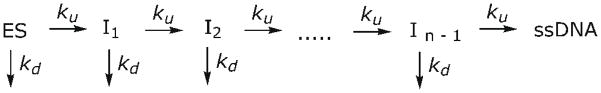Abstract
Helicases utilize the energy of ATP hydrolysis to unwind double-stranded DNA while translocating on the DNA. Mechanisms for melting the duplex have been characterized as active or passive, depending on whether the enzyme actively separates the base pairs or simply sequesters single-stranded DNA that forms due to thermal fraying. Here we show that Dda translocates unidirectionally on single-stranded DNA at the same rate at which it unwinds double-stranded DNA in both ensemble and single molecule experiments. Further, the unwinding rate is largely insensitive to the duplex stability and to applied force. Thus, Dda transduces all of its translocase activity into DNA unwinding activity so that the rate of unwinding is limited by the rate of translocation and the enzyme actively separates the duplex. Active and passive helicases have been characterized by dividing the velocity of DNA unwinding in base pairs per second (Vun) by the velocity of translocation on single-stranded DNA in nucleotides per second (Vtrans). If the resulting fraction is 0.25, then a helicase is considered to be at the lower end of the “active” range. In the case of Dda, the average DNA unwinding velocity was 257 ± 42 bp/s and the average translocation velocity was 267 ± 15 nucleotides/s. The Vun/Vtrans value of 0.96 places Dda in a unique category of being an essentially “perfectly” active helicase.
Keywords: molecular motor, single-molecule, stopped-flow, DNA, fluorescence
Introduction
Helicases are enzymes that unwind double-stranded DNA (dsDNA) to provide the single-stranded DNA (ssDNA) intermediates necessary for virtually every phase of nucleic acid metabolism, reviewed in1–3. They are ubiquitous, and most organisms encode multiple helicases for involvement in different cellular processes. Canonical helicases use the energy of NTP hydrolysis to fuel unidirectional translocation on nucleic acids (NA) and concomitant unwinding of duplex NA.
Whether the helicase physically interacts with and melts the dsNA, an active mechanism, or the helicase traps partially melted NA, a passive mechanism, is a fundamental characteristic of helicase catalyzed NA unwinding4. In an active mechanism, the helicase physically interacts with the dsNA resulting in unzipping of the duplex. For a passive helicase, the activation energy required to unwind dsNA is high, resulting in the rate limiting step being unwinding. Therefore the ratio between unwinding and translocation rates can be used to determine whether a helicase is active or passive5. An active helicase will unwind dsDNA and translocate on ssDNA at similar rates. A passive helicase will display a lower rate for unwinding than translocation; of course, a helicase may display intermediate behavior that can be quantified by the strength of the destabilizing energy melting the DNA. Since increasing the GC content of a duplex increases the activation energy required only if unwinding is rate limiting, comparison of the rates of unwinding with high and low GC content duplexes can be used to determine whether a helicase is active or passive. For a passive helicase, the unwinding rate will decrease with increasing GC content in the NA.
Dda is a helicase from bacteriophage T4 involved in early events in DNA replication6. It is not essential for growth of the phage but is involved in origin dependent replication and aids in recombination by increasing the rate of branch migration catalyzed by the T4 recombinase, UvsX7. It has been suggested to translocate unidirectionally, but the rate of translocation has not been measured. Jongeneel et al.8 investigated the directionality of translocation using partially duplex DNA substrates with either a 3′- or 5′-ssDNA adjacent to the duplex. Dda could unwind substrates with a 5′-ssDNA overhang but not a 3′-ssDNA overhang, suggesting 5′-to-3′ translocation. Raney and Benkovic9 studied the effect of a streptavidin block on the kinetics of ATP hydrolysis by Dda. A 5′-streptavidin block had no effect on the ATPase kinetics while a 3′-streptavidin block reduced the KM for DNA, suggesting translocation in the 5′-to-3′ direction. Morris and Raney10 showed that Dda is able to displace streptavidin from 3′-biotinylated oligonucleotides but not from 5′-biotinylated oligonucleotides, again suggesting translocation in the 5′-to-3′ direction. The kinetics of unwinding and protein displacement by Dda have been extensively studied. Dda does not form stable oligomers11, and has been shown to unwind DNA as a monomer12, albeit with low processivity13. The processivity increases when multiple Dda molecules are bound to the substrate14, although the rate of unwinding remains similar13. Dda displaces proteins from ssDNA10,15 and from dsDNA16. Dda interacts primarily with the translocase strand17 however, it also appears to interact with the displaced strand during specific steps of the unwinding mechanism18.
In this report, we determined the kinetics of Dda’s interactions with ssDNA. We show that the unidirectional translocation rate on ssDNA is very similar to the rate of unwinding of dsDNA, indicating that Dda is an active helicase. This is further supported by the result that Dda is insensitive to the GC content of short duplexes during unwinding and to the applied force during unwinding under single molecule conditions.
Results
Dda associates with DNA at near diffusion controlled rates
For Dda helicase, the rate of unwinding13,18 and protein displacement10,15,16 as well as the equilibrium binding constants for various DNA substrates11,12,14 have been well studied. However, the rates at which Dda binds to, dissociates from, and translocates on ssDNA have not been investigated.
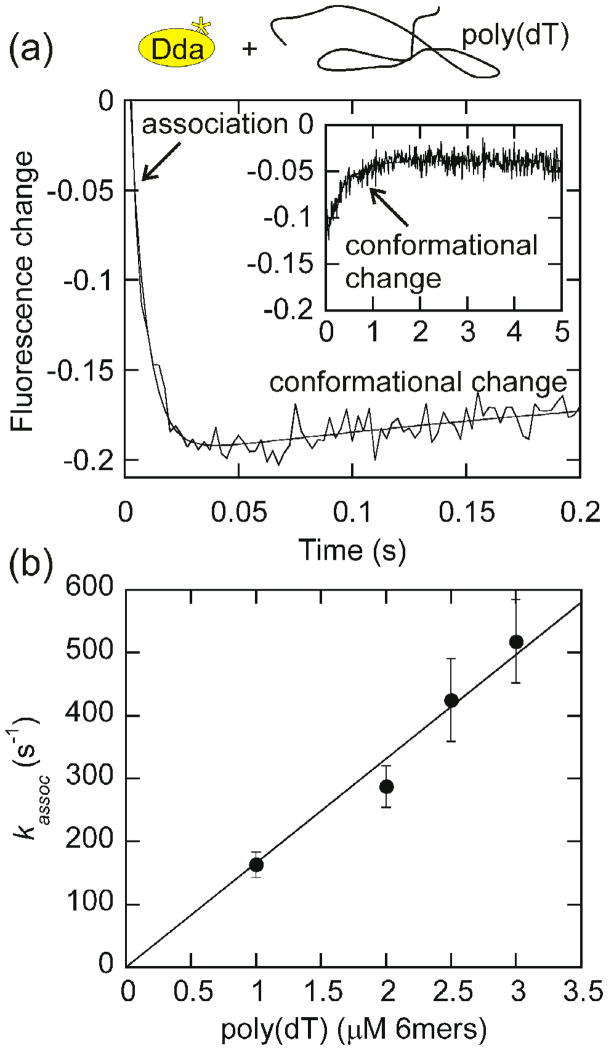
When Dda is rapidly mixed with ssDNA, the fluorescence change of tryptophan is biphasic and can be fit with a double exponential as shown in Fig. 1a. The second phase of the reaction is independent of the concentration of DNA, while the rate of the first phase increases as the DNA concentration increases, indicating that the first phase is association of DNA and Dda (Table S1). The slower second phase likely represents a slow isomerization. The rates of Dda association with increasing concentrations of poly(dT) are plotted in Fig. 1b. The second order association constant obtained from the slope of the line is 1.66 × 108 M−1s−1 which indicates that this rate is close to diffusion controlled (108–109 M−1s−1).
Fig. 1. Measurement of the rate of Dda association with DNA.
(a) The fluorescence change for 200 nM Dda association with 6 μM poly(dT), in nucleotides, is plotted. Data was fit to a double exponential. The observed rate of association (first phase) is 163 ± 20 s−1 at 6 μM poly(dT), in nucleotides (1 μM poly(dT), in binding sites). The second phase (shown in inset) remains constant as the poly(dT) concentration increases (Table S1) and likely represents a slow conformational change. (b) The rate of association (first phase) increases linearly with increasing poly(dT) concentration. Data was plotted with the poly(dT) concentration in 6mers since Dda is known to have a 6 nt binding site size15 and was fit to a line using Kaleidagraph software. The slope of the line, 1.66×108 M−1s−1, represents the association rate constant.
Measurement of the rate of Dda dissociation from circular DNA
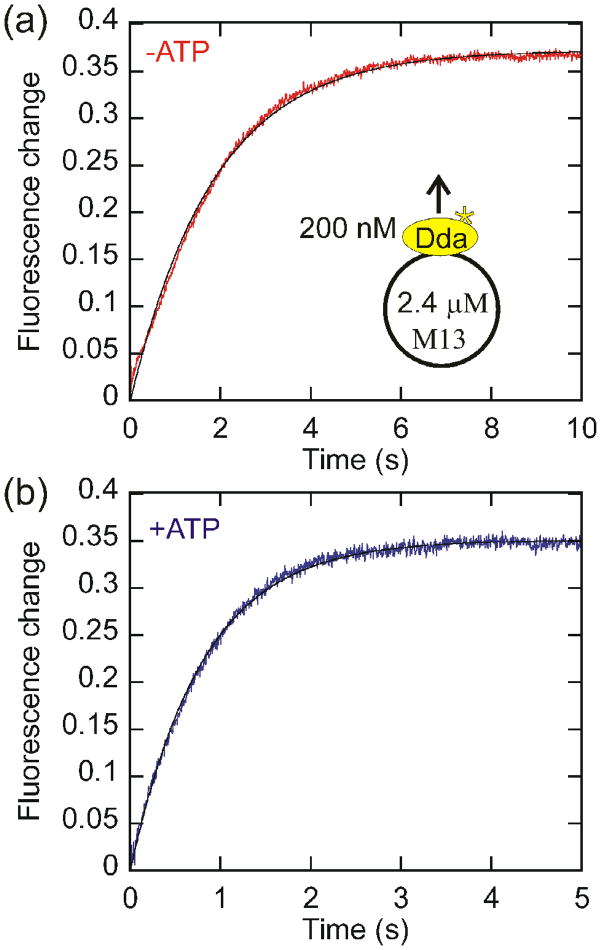
In order to determine the spontaneous dissociation rate in both the presence and absence of ATP, and therefore the effect of ATP binding and hydrolysis on the observed dissociation rate, a circular DNA species was used. This prevents Dda from translocating off the end of an oligonucleotide and affecting the observed dissociation rate. The fluorescence of Dda is significantly different when bound to DNA vs. heparin so heparin serves an effective trap (Fig. S1). Dda was pre-incubated with M13 ssDNA and rapidly mixed with heparin in the absence (Fig. 2a) or presence (Fig. 2b) of ATP. The rapid fluorescence increase is dependent only on the presence of DNA (Table S2), suggesting that it represents dissociation of Dda from DNA. The fluorescence change was fit to a single exponential. Dda dissociates from circular DNA at a rate of 1.12 ± 0.04 s−1 in the absence of ATP and 1.94 ± 0.03 s−1 in the presence of ATP. This suggests that the binding and hydrolysis of ATP causes only a slight increase in the rate of dissociation from internal sites on ssDNA. The dissociation rate was independent of the heparin concentration, indicating that heparin binds to Dda rapidly and the concentration of heparin used is sufficiently high for the rate to be independent of heparin concentration (Table S2).
Fig. 2.
Measurement of the dissociation rate of Dda from circular DNA. The intrinsic dissociation rate of Dda (200nM) from M13 ssDNA (2.4 μM nucleotides) was measured in the absence (a) and presence (b) of ATP by rapidly mixing with heparin. The obtained dissociation rate constants were 1.12 ± 0.04 s−1 in the absence of ATP and 1.94 ± 0.03 s−1 in the presence of ATP.
Measurement of the rate of Dda dissociation from varying length oligonucleotides
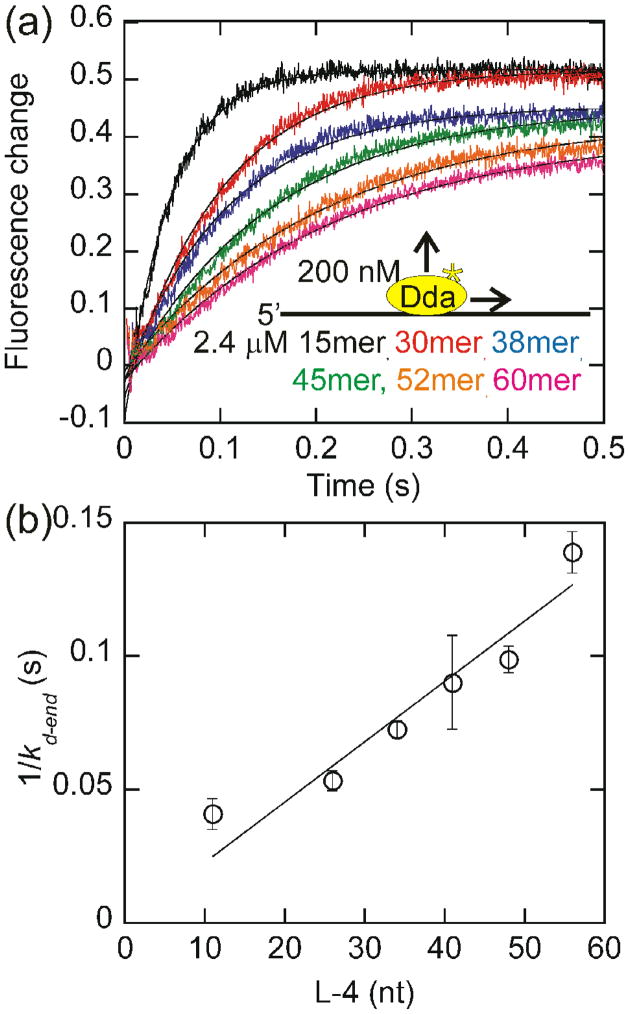
In the presence of ATP, the dissociation rate decreases as the length of the DNA increases (Fig. 3A). The rates of dissociation are faster for oligonucleotides than circular DNA and increase as the length of the substrate decreases, presumably due to translocation off the end of the substrate. As the length of the oligonucleotide increases, the helicase has a longer distance to translocate, assuming that Dda binds randomly to each substrate. Since the dissociation rate is inversely proportional to the length of the oligonucleotide (Equation 1), translocation is unidirectional, and the rate of translocation on ssDNA (Vtrans) is equal to one half the inverse slope of the 1/kd-end vs. L-4 plot (Fig. 3b). The rate of Dda dissociation from linear DNA is faster in the presence of ATP than in the absence due to translocation off the end of the oligonucleotide (Fig. S2). For very short oligonucleotides that are close to the length of the binding site size (6 nucleotides)15, the relationship between the dissociation rate constant and the DNA length is nonlinear. The ends of the substrate appear to affect the observed dissociation rate constant for very short DNA strands (Figs. 3b, 4b, and 5b). For this reason, oligonucleotides less than 30 nucleotides in length are not included in the fits.
Fig. 3.
Measurement of the rate of dissociation of Dda from oligonucleotides of increasing length. (a) The change in fluorescence of Dda (200 nM) as it dissociates from 2.4 μM oligonucleotide of increasing length in the presence of ATP is plotted. The rate constants obtained from single exponential fits were 26.3 ± 1.1 s−1, 22.6 ± 1.8 s−1, 16.4 ± 0.8 s−1, 13.6 ± 2.8 s−1, 12.1 ± 0.6 s−1, and 8.7 ± 0.4 s−1 for dissociation from the 15mer, 30mer, 38mer, 45mer, 52mer, and 60mer, respectively. (b) The inverse of the rate constant for dissociation from the end of the oligonucleotide, kd-end is plotted versus the length of the substrate, adjusted for binding site size. Only oligonucleotides of at least 30 nucleotides in length are included in the fit. One-half the inverse slope of the resulting line provides a rate of translocation by Dda of 252 ± 11 nucleotides/s. The fit was constrained through the origin to constrain kd-in to the value measured in Fig. 2b.
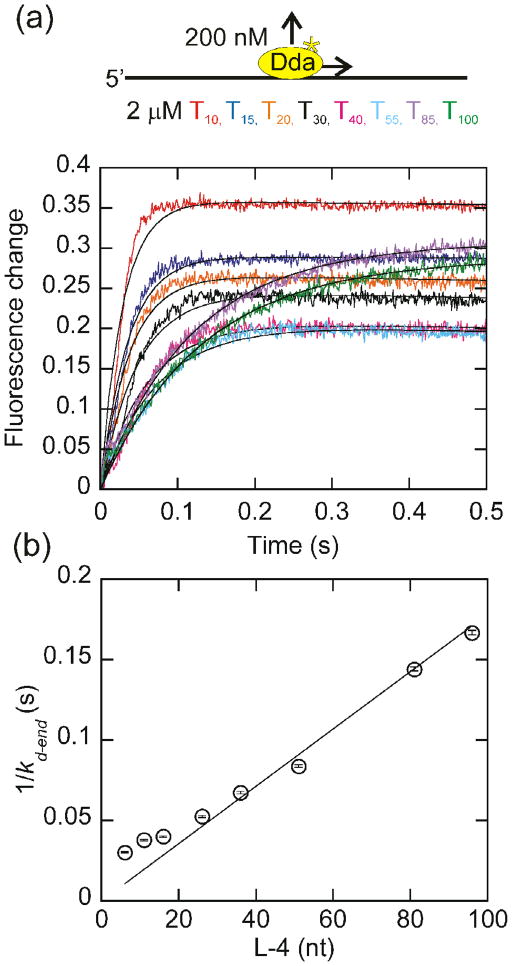
Fig. 4.
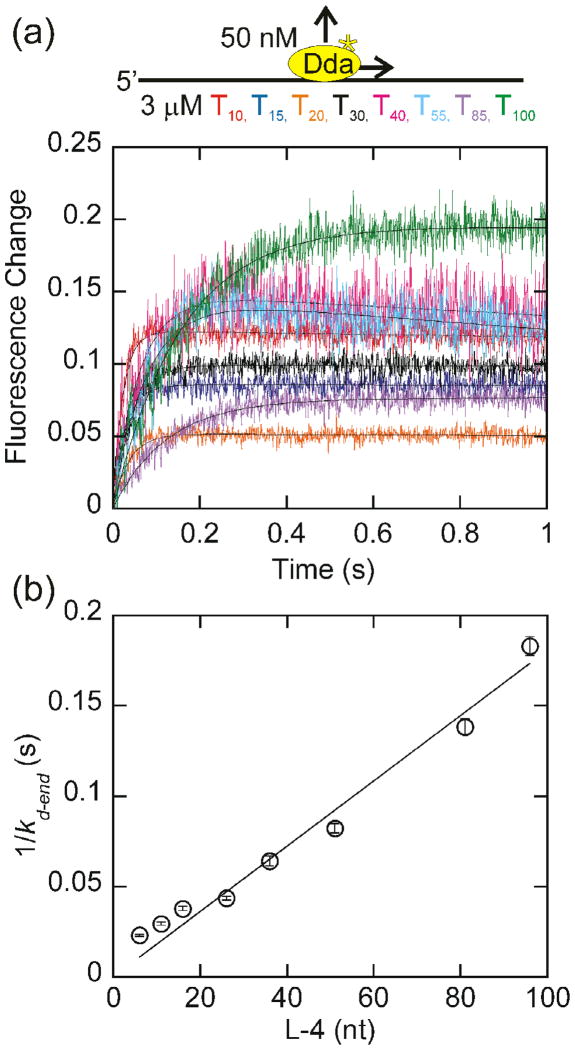
Change in tryptophan fluorescence as a function of oligonucleotide length with homopolymeric oligonucleotides. (a) The change in fluorescence of 200 nM Dda as it dissociates from 2 μM (in nucleotides) dT10, dT15, dT20, dT30, dT40, dT55, dT85, and dT100 is plotted. The observed dissociation rate constants, obtained by fitting to a single exponential were 35.0 ± 0.4 s−1, 28.3 ± 0.3 s−1, 26.8 ± 0.3 s−1, 22.4 ± 0.3 s−1, 16.6 ± 0.2 s−1, 13.7 ± 0.2 s−1, 8.7 ± 0.1 s−1, and 7.8 ± 0.1 s−1 for dissociation from dT10, dT15, dT20, dT30, dT40, dT55, dT85, and dT100, respectively. (b) The inverse of the rate constant for dissociation from the end of the oligonucleotide, kd-end, which is equal to the observed dissociation rate constant minus the intrinsic dissociation rate constant (Fig. 2b) is plotted versus the length of the substrate, adjusted for binding site size. Only oligonucleotides of at least 30 nucleotides in length are included in the fit. One-half the inverse slope of the resulting line provides a translocation rate of 264 ± 10 nucleotides/s. The fit was constrained through the origin to constrain kd-in to the value measured in Fig. 2b.
Fig. 5.
Change in tryptophan fluorescence as a function of oligonucleotide length with homopolymeric oligonucleotides under conditions where the substrate concentration is in vast excess. (a)The change in fluorescence of 50 nM Dda as it dissociates from 3 μM (in nucleotides) dT10, dT15, dT20, dT30, dT40, dT55, dT85, and dT100 is plotted. The observed dissociation rate constants, obtained by fitting to a single exponential were 44.1 ± 1.0 s−1, 34.7 ± 1.0 s−1, 27.2 ± 0.8 s−1, 23.8 ± 0.5 s−1, 16.4 ± 0.7 s−1, 13.0 ± 0.4 s−1, 8.1 ± 0.2 s−1, 6.3 ± 0.2 s−1 for dissociation from dT10, dT15, dT20, dT30, dT40, dT55, dT85, and dT100, respectively. (b) The inverse of the rate constant for dissociation from the end of the oligonucleotide, kd-end, which is equal to the observed dissociation rate constant minus the intrinsic dissociation rate constant (Fig. 2b) is plotted versus the length of the substrate, adjusted for binding site size. Only oligonucleotides of at least 30 nucleotides in length are included in the fit. One-half the inverse slope of the resulting line provides a translocation rate of 287 ± 13 nucleotides/s. The fit was constrained through the origin to constrain kd-in to the value measured in Fig. 2b.
The linear relationship between the observed dissociation rate constant and the oligonucleotide length suggests that Dda translocates unidirectionally. For unidirectional translocation, the dissociation rate is inversely proportional to the length of the DNA19. If the helicase were to translocate bidirectionally, the dissociation rate would be inversely proportional to the square of the length19,20. The data indicates that Dda translocates unidirectionally at a rate of 252 ± 11 nucleotides/s. Although these results do not indicate the direction of translocation, these results are consistent with previous results which suggest that Dda translocates in the 5′-to-3′ direction8–10.
Measurement of the rate of Dda dissociation from homopolymeric oligonucleotides
In order to ensure that there are no sequence dependent effects on the translocation rate, a similar experiment was performed with homopolymeric DNA (Fig. 4). The resulting translocation rate of 264 ± 10 nucleotides/s is very similar to the rate obtained with mixed sequence oligonucleotides, indicating that Dda is not sensitive to the sequence of the substrate. As with the mixed sequence oligonucleotides, the relationship between the dissociation rate constant and the DNA length is not linear for oligonucleotides less than 30 nucleotides in length. This could be due to the ends of the DNA affecting the observed dissociation rate. Since dissociation is fastest from short oligonucleotides, any affects due to the ends of the oligonucleotides will have the most impact on the observed dissociation rate from short oligonucleotides. Since the rate constant for dissociation from the end of the oligonucleotide is proportional to the length of the substrate for oligonucleotides of at least 30 nucleotides, only these substrates are included in the fit.
Although the number of Dda binding sites is in excess of the Dda concentration in these experiments, more than one Dda molecule can be bound to the ssDNA at the concentrations in these experiments. So, the concentration of DNA was increased to decrease the likelihood of multiple enzymes binding to each DNA substrate. With a 10-fold excess of enzyme binding sites, the translocation rate is 287 ± 13 nucleotides/s (Fig. 5b) using the data from oligonucleotides of at least 30 nucleotides in length. This is in close agreement with the translocation rates obtained earlier, indicating that the multiple Dda molecules were not influencing each other in the translocation experiments.
Measurement of the rate of Dda translocation on fluorescein labeled oligonucleotides
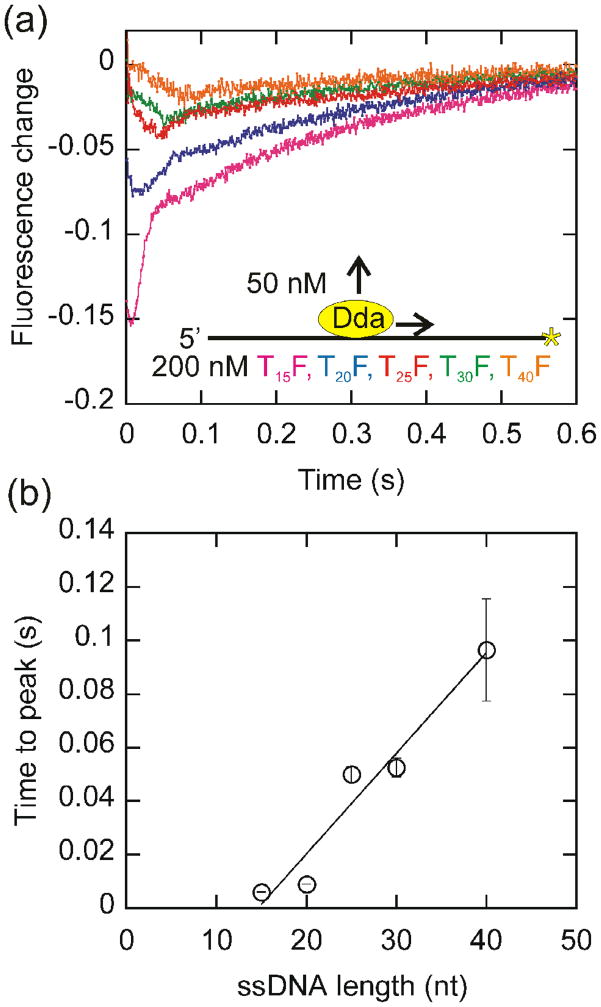
We further investigated the ability of Dda to translocate in a unidirectional manner using a fluorescence assay developed by Dillingham20. Dda was pre-incubated with 3′-fluorescein labeled oligonucleotides. Upon mixing with ATP, an initial decrease in fluorescence was followed by an increase (Fig. 6a). The initial decrease was interpreted to be the result of Dda translocation toward the fluorophore. The subsequent increase in fluorescence most likely results from Dda translocating off the end of the oligonucleotide. Re-initiation of translocation was prevented by the addition of poly(dT). The time to maximal fluorescence quenching is proportional to the length of the oligonucleotide, indicating that the initial decrease in fluorescence corresponds to translocation (Fig. 6b). The inverse of the slope of the line is 266 ± 37 nucleotides/s which is consistent with the translocation rate determined from the dissociation experiments. The average translocation rate obtained for Dda from the data in Figs. 3–6 is 267 ± 15 nucleotides/s. This rate is nearly identical to the published unwinding rate for Dda, 273 ± 43 bp/s13,18. Since the translocation rate is same as the published unwinding rate, Dda is an active helicase using the practical definition recently put forth by Manosas5.
Fig. 6.
Translocation of Dda along fluorescein labeled oligonucleotides of various lengths. (a) The change in the fluorescence of 3′-fluorescein labeled oligonucleotides of varying length was monitored after a 515 nm cutoff filter in the presence of ATP. The initial decrease in fluorescence was determined to correspond to the translocation of Dda. (b) The time to the maximal fluorescence change is plotted versus the length of the oligonucleotide. The translocation rate was determined to be 266 ± 37 nucleotides/s by the inverse slope of a linear fit.
Dda unwinding is not affected by the GC content of the duplex
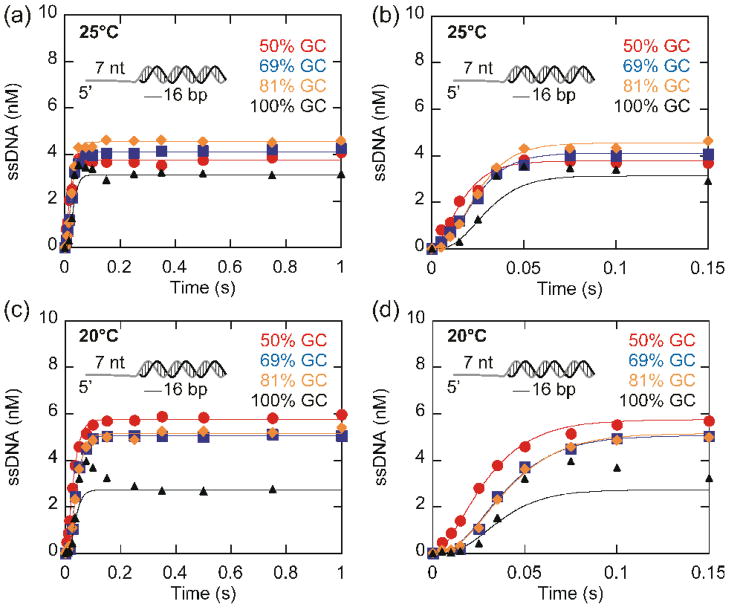
Active vs. passive helicases mechanisms can be evaluated by the comparing the rate of unwinding of substrates with increasing GC content. The rates for unwinding by passive helicases are more sensitive to increasing GC content than active helicases. The rate of unwinding by Dda of a 16 base pair duplex of varying GC content was investigated in order to determine whether the rate of unwinding by Dda is affected by the stability of the duplex. Unwinding reactions were performed at 25 °C (Fig. 7a and b) and 20 °C (Fig. 7c and d). When the GC content of the duplex is increased from 50% to 100%, a noticeable increase in the lag phase occurs and the number of steps required to unwind the substrate increases from 2 to 4 (Fig. 7b and d). The increased number of steps needed to melt the duplex results from the increased stability of the high GC substrates relative to the low GC substrates. For each substrate, the data was fit to an n-step sequential mechanism (Scheme 1)21, with the minimum number of steps (n) required to provide a fit of the data. The resulting n values were 2 for the 50% GC substrate, 3 for the 69% and 81% GC substrates, and 4 for the 100% GC substrate. Although the number of steps required to unwind the duplex increases when the stability of the duplex increases, the rate of unwinding (Vun) remains relatively constant (Table 1) at both 20 °C and 25 °C. If Dda unwinding was affected by the GC content of the duplex, increasing GC content would result in a decrease in the unwinding rate, which was not observed. This indicates that Dda is not affected by the GC content of the duplex. Table 2 shows the translocation and unwinding rates obtained under various conditions. The average translocation and unwinding rates are nearly identical, also indicating that Dda is an active helicase.
Fig. 7.
Unwinding of 16 base pair duplexes of varying stability by Dda. (a–b) Unwinding of 10 nM partial duplex substrates of varying stability by 100 nM Dda at 25°C. Data were fit using KinTek Explorer56 to Scheme 1. The rates of unwinding were 205 ± 26 bp/s, 232 ± 16 bp/s, 231 ± 10 bp/s, 234 ± 27 bp/s for unwinding of 50%, 69%, 81%, and 100% GC substrates, respectively. (c–d) Unwinding of the same substrates at 20 °C. The rate constants for unwinding were 143 ± 8 bp/s, 151 ± 9 bp/s, 148 ± 8 bp/s, 185 ± 21 s−1 for unwinding of 50%, 69%, 81%, and 100% GC substrates, respectively.
Scheme 1.
Table 1.
The unwinding rate is insensitive to the GC content of the duplex.
Table 2.
The unwinding and translocation rates are the same for Dda.
The substrate containing 100% GC in the duplex exhibited an additional phase in the unwinding curve. The unusual peak in the reaction progress curve that appears at around 50 ms (triangles, Fig. 7b) indicates formation of an intermediate. A similar intermediate was previously described for unwinding of substrates containing 24 and 28 base pairs, but not for substrates containing 16 or 20 base pairs18. The intermediate was interpreted to be the result of several fast steps in the unwinding reaction followed by a rate limiting slow step. The intermediate only appeared for those substrates that resulted in lag phases requiring more than 3 kinetic steps. The 16 base pair and 20 base pair substrates described previously required only three kinetic steps to fit the lag phase because these substrates did not have relatively high GC content. In the current report, three of the substrates used here result in unwinding curves that can be described by kinetic mechanisms of 2 steps (50% GC) or 3 steps (69% GC and 81% GC). However, the unwinding curve for the substrate containing 100% GC content is best described by a model that includes 4 kinetic steps. Hence, the appearance of the intermediate is consistent with the increased number of steps needed to unwind the substrate containing 100% GC content, just as reported previously for unwinding of 24 and 28 base pair duplexes18. The increase in number of steps needed for unwinding is a result of the increased stability of the duplex which is determined by the GC content as well as the length of the duplex.
Single molecule unwinding and translocation
Single molecule helicase assays were conducted with magnetic tweezers using a 1.2 kb DNA hairpin specifically attached between a glass surface and a micron-sized magnetic bead22 as a substrate. The hairpin contains a dsDNA stem of 1139 base pairs, ending with a four T loop on one side. At the other end is a fork; one arm of the fork is a 5′-biotinylated 76 base ssDNA. The other contains 10 nucleotides of ssDNA and 150 bases of duplex DNA labeled with digoxigenin on the 3′ end. The magnetic field gradient generated by two permanent magnets produces a strong vertical force whose magnitude is controlled by adjusting the magnet’s position relative to the sample. Dda unwinding of the fork junction results in a gradual increase in the molecule extension, corresponding to the gain of two bases for each base pair unwound. The helicase concentration was very low (about 250 pM) so that each unwinding event is the result of a single enzyme action.
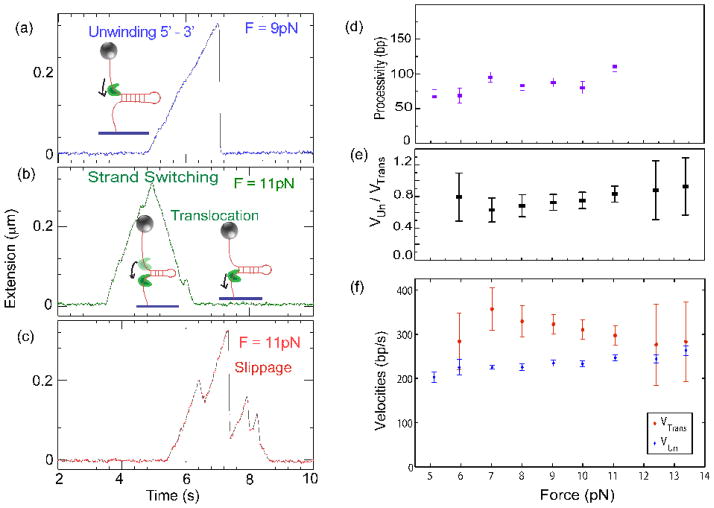
In the presence of saturating ATP, unwinding bursts are observed (Fig. 8a–c). They all start with an unwinding phase of a few hundred base pairs. After the unwinding phase two scenarios are possible: a quasi-instantaneous rezipping of the hairpin due to the helicase dissociation (Fig. 8a), or a slow rezipping, with a rate comparable to the unwinding rate (Fig. 8b). Combinations of these different phases occurred frequently. This is dependent on the force used to pull the hairpin; slow rezipping is absent at 5 pN, and at forces higher than 8 pN, rezipping constitutes about 10% of the total events observed. The unwinding speed is uniform, and unwinding events are often interrupted by partial rezipping corresponding to the helicase slipping backwards by tens to hundreds of base pairs as evidenced by a rapid re-annealing of the hairpin fork (Fig. 8c). The helicase resumes unwinding after such slippage events. Slippages are observed even at very low helicase concentrations, implying that the same enzyme is involved in both events, since dissociation followed by rapid binding is not possible at these low concentrations. During this slippage the enzyme keeps a contact with DNA since it can regain its grip on ssDNA and resume unwinding. Such behaviour is easier to understand for a hexameric enzyme such as the T7 gp4 helicase which encircles ssDNA23 than for a monomeric enzyme like Dda, especially since other non-hexameric helicases, such as UvrD and RecQ rarely slip during unwinding24. During its course of unwinding, Dda may also interact with the displaced DNA strand, resulting in a strand-switching event, characterized by translocation reducing the hairpin reannealing as observed previously in UvrD24. The hairpin fork then does not re-anneal instantaneously, but refolds in the wake of the translocating enzyme, with a rate equal to the translocation speed of the helicase (Fig. 8b). These events enable us to study Dda translocation in addition to its unwinding activity. In all these events, the translocation speed is nearly equal to the unwinding speed.
Fig. 8.
Single molecule unwinding and translocation. One end of a DNA hairpin substrate was attached to a glass coverslip, and the other end was attached to a magnetic bead. Magnets were used to pull the bead at a constant force. The force applied was low enough to prevent mechanical unzipping (below 15pN). This geometry enabled us to follow the enzyme dynamics as a function of the applied force. (a) A typical unwinding event presenting a regular speed of unwinding by Dda in the 5′-to-3′ direction, followed by enzyme dissociation and rapid re-annealing of the hairpin (at 9 pN, 1 base pair ~ 0.9nm). (b) Dda, during its course of unwinding, can sometimes switch strands and just translocate in the 5′-to-3′ direction. While the helicase is unwinding the 1200 base pair hairpin, its extension increases. Dda switches strands after unwinding about 300 base pairs (at 11 pN, 1 base pair unwound ~ 0.98 nm extension increase). The helicase then translocates and the hairpin rezips in its wake, at a rate limited by the translocation speed of the enzyme. Note that the rate of decrease in extension (translocation rate) is equivalent to the rate of increase of extension (unwinding rate). (c) An event demonstrating repeated unwinding due to slippages. In our experimental conditions, slippages occurred frequently. Dda processivity (d) unwinding rate (f), and translocation rate (f) versus applied force, measured in single molecule unwinding assays. (e) Shows the dependence on force of the ratio of the unwinding speed to the translocation speed. Notice that the translocation rate is obtained whenever Dda switches strands; as those events are short and sparse at low force, the error bars associated are rather large, and it was not possible to measure this parameter at 5pN.
An important question which arises is whether the helicase activity is assisted by force applied to the hairpin fork. For a passive helicase, the unwinding speed would increase with the applied force because the force effectively lowers the energy barrier for the helicase to open the double helix. In the case of Dda, we find that the unwinding speed is insensitive to the force applied. For forces ranging from 5 to 13pN the unwinding speed remains close to 250 bp/s, within experimental and statistical errors (Fig. 8f), indicating that Dda is an active helicase. As in ensemble experiments, the ratio between the unwinding and the translocation rates remains close to 1 (considering experimental errors) for the whole range of forces studied (Fig. 8e). The apparently lower unwinding rate might be a systematic effect of undetected small slippage events and the substantial error in the translocation rate. Dda is not a very processive helicase, and at the single molecule level, the different unwinding bursts of Dda have an exponentially distributed processivity. Interestingly, just like the unwinding speed, the processivity is insensitive to the force used to pull the hairpin, as shown in Fig. 8d. The above observations certify that Dda falls in the class of the so-called ‘active’ helicases5.
Discussion
The mechanisms for helicase-catalyzed DNA unwinding are beginning to be understood at the molecular level. One of the ways that helicases have been characterized is in terms of active vs. passive mechanisms. An active helicase interacts with the DNA, resulting in destabilization of the duplex. A passive helicase traps localized melting that occurs through thermal fluctuations4. Most helicases are not completely active or passive, but display activity between these two extremes. One way to quantify this property is to specify the level of destabilizing energy involved in the interaction of the helicase with the DNA fork. A recent report provides a practical guide for distinguishing between active and passive helicases5. The rate of translocation (Vtrans) on ssDNA can be measured via several approaches including bulk fluorescence methods20,25–29 and single molecule methods24,30–37. This rate can then be compared to the rate of unwinding dsDNA (Vun). Those helicases that have a ratio of Vun/Vtrans that is close to 1 are considered to be highly active. Helicases that exhibit a low ratio are considered passive. The ratio that corresponds to an active helicase has been suggested as > 0.25, based on data from a few helicases for which rates are available as well as a theoretical analysis5,38. For example, E. coli RecQ is active (Vun/Vtrans~0.9) and T4 gp41 is passive (Vun/Vtrans~0.08)5. Helicases need not be completely active or passive; for example, for a helicase with Vun/Vtrans=0.25, melting of dsDNA may be partially rate limiting, while for a helicase with Vun/Vtrans=1, the helicase physically separates the duplex and a component of the ATPase cycle or translocation is rate limiting.
The average rate of Dda translocation on ssDNA (267 ± 15 nucleotides/s) was calculated by comparing the rate of Dda dissociation from oligonucleotides of various lengths to that from circular DNA. The translocation rate was also confirmed by measuring the time to the maximal decrease in fluorescence of 3′-fluorescein labeled oligonucleotides and by single molecule techniques. The rate of translocation on ssDNA by Dda is essentially the same as the reported rate of unwinding13,18 as well as that measured in Fig. 7 (average rate is 257 ± 42 bp/s). This rate is also comparable to the rate of unwinding by single molecules of Dda (Fig. 8f). For Dda, the ratio of Vun/Vtrans is 0.96. The unwinding and translocation rates are nearly identical, and well within the experimental error, making this essentially a “perfectly active” helicase. The close match between the unwinding and translocation rates indicates that unwinding of dsDNA by Dda is limited by the rate of translocation on ssDNA. These results are consistent with previous reports showing that when multiple Dda molecules are bound to a single unwinding substrate, the trailing molecules and the leading molecules move at similar rates13, meaning that DNA unwinding due to the lead molecule occurs at the same rate as translocation on ssDNA of the trailing molecules. These results suggest that the rate limiting step for unwinding by Dda is a component of translocation, not of unwinding, per se.
Other helicases with ratios that approach 1 are RecQ and UvrD. In the case of UvrD, the enzyme forms oligomeric species during unwinding, making direct comparisons between translocation and unzipping less straightforward. The Lohman laboratory found that RecBC can simultaneously translocate in the 5′-to-3′ direction at the same rate as the 3′-to 5′ direction37. However, the rate of unwinding under the same conditions is slower than translocation on one (Vun/Vtrans~0.39) or both (Vun/Vtrans~0.54) strands37.
The fact that some helicases translocate on ssDNA much faster than they unzip dsDNA indicates that helicase mechanisms can be much more complex than acting simply as ‘snow plows’. The presence of a complementary strand of NA slows down a passive helicase, resulting in a significantly slower unwinding rate than translocation rate. For example, hexameric helicaseT4 gp41 unwinds dsDNA at the relatively slow rate of ~ 30 bp/s when acting alone33,39. However, translocation on ssDNA occurs at a rate of ~ 400 nt/s33, which is close to the rate of T4 DNA replication. The rate of gp41 is enhanced by more than ten-fold when coupled to the T4 polymerase in vitro40. Therefore unwinding and translocation are similar, when the helicase is coupled to the polymerase. The T7 polymerase interacts with T7 gene 4 helicase, greatly enhancing the unwinding rate. T7 gene 4 translocates on ssDNA at 130 nt/s41 but unwinds DNA at 10–50 bp/s42. When coupled to the T7 polymerase, gene 4 helicase unwinds DNA at rates of 90–130 bp/s43. Therefore, some helicases require a second motor protein in order to fully utilize translocation activity for DNA unwinding activity. Dda does not appear to require accessory proteins to unwind dsDNA at an equivalent rate to that at which it translocates on ssDNA.
An alternative way to distinguish active vs. passive helicases is to measure unwinding with substrates of different GC content. Since an AT base pair is about half as stable as a GC base pair5, the unwinding rate of a passive helicase will decrease with increasing GC content of the duplex. However, an active helicase will be unaffected. RecQ, an active helicase, is somewhat sensitive to the GC content of the duplex with the VunGC/VunAT~0.75. The Patel lab has shown that the rate of unwinding GC rich NA is 6-fold less than AT rich NA for T7 helicase and NS3/4a on RNA. The difference decreases to 3-fold for NS3/4a on DNA substrates and for NS3h on both DNA and RNA42. Dda is the first helicase which has been shown to have both Vun/Vtrans~1, and VunGC/VunAT~1 (Table 2), further emphasizing its characteristics as a ‘perfectly’ active helicase.
The fact that Dda is insensitive to the GC content is consistent with a short step-size that occurs with high force, similar to the chromatin remodeling enzyme, RSC44. Force production is consistent with a role in which Dda removes proteins from the DNA10,16,45–47. Numerous helicases have been found that displace other proteins from DNA. Rrm3 removes proteins bound to DNA in vivo during DNA replication in yeast48. Rep helicase was found to push DnaB helicase during replication in E. coli49. Srs2 regulates recombination by displacing recombinase filaments50, and Pif1 displaces telomerase to regulate telomere length51. In some cases, such as with Srs2, a specific protein-protein interaction between the helicase and the displaced protein induces allosteric changes in the protein that lead to its dissociation from DNA50. Dda can displace streptavidin from biotin-labeled DNA10,15, and has been observed to readily displace DNA-binding proteins from its path during translocation and DNA unwinding16, suggesting that force production occurs when the enzyme moves.
Single molecule Dda unwinding events reveal interesting insights on its activity and interaction with the DNA substrate. In addition to the translocation and unwinding rates obtained from single molecule experiments being similar to those from ensemble experiments, the Vun/Vtrans ratio is close to one, and remains constant as the applied force varies, also indicating that Dda is an active helicase. The slippage events provide evidence that Dda keeps a loose contact with its substrate even during a fast rezipping. This may occur through an interaction with the displaced strand as suggested18, which would facilitate the strand switching observed in some single molecule experiments. Interaction with both strands of the duplex would appear to be advantageous for active helicases.
DNA unwinding requires a step in which the strands are actually separated (melting) as well as a step in which the enzyme moves along the DNA (translocation). It is possible that translocation and unwinding occur concomitantly as a result of movement of the enzyme, but it is also possible that translocation and unwinding occur separately. If the steps are separate, then melting must occur faster, so as to not be rate-limiting. Regardless of the specific mechanism, it would appear that the overall rate for unwinding is limited by a step within the ATPase cycle. For example, ATP hydrolysis, dissociation of ADP or Pi, or a conformational change of the enzyme might limit the overall rate. The rate limiting step for some helicases such T7 gene 4 helicase52, Rho transcription termination factor53 and HCV NS3 helicase54 has been proposed to occur with release of Pi.
The fact that Dda separates the duplex in an active manner predicts that protein-nucleic acid interactions facilitate melting. Evidence for some interaction with the displaced strand was provided when an intermediate in the unwinding reaction was discovered18. The intermediate was explained as a consequence of a slow step for dissociation of the displaced strand after 2–3 steps of unwinding. In the current work, we observed a similar intermediate during unwinding of a substrate containing high GC content. The appearance of this intermediate was not observed with substrates that required only 2–3 steps prior to melting. It was only observed with the high GC content substrate, supporting the conclusion that Dda interacts with both strands, leading to the appearance of the intermediate. The ability of the helicase to switch strands in single molecule experiments and to reinitiate unwinding after slipping also indicate that there may be an interaction between Dda and the displaced strand. In fact, interaction with the displaced strand may actually slow down a helicase rather than enhance activity. In the case of the Pif1 helicase, a DNA:RNA hybrid is unwound faster than a DNA:DNA hybrid. The mechanism for this preference is not known, but it could be due to specific interactions between Pif1 and RNA that activate the helicase, or it could be due to specific interactions between Pif1 and DNA that slow down the helicase. Hence, interaction with the displaced strand may increase or decrease the overall rate of DNA unwinding, depending on the specific mechanism of a helicase.
Thus far, structural studies have not captured the intermediates revealing the specific molecular mechanism(s) for the melting step in helicase-catalyzed DNA unwinding. So, the specific molecular events that occur between a helicase and duplex DNA remain unknown. The fact that Dda helicase moves along ssDNA at the same rate as it unwinds dsDNA indicates that when the enzyme encounters the duplex, its forward motion is not impeded. This could be due to specific interactions that occur between the enzyme and the first base pair. Binding interactions between the helicase and the DNA must lower the activation barrier for melting of the duplex5. Such interactions might include stacking with amino acid residues or hydrogen bonding with the bases, or perhaps distortion of the duplex through kinking of the DNA. For Dda, these interactions occur in a manner that does not limit the overall rate of forward movement. The conformational changes associated with forward motion might sterically split the duplex, assuming appropriate amino acid residues are in place. For example, the structures of other superfamily 1B helicases as well as several DExH helicases contain a hairpin structure referred to as a ‘pin’ that is believed to split the duplex during movement. A combination of binding energy through specific interactions between the enzyme and the base pairs coupled with force production through translocation is likely responsible for the “perfectly active” mechanism of Dda helicase.
Materials and Methods
Materials
Oligonucleotides were purchased from Integrated DNA Technologies and purified as described15. DNA sequences were: 60mer – 5′-TAACGTATTCAAGATACCTCGTACTCTGTACTGACTGCGATCCGACTGTCCTGCATGATG-3′, 52mer – 5′-T24CGCTGATGTCGCCTGGTACGTCGCTGCC-3′, 45mer – 5′-ACCTCGTACTCTGTACTGACTGCGATCCGACTGTCCTGCATGATG-3′, 38mer – 5′-T8GTACTAGTTAACTAGTACCGCTGATGTCGC-3′, 30mer – 5′-CTGACTGCG ATCCGAGTGTCCTGCATGATG-3′, 15mer – 5′-CTGTCCTGCATGATG-3′, 50%GC – T7 CGCTATTGTCTACTGG, 69%GC – T7 CGCTGATGTCGCCTGG, 81%GC – T7 CGCTGCGGTCGCCTGG, 100%GC – T7 CGGGCGGCGGGGCCCG. A 5′-biotinylated 1200 base pair hairpin labelled with several digoxigenins at 3′-end was synthesized as described previously55. Dda was overexpressed and purified as described11.
Association experiments
All concentrations indicated are after mixing. Dda (200 nM) association with poly(dT) in reaction buffer (25 mM Hepes pH 7.5, 10 mM KOAc, 0.1 mM EDTA, 2 mM β-mercaptoethanol) was measured by rapidly mixing in a SX.18MV stopped flow reaction analyzer (Applied Photophysics) at 25 °C. The change in tryptophan fluorescence over time was monitored after a 320 nm cutoff filter (Newport Optical Filter #FSQ-WG320) with excitation at 280 nm through 1 mm slits.
Dissociation experiments
Dda was pre-incubated with ssDNA (at the concentrations indicated in the figure legends) in reaction buffer with 10 mM Mg(OAc)2. All ssDNA concentrations are in nucleotides so that the number of nucleotides, and therefore, Dda binding sites, remains constant as the length of the substrate varies. The reactions were initiated at 25 °C by mixing with heparin (12 mg/ml) in the presence or absence of a saturating concentration of ATP (600 μM) (Table S3). The change in tryptophan fluorescence was monitored after a 320 nm cutoff filter (Newport Optical Filter #FSQ-WG320) with excitation at 280 nm through 1 mm slits (200 nM Dda) or 5 mm slits (50 nM Dda).
Determination of rate of translocation of Dda on DNA
For a model where a nucleic acid translocase needs to bind to only one nucleotide, the number of binding sites is one more than the length of the oligonucleotide (L)28. The length of the ssDNA is inversely proportional to the rate constant for dissociation from the end of the oligonucleotide (kd-end) for an enzyme that translocates unidirectionally on ssDNA19. The translocation rate (Vtrans) can be calculated from these parameters using equation (1)19, where kd-obs is the observed dissociation rate constant and kd-in is the intrinsic rate constant for dissociation from circular DNA.
| (1) |
However, Dda is known to bind 6 nucleotides15. Assuming all 6 nucleotides are required for binding, there are only N = L − B + 1 = L − 528 binding sites (N), for binding site size B. The distance to translocate to reach the end of the oligonucleotide, assuming random binding, is (L-4)/2, and Vtrans is one-half the inverse slope of a 1/kd-end versus L-4 plot based on equation (2).
| (2) |
Translocation experiments using fluorescein fluorescence
200 nM 3′-fluorescein labeled oligonucleotides were preincubated with 50 nM Dda in reaction buffer. The reaction was initiated by mixing with 5 mM ATP, 10 mM Mg(OAc)2, and 5 μM poly(dT) at 25 °C. The change in fluorescein fluorescence was monitored after a 515 nm cut-off filter (Newport Optical Filter #51294) with excitation at 495 nm through 1 mm slits.
DNA unwinding reactions
Oligonucleotides were radiolabeled as described11. Unwinding reactions were performed in reaction buffer with 0.1 mg/ml BSA using a Kintek rapid chemical quench-flow instrument maintained at 25 °C. All concentrations are after mixing. 10 nM radiolabeled substrate was incubated with 100 nM Dda for 3 minutes preceding the initiation of the reaction. 5 mM ATP, 10 mM Mg(OAc)2, 300 nM DNA trap, complementary to the displaced strand, and 5 μM poly(dT), protein trap to prevent rebinding of Dda to the substrate after dissociation, were added to initiate the reaction. Reactions were quenched with 400 mM EDTA, analyzed on a 20% polyacrylamide gel, visualized with a Typhoon Trio phosphorimager (GE Healthcare) and quantified using ImageQuant software(GE Healthcare). Data were fit using KinTek Explorer56 to Scheme 1. The enzyme-substrate (ES) complex is converted to product in n identical, sequential steps, defined by the rate constant ku. At each step, the helicase can also dissociate from the NA with a rate constant of kd. kd was allowed to float because increasing the GC content of the duplex may increase the dissociation rate by increasing the force opposing the helicase, consistent with the observed greater processivity for translocation than for unwinding. 8 base pairs of the 69% GC substrate are known to spontaneously melt based on previous unwinding studies with methylphosphonate modified substrates18. The rate of unwinding (Vun) was calculated using equation (3).
| (3) |
The kinetic step size (m) is defined by equation (4).
| (4) |
L is the length of the duplex. L0 is the number of base pairs that spontaneously melt due to thermal fluctuations, and are not unwound by the helicase18. The number of steps required for the helicase to unwind the duplex is n.
Single molecule experiments
Streptavidin coated MyOne (Invitrogen) magnetic beads were pulled by the magnetic tweezers formed by a pair of permanent magnets55 whose position relative to an antidigoxigenin coated coverslip was monitored, providing a means to change the force exerted on the hairpin over a range of 10−2 – 30 pN. The extension of typically 50 molecules was measured using the ring diffraction image of the bead57 at a temperature of 28°C by 250 pM Dda in a buffer consisting of 20mM Tris-HCl pH 7.6, 25mM NaCl, 3mM MgCl2, 2% BSA, 0.5mM dithiothreitol, and 1mM ATP. A constant force below the threshold for mechanically unzipping the DNA was maintained. Linear fits of the change in extension over time provided unwinding rates. Translocation velocities were obtained in a similar manner from translocation events. The rate of change in extension of the substrate was converted to base pairs using the force-extension curve of ssDNA of a well-defined length in bases in identical buffer conditions. Sometimes, Dda displaced the digoxigenin from the DNA; therefore the experiments were repeated several times in order to collect a large number of unwinding and translocation events. The distribution of unwinding/translocation velocity was computed from 250 – 850 events, depending on the conditions, and presented a Gaussian shape. The errors in the unwinding rate were obtained as a sum of the statistical error and the error in the extension of ssDNA. The mean processivity of the helicase was calculated from the distribution of the maximum number of base pairs unwound in each single burst of activity. The histograms of these events were fit to an exponential whose characteristic length leads to the processivity.
Supplementary Material
Highlights.
Active helicases tightly couple translocation to unwinding while passive do not.
Unlike most helicases, Dda translocates at nearly the same rate as it unwinds DNA.
Dda is relatively insensitive to the GC content of the duplex.
Unwinding by Dda is not affected by a destabilizing force on the duplex.
Dda uses all of its translocase capability for DNA unwinding.
Acknowledgments
We would like to thank Carrie Brown and Clint Langston for technical assistance. This work was supported by National Institutes of Health Grant R01 GM098922 (K.D.R.), National Institutes of Health Grant #P20 RR-16460 from the IDeA Networks of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources (D.L.M.), ANR, Human Frontier Science Program grant (RGP003/2007 to V.C.), the ERC grant ‘MagRepS’ 267862 (V.C. and D.B.), the UAMS graduate student research fund (A.K.B.), and Award Number UL1RR029884 from the National Center For Research Resources (to C. Lowery).
Abbreviations used
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- NA
nucleic acid
- Vtrans
translocation velocity
- Vun
unwinding velocity
- bp
base pairs
- nt
nucleotides
- BSA
bovine serum albumin
Footnotes
Supplementary data to this article can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dillingham MS. Superfamily I helicases as modular components of DNA-processing machines. Biochem Soc Trans. 2011;39:413–423. doi: 10.1042/BST0390413. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante C, Cheng W, Mejia YX. Revisiting the Central Dogma One Molecule at a Time. Cell. 2011;144:480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher RL, Branagan AM, Morrical SW. Coordination of DNA replication and recombination activities in the maintenance of genome stability. J Cell Biochem. 2011;112:2672–2682. doi: 10.1002/jcb.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohman TM. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 5.Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gauss P, Park K, Spencer TE, Hacker KJ. DNA helicase requirements for DNA replication during bacteriophage T4 infection. J Bacteriol. 1994;176:1667–1672. doi: 10.1128/jb.176.6.1667-1672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodadek T, Alberts BM. Stimulation of protein-directed strand exchange by a DNA helicase. Nature. 1987;326:312–314. doi: 10.1038/326312a0. [DOI] [PubMed] [Google Scholar]
- 8.Jongeneel CV, Formosa T, Alberts BM. Purification and characterization of the bacteriophage T4 dda protein. A DNA helicase that associates with the viral helix-destabilizing protein. J Biol Chem. 1984;259:12925–12932. [PubMed] [Google Scholar]
- 9.Raney KD, Benkovic SJ. Bacteriophage T4 Dda Helicase Translocates in a Unidirectional Fashion on Single-stranded DNA. J Biol Chem. 1995;270:22236–22242. doi: 10.1074/jbc.270.38.22236. [DOI] [PubMed] [Google Scholar]
- 10.Morris PD, Raney KD. DNA Helicases Displace Streptavidin from Biotin-Labeled Oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 11.Morris PD, Tackett AJ, Babb K, Nanduri B, Chick C, Scott J, Raney KD. Evidence for a functional monomeric form of the bacteriophage T4 Dda helicase:Dda foes not form stable oligomeric structures. J Biol Chem. 2001;276:19691–19698. doi: 10.1074/jbc.M010928200. [DOI] [PubMed] [Google Scholar]
- 12.Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc Natl Acad Sci USA. 2002;99:14722–7. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eoff RL, Raney KD. Kinetic Mechanism for DNA Unwinding by Multiple Molecules of Dda Helicase Aligned on DNA. Biochemistry. 2010;49:4543–4553. doi: 10.1021/bi100061v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- 15.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 16.Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tackett AJ, Morris PD, Dennis R, Goodwin TE, Raney KD. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 18.Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat Struct Mol Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 19.Young MC, Kuhl SB, von Hippel PH. Kinetic Theory of ATP-driven Translocases on One-dimensional Polymer Lattices. J Mol Biol. 1994;235:1436–1446. doi: 10.1006/jmbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- 20.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–51. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 21.Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J. 2003;85:2224–39. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lionnet T, Dawid A, Bigot S, Barre FX, Saleh OA, Heslot F, Allemand JF, Bensimon D, Croquette V. DNA mechanics as a tool to probe helicase and translocase activity. Nucleic Acids Research. 2006;34:4232–4244. doi: 10.1093/nar/gkl451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun B, Johnson DS, Patel G, Smith BY, Pandey M, Patel SS, Wang MD. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–135. doi: 10.1038/nature10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 26.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomko EJ, Ja H, Park J, Maluf NK, Ha T, Lohman TM. 5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocases. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajagopal V, Gurjar M, Levin MK, Patel SS. The Protease Domain Increases the Translocation Stepping Efficiency of the Hepatitis C Virus NS3-4A Helicase. J Biol Chem. 2010;285:17821–17832. doi: 10.1074/jbc.M110.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matlock DL, Yeruva L, Byrd AK, Mackintosh SG, Langston C, Brown C, Cameron CE, Fischer CJ, Raney KD. Investigation of Translocation, DNA Unwinding, and Protein Displacement by NS3h, the Helicase Domain from the Hepatitis C Virus Helicase. Biochemistry. 2010;49:2097–2109. doi: 10.1021/bi901977k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 31.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 32.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-Molecule Studies Reveal Dynamics of DNA Unwinding by the Ring-Shaped T7 Helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc Natl Acad Sci USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD Enzyme Switches Lead Motor Subunits in Response to [chi] Recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA Helicase Dismantles RecA Filaments by Reeling in DNA in Uniform Steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CG, Bradford C, Lohman TM. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat Struct Mol Biol. 2010;17:1210–1217. doi: 10.1038/nsmb.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Betterton MD, Julicher F. Opening of nucleic-acid double strands by helicases: Active versus passive opening. Phys Rev E. 2005;71:011904. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- 39.Raney KD, Carver TE, Benkovic SJ. Stoichiometry and DNA Unwinding by the Bacteriophage T4 41:59 Helicase. J Biol Chem. 1996;271:14074–14081. doi: 10.1074/jbc.271.24.14074. [DOI] [PubMed] [Google Scholar]
- 40.Dong F, Weitzel SE, von Hippel PH. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc Natl Acad Sci USA. 1996;93:14456–14461. doi: 10.1073/pnas.93.25.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DE, Narayan M, Patel SS. T7 DNA Helicase: A Molecular Motor that Processively and Unidirectionally Translocates Along Single-stranded DNA. J Mol Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 42.Donmez I, Rajagopal V, Jeong YJ, Patel SS. Nucleic Acid Unwinding by Hepatitis C Virus and Bacteriophage T7 Helicases Is Sensitive to Base Pair Stability. J Biol Chem. 2007;282:21116–21123. doi: 10.1074/jbc.M702136200. [DOI] [PubMed] [Google Scholar]
- 43.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bedinger P, Hochstrasser M, Victor Jongeneel C, Alberts BM. Properties of the T4 bacteriophage DNA replication apparatus: The T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983;34:115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- 46.Bedrosian CL, Bastia D. Escherichia coli replication terminator protein impedes simian virus 40 (SV40) DNA replication fork movement and SV40 large tumor antigen helicase activity in vitro at a prokaryotic terminus sequence. Proc Natl Acad Sci USA. 1991;88:2618–2622. doi: 10.1073/pnas.88.7.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yancey-Wrona JE, Matson SW. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azvolinsky AS, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guy CP, Atkinson J, Gupta MK, Mahdi A, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman J, Dillingham MS, Lloyd RG, McGlynn P. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36:654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antony E, Tomko Q, Xiao L, Krejci L, Lohman TM, Ellengerger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 52.Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G. Mechanochemistry of T7 DNA Helicase. J Mol Biol. 2005;350:452–475. doi: 10.1016/j.jmb.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 53.Adelman JL, Jeong YJ, Liao JC, Patel G, Kim DE, Oster G, Patel SS. Mechanochemistry of Transcription Termination Factor Rho. Mol Cell. 2006;22:611–621. doi: 10.1016/j.molcel.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 54.Wang Q, Arnold JJ, Uchida A, Raney KD, Cameron CE. Phosphate release contributes to the rate-limiting step for unwinding by an RNA helicase. Nucleic Acids Res. 2010;38:1312–1324. doi: 10.1093/nar/gkp1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat Chem Biol. 2009;5:904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 57.Gosse C, Croquette V. Magnetic Tweezers: Micromanipulation and Force Measurement at the Molecular Level. Biophysical Journal. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.