Abstract
Viral envelope proteins mediate interactions with host cells, leading to internalization and intracellular propagation. Envelope proteins are glycosylated and are known to serve important functions in masking host immunity to viral glycoproteins. However, the viral infectious cycle in cells may also lead to aberrant glycosylation that may elicit immunity. Our knowledge of immunity to aberrant viral glycans and glycoproteins is limited, potentially due to technical limitations in identifying immunogenic glycans and glycopeptide epitopes. This work describes three different complementary methods for high-throughput screening and identification of potential immunodominant O-glycopeptide epitopes on viral envelope glycoproteins: (i) on-chip enzymatic glycosylation of scan peptides, (ii) chemical glycopeptide microarray synthesis, and (iii) a one-bead-one-compound random glycopeptide library. We used herpes simplex virus type 2 (HSV-2) as a model system and identified a simple O-glycopeptide pan-epitope, 501PPA(GalNAc)TAPG507, on the mature gG-2 glycoprotein that was broadly recognized by IgG antibodies in HSV-2-infected individuals but not in HSV-1-infected or noninfected individuals. Serum reactivity to the extended sialyl-T glycoform was tolerated, suggesting that self glycans can participate in immune responses. The methods presented provide new insight into viral immunity and new targets for immunodiagnostic and therapeutic measures.
INTRODUCTION
Envelope viruses encode one or more membrane proteins that mediate specific binding to host cell ligands and direct early events of membrane fusion and internalization (32). These envelope proteins are often glycosylated, and glycosylation is carried out in the endoplasmic reticulum (ER)-Golgi secretory pathway by host cell-encoded glycosyltransferases (16). Use of the host cell glycosylation machinery for production of viral glycoproteins may ensure that glycan structures are compatible with the host immune system. In this respect, glycans on envelope proteins may provide an immunological shield facilitating evasion from host immunity (39). Large N-linked glycans on influenza virus hemagglutinin and the HIV-1 glycoprotein gp120 have especially been linked to immune evasion (9). However, glycans found on viral glycoproteins, such as high-mannose N-glycans and GalNAc O-glycans (32, 39), are often immature, and they may interact with anti-carbohydrate antibodies and innate immune lectin receptors. The latter may aid in induction of immunity or, in fact, facilitate virus infection.
In addition, viral infection may alter the host cell glycosylation machinery by disruption of the ER-Golgi organization and/or induction of changes in the expression of the required glycosyltransferase repertoire (30, 36). Generally, our knowledge of humoral immunity to aberrant viral glycans and glycoproteins is limited. A major reason for this may be due to technical limitations in identifying and characterizing immunity to complex glycan structures, particularly with combined glycopeptide epitopes. Recently, we described the existence of cancer cell-produced immunodominant O-glycopeptide epitopes that comprise short peptide sequence motifs and aberrant O-glycans (4, 34, 42). Detection of immunity to such combined glycopeptide epitopes requires the use of related glycoforms of the relevant glycoproteins in immune assays. We believe this is a major limiting factor in detection and appreciation of immunity to viral envelope glycoproteins.
In this work, we developed the following different strategies to display (glyco)peptide libraries covering the envelope glycoprotein gG-2 of herpes simplex virus type 2 (HSV-2), which is known to express O-glycans (13, 33): (i) enzymatic on-chip glycosylation of scan peptides, (ii) chemical synthesis of glycopeptide analogues of scan peptides displayed on a microarray chip, and (iii) a one-bead-one-compound (OBOC) random glycopeptide library for epitope discovery and characterization (see Table S1 in the supplemental material for a concise strategy summary). Applying these techniques, we analyzed sets of HSV-2-positive (n = 44) and HSV-2-negative (n = 40) clinically validated sera for the presence of IgG antibodies reactive to displayed peptides and glycopeptides of the gG-2 protein. We identified one prominent glycopeptide pan-reactive epitope that was broadly recognized by serum IgG of HSV-2-seropositive individuals but not by that of seronegative individuals. These data demonstrate that antibodies to combined glycopeptide epitopes are present in serum from HSV-2-infected patients as a result of the infection. In addition to immunity, such glycopeptide epitopes may be of importance for epitope spreading and autoimmune disease in long-term infections (29).
MATERIALS AND METHODS
Synthetic peptide microarrays.
Peptides were prepared by automated peptide synthesis on a Syro II peptide synthesizer (MultiSynTech, Witten, Germany) by a modified 9-fluorenylmethoxy carbonyl (Fmoc)–solid-phase peptide synthesis (SPPS) methodology protocol (see the supplemental Methods in the supplemental material for details) (5). Helix pomatia agglutinin (HPA)-purified mgG-2 was prepared as previously described (37). Protein and peptide products were immobilized on N-hydroxysuccinimide (NHS)-activated hydrogel-coated MPX16 or MPX48 glass slides with a Teflon mask (Schott Nexterion SlideH) with a BioRobotics MicroGrid II spotter (Genomics Solution) using Stealth 3B Microspotting pins (ArrayIt). Approximately 6 nanoliters of glycopeptide/protein in print buffer (150 mM phosphate, 0.005% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate},0.03% NaN3; pH 8.5) was deposited in 16 identical subarrays in a 28-by-28 subgrid at a 0.21-mm pitch between spots or in 48 identical subarrays in a 12-by-12 subgrid at a 0.19-mm pitch between spots per slide format. After printing, the slides were incubated at 70% humidity for 60 min, and the remaining NHS groups were deactivated in blocking buffer (50 mM ethanolamine in 50 mM borate buffer; pH 9.2) for 1 h and then rinsed in Millipore water and spun dry.
On-slide enzymatic glycosylation (synthetic approach 1).
Blocked slides were fitted with adhesive superstructures from which the internal frame for subarray compounding was cut out to form one single-slide-sized well. The well was filled with 1.5 ml of glycosylation mixture (20 mM UDP-GalNAc, 25 μg GalNAc-transferase 2 [GalNAc-T2], 25 mM cacodylic acid sodium [pH 7.4], 10 mM MnCl2, 0.25% Triton X-100), covered with an adhesive slip, placed in a humid chamber, and incubated for 24 h at 37°C. Slides were then washed with phosphate-buffered saline (PBS)-Tween (PBST) (0.5%) (5 min, shaking) and PBS (5 min, shaking) and submerged in sodium citrate buffer (100 mM, pH 2.5) for 15 min while rocking. Slides were again washed with PBS-Tween and PBS, rinsed thoroughly with water, dried by centrifugation, and immediately used in the binding assay described below.
Chemical glycopeptide synthesis (synthetic approach 2).
Site-directed addition of various O-linked mono- or disaccharide units at predefined Ser or Thr units was accomplished by using glycosylated amino acid precursors directly in the peptide synthesis procedure. Thus, the Fmoc-protected glycoamino acids Tn (GalNAcα)-Ser/Thr, Glcβ-Thr, Manα-Thr, GalNAcα-Ser, and Core1 (Galβ1-3GalNAcα)-Thr (with hydroxyl groups of glycan haptens in acetylated form; Sussex Research, Ottawa, Canada) were used in place of corresponding nonglycosylated amino acids at selected positions during peptide synthesis (5). For further details, see the supplemental Methods in the supplemental material.
Binding assay and scanning.
Sera from a total of 84 individuals were collected and used to probe chips displaying the HSV-2 (glyco)peptide microarrays. The serological status of each patient had previously been determined as follows using commercially available enzyme-linked immunosorbent assays (ELISAs) (FocuSelect 1 and 2 IgG): 22 coinfected HSV-1/HSV-2 individuals and 22 HSV-2-seropositive, 20 HSV-1-seropositive, and 20 seronegative individuals. A sample from one patient with frequent relapses of HSV-2 meningitis was also included in the study. Serum samples for microarray analysis were diluted (1:60 or 1:180) into staining buffer (SB; 0.5 M NaCl, 3 mM KCl, 1.5 mM KH2PO4, 6.5 mM Na2HPO4, 1% bovine serum albumin [BSA], 1% Triton X-100; pH = 7.4). For primary and secondary staining, biotinylated lectins (1 to 10 μg/ml; Vector Labs), goat anti-human IgG (Fc specific, 10 μg/ml; Sigma-Aldrich), and streptavidin-Cy3 (2.5 μg/ml; Invitrogen) were diluted in SB. MPX16 slides were fitted with adhesive superstructures (Schott Nexterion), and subarray wells were filled with 60 μl each of sample solution (primary or secondary alike). For the MX48 Teflon-coated slides, the sample solution (5 μl/well) was applied directly onto the slide. All incubation steps were performed in a humid chamber at room temperature for 1 h and were separated by two wash steps in PBS-Tween. After a final wash in PBS, slides were rinsed in water, dried by centrifugation (200 × g), and scanned, and images were analyzed and quantified. The slides were scanned with a ProScanArray microarray scanner (Perkin Elmer) equipped with 3 lasers for excitation at 488 nm, 543 nm, or 633 nm. The scanned images were analyzed with ScanArray Express software. For Cy3 fluorescence, 543 nm (excitation) and 570 nm (emission) were used. Spots were identified using automated spot finding with manual adjustments for occasional irregularities. The mean value of relative fluorescence intensity was used, and the spot intensities were determined by subtracting the median pixel intensity of the local background from the average pixel intensity within the spots. Quadruplicate spots were averaged. The quality control covered intra- and interchip quality analyses of replicates, and the coefficient of the variation (CV) was generally <10%. Patient samples with a relative fluorescence value higher than two standard deviations over the mean of the control group were designated positive.
Random OBOC library staining and sequence deconvolution (synthetic approach 3).
The combinatorial OBOC glycopeptide random synthesis was performed in a custom-made Teflon cylinder reactor with 20 wells as described previously (28) (for details, see the supplemental material). Prior to analysis of HSV-2-positive sera, bead slurry aliquots in methanol (4.0 ml, approximately 10,000 beads/ml) were drained, incubated in PBST for 15 min, and drained again prior to staining. Beads were placed in a fitted polyethylene syringe and covered with a pool of sera from three HSV-2-positive patients (1:100 dilution in SB), and then a plunger was inserted to seal. After 1 h of incubation at room temperature, the beads were drained and washed with PBS-Tween. Next, the beads were covered with secondary antibody (goat anti-human-IgG-alkaline phosphatase [AP]; 1:500 dilution in SB), the plunger was inserted again to seal, and the bead suspension was incubated for 1 h at room temperature. Beads were then drained, washed with PBS-Tween, and covered with the BCIP (5-bromo-4-chloro-3-indolylphosphate)/nitroblue tetrazolium (NBT) ready-to-use substrate (KemEnTec Diagnostics) for color development, the plunger was inserted to seal again, and the bead suspension was incubated for 10 min. Hereafter, the beads were drained and washed with methanol to stop the enzymatic colorimetric reaction. Finally, the beads were resuspended in PBS and spread out in a petri dish and purple-colored beads were singled out manually from the suspension with a micropipette. Each of the isolated beads were portioned out in an Eppendorf tube (one bead/tube), individually washed with methanol (50 μl), and incubated in 50 μl of a reduction cocktail (6.6 mM NaBH4 and 6.0 mM I2 in tetrahydrofuran [THF]) for 20 min. The liquid was removed, and each bead was washed with methanol (50 μl) and then incubated in 5 μl of a cleavage cocktail (trifluoroacetic acid [TFA]-H2O-triethylsilane (TES) [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], 95:3:2 ratio) for 30 min, whereupon liquids were removed under a stream of nitrogen and the resulting residue containing the glycopeptides was stored at 4°C until mass spectrometry (MS) analysis (see the supplemental Methods in the supplemental material).
RESULTS
Chemoenzymatic synthesis of a gG-2 glycopeptide library and analysis of HSV-2 patient sera (synthetic strategy 1).
We first prepared a 20-mer peptide library covering gG-2 with a 10-amino-acid overlap (69 peptides) (see the full list in Table S2 in the supplemental material) via standard Fmoc-SPPS using a parallel robotic peptide synthesizer handling 96 structures per batch (Fig. 1A). The target peptides were designed with N-cap acetylation to allow for enrichment of full-length target sequences directly via microarray printing on an amine-reactive NHS-activated glass surface (5) (see Fig. S1 in the supplemental material). Only peptides containing Ser and/or Thr residues were addressed at this stage (i.e., 64 structures). After matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry confirmation of target sequences, crude peptides were covalently immobilized onto amine-reactive N-hydroxysuccinimide (NHS)-activated glass slides with a robotic microarray pin printer. The use of peptide concentrations of >200 μM ensured saturation of the reactive NHS-esters under the area covered by each spot. The peptide chips were designed to present either 16 or 48 identical subarrays per glass slide (see Materials and Methods).
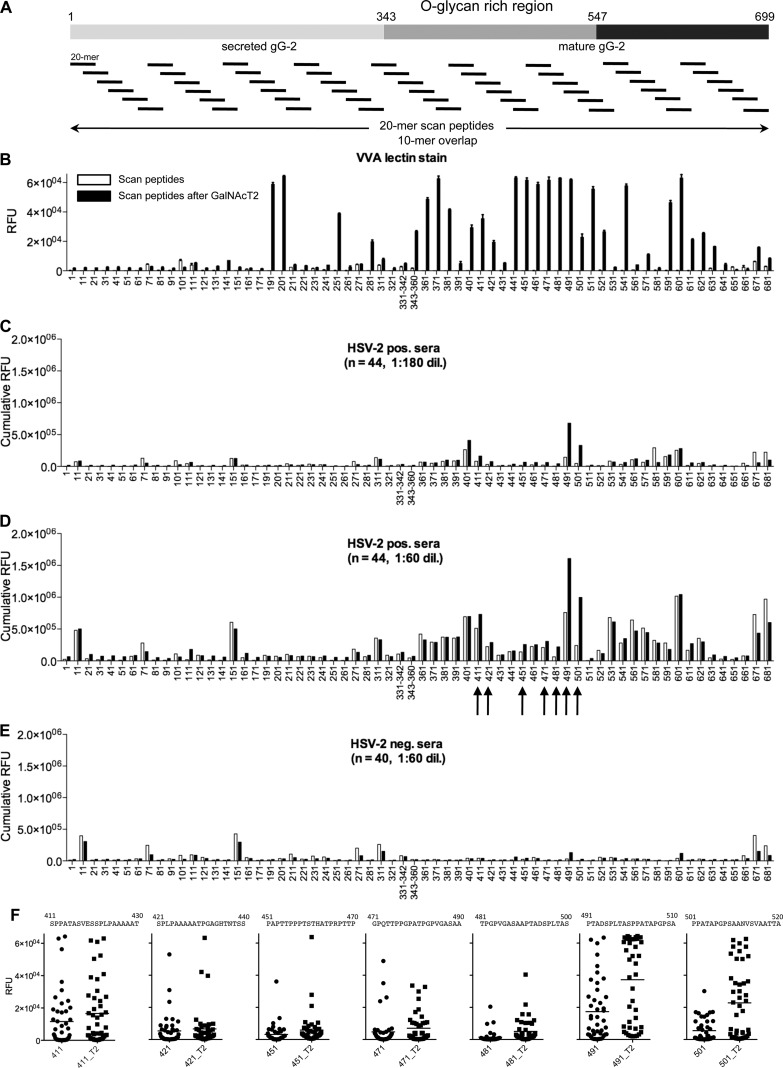
Fig 1.
Chemoenzymatic on-slide glycosylation of gG-2 protein scan peptides with recombinant GalNAc-T2 (synthetic strategy 1). (A) Schematic representation of the whole gG-2 protein (with the O-glycan-rich region, amino acids 343 to 547, within the mature gG-2 protein highlighted) and of the design of the synthetic 20-mer peptides, with a 10-mer overlap, spanning across the protein. (B) VVA lectin staining (5 μg/ml) results before (white columns) and after (black columns) on-chip enzymatic glycosylation. (C and D) Cumulative RFU results of staining of 44 HSV-2-positive sera (HSV-2 positive, n = 22; HSV-1/-2 positive, n = 22) at 1:180 (C) and 1:60 (D) dilutions before (white columns) and after (black columns) on-chip enzymatic glycosylation. (E) Cumulative RFU results of staining of 40 HSV-2-negative sera (HSV-1 positive, n = 20; HSV-1/-2 negative, n = 20) at a 1:60 dilution before (white columns) and after (black columns) on-chip enzymatic glycosylation. (F) Dot plots of selected scan peptides from panel D (highlighted by black arrows) that show increments of cumulative RFU after on-chip GalNAc-T2 (T2) glycosylation relative to the values for the naked peptide. Only scan peptides containing Ser and/or Thr were included on the microarray. The microarray data used for each scan peptide are the averages of four replicates. The error bars in panel B are the standard deviations. See also Table S2 in the supplemental material.
Next, a recombinant soluble polypeptide GalNAc-transferase 2 (GalNAc-T2) was applied to introduce O-linked GalNAc residues by on-chip enzymatic glycosylation.
The on-slide GalNAc glycosylation was evaluated by reactivity with the lectin VVA (isolectin B4) as shown in Fig. 1B. Several regions of the gG-2 protein were glycosylated by GalNAc-T2, and in particular, the region between amino acids 441 and 511 (441-511) had multiple reactive peptides. No significant interaction of the lectin with peptides prior to enzymatic treatment was recorded. While the lectin-binding patterns clearly demonstrate that GalNAc residues have been introduced, the assay does not allow quantification of efficiency in terms of partial or complete glycosylation and the actual sites of glycosylation in the 20-mer peptides are not determined.
To obtain an overview of IgG serum antibody reactivity, we produced cumulative relative fluorescence unit (RFU) data sets for all seropositive and seronegative individuals tested on the microarray (Fig. 1C to E). Reactivities of HSV-2-positive sera (n = 44) toward selected sequences were also displayed as dot plots to address individual sensitivities (Fig. 1F). We found that seropositive individuals exhibited substantial immunoreactivity with several peptides. However, one particular immunoreactive region (amino acids 491 to 520 [491-520]) showed increased cumulative reactivity and sensitivity with the GalNAc glycopeptide compared to those with unglycosylated peptides while retaining type specificity (i.e., 491PTADSPLTASPPATAPGPSAANVSVAATTA520, with potential glycosylation sites underlined) (Fig. 1C and D). Seronegative individuals generally showed limited or no reactivity to peptides and glycopeptides alike (Fig. 1E).
Chemical site-specific glycosylation of a gG-2 glycopeptide library and analysis of HSV-2 patient sera (synthetic strategy 2).
In order to evaluate the discovery success of the chemoenzymatically produced glycopeptide library, we also built a complementary chemically synthesized library that features single-site GalNAc glycopeptide analogues of scan peptides. We prepared all the 20-mer single-glycosylation site analogues required in order to cover, one at a time, all the possible glycosylation sites within positions 4 to 16 in each original naked scan peptide (Fig. 2A). Glycopeptide sequences are shown in Table S2 in the supplemental material.
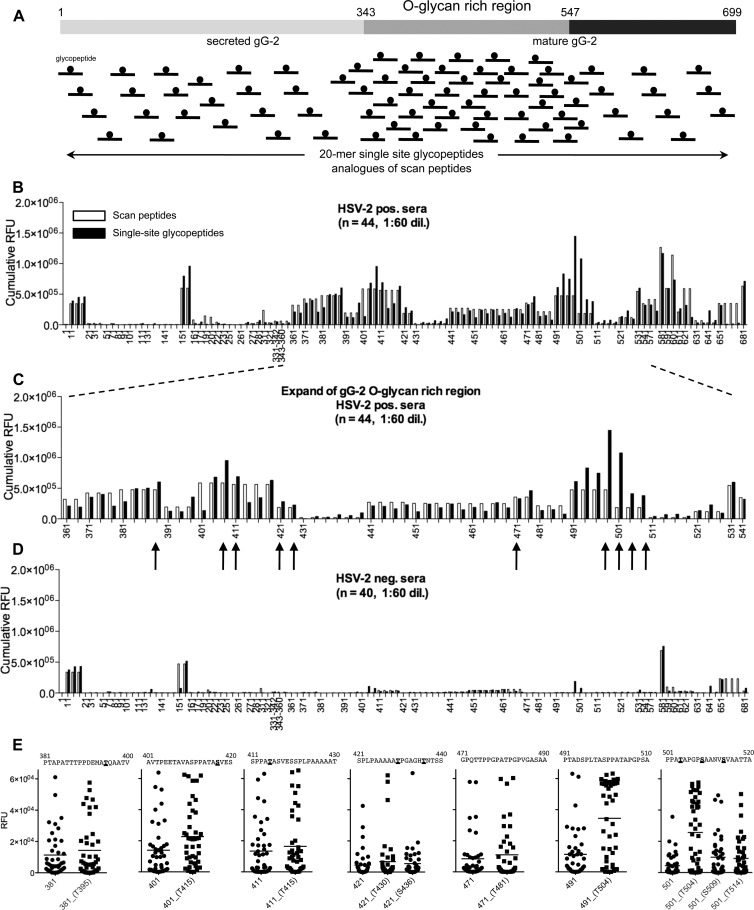
Fig 2.
Single-glycosylation-site glycopeptide analogues of gG-2 protein scan peptides produced by chemical synthesis (synthetic approach 2). (A) Schematic representation of the whole gG-2 protein (with the O-glycan-rich region, amino acids 343 to 547, within the mature gG-2 protein highlighted) and the design of the synthetic 20-mer single-site glycopeptide analogues of scan peptides. Only 20-mer glycopeptide analogues with Ser and/or Thr within positions 4 to 16 were synthesized for each scan peptide. (B and C) Cumulative RFU results of staining of 44 HSV-2-positive sera (HSV-2 positive, n = 22; HSV-1/-2 positive, n = 22) at a 1:60 dilution of scan peptides (white columns) and their respective single-site glycopeptide analogues (black columns); panel C shows the expansion of the O-glycan-rich region. (D) Cumulative RFU results of staining of 40 HSV-2-negative sera (HSV-1 positive, n = 20; HSV-1/-2 negative, n = 20) at a 1:60 dilution of scan peptides (white columns) and their respective single-site glycopeptide analogues (black columns). (E) Dot plots of selected glycopeptides from panel C (highlighted by black arrows) that show increments of cumulative RFU relative to the values for the naked peptide; sites of glycosylation are underlined and in boldface. The microarray data used for each scan peptide glycopeptide are the averages of four replicates. See also Table S2 in the supplemental material for the full peptide list.
Analysis of the cumulative serum IgG response from HSV-2-seropositive individuals to the complete GalNAc glycopeptide scan, arrayed along with the parent naked scan peptides, resulted in a reactivity profile similar to that with the enzymatic approach (Fig. 2B to D). Enhanced cumulative signals from glycopeptides were found again in the region of amino acids 491 to 520, namely, the fourth analog of scan peptide 491-510 (i.e., PTADSPLTASPPATAPGPSA) and the first of peptide 501-520 (i.e., PPATAPGPSAANVSVAATTA). Since the current chemical approach allowed control over the topology of glycosylation, it was possible to gain further insights into the binding epitope and narrow it down to 501PPATAPGPSA510, with glycosylation at Thr504. The reactivities of the HSV-2-positive sera (n = 44) toward selected sequences are also displayed as dot plots to address individual sensitivities (Fig. 1E). The HSV-2-negative sera were all negative except a few cases devoid of type specificity (Fig. 2D).
Characterization of the 501PPATAPGPSA510 immunodominant epitope.
Cumulative RFU analysis of the two glycopeptide libraries suggested that the 501PPATAPGPSA510 region with glycosylation at Thr504 contained an immunodominant GalNAc glycopeptide epitope recognized in particular by HSV-2-seropositive individuals. To further map the epitope with respect to peptide sequence and glycan structure requirements, we synthesized and tested four independent (glyco)peptide libraries: (i) a single-site glycosylation walk (Fig. 3A), (ii) an alanine walk of glycopeptides with GalNAc at Thr504 (Fig. 3B), (iii) a stepwise epitope walk around GalNAc-glycosylated Thr504 (Fig. 3C), and (iv) (glyco)peptide isoforms with Ser in place of Thr504 (both naked and GalNAc functionalized) along with different O-glycans at Thr504 (Fig. 3D). Data in Fig. 3 are shown for serum number 63 as a representative example; microarray data concerning the Ala walk and (glyco)peptide isoforms with a complete serum set are shown in Fig. S2 and S3 in the supplemental material, respectively.
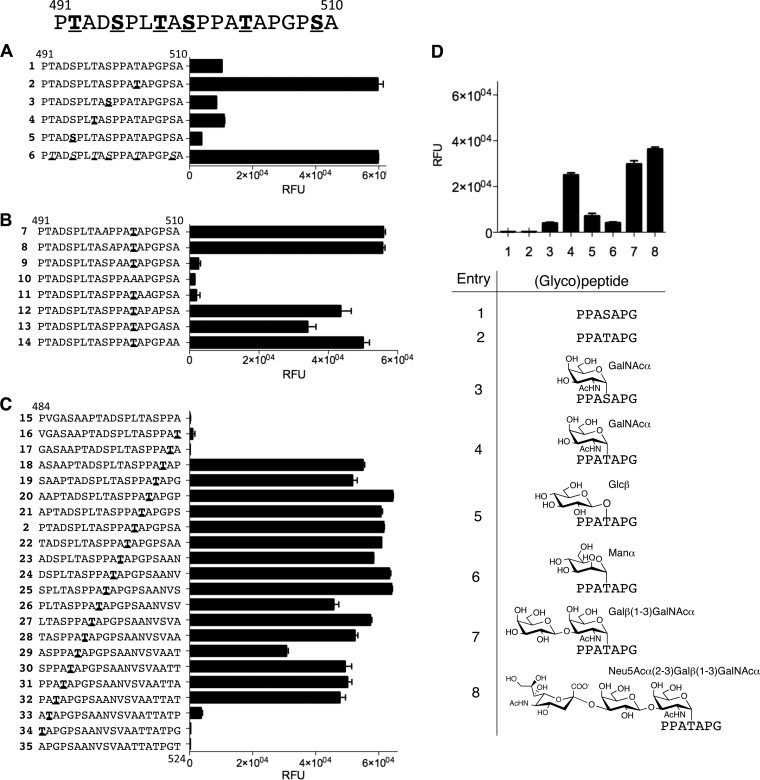
Fig 3.
Epitope mapping on a microarray of the immunodominant sequence in the region of amino acids 491 to 510. (A) Glycopeptide walk of GalNAc attachment sites, including scan peptide 1 (491-510), four single-site synthetic analogues (2 to 5) (glycosylation sites are underlined and in boldface), and glycopeptide 6, produced by GalNAc-T2 glycosylation of glycopeptide 1 in solution (potential glycosylation sites are underlined and italicized). MALDI-TOF analysis confirmed five out of six possible glycosylation sites. (B) Alanine mutation walk within the region of amino acids 500 to 510 (the alanine mutation site is italicized, and the glycosylation sites are underlined and in boldface). (C) Single-amino-acid epitope walk over the region of amino acids 484 to 524 with fixed glycosylation at Thr504 (glycosylation sites are underlined and in boldface). (D) Analysis of different glycoisoforms with focused 7-mer peptide 501PPAXAPG507 analogues (T/S = Ser or Thr). Microarray data presented for a representative patient serum (number 63; HSV-1/-2 positive) at a 1:60 dilution. Each value is the average of four replicates, with the standard deviation shown by error bars. See also Fig. S2 and S3 in the supplemental material for the full data set.
The single-site glycosylation walk identified Thr504 as the immunodominant glycopeptide epitope required for serum reactivity (Fig. 2D and 3A). The Ala scan across the 500-to-510 amino acid segment (Fig. 3B) confirmed that mutation of GalNAc-Thr504 abolished binding completely. Furthermore, both Pro502 and Pro506 proved to be essential for binding. Binding to the glycoepitope was somewhat reduced in a subfraction of sera by mutation of Pro508, whereas Pro501 was clearly not part of the binding epitope, as its replacement provided binding profiles essentially identical to those of parent natural glycopeptides (see data from the entire serum screen in Fig. S2 in the supplemental material). Likewise, an Ala mutation of any other amino acids did not alter the binding profiles. The epitope walk with 20-mer glycopeptides with a single GalNAc at Thr504 across the region of amino acids 484 to 524 narrowed the epitope to 502PA(GalNAc)TAP506 (Fig. 3C). Interestingly, microarray analysis of synthetic glycopeptides spanning over amino acids 491 to 510 with different arrangements of multiple glycosylation showed that HSV-2-seropositive serum number 4 strongly reacted with glycopeptides having GalNAc attached to both Thr498 and Ser500 while GalNAc at Thr504 did not react (see Fig. S3 in the supplemental material).
To define the epitope requirement for the O-glycan structure, we tested reactivity to a panel of different O-glycans attached at Thr504 (501PPATAPG507) (Fig. 3D). First, we established that the epitope was dependent on GalNAc-Thr504, as the same glycopeptide with GalNAc-Ser504 did not react. Second, replacing GalNAc with other monosaccharides, such as βGlc and αMan, showed scanty reactivities (Fig. 3D) (see data of the entire serum collection in Fig. S4 in the supplemental material). Interestingly, the extended O-glycoforms Galβ(1-3)GalNAc (T) and Neu5Acα(2-3)Galβ(1-3)GalNAc (ST) were both reactive with HSV-2-positive sera. This demonstrates for the first time that sialylated O-glycans present on many other glycoproteins and generally considered immunologically silenced (4) can, in fact, give rise to specific antibodies in the context of its peptide backbone.
In addition, chemical glycosylation within the O-glycan-rich region enables, in greater detail, the identification of additional glycopeptide epitopes reacting with the HSV-2-positive sera. Indeed, several glycopeptide epitopes were defined in this way, but they had low sensitivities and did not add to the diagnostic value of the pan-epitope 502-507. This can be appreciated from the first five dot plots of Fig. 2E, showing examples in which the mean RFU values of glycopeptides were slightly higher than those of parent naked peptides. The biological relevance of these glycopeptides requires further studies.
High-throughput OBOC random bead library for glycopeptide confirmation (synthetic strategy 3).
To further establish the significance of the PA(GalNAc)TAP pan-epitope, we used a screening approach based on a one-bead-one-compound (OBOC) random O-glycopeptide library (see Table S1 in the supplemental material) and created about 16,000 structurally different glycopeptides representing permutations of the epitope 502XX(GalNAc)TXXX507 (Fig. 4). The library was prepared as previously described and contained control epitopes derived from a mucin 1 (MUC1) tandem repeat (21). The amino acids relevant to the HSV epitope (P, A, and G) and additional amino acids relevant to the MUC1 epitopes (S, R, and D) were randomly introduced by split-mix synthesis at five variable positions located at X−2 to X+3 relative to the fixed αGalNAc-O-Thr (X0). Thus, the epitope PATAPG was represented in the myriad of peptides synthesized. An aliquot, representing the library sequence variations in 5-fold excess, was incubated with a pool of 3 HSV-2-positive serum samples (1:100 dilution), followed by incubation with alkaline phosphatase-conjugated anti-human-IgG antibody. Upon colorimetric development with ready-to-use BCIP/NBT substrate solution, beads that stained positive were singled out, the hit glycopeptides were released in solution, and their structure was elucidated by MS/MS mass spectrometry analysis (Fig. 4) (for mass spectrometry data, see Table S3 in the supplemental material). Out of the 16,000 structurally different glycopeptides, all isolated hits contained the PA(GalNAc)TAP motif. Remarkably, no other positive beads were stained, indicating that the bead selection approach was highly specific and selective for glycopeptide-specific serum antibodies. A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) demonstrated a full conservation of the motif among clinical HSV-2-confirmed isolates.
Fig 4.
One-bead-one-compound (OBOC) approach for the identification of glycopeptide epitopes (synthetic strategy 3). Design of a 13-mer random OBOC library with a fixed GalNAc-Thr at position 6 and with randomized amino acid residues at positions X−2 to X+3 linked to a cleavable linker as shown. The library was sequentially screened with a pool of HSV-2-positive (n = 3) sera (1:100 dilution), and the sequences of the isolated hits were deconvoluted with mass spectrometry. The glycosylated amino acids are underlined and in boldface, and the hit frequency of each sequence is shown in parentheses in the table.
The 501PPA(GalNAc)TAPG507 glycopeptide serves as a biomarker of HSV-2 infection.
We next tested the biomarker value of the identified epitope 502PA(GalNAc)TAPG507 by detection of HSV-2-associated IgG immunity. We compared the original 20-mer glycopeptide 491-510 with the bead-identified 13-mer AAA502PATAPG507AHGV glycopeptide, each with a GalNAc residue at Thr504 along with the naked 7-mer glycopeptide 501-507 (Fig. 5). Thirty-two (73%) out of the 44 HSV-2-seropositive patients tested, all with reactivity to HPA-purified gG-2 (data not shown), gave a microarray signal more than two standard deviations above the mean of the sera from the HSV-2-seronegative patients (n = 40). With the same threshold, only one HSV-1-positive serum (number 27) would have been mistaken as HSV-2 seropositive. The reactivity of HSV-1-positive serum number 27 responded poorly to concentration gradients and thus could not be diluted out, hinting to interactions of an unspecific nature. The 20-mer glycopeptide 491-510 (Fig. 5A) and the short 7-mer glycopeptide 501-507 (Fig. 5B) were able to identify essentially the same HSV-2-positive patients. The 7-mer at times gave slightly lower signals, yet this was offset by extremely low reactivity among the HSV-2-negative control groups. Moreover, the bead-identified glycopeptide epitope gave, as expected, a type-specific HSV-2 antibody serum reactivity identical to that of the 7-mer (Fig. 5C). Since all other sequence permutations built by the bead library were unreactive, these data demonstrated that this glycoepitope was unique.
Fig 5.
Visualization of biomarker values for pan-epitope 501-507. (A) Synthetic glycopeptide 491-510, with GalNAc glycosylation at Thr504. (B) Synthetic glycopeptide matching the sequence of the most frequent hit from the OBOC library screen. (C) Synthetic glycopeptide 501-507, with GalNAc glycosylation at Thr504. (D) Naked peptide 501-507. Glycosylation sites are underlined and in boldface. Microarray analysis of the full HSV-2-positive and -negative serum set (n = 84) at a 1:60 dilution. Each value is the average of four replicates, with the standard deviation shown by error bars.
We also performed competitive inhibition experiments with different concentrations of the naked or Tn-glycosylated pan-epitope (501-507) or free GalNAc (ranging from 0.1 to 200 μM with 3-fold increments). Data further corroborated the specific nature of the glycopeptide pan-epitope, proving that it is not an array artifact (see Fig. S5 in the supplemental material).
The 502PA(GalNAc)TAPG507 glycopeptide epitope is also recognized by CSF antibodies from a patient with recurrent HSV-2 meningitis.
Next, we compared antibody reactivities to the gG-2 glycopeptides in serum and cerebrospinal fluid (CSF) samples simultaneously drawn from a patient with frequent relapses of HSV-2 meningitis (3). Since the compartmentalized intrathecal antibody response during viral meningitis may be oligoclonal, we compared glycopeptide epitope reactivity of CSF with that of serum. Both body fluids were diluted to identical IgG concentrations of 5 μg/ml to correct for the effects of the blood-CSF gradient for antibodies. This patient showed a remarkably stronger CSF reactivity (>5-fold that of serum) to the 491PTADSPLATSPPA(GalNAc)TAPGPSA510 epitope, whereas no reactivity to the unglycosylated peptide was found (Fig. 6). In order to evaluate the intensity of such an antibody signal in CSF, reactivity toward HPA-purified mgG-2 (37), also printed on a microarray, was evaluated. Although the entire extracellular region of the protein was used as a target antigen, the signal was only approximately half of the single-glycopeptide signal.
Fig 6.
IgG antibody reactivities of serum and cerebrospinal fluid (CSF) from a patient with recurrent HSV-2 meningitis toward the peptide 491-510 (1), its GalNAc-glycosylated analogue at Thr504 (2), HPA-purified mgG-2 (3), and the type-specific peptide 555-572 (4). Each value is the average of four replicates, with the standard deviation shown by error bars.
DISCUSSION
The development of HSV-type-specific serological tests based on envelope glycoproteins has had a major impact on management of HSV infection and is the method of choice for a point-of-care test (1). The only FDA-approved assay is the gG-2 protein-based assay with acceptable specificity. There has also been considerable interest in developing subunit vaccines against HSV, but clinical trials have been less successful. To improve vaccine development, identification of additional targets has been pursued on the protein level (20); however, we also believe that a better understanding of the immunogenicity of posttranslationally modified HSV glycoproteins (including gG-2) is a necessity for better vaccine design.
To address this, we have presented a set of complementary synthesis and screening strategies to discover antibodies directed to virus-associated glycopeptide epitopes in infected patients. The first strategy utilized on-chip enzymatic glycosylation in an attempt to mimic the initial step of mucin glycosylation in the infected cell. In this way, we reasoned that we could generate glycopeptide libraries with natural patterns of glycosylated and nonglycosylated Ser and Thr residues, thus more closely resembling physiological epitopes. The approach shortcuts the need to test every possible permutation of glycosylated/nonglycosylated Ser/Thr residues by means of synthetic glycopeptides in the initial microchip serum screening. We chose to use GalNAc-T2, one of the most ubiquitously expressed of the 20 human GalNAc-T isoenzymes (2, 15, 38), as it is strongly expressed in the CHO and HEK293 cells commonly used for recombinant expression of glycoproteins as well as for propagation of certain viruses, including HSV (22). GalNAc-T isoforms have different peptide substrate specificities, and there is good correlation between in vitro specificity and in vivo function (2, 5). This allows for use of specific GalNAc-Ts to probe O-glycosylation sites in proteins and build O-glycopeptide libraries by enzymatic glycosylation. Application of multiple GalNAc-T's, reflecting the repertoire of isoforms expressed in a given host cell propagating the virus in question, may be used to produce a display of the complete set of natural O-glycosylation sites. Although we utilized only GalNAc-T2 in this study, other enzymes have been proven to be highly efficient on the microarray screening platform (4) and further enzymatic diversification is possible (8). However, the GalNAc-T2 enzyme did extensively glycosylate the region on gG-2 that was previously identified as a mucin-like region (33), and its glycosylation corresponded very well with the prediction program NetOglyc algorithm (v3.1) (19).
Enzymatic on-chip glycosylation can rapidly build a glycopeptide library useful for screening of serum reactivity to glycosylated peptides, but it has certain limitations due to substrate specificities and slower reaction kinetics on solid-phase surfaces compared to those in homogenous solutions (7). Therefore, the second complementary strategy, chemical synthesis, hinges on the use of glycosylated Ser and Thr building blocks able to enter the Fmoc-SPPS cycle, thereby generating glycopeptides of defined topology. This is necessary not only to confirm the structural identity of the reactive glycoepitopes identified by the enzymatic screening method but also to analyze the relevance of each glycan's individual structural elements. Synthesis of large O-linked glycans, such as hexasaccharide selectin ligands, has been demonstrated via both chemical and enzymatic synthesis (24, 40), but usually they require extensive purification strategies for each target structure. Our concept allows for direct on-slide enrichment of large libraries of synthetic products derived from the SPPS blocks and offers a comprehensive tool to rapidly study complete sets of sequences from target proteins without tedious purification protocols. While this approach was possible for shorter neutral O-glycan structures, such as GalNAc and Galβ1-3GalNAc, chemical synthesis becomes costly with increased complexity and number of O-glycans and length of the peptide sequence. Although certain glycans, including sialylated structures such as STn (Neu5Acα2-6GalNAc) and ST (Neu5Acα2-3Galβ1-3GalNAc), are extremely difficult to incorporate by Fmoc synthesis, several elegant examples exist for a limited number of peptide libraries (10, 31).
The third strategy, based on the generation of combinatorial OBOC glycopeptide libraries, provided characterization of the structural variation in the underlying peptide backbone of glycopeptide epitopes. Although the significance of amino acid variation may be studied to some extent by the enzymatic approach, the use of combinatorial libraries allows an open-ended discovery tool for identification of O-GalNAc peptide epitopes. Also, we believe that random OBOC glycopeptide libraries, if produced with wider structural variations by the inclusion of all natural amino acids and/or by introducing design-driven structural constrains, may be able to spearhead the identification of lead glycopeptide epitopes. The refinement of such lead structures and assessment of their biomarker potential may accordingly be pursued by the previous two strategies.
Rewardingly, all three strategies presented here could decipher the immune response to a prototypic envelope glycoprotein, gG-2 of HSV-2, and identify the same pan-reactive glycopeptide epitope. Previous peptide epitope mapping of the envelope gG-2 protein (25–27) showed various immunodominant epitopes, including the nonglycosylated peptide of our pan-epitope, indicating that this part of gG-2 may constitute an antigenic domain (26). However, presence of GalNAc on the Thr504 residue strongly increased the reactivity of the epitope and permitted reduction of the peptide chain to 7 amino acid residues without compromising epitope reactivity. This should be compared with the considerably higher peptide lengths required for even a weak reactivity of the nonglycosylated peptides presented previously (26).
The significance of the glycoepitope is also illustrated by the detection of a prominent intrathecal IgG response to this epitope in the central nervous system (CNS) compartment in patients with recurrent meningitis. A 5-fold-stronger response was found in the CSF than in the serum, even when bias by blood-brain barrier effects was circumvented by adjusting the samples to identical IgG concentrations. We interpret this finding as an indication of a particularly strong presentation of the glycoepitope within the CNS. This may point toward a remarkably selective clonal expansion of glycopeptide-specific IgG antibodies in the CSF, signifying that such glycoepitopes may be used as diagnostic tools in the search for biomarkers of recurrent HSV-2 meningitis and possibly other compartmentalized infections.
Interestingly, we showed that extended glycoforms, such as the sialyl-T of the pan-epitope, can be tolerated even though the entire trisaccharide was not part of the epitope. Sialyl-T glycans are considered biosynthetic self molecules, and they are present on various proteins (41) but do not elicit immune responses in humans per se (6) or within context of its peptide carrier on tumor proteins (4). However, with a favorable glycan presentation protruding away from its peptide carrier, penultimate glycans directly linked to the protein can potentially still be recognized by IgG antibodies. This recognition may persist despite capping with a relative large sialic acid moiety. Although our experiments regard glycopeptides, it is desirable to identify the exact glycoform and location on the native protein. Nevertheless, this will further add to the complexity of events for stimulated immunity to both natural and aberrant truncated N- and O-glycoproteins and deserves additional attention. Despite the fact that truncated O-glycans have been found on envelope glycoproteins expressed on the viral cycle (17, 32), vaccine clinical trials with recombinant HSV-2 envelope glycoproteins have failed (18), possibly due to incompatibility or absence of appropriate posttranslational glycosylation (11, 35). Other viral envelope glycoproteins, including hepatitis C virus (14), respiratory syncytial viruses (12), and the Filoviridae virus family, including Ebola virus (23) and the human herpesviruses, all contain densely glycosylated mucin-like peptide domains with multiple, often tightly clustered, short O-glycans. We believe it is crucial to deepen our understanding of immune responses triggered by glycosylated envelope proteins, as our data clearly demonstrate that the first example of a potent immunodominant glycopeptide epitope eliciting type-specific IgG antibodies can also be found on viral envelope proteins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Benzon Foundation, The Danish Agency for Science, Technology and Innovation (FTP), EU FP7/2007-2013-EuroGlycoArrays 215536, and the University of Copenhagen Programme of Excellence.
Henrik Clausen is acknowledged for helpful advice and discussions, and Maya Bonde Haaland is acknowledged for proofreading the manuscript.
Footnotes
Published ahead of print 4 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ashley-Morrow R, Krantz E, Wald A. 2003. Time course of seroconversion by HerpeSelect ELISA after acquisition of genital herpes simplex virus type 1 (HSV-1) or HSV-2. Sex. Transm. Dis. 30:310–314 [DOI] [PubMed] [Google Scholar]
- 2. Bennett EP, et al. Control of mucin-type O-glycosylation—a classification of the polypeptide GalNAc-transferase gene family. Glycobiology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergstrom T, et al. 1990. Primary and recurrent herpes simplex virus type 2-induced meningitis. J. Infect. Dis. 162:322–330 [DOI] [PubMed] [Google Scholar]
- 4. Blixt O, et al. 2011. Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 13:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blixt O, et al. 2010. A high-throughput O-glycopeptide discovery platform for seromic profiling. J. Proteome Res. 9:5250–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blixt O, et al. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blixt O, Norberg T. 1998. Solid-phase enzymatic synthesis of a sialyl Lewis X tetrasaccharide on a Sepharose matrix. J. Org. Chem. 63:2705–2710 [DOI] [PubMed] [Google Scholar]
- 8. Blixt O, Razi N. 2006. Chemoenzymatic synthesis of glycan libraries. Methods Enzymol. 415:137–153 [DOI] [PubMed] [Google Scholar]
- 9. Bolmstedt A, et al. 1996. Influence of N-linked glycans in V4-V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:213–220 [DOI] [PubMed] [Google Scholar]
- 10. Brocke C, Kunz H. 2002. Synthesis of tumor-associated glycopeptide antigens. Bioorg. Med. Chem. 10:3085–3112 [DOI] [PubMed] [Google Scholar]
- 11. Cohen J. 2010. Immunology. Painful failure of promising genital herpes vaccine. Science 330:304. [DOI] [PubMed] [Google Scholar]
- 12. Collins PL, Mottet G. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73(Part 4):849–863 [DOI] [PubMed] [Google Scholar]
- 13. Dall'Olio F, Malagolini N, Campadelli-Fiume G, Serafini-Cessi F. 1987. Glycosylation pattern of herpes simplex virus type 2 glycoprotein G from precursor species to the mature form. Arch. Virol. 97:237–249 [DOI] [PubMed] [Google Scholar]
- 14. Falkowska E, Kajumo F, Garcia E, Reinus J, Dragic T. 2007. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J. Virol. 81:8072–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerken TA, et al. 2011. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 286:14493–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gill DJ, Clausen H, Bard F. 2011. Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21:149–158 [DOI] [PubMed] [Google Scholar]
- 17. Hansen JE, et al. 1996. Sensitivity of HIV-1 to neutralization by antibodies against O-linked carbohydrate epitopes despite deletion of O-glycosylation signals in the V3 loop. Arch. Virol. 141:291–300 [DOI] [PubMed] [Google Scholar]
- 18. Johnston C, Koelle DM, Wald A. 2011. HSV-2: in pursuit of a vaccine. J. Clin. Invest. 121:4600–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Julenius K, Molgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164 [DOI] [PubMed] [Google Scholar]
- 20. Kalantari-Dehaghi M, et al. 2012. Discovery of potential diagnostic and vaccine antigens in herpes simplex virus-1 and -2 by proteome-wide antibody profiling. J. Virol. 86:4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kracun SK, et al. 2010. Random glycopeptide bead libraries for seromic biomarker discovery. J. Proteome Res. 9:6705–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurt-Jones EA, et al. 2004. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc. Natl. Acad. Sci. U. S. A. 101:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JE, Saphire EO. 2009. Neutralizing ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it. Curr. Opin. Struct. Biol. 19:408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leppanen A, Penttila L, Renkonen O, McEver RP, Cummings RD. 2002. Glycosulfopeptides with O-glycans containing sialylated and polyfucosylated polylactosamine bind with low affinity to P-selectin. J. Biol. Chem. 277:39749–39759 [DOI] [PubMed] [Google Scholar]
- 25. Levi M, Rudén U, Wahren B. 1996. Peptide sequences of glycoprotein G-2 discriminate between herpes simplex virus type 2 (HSV-2) and HSV-1 antibodies. Clin. Diagn. Lab. Immunol. 3:265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liljeqvist JA, et al. 1998. Localization of type-specific epitopes of herpes simplex virus type 2 glycoprotein G recognized by human and mouse antibodies. J. Gen. Virol. 79(Part 5):1215–1224 [DOI] [PubMed] [Google Scholar]
- 27. Marsden H, MacAulay K, Murray J, Smith I. 1998. Identification of an immunodominant sequential epitope in glycoprotein G of herpes simplex virus type 2 that is useful for serotype-specific diagnosis. J. Med. Virol. 56:79–84 [DOI] [PubMed] [Google Scholar]
- 28. Meldal M, Svendsen I, Breddam K, Auzanneau FI. 1994. Portion-mixing peptide libraries of quenched fluorogenic substrates for complete subsite mapping of endoprotease specificity. Proc. Natl. Acad. Sci. U. S. A. 91:3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller SD, et al. 1997. Persistent infection with Theiler's virus leads to CNS autoimmunity via epitope spreading. Nat. Med. 3:1133–1136 [DOI] [PubMed] [Google Scholar]
- 30. Nystrom K, et al. 2007. Virus-induced transcriptional activation of host FUT genes associated with neo-expression of Ley in cytomegalovirus-infected and sialyl-Lex in varicella-zoster virus-infected diploid human cells. Glycobiology 17:355–366 [DOI] [PubMed] [Google Scholar]
- 31. Ohyabu N, et al. 2009. An essential epitope of anti-MUC1 monoclonal antibody KL-6 revealed by focused glycopeptide library. J. Am. Chem. Soc. 131:17102–17109 [DOI] [PubMed] [Google Scholar]
- 32. Olofsson S, Hansen JE. 1998. Host cell glycosylation of viral glycoproteins—a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30:435–440 [DOI] [PubMed] [Google Scholar]
- 33. Olofsson S, Lundström M, Marsden H, Jeansson S, Vahlne A. 1986. Characterization of a herpes simplex virus type 2-specified glycoprotein with affinity for N-acetylgalactosamine-specific lectins and its identification as g92K or gG. J. Gen. Virol. 67(Part 4):737–744 [DOI] [PubMed] [Google Scholar]
- 34. Sorensen AL, et al. 2006. Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology 16:96–107 [DOI] [PubMed] [Google Scholar]
- 35. Stansell E, Canis K, Haslam SM, Dell A, Desrosiers RC. 2011. Simian immunodeficiency virus from the sooty mangabey and rhesus macaque is modified with O-linked carbohydrate. J. Virol. 85:582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sugrue RJ. 2007. Viruses and glycosylation: an overview. Methods Mol. Biol. 379:1–13 [DOI] [PubMed] [Google Scholar]
- 37. Svennerholm B, Olofsson S, Jeansson S, Vahlne A, Lycke E. 1984. Herpes simplex virus type-selective enzyme-linked immunosorbent assay with Helix pomatia lectin-purified antigens. J. Clin. Microbiol. 19:235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tarp MA, et al. 2007. Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology 17:197–209 [DOI] [PubMed] [Google Scholar]
- 39. Vigerust DJ, Shepherd VL. 2007. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vohra Y, Buskas T, Boons GJ. 2009. Rapid assembly of oligosaccharides: a highly convergent strategy for the assembly of a glycosylated amino acid derived from PSGL-1. J. Org. Chem. 74:6064–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wahrenbrock MG, Varki A. 2006. Multiple hepatic receptors cooperate to eliminate secretory mucins aberrantly entering the bloodstream: are circulating cancer mucins the “tip of the iceberg”? Cancer Res. 66:2433–2441 [DOI] [PubMed] [Google Scholar]
- 42. Wandall HH, et al. 2010. Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 70:1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.