Abstract
The role of peroxisome proliferator-activated receptor β/δ (PPARβ/δ) in Harvey sarcoma ras (Hras)-expressing cells was examined. Ligand activation of PPARβ/δ caused a negative selection with respect to cells expressing higher levels of the Hras oncogene by inducing a mitotic block. Mitosis-related genes that are predominantly regulated by E2F were induced to a higher level in HRAS-expressing Pparβ/δ-null keratinocytes compared to HRAS-expressing wild-type keratinocytes. Ligand-activated PPARβ/δ repressed expression of these genes by direct binding with p130/p107, facilitating nuclear translocation and increasing promoter recruitment of p130/p107. These results demonstrate a novel mechanism of PPARβ/δ cross talk with E2F signaling. Since cotreatment with a PPARβ/δ ligand and various mitosis inhibitors increases the efficacy of increasing G2/M arrest, targeting PPARβ/δ in conjunction with mitosis inhibitors could become a suitable option for development of new multitarget strategies for inhibiting RAS-dependent tumorigenesis.
INTRODUCTION
Targeting peroxisome proliferator-activated receptors (PPARs) for the prevention and treatment of diseases is of great interest due to their ability to modulate many physiological functions (1, 20, 34, 46, 47). PPARβ/δ ligands can increase the serum high-density-lipoprotein cholesterol concentration, improve insulin resistance, increase fatty acid catabolism, and exert potent anti-inflammatory activities (1, 20, 30, 34, 35, 42). There is also a potential for targeting PPARβ/δ for the prevention and treatment of cancer. However, the role of PPARβ/δ in cancer remains controversial (reviewed in references 45 to 47, 49, and 50). The first evidence suggesting that PPARβ/δ modulates skin carcinogenesis was provided by the observation that Pparβ/δ-null mice exhibited enhanced epidermal hyperplasia in response to the tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) (48). Consistent with this phenotype, exacerbated skin tumorigenesis was also found in Pparβ/δ-null mice following a two-stage chemical carcinogenesis bioassay (31). Subsequent studies demonstrated that ligand activation of PPARβ/δ inhibits chemically induced tumorigenesis (3, 4, 68). One mechanism that may underlie these PPARβ/δ-dependent chemopreventive effects is modulation of epidermal cell proliferation through inhibition of protein kinase C α/mitogen-activated protein kinase (PKCα/MAPK) (31, 33). Additionally, ligand activation of PPARβ/δ induces terminal differentiation (3, 32, 61), which influences cell proliferation and skin tumorigenesis.
Neoplastic conversion of normal cells to benign tumors and progression of benign tumors to adenomas and carcinomas is associated with overexpression, amplification, and homozygosity of oncogenic Hras (37). Chemicals can cause mutations in Hras in mouse skin tumors (5), and examination of skin tumors produced by chemical carcinogens or UV light revealed that nearly all tumors contain an activated Hras oncogene (2, 12). Targeted introduction of oncogenic Hras into the epidermis of experimental animals can replace the initiation step resulting from exposure to a mutagenic chemical such as 7,12-dimethylbenz[a]anthracene (DMBA) in a two-stage chemical carcinogenesis model (7), and introduction of an Hras oncogene into normal mouse keratinocytes can also produce benign papillomas when grafted onto nude mice (19). Thus, there is strong evidence suggesting that activating the Hras oncogene through mutagenesis contributes to the mechanisms leading to neoplastic transformation during skin carcinogenesis.
Previous work demonstrated that activation of PPARβ/δ attenuated skin tumorigenesis in a 2-stage chemical carcinogenesis bioassay and inhibited proliferation of cells with a mutation in the Hras gene (3). This suggests that PPARβ/δ could inhibit tumorigenesis through inhibition of oncogenic Hras signaling, which was examined in these studies.
MATERIALS AND METHODS
Virus production.
The Hras retrovirus was generated from ψ2 producer cells as described previously (53). The virus titer was determined to be between 1 × 107 and 2 × 107 transforming units/ml by the use of an NIH 3T3 focus-forming assay.
Plasmids.
pCMV-p130 and pCMV-E2F4 vectors were purchased from Origene (Rockville, MD). The pCMV-p107 vector was a kind gift from Liang Zhu (69). The FLAG-p107, pGEX-2T-p107, and pGEX-2T-p130 vectors were kindly provided by Xavier Graña (28). pSG5-PPARβ/δ has been described previously (18). pcDNA-FLAG-PPARβ/δ was constructed by digesting pTNT-FLAG-PPARβ/δ (kindly provided by Gary Perdew of Penn State University) with KpnI and NotI followed by ligation into KpnI- and NotI-linearized pcDNA3.1 vector. The mouse Cdk1 promoter from −205 to +57 was cloned using the following primers: forward primer, 5′-ATAGGTACCGGAAGGAAAACAGAGCTCAAGAG-3′; reverse primer, 5′-ATACTCGAGCACACCGCAGTTCCGG-3′. The PCR product was digested with KpnI and XhoI and ligated into KpnI- and XhoI-linearized pGL4.20 vector (Promega, Madison, WI). Site-directed mutagenesis was performed to produce mutations in the following regions of the mouse Cdk1 promoter reporter construct (pGL4.20-mCdk1-promoter): for the distal E2F binding site, the sense primer was 5′-GTTTCCGCTCCCTTTCGTAATCTGCGCTCCCAGGC-3′ and the antisense primer was 5′-GCCTGGGAGCGCAGATTACGAAAGGGAGCGGAAAC-3′; for the proximal E2F binding site, the sense primer was 5′-GATCCCGGGAGCTTTAATATTGCGAGTTTGAAACTGC-3′ and the antisense primer was 5′-GCAGTTTCAAACTCGCAATATTAAAGCTCCCGGGATC-3′; and for the CHR binding site, the sense primer was 5′-CTTTACCGCGGCGAGTCGACAACTGCTGGCACTCGG-3′ and the antisense primer was 5′-CCGAGTGCCAGCAGTTGTCGACTCGCCGCGGTAAAG-3′. Mutations were confirmed by direct sequencing, and the following mutant reporter constructs were obtained: a mouse Cdk1 promoter reporter construct with a mutant distal E2F binding site (pGL4.20-mCdk1-distal E2F mutant), a mouse Cdk1 promoter reporter construct with a mutant proximal E2F binding site (pGL4.20-mCdk1-proximal E2F mutant), and a mouse Cdk1 promoter reporter construct with a mutant CHR binding site (pGL4.20-mCdk1-CHR mutant).
Cell culture.
Primary keratinocytes from newborn wild-type and Pparβ/δ-null mice were prepared and cultured as previously described (13). Keratinocytes were infected with the Hras retrovirus for 2 days at an estimated multiplicity of infection (MOI) of 1 to 12. Cells were subsequently cultured in control medium or medium containing GW0742 for up to 9 days postinfection. Keratinocytes of the 308 cell line that contain an activated mutation in Hras (56, 65) were cultured as described previously (3, 4).
Cell proliferation assays.
Wild-type and Pparβ/δ-null keratinocytes were infected with a retrovirus carrying Hras at an estimated MOI of 3 for 2 days. Keratinocytes were then treated with 1 μM GW0742 for another 4 days or left untreated. Cell numbers were quantified using a Z1 Beckman Coulter particle counter. Alternatively, cells were seeded in 96-well plates and, after 72 h of treatment with or without 1 μM GW0742, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)—phosphate-buffered saline (PBS) was added to each well at the final concentration of 0.5 mg/ml and cells were incubated at 37°C for 1 h. The medium was removed to quantify optical density with a spectrophotometer at 560 nm.
Immunofluorescence analysis.
HRAS-expressing wild-type and Pparβ/δ-null keratinocytes treated with or without 1 μM GW0742 were cultured in chamber slides to ensure complete attachment. Cells were fixed in 2% formaldehyde–PBS for 15 min at room temperature followed by permeabilization with 100% methanol for 10 min at −20°C. Cells were then washed with PBS and incubated overnight with an anti-phospho-histone 3 (S10) antibody (Cell Signaling, Beverly, MA) at 4°C followed by incubation with an Alexa Fluor 488-conjugated secondary antibody (Cell Signaling, Beverly, MA) and a Cy3-conjugated antitubulin antibody (Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature in the dark. Cells were then washed with PBS before incubation in Hoechst 33342 (1 μg/ml) for 10 min at room temperature. Paraffin-embedded sections from skin tumors were prepared from samples collected for a previously published study from wild-type and Pparβ/δ-null mice, with and without topical application of GW0742 (68). Sections were deparaffinized with xylene and rehydrated with decreasing concentrations of ethanol followed by antigen retrieval by boiling in 10 mM sodium citrate buffer (pH 6.0). Phospho-histone 3 S10 (pH3S10), tubulin, or total DNA (Hoechst) was detected as described above. The mitotic index was calculated as the percentage of cells that gave positive staining results for pH3S10. For every sample, a minimum of 1,000 total cells were examined. Cells were immunostained with antibodies against pH3S10 and β-tubulin and costained with Hoechst to visualize DNA and to identify mitotic cells at different growth phases, and the distribution of cells in various phases of mitosis was determined using a previously described method (38). For every sample, a minimum of 4,000 total cells were examined.
For colocalization analysis, cells were prepared as described above. Anti-p107/p130 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-PPARβ/δ 8095 antibody (18), and anti-E2F4 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) were conjugated with Alexa Fluor 647, Alexa Fluor 488, and Alexa Fluor 568 dye, respectively, using the recommended protocol of the manufacturer (Invitrogen, Carlsbad, CA). Representative photomicrographs were obtained with an Olympus Fluoview 1000 confocal microscope, using a 60× oil objective (numerical aperture [NA], 1.35) at room temperature. Photomicrographs were acquired with FV10-ASW2.0 VIEWER software (Olympus) and deconvolved with Autoquant software (Media Cybernetics, Bethesda, MD).
Flow cytometry analysis.
Cells were stained with bromodeoxyuridine (BrdU) and/or propidium iodide (PI) and analyzed for cell cycle progression as previously described (6, 24). The percentage of cells at each phase of the cell cycle ± standard deviation (SD) was determined with FCS Express software. For anti-HRAS staining analysis, cells were subjected to trypsinization and washed with PBS once before fixation was performed with 2% formaldehyde–PBS for 15 min at room temperature followed by permeabilization with 100% methanol for 10 min at −20°C. Cells were washed with PBS and incubated overnight with an anti-HRAS antibody (C20; Santa Cruz Biotechnology, Santa Cruz, CA) followed by incubation with an Alexa Fluor 488-conjugated secondary antibody (Cell Signaling, Beverly, MA) for 1 h at room temperature in the dark. Approximately 5,000 cells/sample were analyzed by flow cytometry using a Coulter XL-MCL analyzer (Beckman Coulter, Miami Lakes, FL). Pearson's second skewness coefficient, defined as 3 (mean relative HRAS intensity − median relative HRAS intensity)/standard deviation of relative HRAS intensity, was calculated to determine the relative distribution of cells with various levels of HRAS.
DNA microarray analysis.
Wild-type and Pparβ/δ-null keratinocytes were infected with retrovirus carrying Hras at an estimated MOI of 3 for 4 days before treatment with or without 1 μM GW0742 for 24 h. For non-Hras-infected keratinocytes, cells were mock infected for 4 days before treatment with or without 1 μM GW0742 for 24 h. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and purified with an RNeasy minikit (Qiagen, Valencia, CA). RNA (100 ng per sample) was prepared for analysis using a GeneChip Mouse Gene 1.0 ST array (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. The Robust Multichip Average (RMA) approach was used for normalization of microarray data using the R/Bioconductor package as previously described (14). To identify genes that were significantly induced by HRAS, a false-discovery-rate (FDR) cutoff of 0.1 was used. The DAVID algorithm was used to functionally categorize genes involved in different biological process as previously described (25). Principal component analysis (PCA) was performed using the R/Bioconductor package. Data have been deposited in NCBI's Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) (see below).
GSEA.
The log 2-transformed normalized values of the microarray data were used for Gene Set Enrichment Analysis (GSEA) (39, 57). The phenotype data set was constructed by averaging the log 2 values of 62 mitosis-related genes. The Pearson metric was chosen to rank the genes, and the phenotype-permutation option was set to compute the enrichment scores. Sets of E2F target genes were from two published data sets in which E2F target genes were confirmed by chromatin immunoprecipitation (52, 63). The EGR1 gene set was obtained from two confirmed EGR1 target gene databases (15, 58). The Sp1 gene set was obtained from the online Molecular Signature database (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection = TFT). Genes containing a GGGGCGGGGT motif within 2 kb on either side of the transcription start site were considered SP1 target genes. For each analysis, the normalized enrichment score (NES) is indicated together with the corresponding FDR. Transcription factor binding sites were identified using in silico analysis with MatInspector software (Genomatix, Ann Arbor, MI).
Quantitative Western blot analysis.
Cell lysates and supernatants used for Western blot analyses and immunoprecipitations were prepared as previously described (6, 18). An NE-PER nuclear protein extraction kit (Thermo Scientific, Rockford, IL) was used to isolate nuclear and cytosol protein. Equal amounts of cytosol and nuclear protein were used for Western blot analysis. Western blot analysis using radioactive detection methods was performed as previously described (18). The primary antibodies used were as follows: antiphosphoretinoblastoma (anti-phospho-RB; phosphorylated at S780, S795, or S807/811), anti-AURKB, anti-CENP-A, and anti-CHEK1 (Cell Signaling, Beverly, MA), anti-HRAS, anti-cyclin-dependent kinase 1 (anti-CDK1), anti-CDK2, anti-CDK4, anti-cyclin B1, anti-cyclin D1, anti-CKS1/2, anti-NEK2, anti-H2AFZ, anti-BIRC5, anti-RB, anti-E2F1, anti-E2F4, anti-p107, and anti-p130 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-lactic dehydrogenase (anti-LDH; Jackson ImmunoResearch, West Grove, PA), and anti-β-actin (Rockland, Gilbertsville, PA). The anti-PPARβ/δ antibody was previously described (18).
RNA isolation and qPCR analysis.
Total RNA was isolated from cell lines or tumor samples by the use of TRIzol reagent (Invitrogen, Carlsbad, CA). Reverse transcription and quantitative real-time PCR (qPCR) were performed as previously described (44). The relative level of mRNA was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or 18S RNA levels.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as previously described (44) using each of the following antibodies: anti-p107, anti-p130, anti-E2F4, and anti-E2F1 (Santa Cruz Biotechnology, Santa Cruz, CA) and acetylated histone 4 (Millipore, Temecula, CA). Rabbit IgG was used as a negative control. qPCR was performed to determine the relative enrichments of specific proteins in different promoter regions, and ubiquitin C genomic DNA was used for normalization. The following primers were used for ChIP analysis: for the Cdk1 proximal E2F binding site, forward primer 5′-AGCTCAGCTCTGATTGGCTCCTTT-3′ and reverse primer 5′-TTTCAAACTCGCCGCGGTAAAGC-3′; for the Cdk1 distal E2F binding site, forward primer 5′-AAACAGAGCTCAAGAGTCAGTTGGCG-3′ and reverse primer 5′-GCAGAGCGCGAAAGGGAGCGGAAA-3′; for the E2f1 E2F binding site, forward primer 5′-GGCCAATGGAGGAGGCGTT-3′ and reverse primer 5′-TGCAAAGTCCGGGCCACTT-3′; for the Chek1 E2F binding site, forward primer 5′-TTTACGGCAGAGGTGTGCGCTTT-3′ and reverse primer 5′-TTCTCACCAAGCAGTCCTTTGCCA-3′; and for the Ubc sequence, forward primer 5′-CCAGTGTTACCACCAAGAAGGTCA-3′ and reverse primer 5′-CCATCACACCCAAGAACAAGCACA-3′.
ChIP-re-ChIP assay.
For improved cross-linking of proteins that are not directly bound to DNA, a modified cross-linking approach was used (16). Briefly, cells were incubated with 5 mM dimethyl dithiobispropionimidate (DTBP) on ice for 30 min and the reaction was stopped by adding 100 mM Tris-HCl (pH 8.0) and 150 mM NaCl. A second cross-linking was then performed with 1% formaldehyde for 10 min at room temperature and stopped by the addition of glycine to reach a final concentration of 0.125 M. The remaining steps were essentially the same as those of the ChIP protocol described above with the following exceptions. For the first ChIP, sonicated chromatin was incubated with 2 μg of each the following antibodies: control rabbit IgG, anti-PPARβ/δ (8095) antibody (18), or anti-E2F4 that was conjugated to agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). After elution with extraction buffer (0.1 M NaHCO3, 1% sodium dodecyl sulfate [SDS]), samples were diluted with buffer (20 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) and subjected to a second immunoprecipitation with the same antibody (rabbit IgG, anti-PPARβ/δ, or anti-E2F4) for a single pulldown assay or the anti-PPARβ/δ antibody after the first E2F4 antibody pulldown assay.
Immunoprecipitation assay.
Standard immunoprecipitations were performed as previously described (18). For immunoprecipitations involving FLAG-tagged proteins, FLAG matrix gel (Sigma-Aldrich, St. Louis, MO) was used to pull down FLAG-tagged proteins. After three washes, coimmunoprecipitates were eluted with releasing buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100) containing FLAG peptide (Sigma-Aldrich, St. Louis, MO) (100 μg/ml). The eluted precipitates were then resolved with SDS-polyacrylamide gel electrophoresis (SDS-PAGE). For sequential immunoprecipitations, cell lysates were first subjected to anti-FLAG pulldown assays and primary precipitates were eluted as described above. Eluted coimmunoprecipitates were diluted with lysis buffer and subjected to a second immunoprecipitation. For immunoprecipitations involving in vitro-translated proteins, E2F4, PPARβ/δ, and p130 were in vitro translated using a TNT quick coupled transcription/translation kit (Promega, Madison, WI). In vitro-translated proteins were mixed in the absence or presence of 1 μM GW0742 at 30°C for 30 min. The proteins were then diluted with 25 mM MENG buffer supplemented with 150 mM NaCl, 1%Triton X-100, and phosphatase and protease inhibitors before immunoprecipitation was performed as described above.
Luciferase reporter assay.
308 cells or primary keratinocytes that were either mock infected or HRAS expressing were transiently transfected with equal amounts of endotoxin-free pGL4.20, pGL4.20-mCdk1-promoter, pGL4.20-mCdk1-distal-E2F mutant, pGL4.20-mCdk1-proximal-E2F mutant, or pGL4.20-mCdk1-CHR mutant constructs and pCMV-renilla as described above. At 48 h after transfection, cells were treated with dimethyl sulfoxide (DMSO) or 1 μM GW0742 for 24 h. Cells were lysed with 1× passive lysis buffer (Promega, Madison, WI), and luciferase activity was measured with a luminometer.
In vitro kinase assay.
Glutathione transferase-p130 (GST-p130) or GST-p107 protein (200 ng) (see Fig. S4F in the supplemental material) was incubated with 500 ng of recombinant PPARβ/δ (ProteinOne, Rockville, MD) or dilution buffer in the presence of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM EDTA with or without 1 μM GW0742 on ice for 30 min. After this step, 200 ng of a CDK4/cyclin D1 complex (Invitrogen, Carlsbad, CA) and a mixture of 100 μM cold ATP and [γ-32P]ATP was added in the presence of 1× kinase assay buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM EDTA, 2 mM DTT, 40 mM β-glycerophosphate, 20 mM ρ-nitrophenylphosphate, 0.1 mM sodium vanadate, and 0.01% Brij 35). The reaction was performed at 30°C for 15 min, and the reaction was stopped by adding 15 μl of 3× SDS loading buffer. The presence of phosphorylated p130 or p107 was detected by autoradiography, and the presence of total p130 or p107 was detected by Western blot analysis using an anti-GST antibody (Santa Cruz Biotechnologies, Santa Cruz, CA). For this assay, enhanced chemiluminescence (ECL) was used to detect proteins. The presence of recombinant PPARβ/δ was detected by Western blot analysis using an anti-PPARβ/δ antibody (Abcam, Cambridge, MA).
Microarray data accession number.
Microarray data determined in this study have been deposited in NCBI's Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) and are accessible under accession number GSE32498.
RESULTS
Ligand activation of PPARβ/δ induces G2/M arrest, causing selection against high HRAS-expressing cells.
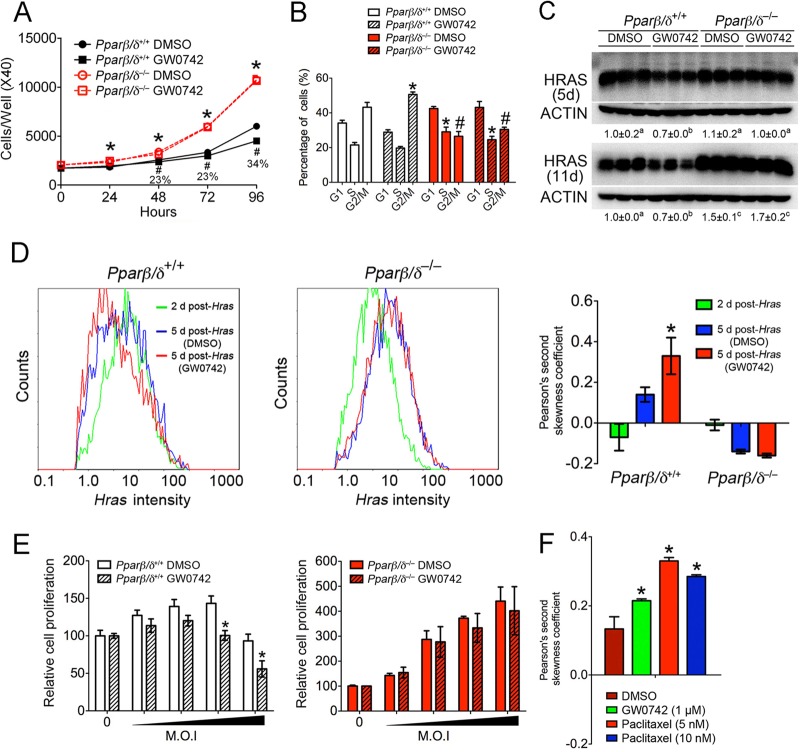
The effect of ligand activation of PPARβ/δ was examined using primary mouse keratinocytes expressing activated HRAS (53). Cell proliferation was greater in HRAS-expressing Pparβ/δ-null cells than in HRAS-expressing wild-type cells, and ligand activation of PPARβ/δ inhibited proliferation of HRAS-expressing wild-type cells (Fig. 1A). This effect was due to a PPARβ/δ-dependent G2/M-phase block (Fig. 1B; see also Fig. S1A to S1C in the supplemental material). This change in proliferation was not due to altered apoptosis (data not shown). Surprisingly, HRAS expression was lower in HRAS-expressing wild-type cells following treatment with GW0742 than in controls, but this effect was not found in HRAS-expressing Pparβ/δ-null cells, whose expression of HRAS was higher than that seen with wild-type cells (Fig. 1C). Since it is unlikely that PPARβ/δ regulates the viral promoter driving HRAS expression, the hypothesis that the reduced expression of HRAS was due to selection against cells expressing higher levels of HRAS was examined. Indeed, ligand activation of PPARβ/δ decreased the percentage of cells with high expression of HRAS and the relative copy numbers of integrated viral Hras DNA in HRAS-expressing wild-type cells; such effects were not found in HRAS-expressing Pparβ/δ-null cells (Fig. 1D; see also Fig. S1E in the supplemental material). This was consistent with the reduced expression of Hras mRNA observed in HRAS-expressing wild-type cells (see Fig. S1F in the supplemental material). Additionally, PPARβ/δ-dependent inhibition of Hras mRNA expression occurred sooner, and the magnitude of this effect was greater, with increasing levels of HRAS (see Fig. S1F in the supplemental material). The efficacy of inhibition of cell proliferation by ligand activation of PPARβ/δ was greater with increased HRAS expression (Fig. 1E). This also shows that there is a range of HRAS expression required to increase cell proliferation and that expression above this range leads to inhibition of proliferation in wild-type keratinocytes, an effect not found in Pparβ/δ-null keratinocytes (Fig. 1E). In addition, as the level of HRAS increased, so did the magnitude of the PPARβ/δ-dependent increase in G2/M arrest (see Fig. S1G in the supplemental material). Collectively, these data suggest that ligand activation of PPARβ/δ selects against cells with higher expression of activated HRAS. Whether the G2/M arrest directly causes selection against cells with higher expression of HRAS was examined by quantifying relative HRAS expression after treatment with a known mitosis inhibitor. Similar to what was observed with ligand activation of PPARβ/δ, inhibition of the G2/M phase with paclitaxel caused selection against cells with higher expression levels of HRAS (Fig. 1F; see also Fig. S1H in the supplemental material). This suggests that G2/M arrest resulting from ligand activation of PPARβ/δ causes selection against cells expressing higher levels of HRAS.
Fig 1.
Ligand activation of PPARβ/δ attenuates cell proliferation by inducing G2/M arrest, causing selection against cells expressing HRAS at high levels. (A and B) HRAS-expressing wild-type and Pparβ/δ-null keratinocytes were treated with or without 1 μM GW0742 for 4 days. Cell numbers were quantified daily. Cell cycle analysis was performed after 3 days of treatment. (C) Western blot analysis of HRAS-expressing keratinocytes 5 days or 11 days postinfection. Expression levels were normalized to β-actin and are presented as fold change relative to control DMSO results. (D) Flow cytometric analysis using anti-HRAS antibody was performed, and Pearson's second skewness coefficient was calculated for HRAS intensity. (E) Cells were infected with an Hras retrovirus at increasing MOI, and after 3 days of culture with or without 1 μM GW0742, an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay was performed. (F) Flow cytometric analysis of HRAS intensity in HRAS-expressing keratinocytes performed as described above for panel D. For all data sets, n = 3 or 4 independent samples per treatment group. Values represent means ± standard errors of the means (SEM). *, significantly different from wild-type vehicle control (DMSO) result, P ≤ 0.05. #, significantly less than GW0742-treated wild-type result, P ≤ 0.05. Values with different superscripts are significantly different, P ≤ 0.05.
Inhibition of mitosis by ligand activation of PPARβ/δ in HRAS-expressing cells.
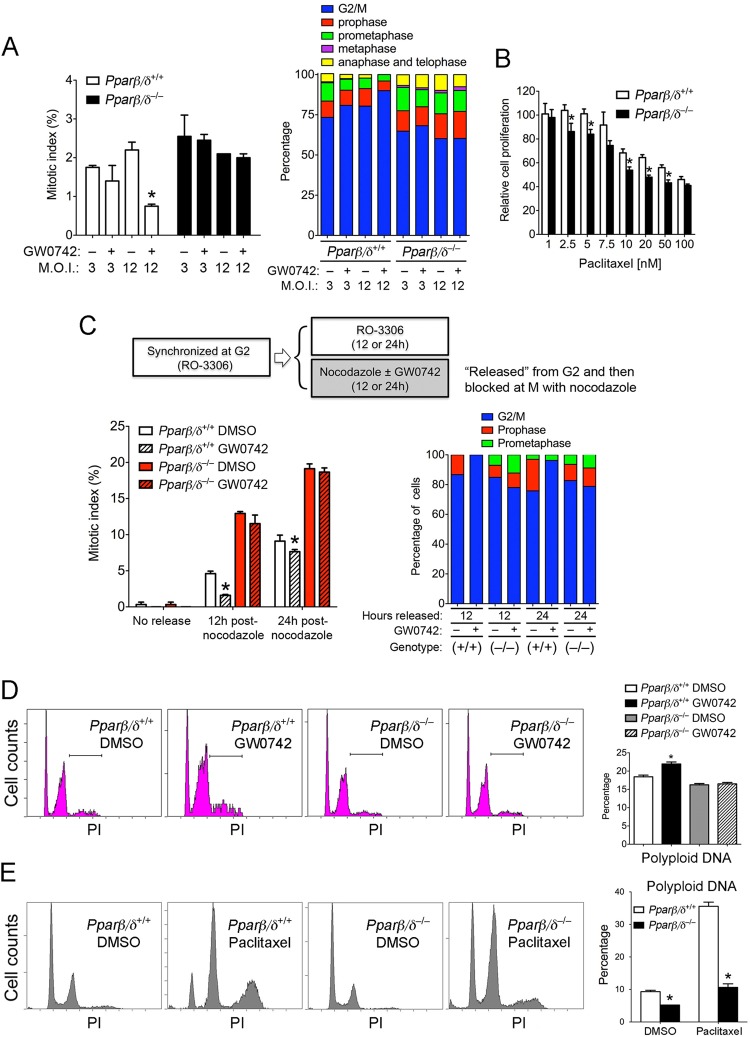
A G2/M-phase arrest could be mediated by a block in mitosis, which was examined in the following experiments by quantifying the mitotic index and comparing the effects of mitosis inhibitors. Ligand activation of PPARβ/δ markedly reduced the mitotic index in HRAS-expressing wild-type cells with a higher level of HRAS, which was reflected by a PPARβ/δ-dependent increase in the numbers of cells at the G2/M boundary and a decrease in the numbers of cells at metaphase, anaphase, and telophase (Fig. 2A). A lower percentage of cells at the G2/M boundary and higher percentages of cells in metaphase, anaphase, and telophase were observed in Pparβ/δ-null counterparts (Fig. 2A). Enhanced sensitivity to paclitaxel (a microtubule stabilizer that blocks mitosis)-induced inhibition of cell proliferation was found in HRAS-expressing Pparβ/δ-null cells compared to wild-type cells (Fig. 2B). This is consistent with a higher level of mitosis (Fig. 2A) and enhanced proliferation in HRAS-expressing Pparβ/δ-null keratinocytes (Fig. 1A). Blocking mitosis at prometaphase with paclitaxel caused a greater increase in the mitotic index in Pparβ/δ-null cells than in wild-type cells (see Fig. S2A in the supplemental material). While paclitaxel effectively blocked HRAS-expressing wild-type cells in prometaphase, a number of HRAS-expressing Pparβ/δ-null cells proceeded to metaphase following treatment with paclitaxel (see Fig. S2B and S2C in the supplemental material). To examine the effect of ligand activation of PPARβ/δ on mitosis entry, the cells were synchronized at the G2 phase with RO-3306 (a CDK1 inhibitor) and then released into nocodazole to a block at the prometaphase in the presence or absence of GW0742. Ligand activation of PPARβ/δ decreased the mitotic index only in HRAS-expressing wild-type cells (Fig. 2C; see also Fig. S2D in the supplemental material). Further, the mitotic index was greater after release from the G2/M boundary in HRAS-expressing Pparβ/δ-null cells compared to wild-type cells (Fig. 2C; see also Fig. S2D in the supplemental material). While the majority of HRAS-expressing wild-type cells released from the G2 block and treated with GW0742 remained in the G2/M boundary, a higher percentage of HRAS-expressing Pparβ/δ-null cells proceeded to prophase and prometaphase compared to controls (Fig. 2C). Since it is known that keratinocytes in the G2/M state can exhibit polyploidy (67), this was examined in HRAS-expressing cells. Ligand activation of PPARβ/δ increased levels of cells with polyploidy DNA concomitantly with an increase in levels of cells at the G2/M block only in HRAS-expressing wild-type cells (Fig. 2D). Similarly, a markedly greater increase in levels of cells with polyploid DNA was found in HRAS-expressing wild-type but not Pparβ/δ-null cells after treatment with paclitaxel (Fig. 2E).
Fig 2.
Ligand activation of PPARβ/δ inhibits mitosis of HRAS-expressing keratinocytes. Wild-type and Pparβ/δ-null keratinocytes were infected with an Hras retrovirus at an estimated MOI of 3 or 12 for 2 days and then treated with or without 1 μM GW0742 for another 3 days. (A) (Left panel) Mitotic index. (Right panel) The distribution of cells in different mitotic phases. (B) HRAS-expressing keratinocytes were treated with the indicated concentration of paclitaxel for 24 h, and cell proliferation was determined. (C) HRAS-expressing keratinocytes were synchronized at the G2 phase by treatment with RO-3306 for 36 h. Cells were then either maintained in RO-3306 or released from the G2-phase block and cultured with nocodazole with or without 1 μM GW0742 for 12 or 24 h. The mitotic index and percentages of cells at the G2/M boundary and in different mitotic phases were determined. (D and E) Representative DNA histograms from HRAS-expressing keratinocytes treated with and without 1 μM GW0742 (D) or 10 nM paclitaxel (E). The percentages of cells with polyploidy DNA are shown in the far right panels. For all data sets, n = 3 independent samples per treatment group. Values represent means ± SEM. *, significantly different from wild-type controls, P ≤ 0.05.
Ligand activation of PPARβ/δ decreases expression of E2F target genes that regulate mitosis in HRAS-expressing keratinocytes.
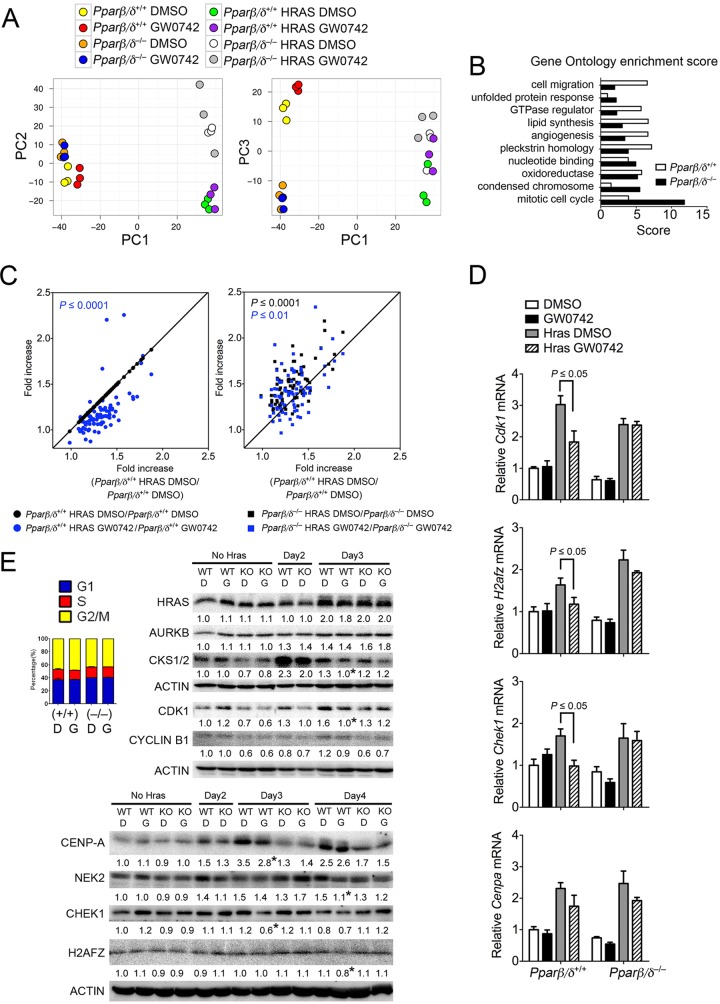
Microarray analysis was performed to identify potential genes that could regulate mitosis through PPARβ/δ. Principal component analysis showed that the major differences in gene expression profiles were due to expression of HRAS (Fig. 3A). Differences between HRAS-expressing wild-type and Pparβ/δ-null cells with respect to gene expression were markedly larger than those seen with control cells, and the effect of ligand activation was also PPARβ/δ dependent (Fig. 3A). Gene ontology analysis showed significant enrichment of HRAS-induced genes that regulate chromosome condensation and mitotic cell cycle in both genotypes, and the enrichment score was much higher in HRAS-expressing Pparβ/δ-null cells than in wild-type cells (Fig. 3B). Eighty-two mitosis-related genes induced by HRAS in either wild-type or Pparβ/δ-null cells were identified by this analysis. Expression of 62 of these genes was repressed by ligand activation of PPARβ/δ in HRAS-expressing wild-type cells but not Pparβ/δ-null cells (Fig. 3C). Further, the fold induction caused by HRAS was greater in Pparβ/δ-null cells than in wild-type cells (Fig. 3C). Changes in expression of 18 genes selected based on microarray and bioinformatic analysis, including Cdk1, H2afz, Chek1, and Cenpa, were verified by qPCR (Fig. 3D and data not shown). Western blot analysis of HRAS-expressing cells also showed PPARβ/δ-dependent repression of CDK1, cyclin B1, H2AFZ, CHEK1, CENPA, and NEK2 by ligand activation, and these effects were not due to changes in cell cycle distribution (Fig. 3E). The PPARβ/δ-dependent repression of cyclin B1 persisted (see Fig. S1D in the supplemental material).
Fig 3.
Ligand activation of PPARβ/δ decreases expression of genes that modulate mitosis in HRAS-expressing keratinocytes. Microarray analysis was performed using control or HRAS-expressing wild-type or Pparβ/δ-null keratinocytes. (A) PCA of normalized microarray data. (B) Differential enrichment of genes involved in different biological processes was determined by gene ontology analysis by the use of DAVID software. The criterion for inclusion in analysis was a minimum of a 1.3-fold change induced by HRAS. (C) Significant (82-gene) enrichment with respect to mitosis and chromosome condensation regulators in HRAS-expressing Pparβ/δ-null keratinocytes was found by DAVID analysis. Ligand activation of PPARβ/δ repressed induction of 62 of the 82 mitosis genes in HRAS-expressing wild-type but not Pparβ/δ-null keratinocytes. (D and E) Confirmation of changes in mRNA (D) and protein (E) expression of mitosis-related genes regulated by ligand activation of PPARβ/δ in HRAS-expressing keratinocytes. qPCR analysis was performed 24 h after treating HRAS-expressing cells with 1 μM GW0742. Levels of protein expression of control cells (No Hras), HRAS-expressing wild-type (WT), and Pparβ/δ-null (KO) cells were compared. Control HRAS-expressing cells were obtained 2 days postinfection (Day2). The effect of ligand activation of PPARβ/δ was examined by treating HRAS-expressing cells 2 days postinoculation with either DMSO (columns D) or 1 μM GW0742 (columns G) for 24 or 48 h (Day3 or Day4, respectively). Expression levels were normalized to β-actin and are presented as fold changes relative to control DMSO results. The inset represents the distribution of cell cycle phases obtained 24 h post-ligand treatment. For all data sets, n = 3 or 4 independent samples. Values represent means ± SEM. *, significantly different from wild-type controls, P ≤ 0.05.
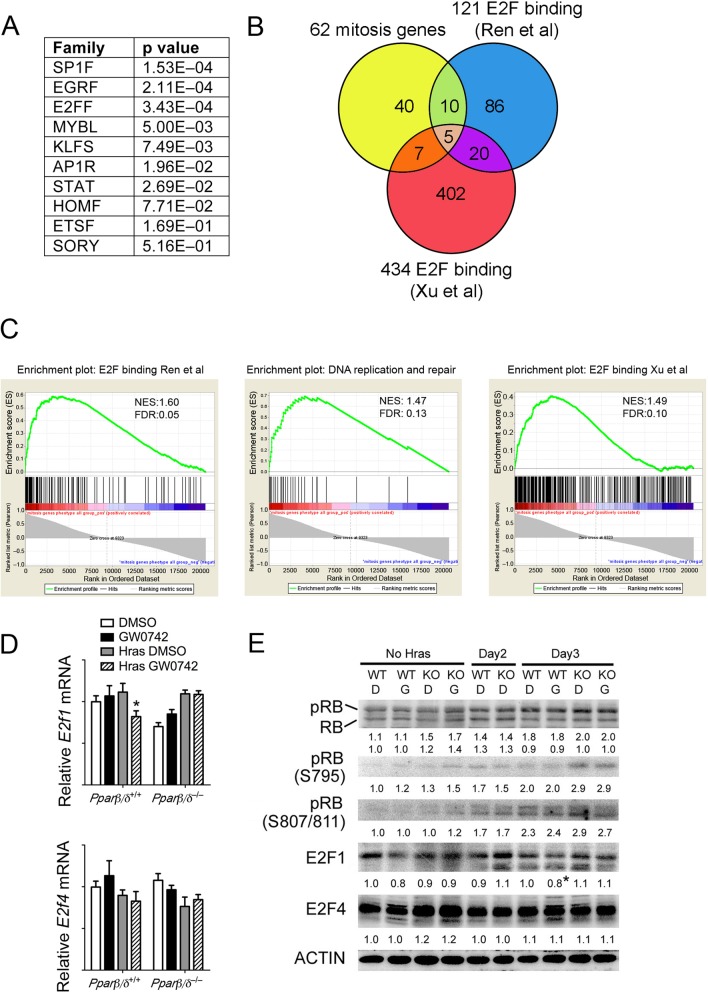
Bioinformatic analysis of the promoters of the 62 mitosis-related genes showed that while PPREs were not found, several common regulatory elements, including E2F, SP1, and EGR, were present in most genes (Fig. 4A). Since E2F is known to regulate mitosis (26, 41), the 62 genes were compared with genes in two ChIP-confirmed E2F target gene databases (52, 63). Twenty-two of these genes were also found to be E2F target genes based on this analysis (Fig. 4B). Gene set enrichment analysis (GSEA) revealed that E2F target genes were regulated similarly to the mitosis-related genes, including genes involved in DNA repair and synthesis (Fig. 4C); SP1 and EGR were ruled out as being central to this regulation (data not shown). E2F1 is an activator E2F that upregulates expression of target genes, whereas E2F4 is a repressor form of E2F that represses expression of target genes (9). PPARβ/δ-dependent repression of E2F1 was observed following ligand activation (Fig. 4D and E), consistent with the fact that E2F1 is autoregulated (29). Expression of E2F4 was not changed in response to ligand activation of PPARβ/δ (Fig. 4D and E). The observed change in expression of mitosis-related genes was not mediated by altered phosphorylation of retinoblastoma (RB), because no change in phospho-RB was observed following ligand activation of PPARβ/δ (Fig. 4E). This suggests that PPARβ/δ-dependent modulation of mitosis-related gene expression occurs downstream of RB.
Fig 4.
PPARβ/δ regulates E2F target genes in HRAS-expressing keratinocytes. (A) Promoter transcription factor binding site analysis was performed to identify common transcription factors that regulate expression of the 62 differentially expressed mitosis-related genes modulated by PPARβ/δ. The P values indicate the relative significance of promoter enrichment for each respective transcription factor. (B) The overlap of 62 mitosis-related genes and two ChIP-validated E2F target gene databases is presented in the Venn diagram. (C) The gene sets described for panel B were examined by gene set enrichment analysis (GSEA). The gene set corresponding to DNA replication and DNA repair was a subset of E2F target genes from the Ren et al. database (52). (D) qPCR validation of microarray data. (E) Western blot analysis of mock-infected (No Hras) or HRAS-expressing keratinocytes treated with or without GW0742 at 2 to 4 days postinfection. Expression levels were normalized to β-actin and are presented as fold change relative to control DMSO results. For all data sets, n = 3 or 4 independent samples. Values represent means ± SEM. *, significantly different from HRAS-expressing wild-type control, P ≤ 0.05.
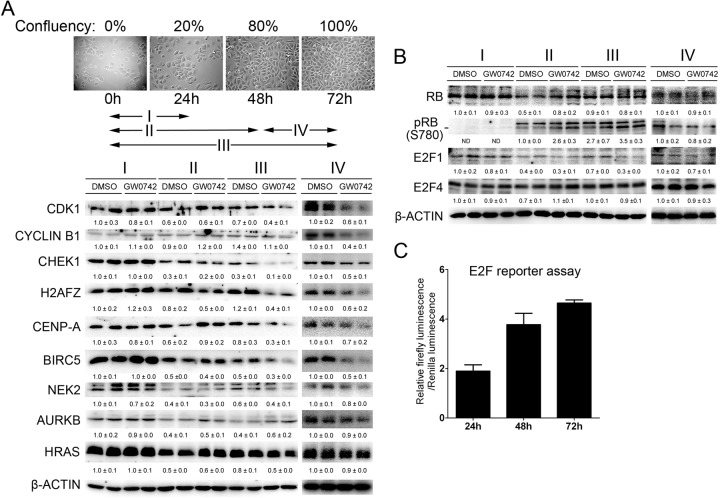
Consistent with the finding that ligand activation of PPARβ/δ in 308 cells causes G2/M arrest (4), repression of E2F target genes that regulate mitosis, including cyclin B1, CHEK1, CDK1, and H2AFZ, was also observed following ligand activation of PPARβ/δ in these cells (Fig. 5A). This is important because 308 cells have an activated Hras mutation, in contrast to the keratinocyte model of viral HRAS transformation. Interestingly, the observed repression was greatest in cells with higher confluence when E2F activity was highest (Fig. 5C). Additionally, expression of HRAS was repressed in response to ligand activation of PPARβ/δ but only after 72 h of treatment, when the cells were at higher confluence (Fig. 5A). Ligand activation of PPARβ/δ in confluent 308 cells for only 24 h caused significant repression of E2F target genes that regulate mitosis with no apparent change in HRAS expression, indicating that reduced HRAS expression does not mediate these changes (Fig. 5A). As found in HRAS-expressing keratinocytes, expression of E2F1 was repressed by ligand activation of PPARβ/δ in 308 cells (Fig. 5B). Moreover, examination of phospho-RB showed that PPARβ/δ-dependent modulation of mitosis genes in 308 cells occurs downstream of RB (Fig. 5B).
Fig 5.
Ligand activation of PPARβ/δ inhibits expression of promitotic proteins in 308 cells. 308 cells were cultured for up to 72 h and treated with GW0742 to activate PPARβ/δ for various periods of time as illustrated by the arrows. (A and B) Western blot analysis of mitosis-related proteins (A) or RB, E2F1, and E2F4 (B) in 308 cells treated as described above. Expression levels were normalized to β-actin and are presented as fold change relative to control DMSO results. (C) 308 cells were cultured for 24, 48, or 72 after transfection with an E2F-luciferase reporter construct. Luciferase activity was measured and normalized to renilla as an internal control. For all data sets, n = 3 independent samples per treatment group. Values represent means ± SEM.
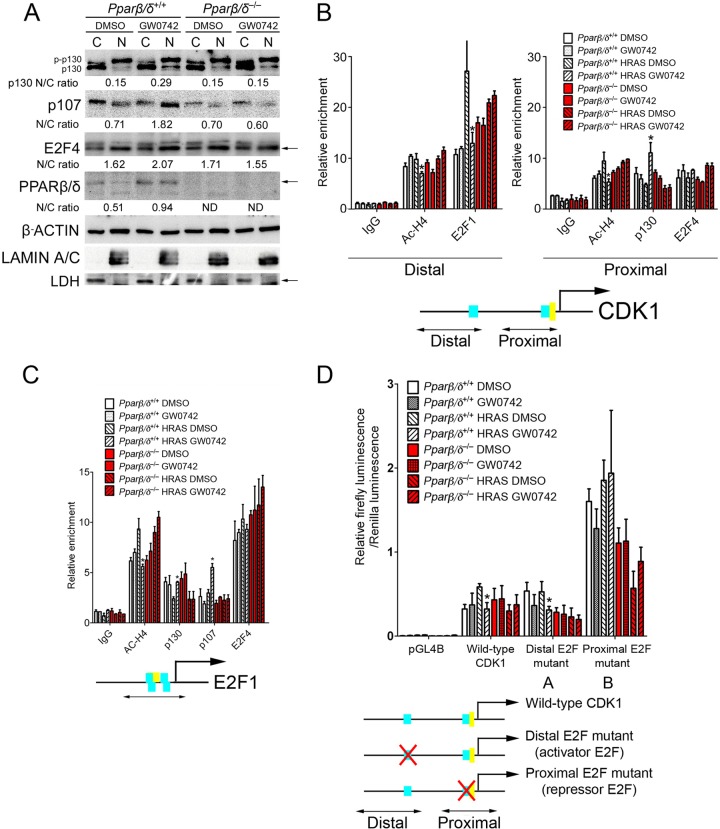
Ligand activation of PPARβ/δ represses CDK1 and E2F1 by increasing recruitment of p107/p130 to E2F4 binding sites.
The molecular mechanisms of repression of E2F target genes following ligand activation of PPARβ/δ were examined next. The decreased expression of E2F target genes could be caused by an increase of repressor E2F4 activity, as the E2F4 repressor is known to form a complex with RB/p107/p130 to repress target gene expression (9). The nucleus-to-cytosol ratio of p130 (hypophosphorylated), p107, E2F4, and PPARβ/δ was increased by ligand activation of PPARβ/δ in 308 cells (see Fig. S3C in the supplemental material) and in HRAS-expressing wild-type but not Pparβ/δ-null cells (Fig. 6A; see also Fig. S3A in the supplemental material). These effects were not observed in mock-infected keratinocytes (see Fig. S3A in the supplemental material). Since the CDK1/cyclin B1 complex is known to be critical for mitosis entry and since ligand activation of PPARβ/δ repressed HRAS-induced CDK1 expression, the promoter occupancy of E2Fs was examined in the distal E2F1 (activator) and proximal E2F4 (repressor) binding sites of the CDK1 promoter (70). Ligand activation of PPARβ/δ caused a reduction in the acetylated histone 4 level in both the E2F1 and E2F4 binding sites and decreased promoter occupancy of E2F1 in the E2F1 binding site in HRAS-expressing wild-type but not Pparβ/δ-null cells (Fig. 6B). While no occupancy of p130 or p107 in the E2F1 binding site was found (data not shown), promoter occupancy of p130 in the E2F4 binding site was increased following ligand activation of PPARβ/δ in HRAS-expressing wild-type but not Pparβ/δ-null keratinocytes (Fig. 6B). E2F4 promoter occupancy was evident on the E2F4 binding site; ligand activation of PPARβ/δ did not alter this occupancy (Fig. 6B). None of these changes were observed in mock-infected keratinocytes (Fig. 6B). Similar effects were also noted in 308 cells (see Fig. S3D in the supplemental material). Combined, these observations are consistent with the notion that ligand activation of PPARβ/δ represses HRAS-induced expression of CDK1 by repressing E2F1 activator activity and also increasing E2F4/p130 repressor activity. While the change in occupancy of E2F4/p130 on the CDK1 promoter following ligand activation of PPARβ/δ is associated with a decrease in expression of CDK1 protein of only 37%, the stoichiometry of transcriptional and translational proteins required to mediate this repression is unknown and could involve multiple biological factors in addition to E2F4/p130 binding.
Fig 6.
Ligand activation of PPARβ/δ represses CDK1 and E2F1 by increasing recruitment of p107/130 to E2F4 binding sites. Wild-type and Pparβ/δ-null cells were mock infected or Hras infected for 2 days. Cells were cultured with or without 1 μM GW0742 for 24 h. (A) Western blot analysis of cytosol (columns C) and nuclear (columns N) extracts from HRAS-expressing keratinocytes. Expression levels were normalized to β-actin. The average ratios of nuclear to cytoplasmic protein (N/C) are shown. (B and C) Promoter occupancy of acetylated histone 4 (Ac-H4), E2F1, p130, p107, and E2F4 was examined by ChIP analysis of the mouse CDK1 (B) or E2F1 (C) promoter. For the CDK1 and E2F1 promoters, the distal E2F1 activator binding site and the proximal E2F4 repressor binding site are depicted as two blue boxes. The CHR binding site is depicted as the yellow box. The relative positions of the PCR products used for ChIP analysis are shown by the lines with double arrows. (D) Promoter analysis of the mouse CDK1 promoter. Mutations in the distal activator E2F1 binding site, the proximal repressor E2F4 binding site, and the proximal CHR binding site are illustrated. For all data sets, n = 3 independent samples. Values represent means ± SEM. *, significantly different from HRAS-expressing wild-type control, P ≤ 0.05.
Ligand activation of PPARβ/δ also caused a reduction in acetylated histone 4 levels and increased promoter occupancy of p130 and p107 in the E2F binding site of the E2f1 promoter (a gene autoregulated by E2F) in HRAS-expressing wild-type but not Pparβ/δ-null keratinocytes (Fig. 6C). E2F4 promoter occupancy was evident in the E2F binding site, and ligand activation of PPARβ/δ did not alter this occupancy (Fig. 6C). None of these changes were observed in mock-infected keratinocytes (Fig. 6C). Similar effects were also noted in 308 cells (see Fig. S3E in the supplemental material). Combined, these observations suggest that ligand activation of PPARβ/δ represses HRAS-induced expression of E2F1 by increasing E2F4/p130/p107 repressor activity.
Since CHEK1 is regulated by E2F1 (8), the effect of PPARβ/δ activation on promoter occupancy of E2F1, E2F4, and p130 was also examined. Ligand activation of PPARβ/δ caused a reduction in acetylated histone 4 levels and decreased promoter occupancy of E2F1 in the E2F binding site on the Chek1 promoter in HRAS-expressing wild-type but not Pparβ/δ-null keratinocytes (see Fig. S3B in the supplemental material). No significant promoter occupancy of E2F4 or p130 with respect to the E2F binding site was detected in HRAS-expressing keratinocytes (see Fig. S3B in the supplemental material).
To further characterize the mechanism by which ligand activation of PPARβ/δ represses CDK1 expression, mutation analysis of the CDK1 promoter was performed. Four CDK1 promoter-luciferase constructs were designed (Fig. 6D). Ligand activation of PPARβ/δ caused repression of the wild-type CDK1 promoter and the distal E2F mutant CDK1 promoter in HRAS-expressing wild-type but not Pparβ/δ-null keratinocytes (Fig. 6D). Basal luciferase activity was significantly higher in both the proximal E2F mutant (Fig. 6D) and the CHR mutant (data not shown), consistent with the finding that E2F4 represses CDK1 expression. However, repression of the CDK1 promoter activity was not found in response to ligand activation of PPARβ/δ with the proximal E2F mutant (Fig. 6D) or the CHR mutant (data not shown). Similar effects were also noted in 308 cells (see Fig. S3F in the supplemental material). These observations suggest that while E2F1 activity is dispensable, E2F4 repressor activity is indispensable for PPARβ/δ-dependent repression of CDK1 expression.
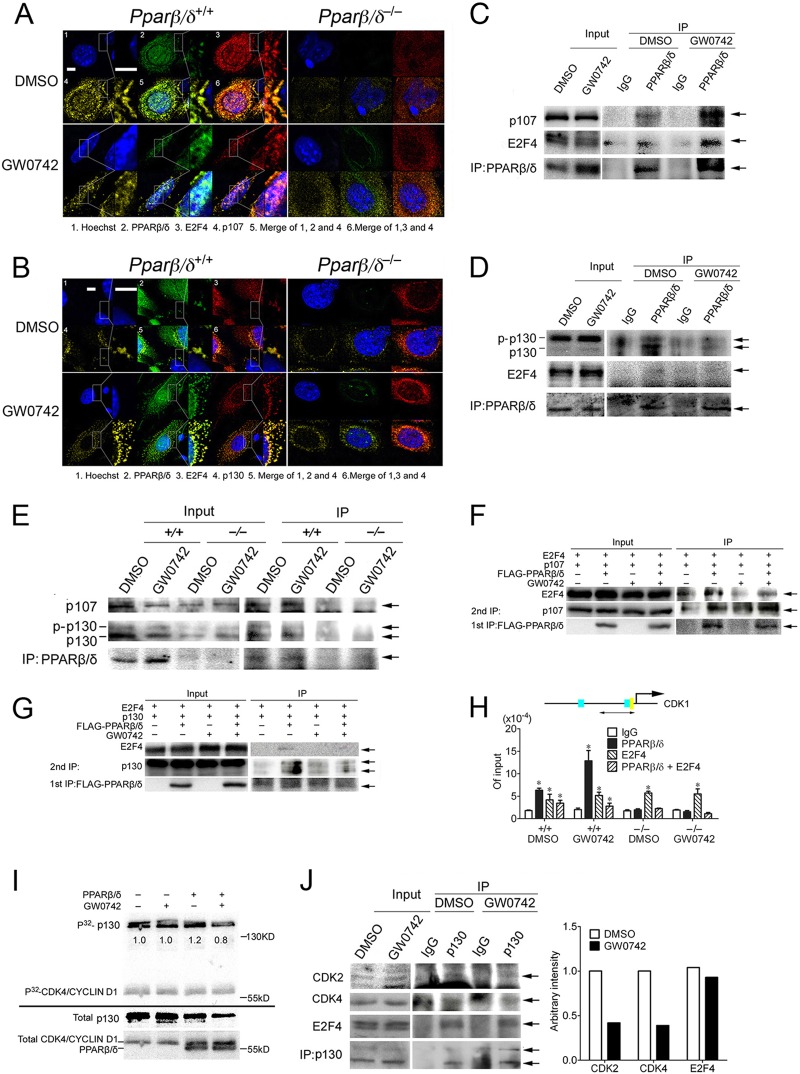
PPARβ/δ interacts with p107 and p130.
Since nuclear translocation of PPARβ/δ in response to ligand activation in HRAS-expressing cells occurred concomitantly with the increased nuclear accumulation of hypophosphorylated p130 and p107 (Fig. 6A), this suggests that PPARβ/δ may physically interact with p130 and p107 to facilitate their translocation. It is already known that E2F4 and p130/p107 physically interact, and indeed, colocalization of p130/p107 and E2F4 was found in both wild-type and Pparβ/δ-null keratinocytes, as shown by confocal microscopy (Fig. 7A and B). In addition, colocalization of PPARβ/δ and p130/p107 was observed in HRAS-expressing wild-type but not Pparβ/δ-null cells (Fig. 7A and B). p107 and E2F4 were coimmunoprecipitated with PPARβ/δ (Fig. 7C), and PPARβ/δ and E2F4 were coimmunoprecipitated with p107 in HEK293T cells (see Fig. S4A in the supplemental material). While E2F4 and both forms of p130 were coimmunoprecipitated with PPARβ/δ, hypophosphorylated p130 was preferentially pulled down (Fig. 7D). The finding that both p130/p107 and E2F4 were coimmunoprecipitated with PPARβ/δ suggests that either (i) PPARβ/δ can physically bind to p130/p107 and E2F4 or (ii) PPARβ/δ can bind to p130/p107 only and E2F4 was coimmunoprecipitated because E2F4 associates with p107/p130. To distinguish between these possibilities, in vitro-translated p130, PPARβ/δ, and E2F4 proteins were used in a coimmunoprecipitation assay. While PPARβ/δ physically interacted with p130 (see Fig. S4B in the supplemental material), no direct interaction between PPARβ/δ and E2F4 was observed with either E2F4 (see Fig. S4C in the supplemental material) or PPARβ/δ pull down (see Fig. S4D in the supplemental material). A direct interaction between p130/p107 and PPARβ/δ was also found in HRAS-expressing primary keratinocytes (Fig. 7E). An interaction between endogenous p130/p107 and PPARβ/δ was also found in HEK293T cells (see Fig. S4E in the supplemental material). Combined, these findings suggest that PPARβ/δ can directly interact with p130/p107 but not E2F4.
Fig 7.
PPARβ/δ binds with p107/p130. (A and B) Confocal immunofluorescence of HRAS-expressing wild-type and Pparβ/δ-null keratinocytes treated with DMSO or 1 μM GW0742 for 24 h. Photomicrographs are shown at higher magnification only for wild-type keratinocytes. Bars, 3 μm. (C to E) IP assays in HEK293T cells transiently transfected with pCMV-p107/pCMV-p130, pCMV-E2F4, and pSG5-PPARβ/δ (C and D) or in HRAS-expressing keratinocytes (E) treated with DMSO or 1 μM GW0742 for 24 h. Arrows indicate p107 (C), hypophosphorylated p130 (D), or p107 or hyper- or hypophosphorylated p130 (E). (F and G) Sequential pulldown assay of HEK293T cells transiently transfected with pCMV-p107 (F) or pCMV-p130 (G), pCMV-E2F4, and pcDNA-FLAG-PPARβ/δ and treated with DMSO or 1 μM GW0742 for 24 h. Arrows indicate p107 (F), p130 (G), or E2F4 or FLAG-PPARβ/δ. (H) ChIP-re-ChIP assay on HRAS-expressing wild-type and Pparβ/δ-null keratinocytes after 24 h of DMSO or 1 μM GW0742 treatment. Relative recruitment of PPARβ/δ, E2F4, or the complex of the two on the repressor E2F4 binding site of the Cdk1 gene was quantified by qPCR. Data represent the proportion of input. (I) An in vitro kinase assay was performed, and the ratio of the phosphorylated p130 to total p130 is shown below each band. (J) Co-IP assays were performed with HRAS-expressing keratinocytes treated with DMSO or 1 μM GW0742 for 24 h. Arrows indicate CDK2, CDK4, E2F4, or p130. The amount of CDK2, CDK4, and E2F4 pulled down was quantified by normalization to the amount of p130 pulled down.
The observation that both p130/p107 and E2F4 were coimmunoprecipitated with PPARβ/δ (Fig. 7C and D) implies that p130/p107, E2F4, and PPARβ/δ may form a complex. A sequential immunoprecipitation approach was used to examine this idea. Indeed, E2F4 was detected in a complex with PPARβ/δ and p107/p130 following sequential immunoprecipitation of PPARβ/δ followed by immunoprecipitation of p107/p130 (Fig. 7F and G), suggesting that this complex exists in a system when the three proteins are overexpressed. To determine if this complex is found on the promoter of the Cdk1 gene, a ChIP-re-ChIP assay was performed using a cross-linker (DTBP) that allows detection of proteins that are not directly bound to chromatin. With this approach, promoter occupancy of PPARβ/δ was detected at the same site in HRAS-expressing wild-type cells and increased in the presence of GW0742 (Fig. 7H) but PPARβ/δ was not detected on the E2F4 repressor sites when formaldehyde was used as the cross-linker (data not shown). Enriched promoter occupancy of E2F4 and PPARβ/δ was found after sequential PPARβ/δ and E2F4 pulldown only in wild-type cells (Fig. 7H). These data suggest that PPARβ/δ, E2F4, and p130/p107 may form a complex on the Cdk1 promoter. Whether this occurs for other E2F target genes remains to be determined.
PPARβ/δ may preferentially interact with hypophosphorylated p130 (Fig. 7D and E), suggesting that the binding of PPARβ/δ to p130 may protect p130 from phosphorylation. Since p130 can be phosphorylated by a CDK4/cyclin D1 complex (22), an in vitro kinase assay was performed to examine this hypothesis. The addition of both PPARβ/δ and GW0742 decreased the phosphorylation of p130 by 33% (Fig. 7I). The decreased phosphorylation of p130 was not due to competition between PPARβ/δ and p130 for CDK4/cyclin D1, because PPARβ/δ was not phosphorylated by CDK4/cyclin D1 (Fig. 7I). To determine whether the observed decrease of phosphorylation of p130 by ligand activation of PPARβ/δ was due to the decreased binding of CDK4/cyclin D1 complex to p130, the interaction of p130 and CDK4 was examined. Ligand activation of PPARβ/δ decreased the association between p130 and CDK4 in HRAS-expressing cells (Fig. 7J). In addition, the interaction between p130 and CDK2, which can also phosphorylate p130 (10), was also decreased by ligand activation of PPARβ/δ (Fig. 7J). No significant change in the phosphorylation of p107 by CDK4/cyclin D1 in the presence of PPARβ/δ and/or GW0742 was observed (see Fig. S4G in the supplemental material).
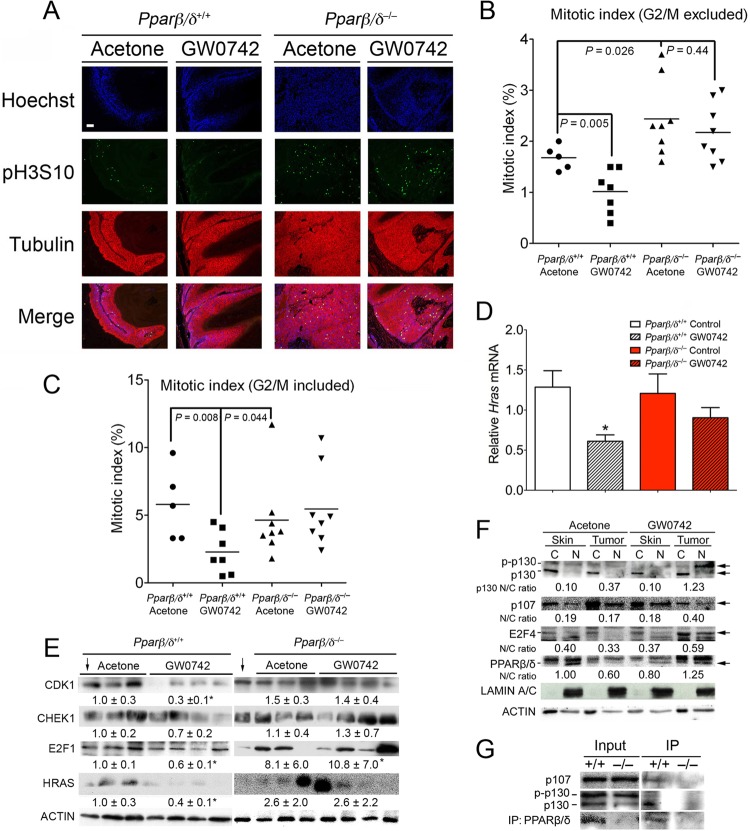
Ligand activation of PPARβ/δ attenuates mitosis in vivo.
To determine if the changes found in HRAS-expressing cells in response to ligand activation of PPARβ/δ also occur in vivo, the mitotic index and expression of Hras were examined in skin tumors obtained from a two-stage bioassay (initiation with DMBA, promotion with TPA). Ligand activation of PPARβ/δ caused a decrease in the mitotic index in skin tumors from wild-type but not Pparβ/δ-null mice (Fig. 8A to C). In addition, the mitotic index in skin tumors from Pparβ/δ-null mice was higher than that seen with wild-type mice (Fig. 8A to C). Consistent with the hypothesis that PPARβ/δ-dependent inhibition of mitosis causes selection against cells expressing higher levels of HRAS, expression of Hras mRNA was lower in skin tumors from wild-type mice treated with GW0742, an effect not found in Pparβ/δ-null mice (Fig. 8D). In addition, ligand activation of PPARβ/δ also decreased the level of proteins that promote mitosis, including CDK1, CHEK1, and E2F1, in skin tumors from wild-type but not Pparβ/δ-null mice (Fig. 8E). Expression of HRAS was also reduced by ligand activation of PPARβ/δ in wild-type mouse skin tumors but not in Pparβ/δ-null mouse skin tumors (Fig. 8E). Consistent with results observed in HRAS-expressing primary keratinocytes and 308 cells (Fig. 6A; see also Fig. S3A and S3C in the supplemental material), ligand activation of PPARβ/δ increased the nucleus-to-cytosol ratio of p130 (hypophosphorylated), p107, E2F4, and PPARβ/δ in skin tumors but not in adjacent nontransformed skin (Fig. 8F). There was also an increase in nuclear accumulation of phosphorylated p130 in skin tumors following ligand treatment (Fig. 8F). There are at least two possibilities to explain why the two forms of p130 increase in numbers when PPARβ/δ is activated. First, even though PPARβ/δ preferentially interacts with hypo-p130, PPARβ/δ can also interact with phosphorylated p130 (Fig. 7D and E). Thus, when PPARβ/δ is activated, nuclear translocation of PPARβ/δ may lead to an increase in both hypo- and phosphorylated p130 levels. The second possibility is that, even though ligand-activated PPARβ/δ decreases phosphorylation of p130 (Fig. 7I), it does not completely prevent p130 from being phosphorylated by CDKs. Thus, nuclear hypo-p130 may be phosphorylated by instances of the CDK2/CDK4 complex that are present in the nucleus and this may account for the increased levels of both forms of p130 observed in the nucleus when PPARβ/δ is activated. An association between PPARβ/δ and p107 and hypophosphorylated p130 was also detected in wild-type skin tumors treated with GW0742 (Fig. 8G). These findings suggest that ligand activation of PPARβ/δ also attenuates mitosis in chemically induced skin tumors with an HRAS mutation through cross talk with E2F signaling.
Fig 8.
Ligand activation of PPARβ/δ selects against higher HRAS-expressing chemically induced skin tumors by inhibiting mitosis. (A) Representative photomicrographs of skin tumors in wild-type or Pparβ/δ-null mouse skin treated with or without GW0742. The mitotic index was calculated from a minimum of 1,000 cells per sample. (B and C) Quantification of the mitotic index for the samples examined in panel A, excluding cells in the G2/M boundary (B) and including cells in the G2/M boundary (C). For panels B and C, representative skin tumors were used (n = 5 to 8). (D) RNA was isolated from skin tumors, and Hras mRNA expression was determined by qPCR and normalized to 18S RNA. For all data sets, n = 6 to 10 independent samples. Values represent means ± SEM. *, significantly different from wild-type controls, P ≤ 0.05. Bar, 100 μm. (E and F) Western blot analysis of CDK1, CHEK1, E2F1, and HRAS (E) or p130, p107, E2F4, and PPARβ/δ (F) in skin and skin tumors. Expression levels of proteins were normalized to β-actin and are presented as the fold change relative to control DMSO (E) or as the average ratio of nuclear to cytoplasmic protein (N/C) (F). The arrows pointing down in panel E indicate the same wild-type control sample used for comparison. (G) Co-IP assay showing an interaction between PPARβ/δ and p107 or p130 in cell lysates from skin tumors treated with GW0742.
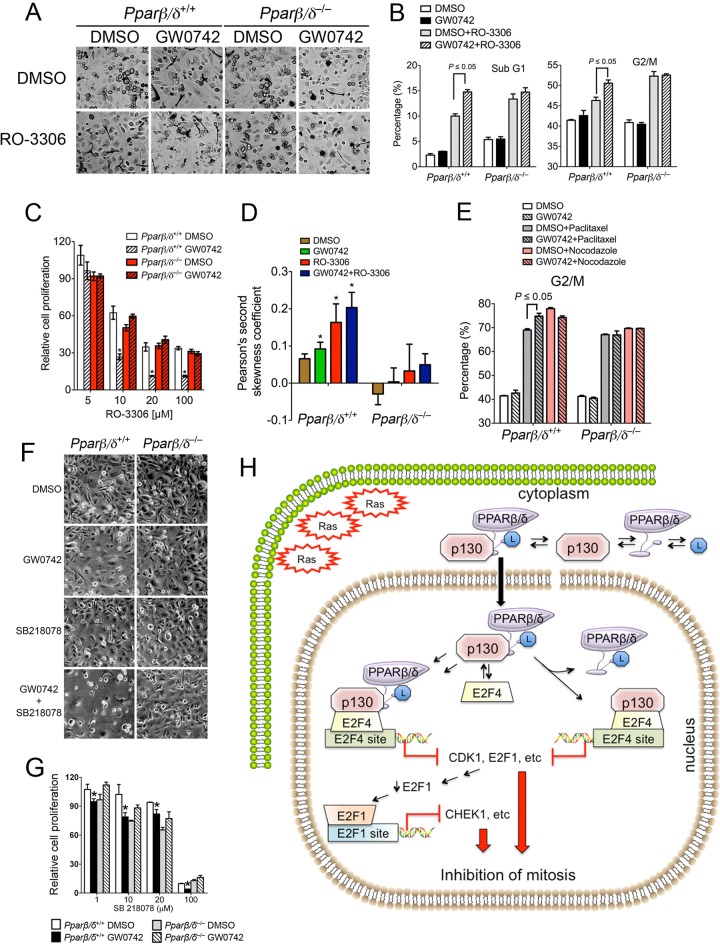
Enhanced sensitivity to pharmacological inhibition of mitosis in HRAS-expressing cells by ligand activation of PPARβ/δ.
Other therapeutics, including RO-3306 (CDK1 inhibitor), paclitaxel (microtubule stabilizer), nocodazole (microtubule destabilizer), and SB218078 (CHEK1 inhibitor), can effectively inhibit growth of transformed cells by blocking progression at the M phase of the cell cycle. Since ligand activation of PPARβ/δ with GW0742 also causes G2/M arrest in HRAS-expressing keratinocytes, the effect on cell proliferation of combining GW0742 with other mitosis inhibitors was examined. RO-3306 increased the percentage of cells in the sub-G1 and G2/M phases of the cell cycle in HRAS-expressing keratinocytes, and this effect was markedly increased by cotreatment with GW0742, an effect not found in similarly treated HRAS-expressing Pparβ/δ-null keratinocytes (Fig. 9A and B). These changes were consistent with the enhanced PPARβ/δ-dependent inhibition of cell proliferation observed in HRAS-expressing wild-type keratinocytes cotreated with RO-3306 and GW0742 (Fig. 9C). Similar to the effect of ligand activation of PPARβ/δ, treatment with RO-3306 caused selection against cells expressing higher levels of HRAS in wild-type keratinocytes (Fig. 9D). Cotreatment of RO-3306 with GW0742 enhanced this selection (Fig. 9D). Paclitaxel also increased the percentage of cells in the G2/M phase of the cell cycle in HRAS-expressing keratinocytes, and this effect was increased by cotreatment with GW0742, an effect not found in HRAS-expressing Pparβ/δ-null keratinocytes (Fig. 9E). In contrast, cotreatment of GW0742 with nocodazole did not increase the effects induced by nocodazole alone (Fig. 9E). Similarly enhanced inhibition of cell proliferation was also found following cotreatment with GW0742 and RO-3306 or paclitaxel in 308 mouse keratinocytes (data not shown). Enhanced PPARβ/δ-dependent inhibition of cell proliferation was also observed in HRAS-expressing wild-type cells cotreated with the CHEK1 inhibitor SB218078 and GW0742 (Fig. 9F and G).
Fig 9.
Ligand activation of PPARβ/δ leads to hypersensitivity to pharmacological inhibition of mitosis in HRAS-expressing keratinocytes. (A to D) HRAS-expressing wild-type or Pparβ/δ-null keratinocytes were treated with or without 1 μM GW0742 for 2 days and then treated with 1 μM GW0742, 10 μM RO-3306, or 10 μM RO-3306 and 1 μM GW0742 for 24 h. (A) Representative photomicrographs after 24 h of cotreatment. A significant increase in cell death was observed in HRAS-expressing wild-type keratinocytes cotreated with GW0742 and RO-3306 but not Pparβ/δ-null keratinocytes. (B) Distribution of cells in the sub-G1 and G2/M phases of the cell cycle. (C) The percentage of viable cells was normalized to the corresponding control in the absence of RO-3306. (D). Flow cytometric analysis using an anti-HRAS antibody was performed, and Pearson's second skewness coefficient of HRAS intensity was calculated. (E) HRAS-expressing keratinocytes were cultured in medium with or without 1 μM GW0742 or treated with 20 nM paclitaxel or nocodazole (100 ng/ml) with or without 1 μM GW0742 for 40 h. The distribution of cells in the G2/M phase of the cell cycle was determined. (F and G) HRAS-expressing wild-type or Pparβ/δ-null keratinocytes were treated with or without 1 μM GW0742 for 2 days and then treated with 1 μM GW0742, 10 μM SB218078, or 10 μM SB218078 and 1 μM GW0742 for 24 h. (F) Representative photomicrographs after 24 h of cotreatment. A significant increase in cell death was observed in HRAS-expressing wild-type keratinocytes cotreated with GW0742 and SB218078 but not Pparβ/δ-null keratinocytes. (G) The percentage of viable cells was normalized to the corresponding control in the absence of SB218078. For all data sets, n = 3 independent samples per treatment group. Values represent means ± SEM. *, significantly different from HRAS-expressing wild-type control, P ≤ 0.05. (H) Mechanism of PPARβ/δ-mediated inhibition of mitosis in HRAS-expressing cells. PPARβ/δ chaperones p130 into the nucleus, where it represses E2F target genes, causing inhibition of mitosis.
DISCUSSION
This report is the first to demonstrate PPARβ/δ-dependent inhibition of cell proliferation in HRAS-expressing cells by increasing G2/M arrest. This is consistent with previous work that showed inhibition of skin carcinogenesis and inhibition of proliferation in keratinocyte cell lines with Hras mutations (3, 4). The current studies revealed that induction of G2/M arrest caused by ligand activation of PPARβ/δ specifically targets cells with higher expression of HRAS. That the PPARβ/δ-dependent induction of G2/M arrest causes selection against cells with higher expression of HRAS is consistent with results showing that cells treated with chemicals that induce G2/M arrest also cause selection against cells expressing higher levels of HRAS. Interestingly, human cancer cell lines expressing oncogenic RAS were previously shown to be more sensitive to mitotic perturbations than normal cells (36), an observation also noted in the present studies. For example, as HRAS activity increased in keratinocytes (as the result of viral transduction) or 308 cells (with an activating mutation in HRAS), the efficacy of GW0742 with respect to induction of G2/M arrest increased. Moreover, ligand activation of PPARβ/δ inhibits mitosis in skin tumors, and this phenotype is also associated with reduced expression of HRAS in these tumors. It was also shown that cells expressing oncogenic RAS exhibit a disadvantage with respect to cell proliferation following knockdown of many mitotic genes (36). This is important because PPARβ/δ-dependent repression of many of these mitosis-related genes was also observed in HRAS-expressing keratinocytes, 308 cells, and skin tumors in the present study.
Among the mitosis-related genes that were repressed by ligand activation of PPARβ/δ in HRAS-expressing cells, Cdk1 and Chek1 are of great interest. While some of these changes were relatively modest, this could have been due to the presence of an endogenous high-affinity agonist that prevents alterations in expression of greater robustness. An active cyclin B1-CDK1 complex is a trigger to enter mitosis, whereas depletion of cyclin B1-CDK1 can cause a block in mitosis concomitantly with repeated rounds of S phase, leading to cells with polyploidies in both fission yeast and human cells (11, 23, 27, 40). CHEK1 is required for spindle checkpoint function (specifically at the tension-sensing branch of the checkpoint), and Chek1−/− cells can exit mitosis in the presence of paclitaxel and undergo endoreduplication, leading to polyploidies (66). Ligand activation of PPARβ/δ decreased expression of CDK1 and CHEK1 in HRAS-expressing cells, and the observed phenotype, including delayed entry into mitosis, retarded exit from mitosis, and increased polyploidy cell numbers, is similar to the phenotype of Cdk1-null and Chek1-null cells. Combined, these observations suggest that the PPARβ/δ-dependent decrease in expression of CDK1 and CHEK1 alone in HRAS-expressing cells may largely underlie the observed mitosis block following ligand activation of PPARβ/δ.
Inhibition of cell cycle kinetics induced by ligand activation of PPARβ/δ could have been due in part to direct regulation of target genes by PPARβ/δ, which was not examined in the present study. However, results from these studies also establish that PPARβ/δ-dependent inhibition of mitosis in cells with an activating Hras mutation can also inhibit cell cycle progression and is mediated by a mechanism (Fig. 9H) that involves (i) PPARβ/δ directly binding with p107/p130 proteins; (ii) translocation of PPARβ/δ to the nucleus in response to ligand activation, leading to increased nuclear hypophosphorylated p130 and p107; (iii) ligand bound-PPARβ/δ maintaining p130 in a hypophosphorylated state; and (iv) heightened nuclear p107/p130 causing increased recruitment of the p130/p107/E2F4 complex to the promoters of mitosis-related genes and inhibition of their transcription, i.e., of genes with repressor E2F4 binding sites (such as Cdk1 and E2f1) that are repressed directly by this complex. Because of this PPARβ/δ-dependent downregulation of E2F1 expression, decreased E2F1 recruitment to promoters of other genes preferentially regulated by activator E2Fs (such as Chek1) is secondarily influenced by this regulation. Thus, it is not surprising that expression of CHEK1 is also downregulated by ligand activation of PPARβ/δ. Alternatively, it remains possible that the PPARβ/δ/p130/p107/E2F4 complexes exhibit differential affinities for binding sites on chromatin or that their presence leads to differences in recruitment of transcriptional corepressors. Further studies are needed to examine these ideas. Since cells with RAS mutations are more sensitive to mitotic perturbations than normal cells, the present studies focused more on the regulation of mitosis genes. However, it is also noteworthy that expression of many E2F target genes involved in DNA replication and DNA repair was also reduced by ligand activation of PPARβ/δ. This change in gene expression is reflected by the decrease in cells in the S phase in HRAS-expressing keratinocytes following ligand activation of PPARβ/δ.
The interaction between p107/p130 and PPARβ/δ is independent of HRAS. Moreover, PPARβ/δ preferentially binds to the hypophosphorylated form of p130 based on data from coimmunoprecipitations. This conclusion is also supported by results showing the more prominent colocalization of p130 and PPARβ/δ in the cytosol of cells, since hypophosphorylated p130 was found primarily in the cytoplasm. In the presence of ligand, PPARβ/δ may inhibit phosphorylation of p130 by CDK4. This suggests that, when p130 is shuttled to the nucleus via random nuclear translocation of PPARβ/δ under normal conditions, p130 is phosphorylated by the CDK4/cyclin D1 complex present in the nucleus, thus losing its repressor activity. This also suggests that ligand activation of PPARβ/δ is essential for repression of mitosis genes, because it maintains p130 in a hypophosphorylated state and chaperones hypophosphorylated p130 into the nucleus in cells with activated HRAS signaling.
Nuclear translocation of PPARβ/δ in HRAS-expressing cells following ligand activation is central to the inhibition of mitosis genes. Nuclear translocation of PPARβ/δ and increased nuclear p107 and p130 levels in normal keratinocytes are typically not observed. In contrast, increased nuclear translocation of PPARβ/δ and concurrent increases of p107 and p130 levels in HRAS-expressing keratinocytes and skin tumors illustrate the essential nature of nuclear translocation of PPARβ/δ following ligand activation in the presence of activated HRAS signaling. The increase in both cytosolic and nuclear PPARβ/δ levels following ligand activation in control keratinocytes is most likely due to the stabilization of the receptor rather than to an increase of protein synthesis and nuclear translocation for the following reasons. First, no increase in the level of PPARβ/δ mRNA was observed following ligand activation. Second, ligand activation of PPARβ/δ is known to prevent its ubiquitin-mediated degradation, thus increasing its half-life (17). However, the mechanism of nuclear translocation of PPARβ/δ in HRAS-expressing keratinocytes, skin tumors, and confluent 308 cells following ligand activation remains unclear. It is possible that increased HRAS activity activates downstream kinases and alters the phosphorylation status of PPARβ/δ, leading to its nuclear translocation.
Increasing G2/M arrest of cells expressing high levels of HRAS can be achieved by ligand activation of PPARβ/δ, and cotreatment of a PPARβ/δ ligand with various mitosis inhibitors enhances the efficacy of increasing G2/M arrest. This supports the hypothesis that combining ligand activation of PPARβ/δ with mitosis inhibitors is a feasible approach for treating tumors that express higher levels of RAS. Indeed, oncogenic RAS signaling is increased in a number of human cancers, including lung, colon, pancreas, and melanoma (55). While the role of PPARβ/δ in some cancers remains controversial (reviewed in references 45 to 47, 49, and 50), the body of evidence suggesting that PPARβ/δ protects against cancer is increasing. For example, a recent compelling study demonstrated that colorectal cancer patients with relatively low expression of PPARβ/δ in the primary tumor were ∼4 times as likely to die from this disease as patients with relatively higher expression of PPARβ/δ in their primary tumors (64). It is also not disputed that ligand activation of PPARβ/δ inhibits chemically induced skin carcinogenesis (3, 4, 32, 68). Moreover, preclinical and clinical studies have also shown that ligand activation of PPARβ/δ inhibits or prevents metabolic syndrome, obesity, dyslipidemias, glucose intolerance, and chronic inflammation, characteristics that are positively associated with cancer (43, 51, 59, 60, 62). Since targeting single molecules for chemoprevention and chemotherapy has not proven highly effective (21) due in part to the genetic heterogeneity associated with diseases (54), targeting PPARβ/δ in conjunction with mitosis inhibitors could become a suitable option for development of new multitarget strategies for inhibiting RAS-dependent tumorigenesis.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Andrew Billin and Timothy Willson for providing the GW0742 and the Center for Quantitative Cell Analysis and the Genomic Core Facility at the Huck Institutes of Life Sciences of The Pennsylvania State University for their technical support with flow cytometry and data analysis.
This work was supported by National Institutes of Health grants CA124533, CA126826, CA141029, CA140369, and AA018863 to J.M.P. and CA122109 and CA117957 to A.B.G. and by grants from the National Cancer Institute Intramural Research Program (ZIABC005561, ZIABC005562, and ZIABC005708) to F.J.G.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print 2 April 2012
REFERENCES
- 1. Akiyama TE, Meinke PT, Berger JP. 2005. PPAR ligands: potential therapies for metabolic syndrome. Curr. Diab. Rep. 5:45–52 [DOI] [PubMed] [Google Scholar]
- 2. Balmain A, Ramsden M, Bowden GT, Smith J. 1984. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature 307:658–660 [DOI] [PubMed] [Google Scholar]
- 3. Bility MT, et al. 2008. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits chemically-induced skin tumorigenesis. Carcinogenesis 29:2406–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bility MT, Zhu B, Kang BH, Gonzalez FJ, Peters JM. 2010. Ligand activation of peroxisome proliferator-activated receptor-β/δ and inhibition of cyclooxygenase-2 enhances inhibition of skin tumorigenesis. Toxicol. Sci. 113:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bizub D, Wood AW, Skalka AM. 1986. Mutagenesis of the Ha-ras oncogene in mouse skin tumors induced by polycyclic aromatic hydrocarbons. Proc. Natl. Acad. Sci. U. S. A. 83:6048–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borland MG, et al. 2011. Stable over-expression of PPARβ/δ and PPARγ to examine receptor signaling in human HaCaT keratinocytes. Cell Signal. 23:2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown K, et al. 1986. v-ras genes from Harvey and BALB murine sarcoma viruses can act as initiators of two-stage mouse skin carcinogenesis. Cell 46:447–456 [DOI] [PubMed] [Google Scholar]
- 8. Carrassa L, Broggini M, Vikhanskaya F, Damia G. 2003. Characterization of the 5′ flanking region of the human Chk1 gene: identification of E2F1 functional sites. Cell Cycle 2:604–609 [PubMed] [Google Scholar]
- 9. Chen HZ, Tsai SY, Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9:785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng L, Rossi F, Fang W, Mori T, Cobrinik D. 2000. Cdk2-dependent phosphorylation and functional inactivation of the pRB-related p130 protein in pRB(−), p16INK4A(+) tumor cells. J. Biol. Chem. 275:30317–30325 [DOI] [PubMed] [Google Scholar]
- 11. Correa-Bordes J, Nurse P. 1995. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell 83:1001–1009 [DOI] [PubMed] [Google Scholar]
- 12. Daya-Grosjean L, Robert C, Drougard C, Suarez H, Sarasin A. 1993. High mutation frequency in ras genes of skin tumors isolated from DNA repair deficient xeroderma pigmentosum patients. Cancer Res. 53:1625–1629 [PubMed] [Google Scholar]
- 13. Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. 1995. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 254:3–20 [DOI] [PubMed] [Google Scholar]
- 14. Do JH, Choi DK. 2006. Normalization of microarray data: single-labeled and dual-labeled arrays. Mol. Cells 22:254–261 [PubMed] [Google Scholar]
- 15. Fu M, et al. 2003. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene 315:33–41 [DOI] [PubMed] [Google Scholar]
- 16. Fujita N, Wade PA. 2004. Use of bifunctional cross-linking reagents in mapping genomic distribution of chromatin remodeling complexes. Methods 33:81–85 [DOI] [PubMed] [Google Scholar]
- 17. Genini D, Catapano CV. 2007. Block of nuclear receptor ubiquitination. A mechanism of ligand-dependent control of peroxisome proliferator-activated receptor δ activity. J. Biol. Chem. 282:11776–11785 [DOI] [PubMed] [Google Scholar]
- 18. Girroir EE, et al. 2008. Quantitative expression patterns of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) protein in mice. Biochem. Biophys. Res. Commun. 371:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Greenhalgh DA, Welty DJ, Player A, Yuspa SH. 1990. Two oncogenes, v-fos and v-ras, cooperate to convert normal keratinocytes to squamous cell carcinoma. Proc. Natl. Acad. Sci. U. S. A. 87:643–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grimaldi PA. 2005. Regulatory role of peroxisome proliferator-activated receptor δ (PPARδ) in muscle metabolism. A new target for metabolic syndrome treatment? Biochimie 87:5–8 [DOI] [PubMed] [Google Scholar]
- 21. Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674 [DOI] [PubMed] [Google Scholar]
- 22. Hansen K, et al. 2001. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 20:422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayles J, Fisher D, Woollard A, Nurse P. 1994. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78:813–822 [DOI] [PubMed] [Google Scholar]
- 24. He P, et al. 2008. Effect of ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in human lung cancer cell lines. Toxicology 254:112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 26. Ishida S, et al. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Itzhaki JE, Gilbert CS, Porter AC. 1997. Construction by gene targeting in human cells of a “conditional” CDC2 mutant that rereplicates its DNA. Nat. Genet. 15:258–265 [DOI] [PubMed] [Google Scholar]
- 28. Jayadeva G, et al. 2010. B55α PP2A holoenzymes modulate the phosphorylation status of the retinoblastoma-related protein p107 and its activation. J. Biol. Chem. 285:29863–29873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson DG, Ohtani K, Nevins JR. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8:1514–1525 [DOI] [PubMed] [Google Scholar]
- 30. Kilgore KS, Billin AN. 2008. PPARβ/δ ligands as modulators of the inflammatory response. Curr. Opin. Invest. Drugs 9:463–469 [PubMed] [Google Scholar]
- 31. Kim DJ, et al. 2004. Peroxisome proliferator-activated receptor β (δ)-dependent regulation of ubiquitin C expression contributes to attenuation of skin carcinogenesis. J. Biol. Chem. 279:23719–23727 [DOI] [PubMed] [Google Scholar]
- 32. Kim DJ, et al. 2006. PPARβ/δ selectively induces differentiation and inhibits cell proliferation. Cell Death Differ. 13:53–60 [DOI] [PubMed] [Google Scholar]
- 33. Kim DJ, et al. 2005. Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits epidermal cell proliferation by down-regulation of kinase activity. J. Biol. Chem. 280:9519–9527 [DOI] [PubMed] [Google Scholar]
- 34. Lee CH, Olson P, Evans RM. 2003. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology 144:2201–2207 [DOI] [PubMed] [Google Scholar]
- 35. Lee CH, et al. 2006. PPARδ regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. U. S. A. 103:3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo J, et al. 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137:835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malumbres M, Barbacid M. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3:459–465 [DOI] [PubMed] [Google Scholar]
- 38. Moffat J, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298 [DOI] [PubMed] [Google Scholar]
- 39. Mootha VK, et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273 [DOI] [PubMed] [Google Scholar]
- 40. Moreno S, Nurse P. 1994. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature 367:236–242 [DOI] [PubMed] [Google Scholar]
- 41. Müller H, et al. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oliver WR, Jr, et al. 2001. A selective peroxisome proliferator-activated receptor δ agonist promotes reverse cholesterol transport. Proc. Natl. Acad. Sci. U. S. A. 98:5306–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pais R, Silaghi H, Silaghi AC, Rusu ML, Dumitrascu DL. 2009. Metabolic syndrome and risk of subsequent colorectal cancer. World J. Gastroenterol. 15:5141–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Palkar PS, et al. 2010. Cellular and pharmacological selectivity of the PPARβ/δ antagonist GSK3787. Mol. Pharmacol. 78:419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peters JM, Foreman JE, Gonzalez FJ. 2011. Dissecting the role of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in colon, breast and lung carcinogenesis. Cancer Metastasis Rev. 30:619–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peters JM, Gonzalez FJ. 2009. Sorting out the functional role(s) of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in cell proliferation and cancer. Biochim. Biophys. Acta 1796:230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peters JM, Hollingshead HE, Gonzalez FJ. 2008. Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clin. Sci. (London) 115:107–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peters JM, et al. 2000. Growth, adipose, brain and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor β/δ. Mol. Cell. Biol. 20:5119–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peters JM, Shah YM, Gonzales FJ. 2012. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 12:181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peters JM, Morales JL, Gonzales FJ. 2011. Modulation of gastrointestinal inflammation and colorectal tumorigenesis by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ). Drug Discov. Today 8:e85–e93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prizment AE, Flood A, Anderson KE, Folsom AR. 2010. Survival of women with colon cancer in relation to precancer anthropometric characteristics: the Iowa Women's Health Study. Cancer Epidemiol. Biomarkers Prev. 19:2229–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ren B, et al. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roop DR, et al. 1986. An activated Harvey ras oncogene produces benign tumours on mouse epidermal tissue. Nature 323:822–824 [DOI] [PubMed] [Google Scholar]
- 54. Schadt EE. 2009. Molecular networks as sensors and drivers of common human diseases. Nature 461:218–223 [DOI] [PubMed] [Google Scholar]
- 55. Schubbert S, Shannon K, Bollag G. 2007. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7:295–308 [DOI] [PubMed] [Google Scholar]
- 56. Strickland JE, et al. 1988. Development of murine epidermal cell lines which contain an activated rasHa oncogene and form papillomas in skin grafts on athymic nude mouse hosts. Cancer Res. 48:165–169 [PubMed] [Google Scholar]
- 57. Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Svaren J, et al. 2000. EGR1 target genes in prostate carcinoma cells identified by microarray analysis. J. Biol. Chem. 275:38524–38531 [DOI] [PubMed] [Google Scholar]
- 59. Terzić J, Grivennikov S, Karin E, Karin M. 2010. Inflammation and colon cancer. Gastroenterology 138:2101–2114 e2105 [DOI] [PubMed] [Google Scholar]
- 60. Tsugane S, Inoue M. 2010. Insulin resistance and cancer: epidemiological evidence. Cancer Sci. 101:1073–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Westergaard M, et al. 2001. Modulation of keratinocyte gene expression and differentiation by PPAR-selective ligands and tetradecylthioacetic acid. J. Invest. Dermatol. 116:702–712 [DOI] [PubMed] [Google Scholar]
- 62. Wolin KY, Carson K, Colditz GA. 2010. Obesity and cancer. Oncologist 15:556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu X, et al. 2007. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 17:1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang L, et al. 2011. Biological function and prognostic significance of peroxisome proliferator-activated receptor δ in rectal cancer. Clin. Cancer Res. 17:3760–3770 [DOI] [PubMed] [Google Scholar]
- 65. Yuspa SH, Morgan DL. 1981. Mouse skin cells resistant to terminal differentiation associated with initiation of carcinogenesis. Nature 293:72–74 [DOI] [PubMed] [Google Scholar]
- 66. Zachos G, et al. 2007. Chk1 is required for spindle checkpoint function. Dev. Cell 12:247–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zanet J, et al. 2010. A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS One 5:e15701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu B, et al. 2010. Chemoprevention of chemically induced skin tumorigenesis by ligand activation of peroxisome proliferator-activated receptor-β/δ and inhibition of cyclooxygenase 2. Mol. Cancer Ther. 9:3267–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhu L, et al. 1993. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 7:1111–1125 [DOI] [PubMed] [Google Scholar]
- 70. Zhu W, Giangrande PH, Nevins JR. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23:4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.