infectious agents has been the central duty of clinical microbiology laboratories. Technological advances, such as antigen-specific latex agglutination (7, 23) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (18, 26), allowed the earlier identification of bacterial pathogens but were difficult to extend to mixed, uncultured, or environmental samples (2, 8). In this respect, nucleic acid amplification and related non-culture-based techniques offer the distinctive edge of being highly sensitive and specific to even fastidious and uncommon organisms, provided that the proper target is selected (14, 31).
The availability of high-throughput DNA sequencing technologies has ignited an explosion of genome data: 1,710 complete and 2,294 draft bacterial genomes have been made available, and more than 2,000 bacterial genome projects are in progress. They represent a wide diversity of clinically important bacteria, from well-known virulent species such as Mycobacterium tuberculosis (5), Staphylococcus aureus (13) and Bacillus anthracis (21) to various pathogenic species that have been recognized or described only more recently (25, 30). This wealth of information has major implications for the clinical microbiologist. First, the selection of molecular targets is no longer limited to the small number of classical phylogeny genes, as the more than 4,000 genomes offer a wide repertoire of potential gene targets for diagnostic, epidemiological, and phylogenetic studies. Second, and perhaps more importantly, the massive amount of genome data requires considerable bioinformatics expertise on the clinical microbiologist's part to unveil its full potential (24).
Recognizing such, we had proposed a novel pangenomic analysis approach to select highly specific targets for multiplex PCR identification and the detection of bacterial pathogens (10), and subsequently we developed an improved version of the algorithm, ssGeneFinder (http://147.8.74.24/ssGeneFinder/) (11). Here, we report an automated, web-based platform, ssGeneFinder Webserver, which allows the user to easily generate specific targets for any publicly available genome using our algorithm. We have also applied this ssGeneFinder Webserver to identify specific gene targets for the enteric pathogen Salmonella enterica serovar Typhi, a bacterium of both clinical and public health interest, and atypical strains which are difficult to identify are well reported (15, 29). Furthermore, the sensitivity and specificity of PCR primers designed for amplifying the specific gene targets were validated using typical and atypical strains of S. Typhi, strains of other serovars of S. enterica and other members of the Enterobacteriaceae, and stool samples negative for S. Typhi.
MATERIALS AND METHODS
Algorithm and design of ssGeneFinder Webserver.
ssGeneFinder and its web interface, the ssGeneFinder Webserver, were programmed in Perl and PHP, respectively. The core algorithm was based on the pangenomic analysis approach detailed in our previous publications (10, 11), and the current implementation is shown in Fig. 1. A notable feature of the webserver is its genome archive, which has been maintained with weekly updates using genome data from NCBI GenBank. When using the ssGeneFinder Webserver, the user only needs to specify the target species, nontarget species, and an email address for result notification. There is no need to download or install any software. A fully automated analysis is performed on the server, and the results are sent to the email address when ready.
Fig 1.
ssGeneFinder algorithm as implemented on the ssGeneFinder Webserver.
Identification of S. Typhi-specific targets using ssGeneFinder Webserver.
Eleven S. Typhi genomes were selected as targets (see Fig. S1A and B in the supplemental material), in which two were completely sequenced and the remaining nine were draft genomes (Table 1). Nontarget genomes, including 89 non-Typhi Salmonella serovar, 257 Escherichia, and 29 Shigella genomes (see Table S1 in the supplemental material) in the ssGeneFinder genome archive were specified (see Fig. S1C and D). Default settings on the server were used for the analysis (E value for conserved targets, 1 × 10−40; E value for elimination, 1.0; minimum target sequence length, 50 bp; minimum percentage of genomes in which the targets must be conserved, 95%); the email address of one of the authors was specified to simulate actual use (see Fig. S1E). The identified potential targets were obtained using the link provided in the notification email (see Fig. S1F and G). The standard operating procedure for using the ssGeneFinder Webserver is shown in Fig. S1 in the supplemental material.
Table 1.
List of S. Typhi genomes used in the study
| S. Typhi strain | Sequencing status | GenBank accession no. |
|---|---|---|
| Ty2 | Complete | AE014613a |
| CT18 | Complete | AL513382a |
| AL513383b | ||
| AL513384b | ||
| 404ty | Draft | CAAQ00000000c |
| E00-7866 | Draft | CAAR00000000c |
| E01-6750 | Draft | CAAS00000000c |
| E02-1180 | Draft | CAAT00000000c |
| E98-0664 | Draft | CAAU00000000c |
| E98-2068 | Draft | CAAV00000000c |
| E98-3139 | Draft | CAAZ00000000c |
| J185 | Draft | CAAW00000000c |
| M223 | Draft | CAAX00000000c |
Chromosome sequence.
Plasmid sequence.
Draft genome contigs.
Manual selection of reported S.
Typhi-specific targets. The sequences reported by the ssGeneFinder Webserver were subjected to a BLASTn similarity search against the nucleotide collection (nr/nt), whole-genome shotgun contigs (wgs), and human genomic plus transcript (human G+T) databases. Limited by our computational capacity, BLASTn searches against the nr/nt, wgs, and human G+T databases were not performed by the ssGeneFinder Webserver, and the user had to perform their BLASTn search at the NCBI website (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). By manual inspection of the BLAST results, potentially S. Typhi-specific targets matching other species with an E value of less than 1.0 were rejected to avoid sequence similarities that may cause unintended PCR amplification (see Table 3). PCR primers were designed for the remaining sequences using NCBI Primer-BLAST as previously described (11).
Table 3.
Conserved and potentially specific S. Typhi targets reported by ssGeneFinder Webserver
| Target no. (location on reference assembly and inferred identity) | Target length (bp) | Most significant non-S. Typhi serovar matcha (accession no. [BLAST E value]) | Most significant match from human sequencesb (accession no. [BLAST E value]) | Note |
|---|---|---|---|---|
| Usid000034 (contig01715, bp 759-849; S. Typhi-specific intergenic region) | 91 | Vicugna pacos wgs sequence (ABRR01142651.1 [0.56]) | Homo sapiens chromosome 15 (NT_010194.17 [0.6]) | Excluded from further analysis |
| Usid000040 (contig00298, bp 2635-2730; fimbrial chaperone protein gene) | 96 | Tarsius syrichta wgs sequence (ABRT011036967.1 [2.0]) | No match | Feasible molecular target |
| Usid000041 (contig00298, bp 2842-2970; fimbrial chaperone protein gene) | 129 | E. cloacae SCF1 (CP002272.1 [0.007]) | H. sapiens chromosome 16 (NT_010498.15 [2.1]) | Excluded from further analysis |
| Usid000076 (contig02563, bp 293-384; fimbrial protein gene) | 92 | Antirrhinum hispanicum genomic sequence (AJ300474.1 [0.31]) | H. sapiens chromosome 16 (NT_010498.15 [2.1]) | Excluded from further analysis |
| Usid000077 (contig02563, bp 407-531; fimbrial protein gene) | 125 | Tursiops truncatus wgs sequence (ABRN02054357.1 [0.046]) | H. sapiens chromosome 1 (NT_004487.19 [0.6]) | Excluded from further analysis |
| Usid000078 (contig02563, bp 552-664; fimbrial protein gene) | 113 | Pyrolobus fumarii 1A (CP002838.1 [3.8]) | No match | Feasible molecular target |
| Usid000090 (contig01717, bp 1254-1406; hypothetical protein-coding gene) | 153 | Daubentonia madagascariensis wgs sequence (AGTM011597351.1 [3e−4]) | H. sapiens chromosome 1 (NT_032977.9 [0.049]) | Excluded from further analysis |
| Usid000091 (contig01717, bp 1453-1552; hypothetical protein-coding gene) | 100 | Rattus norvegicus wgs sequence (AABR05012442.1 [0.56]) | H. sapiens chromosome 5 (NW_001838934.1 [0.17]) | Excluded from further analysis |
| Usid000092 (contig01717, bp 1586-1691; hypothetical protein-coding gene) | 106 | Canis lupus familiaris wgs sequence (AAEX03007752.1 [0.046]) | H. sapiens chromosome 1 (NT_032977.9 [2.1]) | Excluded from further analysis |
Using online BLASTn search against the nucleotide collection (nr/nt) and whole-genome shotgun contigs (wgs) with default parameters.
Using online BLASTn search against the human genomic plus transcript (human G+T) database with default parameters.
Bacterial strains and identification.
All clinical isolates were phenotypically identified by conventional biochemical methods and the Vitek system (GNI+) (bioMérieux Vitek, Durham, NC). The 40 S. Typhi isolates included representative strains such as S. Typhi Ty2 and the attenuated Vi-negative vaccine strain S. Typhi Ty21a; also included were 36 stool and blood culture isolates from local tertiary hospitals, further characterized by their positive agglutination reaction with poly(O), poly(H), 9O, Vi, and H-d Salmonella antisera. Two atypical S. Typhi isolates previously published by our group were also included. One was a locally isolated H1-j strain of S. Typhi from a patient with no history of travel to Indonesia or personal contact with Indonesians (15). The strain agglutinated with poly(O), 9O, Vi, and H1-j Salmonella antisera but not with poly(H) antisera. The other strain was an electron microscopy-confirmed aflagellated mutant of S. Typhi; apart from positive agglutination with poly(O), 9O, and Vi Salmonella antisera, the isolate also agglutinated with 2O antiserum but not with poly(H) or any individual H Salmonella antisera (29). To serve as negative controls, 110 non-Typhi Salmonella serovar isolates were included. They included S. enterica serovar Paratyphi A (n = 4), S. enterica serovar Paratyphi B (n = 1), S. enterica serovar Typhimurium (n = 5; including ATCC 14028 and mutants SL3261 and SL7207), S. enterica serovar Enteritidis (n = 12; identified according to the Kauffmann-White scheme), and other Salmonella isolates of various serogroups according to their O antigens, including non-Paratyphi group A Salmonella serovar (n = 6), non-Paratyphi Salmonella serovar group B (n = 29), non-Paratyphi Salmonella serovar group C (n = 12), non-Typhi Salmonella serovar group D (n = 35), Salmonella serovar group E (n = 3), and others (n = 3). One hundred fifteen isolates from the family Enterobacteriaceae, including 77 Escherichia coli isolates, an Escherichia fergusonii isolate, 1 of each of Shigella sonnei and Shigella flexneri, and 35 other isolates, including Klebsiella spp. (n = 13), Enterobacter spp. (n = 9), and other genera (n = 13), were also included as negative controls (Table 2).
Table 2.
List of bacterial isolates used for target verification
| Bacterial species | Strain no. | Source |
|---|---|---|
| S. Typhi | Ty2, Ty21a, PW1957, PW1958, PW1961, PW2084-PW2091, PW2218-PW2224, PW2227-PW2246 | Reference strains and clinical isolates |
| Non-Typhi Salmonella serovar | ATCC 14028, SL3261, SL7207, PW2092-PW2141, PW2159-PW2208, PW2247-PW2253 | Reference strains and clinical isolates |
| E. coli | EB01, EC01-EC59 (AKLT), E01-E17 (JLLT) | Clinical isolates |
| Escherichia fergusonii | EB26 | Clinical isolate |
| Shigella sonnei | JLLT-159558 | Clinical isolate |
| Shigella flexneri | JLLT-169945 | Clinical isolate |
| Klebsiella pneumoniae | EB02, EB04, EB14, EB16, EB20, EB22, EB29, EB30, EB37, EB38 | Clinical isolates |
| Klebsiella oxytoca | EB05, EB24, EB35 | Clinical isolates |
| Proteus mirabilis | EB09, EB10, EB17, EB19, EB23, EB27, EB31, EB34 | Clinical isolates |
| Enterobacter cloacae | EB11, EB12, EB15, EB18, EB21, EB28, EB33, EB36 | Clinical isolates |
| Morganella morganii | EB06, EB32 | Clinical isolates |
| Citrobacter koseri | EB03 | Clinical isolate |
| Enterobacter aerogenes | EB39 | Clinical isolate |
| Kluyvera intermedia | EB13 | Clinical isolate |
| Pantoea agglomerans | EB40 | Clinical isolate |
Extraction of bacterial DNA and PCR amplification of the S. Typhi-specific gene targets.
DNA extraction from bacterial isolates was performed as previously published (10, 32). The PCR mixture (25 μl) for the amplification of S. Typhi-specific gene targets contained purified DNA template (1.0 μl), 1.0 μM each target-specific primer (Sigma-Aldrich, Steinheim, Germany), 2.5 μl 10× PCR buffer II, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphates (dNTPs) (GeneAmp; Applied Biosystems, CA), and 0.675 U Taq polymerase (AmpliTaq Gold; Applied Biosystems, CA). Thermocycling was performed in an automated thermocycler (Veriti 96-well fast thermal cycler; Applied Biosystems, CA) with a hot start at 95.0°C for 10 min, 40 touchdown cycles of 95.0°C for 1 min, annealing for 1 min at temperatures decreasing from 70.0 to 50.5°C (with 0.5°C decremental steps), 72.0°C for 30 s, and a final extension at 72.0°C for 7 min. Analytical gel electrophoresis, DNA sequencing using the reverse primers for amplicon validation, and the assessment of PCR sensitivity were performed as previously described (10). Three culture-negative stool samples were spiked, each with 3 × 103, 3 × 104, and 3 × 104 CFU per sample. Bacterial DNA from the nine spiked samples was extracted and purified for PCR. A pair of pan-Salmonella primers (LPW19677 [5′-CCCAGCCGGTGCCAGAAATG-3′] and LPW19678 [5′-TGGATGTCCGTACTGATGGTGTGT-3′]) was also designed to serve as an amplification positive control for the Salmonella isolates.
Sensitivity and specificity of the assay.
The sensitivity of the PCR was assessed with S. Typhi isolate PW2246 using a previously method published (10). To assess the specificity, in addition to the 110 non-Typhi Salmonella and 115 Enterobacteriaceae isolates, 66 human stool samples that were culture negative for S. Typhi were randomly selected from a clinical microbiology laboratory in a teaching hospital. Samples were stored at 4°C until DNA extraction and purification and processed in lots of about 10 specimens, and one specimen from each lot was spiked with 20 μl S. Typhi suspension in phosphate-buffered saline (PBS) at 0.5 McFarland standard to indicate successful DNA extraction and PCR inhibitor removal.
S. Typhi sequence and analysis data.
All of the genome files used in our sample run, together with the generated reports, targets, and alignments, are available from the ssGeneFinder Webserver page at http://147.8.74.24/tom/ssGeneFinder%20Webserver/browse.php?id=953b7d07e0de86e5b843f70d74865f29.
RESULTS
ssGeneFinder Webserver and functionality of the platform.
As of March 2012, the genome archive of the ssGeneFinder Webserver (version 2.3.2) contained 1,925 complete and 3,316 draft bacterial genomes. Any nonduplicate combinations of target and nontarget genomes could be specified as input. There was no restriction on the number of target and nontarget genomes, but the number of iterative BLASTn searches was fixed at 10. Preoptimized parameters were available as defaults on the ssGeneFinder Webserver. The user's email address, however, is required for the server to issue a notification indicating the completion of the analysis.
The result notification email contained a summary report, a machine log file of the run, and a link to a customized page where the identified potential targets, their multiple alignments generated from the target genomes, and all of the genome files used in the analysis could be downloaded.
Identification of S. Typhi-specific targets.
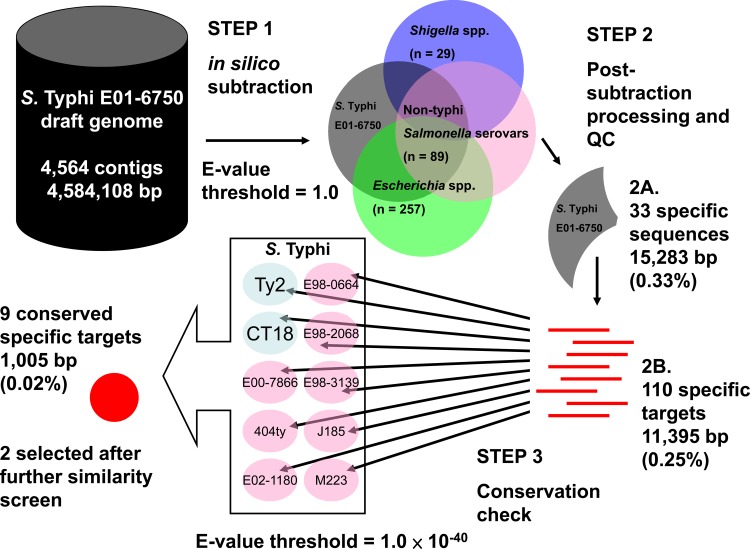
The starting genome selected by the ssGeneFinder Webserver was that of S. Typhi E01-6750. The draft genome contained 4,564 sequence contigs with a total length of 4,584,108 bp, excluding ambiguous nucleotides (Fig. 1, step 1). After 10 iterative rounds of in silico elimination, 33 sequences (total length of 15,283 bp; 0.33% of the starting genome) were found to be specific to S. Typhi E01-6750, i.e., not found in any of the nontarget Salmonella, Shigella, or Escherichia genomes using the default threshold (Fig. 1, step 2A). As ssGeneFinder used sequence masking to improve the efficiency of the iterative search process, the 33 sequences were further subjected to unmasking. In the process, some sequences were broken into fragments, as they contained regional similarities to the nontarget genomes; some fragments became shorter than the minimum target length specified and were eliminated. One hundred ten sequence fragments (total length, 11,395 bp; 0.25% of the starting genome) remained after the process and were used as queries to search against the 11 target S. Typhi genomes to determine if they were conserved (Fig. 1, step 2B). Finally, nine sequence regions (total length, 1,005 bp; 0.02% of the starting genome) were found to be conserved in at least 95% of the target genomes (Fig. 1, step 3) and reported as potential targets to the user (Table 3).
Screening and target selection.
For the online BLASTn search, seven of the nine potential targets were excluded because of sequence similarity to DNA sequences from human and other non-Typhi Salmonella serovar species (E value for elimination, 1.0). Two potential targets (usid000040 and usid000078) had no match to any sequence in the nucleotide collection and human genomic and transcript databases, having an E value below the threshold, and they were subjected to primer design (Table 3).
Primers were designed using Primer-BLAST to amplify the two potential targets: usid000040, an S. Typhi-specific fimbrial chaperone protein gene, with the pair LPW18819 (5′-ACGAGGCAAAGACGAGGGTGA-3′) and LPW18820 (5′-AGCCCTGTTAAGCAAGCGCTTTAG-3′); usid000078, an S. Typhi-specific fimbrial protein gene, with the pair LPW18821 (5′-ACAGCAATCGACGTTGCAATACTT-3′) and LPW18822 (5′-TATACCGCTCGTTCGCCGCT-3′).
PCR detection and identification of S. Typhi.
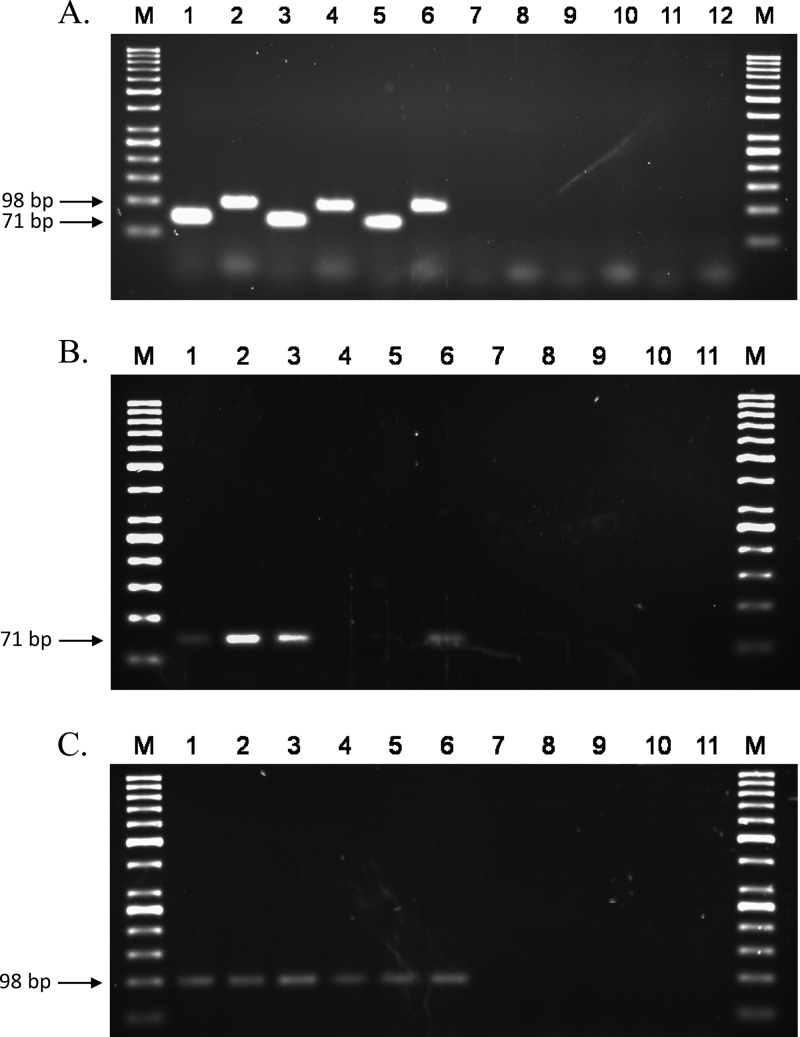
For all of the 40 S. Typhi isolates, both PCR assays produced amplicons of the expected size, 71 bp for target usid000040, the S. Typhi-specific fimbrial chaperone protein gene fragment amplified using LPW18819/LPW18820, and 98 bp for target usid000078, the S. Typhi-specific fimbrial protein gene fragment amplified using LPW18821/LPW18822 (Fig. 2A). The amplicons were confirmed by DNA sequencing unidirectionally using either LPW18820 for usid000040 or LPW18822 for usid000078. The sequences obtained were subjected to a BLASTn search against the genomic region of S. Typhi, and the identities of all amplicons were confirmed. All Salmonella isolates were successfully amplified using the pan-Salmonella primers, and none of the 110 non-Typhi Salmonella serovar isolates gave positive results in either of the S. Typhi assays.
Fig 2.
(A) Specificity of the S. Typhi PCR. Lane M, molecular marker GeneRuler 50-bp DNA ladder; lanes 1, 3, and 5, S. Typhi isolates PW1957 (an H1-j strain of S. Typhi that is uncommonly found outside Indonesia), PW1958 (an S. Typhi variant with 2O and 9O antigens), and ATCC 19430 (S. Typhi Ty2) using primer pair LPW18819/LPW18820; lanes 2, 4, and 6 are similar to lanes 1, 3, and 5, except primer pair LPW18821/LPW18822 was used; lane 7, S. Typhi culture-negative stool sample S01 using primer pair LPW18819/LPW18820; lane 8, S. Typhi culture-negative stool sample S02 using primer pair LPW18821/LPW18822; lanes 9 and 10, Klebsiella pneumoniae isolate EB02 using either primer pair in the order described for lanes 1 and 2 (LPW18819/20 for lane 9 and LPW18821/22 for lane 10); lanes 11 and 12, Pantoea agglomerans isolate EB40. The predicted amplicon sizes for LPW18819/LPW18820 and LPW18821/LPW18822 were 71 and 98 bp, respectively. (B) Sensitivity of the primer pair LPW18819/LPW18820. Lane M, molecular marker GeneRuler 50-bp DNA ladder; lanes 1 to 3, spiked stool samples CE5, BE5, and AE5 at an inoculum of 3.07 × 105 CFU per sample; lanes 4 to 6, spiked stool samples CE4, BE4, and AE4 at an inoculum of 3.07 × 104 CFU per sample; lanes 7 to 9, spiked stool samples CE3, BE3, and AE3 at an inoculum of 3.07 × 103 CFU per sample; lane 10, S. Typhi culture-negative stool sample NS28; lane 11, S. Typhi culture-negative stool sample NS29. For the spiked stool samples, each PCR mixture contained 0.5 μl of purified DNA extract as the template (total of 200 μl of purified extract per sample). The predicted amplicon size was 71 bp. (C) Sensitivity of the primer pair LPW18821/22. Lane M, molecular marker GeneRuler 50-bp DNA ladder; lanes 1 to 3, spiked stool samples CE5, BE5, and AE5 at an inoculum of 3.07 × 105 CFU per sample; lanes 4 to 6, spiked stool samples CE4, BE4, and AE4 at an inoculum of 3.07 × 104 CFU per sample; lanes 7 to 9, spiked stool samples CE3, BE3, and AE3 at an inoculum of 3.07 × 103 CFU per sample; lane 10, S. Typhi culture-negative stool sample NS28; lane 11, S. Typhi culture-negative stool sample NS29. For the spiked stool samples, each PCR mixture contained 0.5 μl of purified DNA extract as the template (total of 200 μl of purified extract per sample). The predicted amplicon size was 98 bp.
Sensitivity and specificity of the PCR.
The S. Typhi PW2246 suspension in PBS had a viable count of 1.53 × 108 CFU per ml. Assuming the complete extraction of bacterial DNA from the spiked stool samples and no loss during the purification process, the detection limit of the primer pair LPW18819/LPW18820 was 1,000 CFU per 25-μl PCR mix (Fig. 2B); that of primer pair LPW18821/LPW18822 was 100 CFU per 25-μl PCR mix (Fig. 2C). There were no false positives for the 110 isolates of non-Typhi Salmonella serovars tested. None of the 115 family Enterobacteriaceae (non-Salmonella) isolates produced PCR amplicons compatible with the predicted sizes, i.e., 71 and 98 bp, for LPW18819/LPW18820 and LPW18821/LPW18822. None of the 66 culture-negative stool samples gave a positive result for either of the primer pairs (Fig. 2).
DISCUSSION
We report a user-friendly web platform, ssGeneFinder Webserver, for the selection of specific genomic targets for the direct PCR detection and identification of bacterial pathogens from cultured isolates and spiked stool samples. To test the concept of using the huge amount of genome sequence data for identifying specific diagnostic targets for identifying bacteria, we used a pangenomic approach to design PCR primers to detect Burkholderia species that were occasionally misidentified by commercial systems and required DNA sequencing for unambiguous differentiation (10). Subsequently, the process was automated and was successfully applied to identifying specific molecular targets for the detection of the E. coli O104:H4 strain implicated in the Europe 2011 outbreak (11). Users can download the required genomes from NCBI GenBank and the ssGeneFinder software from our website (http://147.8.74.24/tom/ssGeneFinder/), and they can run the analysis on their own computers. While such a technique allowed advanced users and sequencing centers to use their unpublished genome data in customized analyses, to run an ssGeneFinder analysis still required computer skills in addition to microbiological knowledge in selecting realistic negatives as nontarget genomes. In the ssGeneFinder Webserver described in this study, all of the bacterial genome data from NCBI GenBank have been downloaded onto the server; all of the operations required for the analysis were converted to a simple, user-friendly interface. Moreover, the user does not even need to install the software on his computer, as the web interface simply required an internet connection and an email account to receive the analysis results.
Accurate identification of S. Typhi has major clinical and public health implications. Using the ssGeneFinder Webserver presented in this paper, we successfully identified serovar-specific targets for S. Typhi, and the sensitivity and specificity of the two selected targets were characterized and confirmed using a panel of 40 S. Typhi isolates, 110 non-Typhi Salmonella isolates, 115 Enterobacteriaceae (non-Salmonella) isolates, and 66 stool samples that cultured negative for S. Typhi. Unlike the nontyphoidal strains, typhoidal Salmonella isolates are associated with serious extraintestinal manifestations of typhoid fever with a mortality of 10% without treatment (4). Typhoidal Salmonella strains may also persist in the gallbladder of asymptomatic carriers and be chronically shed for more than 1 year (17). Although serotyping by the determination of the O, H, and Vi antigens can differentiate among different S. enterica subsp. enterica serovars and remains the gold standard for their routine identification (27), as highlighted by the atypical isolates we and others reported, variations in antigenic expression may prevent accurate serotype determination (15, 29). The molecular typing of the Salmonella serovars has been hindered by the lack of a single marker which can differentiate among all serovars. While 16S rRNA gene sequences have been used to type isolates to the subspecies level (29), molecular serogrouping required the simultaneous interrogation of gene clusters involved in Salmonella antigen biosynthesis, such as the rfb genes for various O antigens (9). DNA microarray-based typing has revealed the presence of groups of serovar-specific genes (20, 22) and multiplex PCR assays (12, 16), and panels (1) intending to differentiate among the different serovars have been published. Although a commercial low-density DNA microarray for the identification of Salmonella serovars has been marketed, it failed to distinguish between some serovars and was not tested using S. Typhi (28). Recently, matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry-based typing has been shown to identify some Salmonella serovars, but again S. Typhi was not included in the study (6). While multiple serodiagnostic tests, such as the Widal test, Multi-Test Dip-S-Ticks, TyphiDot, and TUBEX, are available in certain localities, they do not directly identify the bacterium and have sensitivity and specificity depending greatly on the time of testing and patient's history of S. Typhi and nontyphoidal Salmonella infection (19). None of the above-described tests are applicable to the direct detection of S. Typhi from mixed, uncultured, or environmental samples, such as the stool samples from an S. Typhi carrier and contaminated food. Although this PCR-based assay does not yield an isolate for further microbiological characterization, it can potentially complement current techniques by offering a rapid screening and identification test for diverse specimens suspected to contain S. Typhi. Nevertheless, as the protocol presented did not achieve the sensitivity of enrichment culture, it currently does not replace direct isolation for the detection of S. Typhi; enrichment culture, for instance, has been reported to detect as few as 1 to 10 Salmonella organisms from a stool sample (3).
In the current study, as relatively few genomes of the S. Paratyphi serovars (only one complete genome for each of S. Paratyphi A, B, and C) have been published, they were not included as targets. The PCR assay designed therefore was specific to S. Typhi and did not detect the Paratyphi serovars. This incompleteness of the public genomic sequence repository relative to all pathogenic, commensal, and environmental species certainly limits target selection using the server, although such limitations are expected to be overcome by high-throughput sequencing technologies. Where the 16S rRNA gene is limited by its phylogenetic resolution (33), the ssGeneFinder Webserver provides an effective algorithm to identify specific targets for detection and identification. Being user friendly and efficient in computation, ssGeneFinder is useful in identifying gene targets that were sensitive despite intragroup variation in the target organisms; specific to various levels in classification, including species, isolate, and serovar; and, with further validation, potentially applicable to the direct detection and identification of clinically important bacteria from a wide diversity of uncultured, mixed, and environmental samples.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly supported by the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau, Research Grants Council Grant, University Development Fund, Committee for Conference and Research Grant, The University of Hong Kong, and the Croucher Foundation.
Footnotes
Published ahead of print 21 March 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Arrach N, et al. 2008. Salmonella serovar identification using PCR-based detection of gene presence and absence. J. Clin. Microbiol. 46:2581–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buchwald DS, Blaser MJ. 1984. A review of human salmonellosis. II. Duration of excretion following infection with nontyphi Salmonella. Rev. Infect. Dis. 6:345–356 [DOI] [PubMed] [Google Scholar]
- 4. Cohen ML. 1992. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science 257:1050–1055 [DOI] [PubMed] [Google Scholar]
- 5. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 6. Dieckmann R, Malorny B. 2011. Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 77:4136–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doern GV. 1982. Evaluation of a commercial latex agglutination test for identification of Staphylococcus aureus. J. Clin. Microbiol. 15:416–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferreira L, et al. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fitzgerald C, et al. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45:3323–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ho CC, et al. 2011. Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: a proof-of-concept study. J. Clin. Microbiol. 49:814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ho CC, Yuen KY, Lau SK, Woo PC. 2011. Rapid identification and validation of specific molecular targets for detection of Escherichia coli O104:H4 outbreak strain by use of high-throughput sequencing data from nine genomes. J. Clin. Microbiol. 49:3714–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S, et al. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 44:3608–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuroda M, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 14. Lau SK, et al. 2011. First report of disseminated Mycobacterium skin infections in two liver transplant recipients and rapid diagnosis by hsp65 gene sequencing. J. Clin. Microbiol. 49:3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau SK, et al. 2005. Typhoid fever associated with acute appendicitis caused by an H1-j strain of Salmonella enterica serotype Typhi. J. Clin. Microbiol. 43:1470–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leader BT, Frye JG, Hu J, Fedorka-Cray PJ, Boyle DS. 2009. High-throughput molecular determination of Salmonella enterica serovars by use of multiplex PCR and capillary electrophoresis analysis. J. Clin. Microbiol. 47:1290–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levine MM, Black RE, Lanata C. 1982. Precise estimation of the numbers of chronic carriers of Salmonella typhi in Santiago, Chile, an endemic area. J. Infect. Dis. 146:724–726 [DOI] [PubMed] [Google Scholar]
- 18. Neville SA, et al. 2011. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49:2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsen SJ, et al. 2004. Evaluation of rapid diagnostic tests for typhoid fever. J. Clin. Microbiol. 42:1885–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Porwollik S, et al. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 186:5883–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Read TD, et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86 [DOI] [PubMed] [Google Scholar]
- 22. Scaria J, et al. 2008. Microarray for molecular typing of Salmonella enterica serovars. Mol. Cell. Probes 22:238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steinmetz I, et al. 1999. Rapid identification of Burkholderia pseudomallei by latex agglutination based on an exopolysaccharide-specific monoclonal antibody. J. Clin. Microbiol. 37:225–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tse H. 2010. Computer code: more credit needed. Nature 468:37. [DOI] [PubMed] [Google Scholar]
- 25. Tse H, et al. 2010. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J. Bacteriol. 192:1471–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Versalovic J. (ed). 2011. Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
- 28. Wattiau P, et al. 2008. Evaluation of the Premi Test Salmonella, a commercial low-density DNA microarray system intended for routine identification and typing of Salmonella enterica. Int. J. Food Microbiol. 123:293–298 [DOI] [PubMed] [Google Scholar]
- 29. Woo PC, Fung AM, Wong SS, Tsoi HW, Yuen KY. 2001. Isolation and characterization of a Salmonella enterica serotype Typhi variant and its clinical and public health implications. J. Clin. Microbiol. 39:1190–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Woo PC, et al. 2009. The complete genome and proteome of Laribacter hongkongensis reveal potential mechanisms for adaptations to different temperatures and habitats. PLoS Genet. 5:e1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woo PC, et al. 2010. Internal transcribed spacer region sequence heterogeneity in Rhizopus microsporus: implications for molecular diagnosis in clinical microbiology laboratories. J. Clin. Microbiol. 48:208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woo PC, et al. 2002. Streptococcus sinensis sp. nov., a novel species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40:805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woo PC, et al. 2011. Automated identification of medically important bacteria by 16S rRNA gene sequencing using a novel comprehensive database, 16SpathDB. J. Clin. Microbiol. 49:1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.