Summary
Protein design tests our understanding of protein stability and structure. Successful design methods should allow the exploration of sequence space not found in nature. However, when redesigning naturally occurring protein structures most fixed backbone design algorithms return amino acid sequences that share strong sequence identity with wild-type sequences, especially in the protein core. This behavior places a restriction on functional space that can be explored and is not consistent with observations from nature, where sequences of low identity have similar structures. Here, we allow backbone flexibility during design to mutate every position in the core (38 residues) of a four-helix bundle protein. Only small perturbations to the backbone, 1-2 Å, were needed to entirely mutate the core. The redesigned protein, DRNN, is exceptionally stable (melting point > 140 °C). An NMR and X-ray crystal structure show that the side chains and backbone were accurately modeled (all-atom RMSD = 1.3 Å).
Keywords: Computational Protein Design, de novo Protein Design, Flexible Backbone Protein Design
Introduction
A primary goal of protein design is to create proteins that have sequences, structures and functions not found in nature. This goal can be reached by designing new protein structures from scratch or by modifying sequences and structures of proteins found in nature. The second approach is appealing, because in many cases it should be more likely to succeed, and it is the approach nature typically uses to evolve new functional proteins. There are many examples of naturally occurring protein pairs that are structurally homologous (have the same fold), but have different functions and low sequence identity (< 15%). Recapitulating or expanding on this sequence diversity by design, however, is not straightforward. Most computational methods for protein design are built on side-chain optimization algorithms that work most efficiently with a fixed protein backbone (Gordon et al., 1999). When redesigning naturally occurring proteins with these methods, the computationally optimized sequences often closely resemble the native sequence, especially in the protein core, where >60% sequence identity is common(Desjarlais and Handel, 1999; Kuhlman and Baker, 2000; Pokala and Handel, 2001). It is clear from these studies and from the structural analysis of naturally occurring homologs that to expand sequence diversity it is necessary to allow perturbations to the protein backbone conformation. Even small changes to the backbone (<2 Å), can open large regions of sequence space (Yin et al., 2007). The challenge for protein designers is to identify backbone and sequence perturbations that are energetically favorable.
A variety of strategies have been developed for performing protein design with backbone flexibility (Apgar et al., 2009; Dantas et al., 2007; Davis et al., 2009; Desjarlais and Handel, 1999; Friedland et al., 2008; Fung et al., 2008; Georgiev and Donald, 2007; Grigoryan and Degrado; Havranek and Baker, 2009; Mandell and Kortemme, 2009; Su and Mayo, 1997), however, few have been experimentally validated with high-resolution structures of the design model(Correia et al.; Harbury et al., 1998; Harbury et al., 1995; Hu et al., 2007; Kuhlman et al., 2003; Kuhlman et al., 2002; Murphy et al., 2009; Sammond et al.). Perhaps the most tested approach has been iterative rounds of sequence optimization and backbone refinement with the molecular modeling program Rosetta. Sequence optimization is performed using a simulated annealing protocol that searches for low-energy combinations of side-chain rotamers. Structure refinement uses Monte Carlo sampling of small backbone torsion angle perturbations coupled with gradient-based minimization of dihedral angles. Both stages of optimization use an energy function that rewards tight packing, commonly observed side-chain and backbone torsion angles, favorable hydrogen-bond geometries and low energies of desolvation. This approach has been used to design a protein from scratch, design a protein-binding peptide and design new protein loop conformations (Dantas et al., 2007; Hu et al., 2007; Kuhlman et al., 2003). In this study, we explore whether iterative optimization of sequence and structure with Rosetta can be used to aggressively redesign an entire protein core.
Our specific goal was to mutate every residue in the core of the four-helix bundle protein, CheA phosphotransferase, while maintaining the overall fold and stability of the protein (Figure 1A). Several de novo design and redesign projects have focused on helix bundle proteins(Hecht et al., 1990). From these studies, it is evident that many sequences will adopt collapsed helical structures as long as the amphipathic nature of the helices is preserved and the sequence has significant helical propensity (DeGrado and Nilsson, 1997; Kamtekar et al., 1993). What is more challenging to design are sequences that adopt a specific pre-determined structure and show characteristics of natural helix bundle proteins, such as cooperative thermal unfolding. Many previously reported helical bundle designs formed a molten globule, i.e., an ensemble of collapsed structurally degenerate conformations. In cases where the structure for a design was experimentally determined, it often did not agree with the initial design model (Hill and DeGrado, 2000; Lovejoy et al., 1993; Willis et al., 2000). One striking success story is the accurate de novo design of a symmetric four-helix coiled-coil with a right-handed super-helical twist(Harbury et al., 1998). A key component of this work was optimization of packing energies via backbone refinement as well as sequence design with a reduced amino acid alphabet. Here, we show that flexible backbone design can be used to perturb the structure and sequence of a pre-existing protein with atomic-level accuracy.
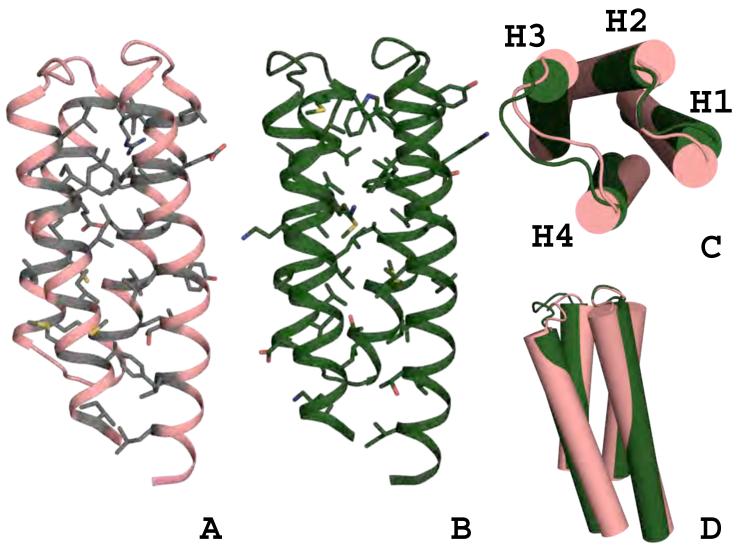
Figure 1.
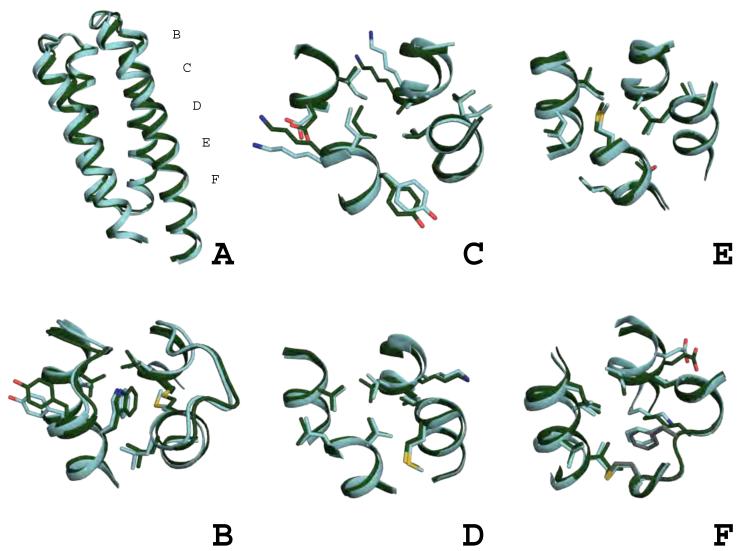
Global comparison of the wild-type template and DRNN design model.
Thirty-eight design positions shown as grey sticks were identified in the wild-type template (A). The final design model for DRNN with the designed positions shown as green sticks (B). DRNN’s backbone and helix crossing angles have been subtly changed by the flexible backbone design procedure (C and D). The helices are labeled H1-H4 in panel C. Panels A, B, and D are in the same orientation and panel C is a top down view of the bundle.
Results
Core Redesign of the CheA Four Helix Bundle
The four-helix bundle CheA phosphotransferase was chosen as the design template (PDB ID: 1TQG] because of its simple up-down helix bundle topology and its moderate size of 105 amino acids. Thirty-eight positions from the CheA X-ray crystal structure were identified as being completely or partially buried and were targeted for mutation (Figure 1A and Figure 2). Our initial hypothesis, based on previous protein redesign experiments, was that the protein backbone would need to be perturbed in order to completely redesign the protein core. To test this hypothesis, four different computational procedures were used to generate designed sequences: (1) fixed backbone design with all amino acid types allowed at each design position (FBAA), (2) fixed backbone design with the native amino acid disallowed at each design position (FBNN), (3) flexible backbone design with all amino acid types allowed at each design position (DRAA) and (4) flexible backbone design with the native amino acid disallowed at each design position (DRNN). In the naming scheme, FB stands for fixed backbone, DR stands for the design and backbone refine strategy of flexible backbone design, AA denotes that all amino acids were allowed during design and NN indicates that only non native amino acids were allowed during design.
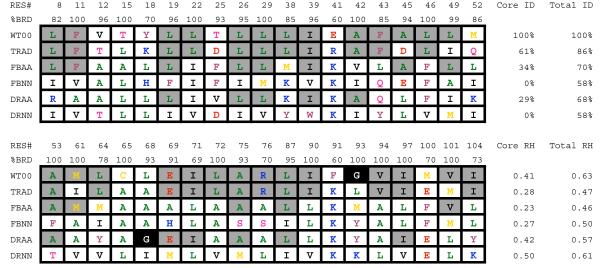
Figure 2.
Comparison of wild-type and designed sequences.
The core sequences for wild-type(WT00), the traditional output from RosettaDesign (TRAD), and the four design experiments FBAA, FBNN, DRAA, and DRNN are shown. The core and total sequence identity and the core and total RosettaHoles scores are given for each sequence. The percent of burial for each core position is shown as %BRD. Residue number is listed as RES#. Gray boxes indicate that a position is conserved between the wild-type sequence and one or more of the designed sequences. The one letter amino-acid codes are colored red (E,D), orange (M,C), green (L,A), blue (K,R,H), black (I,V), pink (N,Q,S,T), plum (F,W,Y), and glycine is shown white on a black background. See also Supplemental Figure 8.
The fixed backbone design protocol used Rosetta’s standard rotamer-optimization method, which uses Monte Carlo sampling of backbone-dependent side-chain rotamers to search for low energy sequences. The flexible backbone protocol used the same sequence-optimization algorithm, but iterated sequence optimization with high-resolution backbone refinement using Monte Carlo sampling and gradient-based minimization of backbone torsion angles. Backbone perturbations with this protocol are generally modest, 1-2 Å. 25,000 independent trajectories were generated for each protocol. As anticipated, in the two approaches where all amino acid types were allowed, FBAA and DRAA, the flexible backbone procedure DRAA generated sequences with lower sequence identity to the wild-type protein. The average sequence identity over the designed positions was 26% in the DRAA protocol and 65% with the fixed backbone protocol. To check if the fixed backbone protocol generated models with lower sequence identity we searched for the best scoring fixed backbone models with less than 50% core identity to the wild-type sequence. Models with Rosetta energies within 6 Rosetta Energy Units of the lowest scoring fixed backbone model were identified that had sequence identities between 40% and 50%. The final FBAA sequence chosen for experimental characterization was selected from this filtered set. Sequence Logos of the 200 lowest energy sequences for each computational protocol illustrate the types of amino acids designed at each position (Supplemental Figure 8).
The RosettaHoles algorithm was used to evaluate packing density in the redesigned proteins compared to wild-type CheA and statistics from high-resolution X-ray crystal structures(Sheffler and Baker, 2009). RosettaHoles explicitly searches for small voids in the protein that are inaccessible to water, and assigns a score to each residue between 0 and 1 that reflects the quality of packing around that residue. RosettaHoles scores closer to 1 indicate fewer voids. Residues in high-resolution crystal structures generally have scores between 0.5 and 1.0 for the entire protein. Models generated with the FBAA and FBNN protocols had RosettaHoles scores between 0.2 and 0.3 for the core residues, while the DRNN and DRAA models had scores between 0.4 and 0.5.
For each of the four protocols, a single sequence was selected for experimental validation (Figure 2). Sequences were selected for experimental testing based on their total Rosetta energy, the quality of packing, correctly predicted secondary structure, performance in ab initio folding experiments and deviation from the wild-type sequence (see methods for more details). In choosing a sequence from the FBAA protocol, we also did not consider sequences that had >50% core sequence identity with the wild-type sequence. For comparison, Figure 2 also shows the lowest scoring sequence generated with the FBAA protocol, labeled as TRAD. The TRAD sequence has 61% identity with the wild-type sequence in the core of the protein.
The computational experiments that incorporated flexible backbone design show subtle but important backbone movements (Figure 1B, 1C, 1D, Figure 3 and Supplemental Figures 3, 4, and 5). The backbone movements generated by this procedure are most often small local changes, with the most variation occurring at loops and termini, larger lever arm movements associated with small backbone dihedrals are limited because gradient based minimization favors collapsed, well-packed protein chain. The designed sequence, DRNN, and the DRNN design model are the most varied from the native sequence and CheA crystal structure (Figure 1B, 1C and 1D) and will be used to illustrate the types of backbone changes due to flexible backbone design. The final DRNN design model has a backbone RMSD of 1.6 Å compared to the CheA crystal structure. The largest backbone deviations between the design model and the crystal structure are seen in loop 3, helix 1, and helix 4. Although its sequence was not varied, loop 3 is pushed away from the center of the helix bundle because of the incorporation of a tryptophan at position 39, previously an isoleucine. Using a global alignment, the backbone RMSD of loop 3 compared to the wild-type protein is 1.9 Å and the all atom RMSD is 2.9 Å. Helix 1 is perturbed by 1.9 Å and helix 4 is perturbed by 2.1 Å (Figure 1C and 1D). The sequence identity of the 38 designed core residues is 0% compared to the native CheA and the total sequence identity is 57%. A diverse set of mutations were predicted for the 38 core design positions, 27 mutations were hydrophobic/aromatic residues mutated to different hydrophobic/aromatic residues, 6 mutations were hydrophobic/aromatic residues mutated to polar residues, 3 mutations were polar residues mutated to hydrophobic/aromatic residues, and 2 mutations were polar amino acids mutated to polar amino acids. In this study, residue positions on the template CheA were classified as buried core positions if they were greater than 50% buried and made significant contacts with residues that were completely buried. This is an intentionally broad definition of the protein core and was intended to capture as much of the protein core as possible, without redesigning the entire protein.
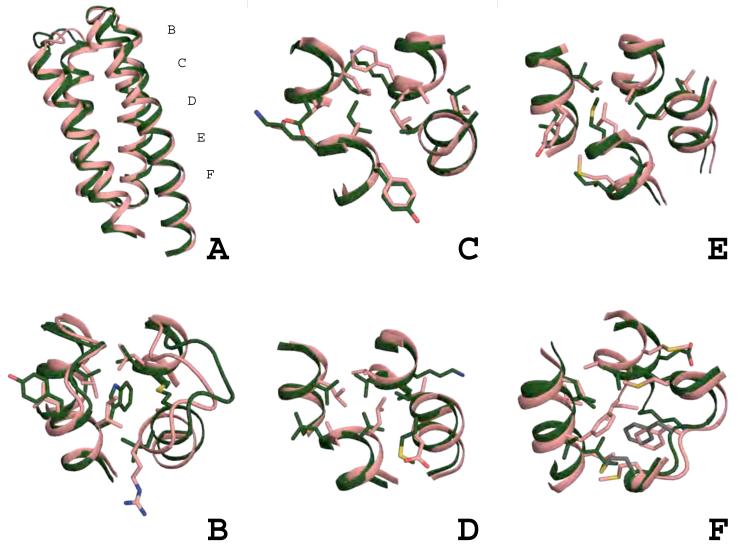
Figure 3.
Comparison of wild-type template and DRNN design model.
The design and the wild-type bundle can be divided into five layers of interacting side-chains. Panel A shows the global view of the side-chain layers. Panels B-F show the layers with wild type in salmon and DRNN in green; positions that were not designed are shown in grey. See also Supplemental Figures 3, 4, & 5.
Protein Expression and Behavior
Three of the designed proteins, FBAA, DRAA and DRNN expressed in E. coli in soluble form at a variety of induction temperatures, 16 °C-37 °C, and produced greater than 33 mg/L of purified protein of culture. The proteins eluted as single peaks from size exclusion chromatography with apparent molecular weights consistent with the expected monomer weights, ~14 kD. In contrast, FBNN was found only in an insoluble form. This behavior was seen at all tested temperatures and IPTG induction concentrations.
Biophysical Characterization of Redesigned CheA
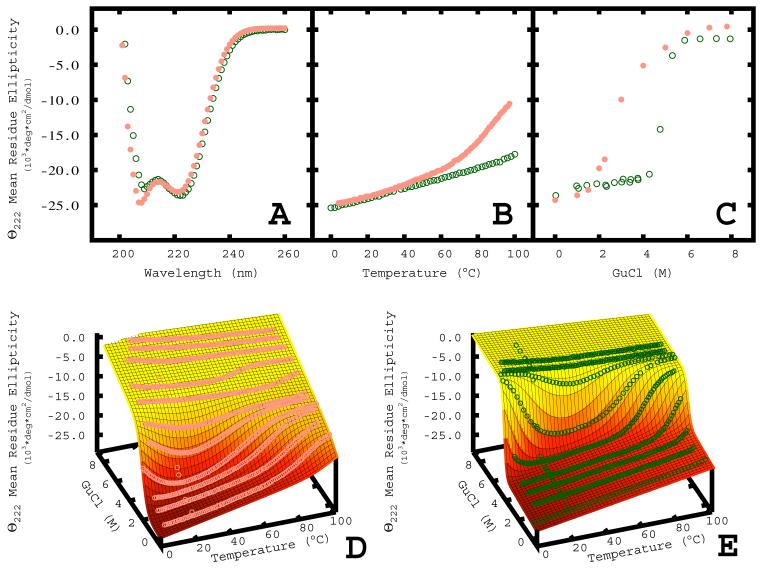
Far-UV circular dichroism experiments confirmed that the designed proteins are primarily α-helical, with strong minima present at 208 nm and 220 nm (Figure 4A and Supplemental Figures 1 and 2). Two of the designed proteins (FBAA, DRNN) did not unfold when subjected to temperatures of up to 97 °C (Figure 4B and Supplemental Figure 1). Chemical denaturation with guanidine hydrochloride (GdnHCl) shows that the designed proteins undergo highly cooperative unfolding events (Figure 4C and Supplemental Figures 1 and 2). To determine accurate values for m, the temperature of the midpoint of unfolding (Tm), ΔH°, ΔCp°, and ΔG°, a Gibbs-Helmholtz surface was constructed by fitting several thermally induced denaturations in the presence of varying amounts of GdnHCl to the Gibbs-Helmholtz equation modified to take into account the effect of denaturant concentration (Table 1, Figure 4D, 4E and Supplemental Figures 1 and 2)(Kuhlman and Raleigh, 1998). The designed proteins are hyperthermostable with Tm values between 96 and 142 °C and ΔG° values for unfolding between 5.5 and 16.2 kcal/mol. Remarkably, the computationally most ambitious design, DRNN, was the most stable. For comparison, the wild-type protein has a ΔG° of unfolding of 3.5 kcal/mol and a Tm of 91°C. The designed proteins have ΔCp° ranging from 0.83 to 1.1 kcal/mol•deg, which are typical values for proteins of this size (Myers et al., 1995). The ΔH° values range from 63 to 128 kcal/mol and the m values range from 1.9 to 3.4 kcal/(mol•M), the wild-type protein has values of 41 kcal/mol and 1.4 kcal/(mol•M) respectively.
Figure 4.
Biophysical characterization of DRNN and wild-type template.
Far-UV Circular Dichroism (A), Thermal Denaturation (B), and Chemical Denaturation (C) of DRNN (green) and wild-type (salmon). Global fits (mesh) of thermal and chemical denaturation data for wild type (D) and DRNN (E) obtained by fitting the data to the Gibbs-Helmholtz equation. All experiments were carried out at 10-20 μM protein concentration in 50 μM sodium phosphate at pH 7.4 and 20°C .
Table 1.
Thermodynamic parameters for wild-type and designed sequences.
| ΔGo (Kcal/mol) |
Tm (°C) |
ΔCp° (Kcal/mol*K) |
ΔH° (Kcal/mol) |
m (Kcal/mol*M) |
|
|---|---|---|---|---|---|
| WT | 3.5 | 91 | 0.61 | 41 | 1.4 |
| FBAA | 14.9 | 144 | 0.83 | 107 | 2.3 |
| DRAA | 5.5 | 96 | 0.90 | 63 | 1.9 |
| DRNN | 16.2 | 142 | 1.08 | 128 | 3.4 |
Values for ΔG°, Tm, ΔCp°, ΔH°, and m were calculated by globally fitting a surface of chemical and thermal melts using the Gibbs-Helmholtz equations. See also Supplemental Figures 1 & 2.
Because DRNN was the most aggressive redesign of CheA and the most stable redesign, we choose it for high-resolution structure determination by NMR and X-ray crystallography.
X-ray Crystal Structure of DRNN
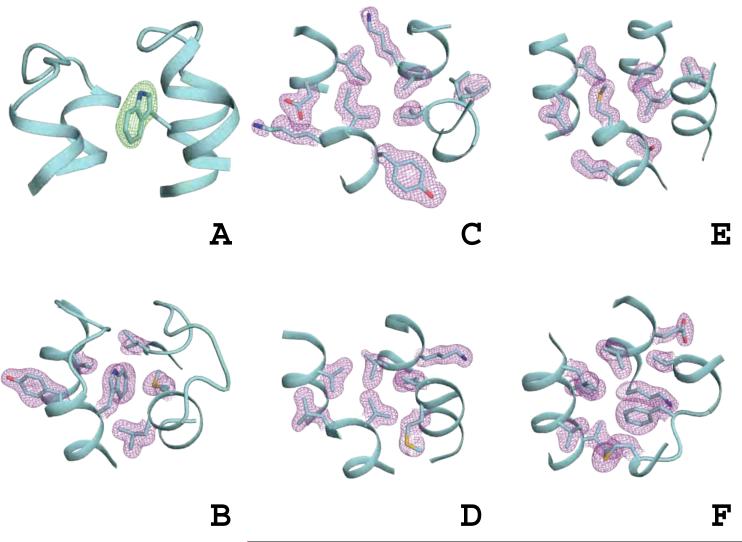
The structure of the designed protein DRNN was determined by X-ray crystallography using diffraction data to a resolution of 1.85 Å. The structure was determined by molecular replacement using the design model with all side-chain atoms removed (to test for potential model bias). In the resulting 2Fo-Fc electron density map almost all of the side chains of the designed residues were clearly defined (Figure 5A). The final model has excellent stereochemical parameters (as determined by Molprobity (Davis et al., 2004)) and also ranks in the ~95th percentile for RosettaHoles packing score, 0.64, in the 1.0-2.0 Å resolution range (Figure 5B-F)(Sheffler and Baker, 2009).
Figure 5.
X-ray crystal structure of DRNN.
A) Fo-Fc electron density (green) for residue W39 after molecular replacement using DRNN without side-chain atoms as the search model. Ribbon-presentation of the DRNN backbone in cyan. B-F) The final 2Fo-Fc density (purple) for molecule A of the DRNN X-ray crystal structure in the five layers used to describe the wild-type and design model; sticks are shown for all design positions and residues 56M and 58F in F. See also Supplemental Table 2
There is strong agreement between the DRNN design model and the experimentally determined structure (Figure 6, Supplemental Figure 9). The all-atom RMSD between the design model and both chains A and B in the asymmetric unit of the experimental structure are 1.5 Å and 1.3 Å, respectively. The 38 core design positions were predicted with good accuracy, 34 positions were observed in the correct rotamer state. Three design positions (Y37, K90, K92) were observed in different rotamer states due to the presence of crystal contacts (K90), or hydrogen bonding with nearby waters (Y37 and K92) that were not included in the design model. Valine 29 was observed in a rotamer different from that in the design model for unknown reasons. The prediction of the backbone of loop 3, which was extensively remodeled, is also highly accurate, with RMSD values of 0.32 Å and 0.38 Å, respectively, over backbone atoms for both chains A and B. Additionally, a hydrogen bond between the side chain of W39 and the backbone carbonyl oxygen of P33 in loop 1 is present in the crystal structure as designed.
Figure 6.
Comparison of DRNN design model and DRNN X-ray crystal structure. The DRNN design model (green) and chain B of the X-ray crystal structure (cyan) shown in a global view (A) and as the five layers that make the bundle core (B-F); positions that were not designed are shown in grey in F.
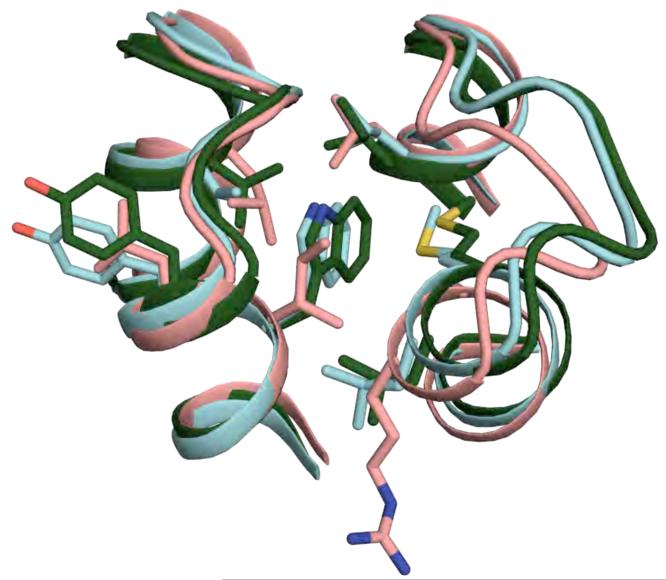
We also compared the DRNN X-ray crystal structure to the 1TQG X-ray crystal structure, the starting template for the flexible backbone design procedure. The DRNN X-ray crystal structure is more similar to the DRNN design model than the starting template (Supplemental Figure 9). The Cα RMSD between the DRNN crystal structure and the DRNN model is 0.8 Å, while the Cα RMSD between the DRNN crystal structure and the 1TQG starting template is 1.7 Å. The structures were further compared by making a histogram of distances between equivalent Cα atoms in the DRNN design model or 1TQG template and the DRNN crystal structure. While 48% of the equivalent Cα atoms were within 0.5 Å of each other when comparing the DRNN model to the DRNN crystal structure, only 29% were within 0.5 Å when comparing the 1TQG template to the DRNN crystal structure. Visually, the most striking comparison is for loop 3, where the DRNN design model is similar to the DRNN crystal structure while loop 3 from the template is more tightly packed against loop 1 (Figure 9).
Figure 9.
Comparison of wild-type template, DRNN design model and crystal structure. The wild-type template (salmon), DRNN design model (green), and the DRNN X-ray crystal structure (cyan) compared in the region of W39 (helix layer B shown in figures 3B, 5B, and 6B). See also Supplemental Figure 9.
NMR Structure of DRNN
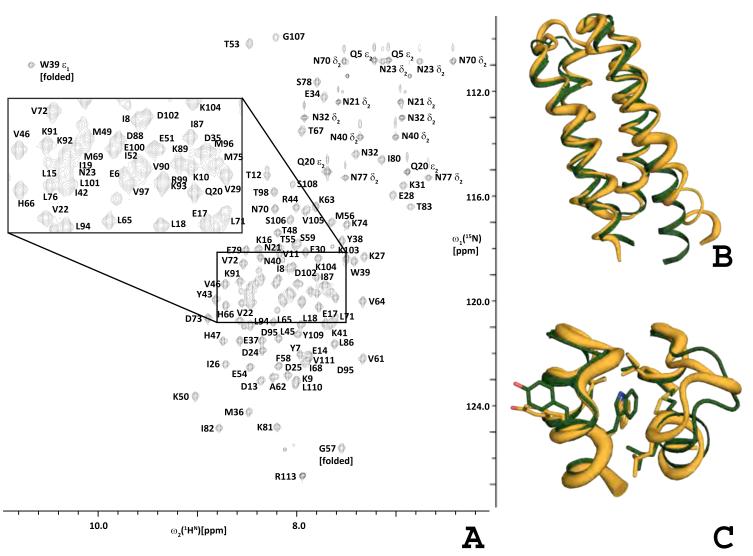
In order to also obtain an NMR solution structure, DRNN, was nominated as a PSI:Biology community outreach target assigned to the Northeast Structural Genomics Consortium (http://www.nesg.org; NESG target ID OR38). The 2D [15N, 1H]-HSQC spectrum of DRNN (Figure 7A) shows that a homogeneous NMR sample containing well-folded DRNN was obtained. Furthermore, the estimated correlation time for isotropic reorientation (τc = 5ns) confirms that DRNN is monomeric in solution.
Figure 7.
2D [15N,1H] HSQC and NMR solution structure of DRNN.
2D [15N,1H] HSQC spectrum (~1 mM protein concentration, 20 mM sodium phosphate, pH 6.5) recorded at 750 MHz 1H resonance frequency. Resonance assignments are indicated using the one-letter code for amino acids (A). Global comparison of the DRNN model (green) and the DRNN solution structure (orange) (B). The region around W39 of the DRNN model and the solution structure (C) (corresponds to layer B in figures 5 and 6) See also Supplemental Figure 7.
A high-quality NMR solution structure was obtained (Supplemental Table 3), which like the crystal structure is similar to the design model: the RMSD calculated for the backbone heavy atoms N, C and C’ between the DRNN design model and the mean coordinates of the 20 conformers representing the solution is 2 Å. Deviations between the design model and NMR structure are, however, primarily observed for the poorly defined conformations of the N-terminus of helix 1 and C-terminus of helix 4 (Figure 7B). Hence, the corresponding RMSD calculated for residues 15-105 only is 1.2 Å. Both the DRNN design model and the DRNN X-ray crystal structure are in excellent agreement with the NMR derived conformational constraints, i.e., only 11 out of 1,406 distance constraints are violated by more than 0.5 Å in the crystal structure or the design model.
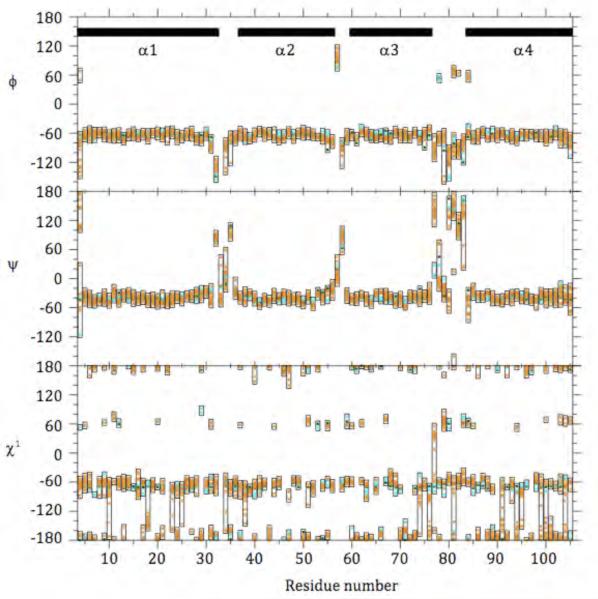
Comparison of χ1-angles in the NMR structure (Figure 8) and the design model reveals that 35 of the 38 designed core residues are in the expected (i.e., designed) rotameric state, and that significantly different rotamer states are observed only for L15, L18, and T53. Notably, the closest agreement between the NMR structure and the design model is observed in the region surrounding W39, with the all heavy atom RMSD calculated for the 19 closest neighbors of W39 being only 1.35 Å (Figure 7C and Supplemental Figure 6).
Figure 8.
Comparison of DRNN NMR Structural Ensemble, DRNN X-ray Crystal Structure, and DRNN design model in φ, ψ, and χ1 space. The values of the ensemble of conformers representing the NMR solution structure are shown in orange with boxes drawn around the observed range. The values observed for the two chains of the X-ray structure are shown in blue, and the values for the design model are shown in green. The black bars at the top indicate the location of the α-helices. See also Supplemental Figure 6.
Discussion
The experimentally determined X-ray and NMR structures of DRNN show that it is possible to use flexible backbone design to aggressively sample sequence space compatible with a naturally occurring protein fold. The redesigned protein DRNN, has zero core sequence identity with the parent CheA but adopts a structure that is similar to CheA with distinct conformational perturbations that were predicted by the design protocol. The remodeling of protein sequences and conformations is a common path used by nature to evolve new functional proteins. Our results suggest that it should be possible to use computational protein design to achieve precise placements of backbone and side-chain atoms as a critical step in building novel binding and active sites. Of the four proteins that were experimentally characterized, only FBNN failed to express in a soluble form in bacteria. This result suggests that there may be a limit to the degree that a sequence can be redesigned without explicit modeling of backbone relaxation, although additional experiments of this type are needed before more general conclusions can be made. Also, if constrained to using a fixed backbone during the design process, protocols that adjust the energy function to soften repulsive forces may be better suited for dramatically redesigning protein cores (Dahiyat and Mayo, 1997; Grigoryan et al., 2007).
The DRNN sequence has exceptional thermostability with a Tm > 140 °C and a free energy of folding of −16 kcal / mol at 25°C. High stability has also been observed in previous computational redesigns of naturally occurring proteins (Dantas et al., 2007; Dantas et al., 2003; Malakauskas and Mayo, 1998; Schweiker and Makhatadze, 2009). In many of these studies the whole protein was redesigned or mutations were dispersed between buried and exposed residues. Our results confirm that high thermostability can be achieved by computational remodeling of just the hydrophobic core. This was also demonstrated in a recent study from Borgo and Havranek (Borgo and Havranek, 2012). Iterative computational cycles of point mutations and backbone relaxation were used to identify small sets of mutations that fill voids in protein cores. The redesigns were stabilized by several kilocalories per mole.
Why is DRNN more stable than the wild-type protein? Possible sources of stability include the incorporation of amino acids with higher intrinsic propensity to form a helix, a burial of more hydrophobic surface area, and a preference for lower energy side-chain rotamers. One of the Rosetta scoring terms used during sequence optimization is based on the probability of observing an amino acid with a particular φ and ψ angle in naturally occurring protein structures. This scoring term accounts for the intrinsic preferences of the amino acids to be in α-helices and β-strands. Interestingly, the value for this score term on average is only slightly more favorable, 1-2%, for DRNN than the wild-type protein. In fact, eighteen of the designed residues in DRNN are β-branched amino acids (valine, threonine or isoleucine), which are typically enriched in β-strand structure(Minor and Kim, 1994). In contrast, ten of the designed positions are β-branched amino acids in the wild-type sequence.
Each amino-acid side chain has intrinsic preferences for the various rotamers that it can adopt. These preferences are highly dependent on the backbone φ and ψ angles of the residue. These preferences are incorporated in the Rosetta scoring function by evaluating the log odds of observing a particular rotamer in the protein database, conditioned on φ and ψ angle. Rosetta uses backbone-dependent rotamer statistics compiled by Dunbrack(Shapovalov and Dunbrack). On average, the rotamers used in DRNN (both in the model and in the crystal structure) are only slightly more favorable, 2-3%, than the rotamers adopted in the wild-type structure (Supplemental Figure 7).
The hydrophobic effect is the primary driving force for protein folding (Dill, 1990) and the burial of more hydrophobic atoms can increase protein stability (Lim et al., 1994; Munson et al., 1996). To evaluate the number of hydrophobic atoms buried in DRNN and wild-type CheA, the solvent accessible surface area of each atom was calculated using a 1.4 Å probe, representative of water solvent. Fourteen additional non-hydrogen hydrophobic atoms were completely buried in DRNN, versus the wild-type CheA and an additional sixteen hydrophobic atoms are greater than 50% buried (Supplemental Table 1). This suggests that the extreme thermostability of DRNN may be partially due to the burial of an additional 27 hydrophobic atoms. However, a similar analysis of the FBAA, FBNN and DRAA design models indicates that there is not a simple correlation between the number of buried hydrophobic atoms and the observed changes in protein stability (Supplemental Table 1). While the FBAA design was nearly as stable as DRNN, in the FBAA design model there is one less buried hydrophobic atom than in the wild type protein. In summary, we have not identified a single metric or characteristic that explains why FBAA and DRNN are more stable than DRAA and the wild-type protein. Like DRNN, the FBAA, FBNN, and DRAA models all have favorable Ramachandran dihedral angles and the side chains are modeled using favorable side chain torsion angles.
In this study we characterized DRNN using both X-ray crystallography and NMR spectroscopy. The X-ray structure is valuable for validating the details of side chain packing in the protein core, while the NMR structure allows one to detect internal dynamics in solution. The NMR spectra obtained for DRNN show that the protein’s global conformation is not affected at room temperature by chemical exchange on the chemical shift timescale (milli- to micro-seconds). In future work, it will be interesting to explore the backbone and side-chain dynamics of DRNN at faster timescales (nanoseconds) and compare results with the wild-type protein and other computationally designed proteins: in a previous study of a designed three-helix bundle, DeGrado and co-workers demonstrated, by measuring NMR spin relaxation parameters, that the side chains in the core of a designed protein were more dynamic on average than is commonly observed for natural proteins(Walsh et al., 2001).
In conclusion, the redesign strategy applied here promises to be valuable for the stabilization of enzymes, ligand-binding proteins, and protein-protein interface partners where preservation of a functional surface or pocket is important. In these cases, our approach can be extended by constraining the relative spatial locations of functionally important residues, while surrounding residues are remodeled in sequence and structural space. Design with backbone flexibility will also be important for repurposing proteins to bind novel substrates and ligands. In this case, constraints can also be used to direct functional residues into desired conformations, while the surrounding sequence and backbone are optimized for the targeted new ligands.
Experimental Procedures
Computational Methods
Fixed Backbone Protein Design Protocol
The fixed backbone protein design protocol used here is the standard fixed backbone design protocol released with Rosetta3.3. The design protocol consists of applying a side-chain packing algorithm, which uses simulated annealing to search rotamer space, using rotamers from the Dunbrack rotamer library and using the Rosetta energy function to evaluate the fitness of sequences(Leaver-Fay et al.).
Flexible Backbone Protein Design Protocol
The redesign sequences were generated using a new protocol within the Rosetta framework. The protocol has two stages, fixed backbone sequence design and fixed sequence backbone and side-chain dihedral optimization. The protocol iterates between these two stages until the energy difference between cycle i and cycle i-1 is less than 1.0 Rosetta Energy Units (REU), in practice this is ~5 redesign simulations for proteins between 100 and 200 residues. The fixed backbone sequence design step uses the standard Rosetta side-chain packing algorithm described above and elsewhere. The fixed sequence backbone and side-chain dihedral optimization employs the Rosetta structure-optimization protocol used in structure prediction and refinement.
Computational Protein Design Experiments
Four different types of computational experiments were performed: (1) fixed backbone design where all amino acids were allowed at design positions (FBAA), (2) fixed backbone design where the native amino acid was not allowed at design positions (FBNN), (3) flexible backbone where all amino acids were allowed at design positions (DRAA) and (4) flexible backbone design where the native amino acid was not allowed at design positions (DRNN).
Core Redesign of the CheA Four-Helix Bundle
To redesign the core residues of the CheA four-helix bundle, 38 positions were identified as buried or partially buried. These positions have at least 15 neighbors each within 10 Å, where a neighbor is defined by the distance between Cβ atoms on residues i and j. Positions identified as core residues were visually inspected to remove any non-buried surface positions with a high number of neighbors. During this visual inspection, all attempts were made to include all partially buried side-chain positions, excluding positions identified as being in a loop by the DSSP algorithm(Kabsch and Sander, 1983). During the design stage, the 38 designable core positions were allowed to change amino-acid identity as described for each type of protein design experiment. An additional seven surface positions were allowed to design and mutate to any amino acid identity. The remaining 60 positions were not allowed to change amino acid identity but were free to change rotamer state. The possible rotamer states for each amino acid type are taken from the Dunbrack backbone dependant rotamer library(Dunbrack, 2002). The 38 core designable positions were given more rotamer freedom, allowing additional sampling of rotamer states, the side-chain chi angles where given 12 extra rotamer states at ± 0.25, 0.50, 0.75, 1.00, 1.25, and 1.50 standard deviations from the most favorable dihedral angles for each rotamer. The seven designable and 60 surface positions were given extra rotamer states at ± 0.5 and 1.0 standard deviation from the most favorable rotamer states. All positions were free to sample φ, ψ, ω, and all dihedral χ angles during backbone and side-chain perturbation and minimization. A total of 25,000 design simulations were performed for each computational protein design experiment.
Selection of Designed Sequences for Experimental Characterization
The 25,000 designed sequences were ranked by their quality of core packing, as measured by RosettaHoles, sequences with scores less than 0.5 (0.4 for FBAA and FBNN) were pruned(Sheffler and Baker, 2009). Sequences where the core design positions were predominately of a single amino-acid type, greater than 50% of the design positions, were pruned. This filter eliminates sequences where the protein core is composed primarily of only a few amino-acid types, mostly alanine and leucine. The 50 lowest-scoring models, based on total Rosetta energy, were evaluated for their secondary structure propensities using the secondary structure prediction server JPRED3(Cole et al., 2008). All 50 design models were predicted to have similar secondary structures compared to the design model and the native CheA. The ten lowest-energy models were subjected to structure prediction using Rosetta’s structure prediction method. This filter evaluates if the designed sequence is predicted to adopt the desired fold, all designed sequences recovered the desired fold. The ten lowest-energy sequences for each experiment were evaluated by eye and one sequence from each experiment was chosen for experimental characterization. It is interesting to note that the sequence chosen from the DRNN experiment was also the lowest-scoring sequence out of the 25,000 designed sequences generated in that experiment.
Experimental Methods
Protein Expression and Purification
A codon-optimized gene for each designed sequence, and a modified version of the wild-type CheA was purchased from Genscript, lowercase letters are due to cloning and capital letters are the designed sequences.
> 1TQG_MOD_WT
mGSHQEYLQQFVDETKEYLQNLNDTLDELEKNPEDMELINEAFRALHTLKEMAETMGFSSMAKLC
HTLENILDKARNSEIKITSDLLDKIKDGVDMITRMVDKIVS
gsylvprgslehhhhhh*
>FBAA
mGSHQEYLQKFADEAKELLQNINDFLKELEKNPEDMEMINKVLRAFHTLKELAETMGFSSMAKMA
HTAANLADKAANSEIKITSDLLDKLKDMADMLTRFVDKLVS
gsylvprgslehhhhhh*
>FBNN
mGSHQEYIQKVADELKEHFQNINDFIKEMEKNPEDMEKVNKIQREFHTAKEIFETMGFSSAAKIA
HTAHNLADKSSNSEIKITSDLIDKLKDYADMLTRFMDKLVS
gsylvprgslehhhhhh*
>DRAA
mGSHDEYRKKAADELKELLQNINDVLDELEKNPEDMEKINKAQRLFHTIKDKAQTMGFSSAAKYA
HTGENIADKAANSEIKITSDLLDKLKDYADMITRELDKYVS
gsylvprgslehhhhhh*
>DRNN
mGSHQEYIKKVTDELKELIQNVNDDIKEVEKNPEDMEYWNKIYRLVHTMKEITETMGFSSVAKVL
HTIMNLVDKMLNSEIKITSDLIDKVKKKLDMVTRELDKKVS
gsylvprgslehhhhhh*
Each gene was supplied as 4 μg of lyophilized DNA in pUC57 vector. The gene of interest was amplified from the parent vector using polymerase chain reaction (PCR), purified using a PCR-clean-up kit from Fermentas, double digested with NdeI and XhoI from NEB, purified again using a PCR-clean-up kit, and finally ligated into a pET-21 b(+) vector from Novagen that had been prepared by double-digesting with NdeI and XhoI and using a Fermentas gel-extraction clean-up kit. The ligation reaction product was transformed into XL-10 Gold cells from Stratagene.
Each protein was expressed in BL21 (DE3) pLysS cells from Stratagene. Cells were grown in LB media with 100 μg/ml ampicillin at 37°C to an OD600 of 0.6 and induced with 0.5 mM IPTG for 12 hours at 16 °C. Cells were centrifuged at 4500 x g for 30 minutes and cell pellets were resuspended in 0.5 M NaCl, 0.2 M Na2HPO4/NaH2PO4 pH 7.0, 10% (v/v) glycerol, 1% (v/v) Triton-X 100, dithiothreitol, and treated with DNAase, RNase, benzamidine, and phenylmethanesulfonylfluoride after three rounds of sonication. The cell lysate was cleared twice by centrifugation at 18,000 x g for 30 minutes. The supernatants were then filtered using a 0.22 μM filter from Millipore. The supernatant was purified by immobilized-metal affinity chromatography using a HisTRAP column from GE Healthcare. The elution was concentrated to 2 mL and further purified by size exclusion chromatography using a Superdex S75 column from GE Healthcare. For the FBNN sequence, induction conditions with IPTG concentrations ranging from 0.1 mM to 0.5 mM and induction ranging from 4 hours to 12 hours were tested. Ultimately, the FBNN sequence did not generate soluble protein.
Circular Dichroism
CD data were collected on a Jasco J-815 CD spectrometer. Far-UV CD scans were collected using a cuvette with a pathlength of 1 mm at concentrations between 10-20 μM protein in 50 μM sodium phosphate at pH 7.4 and 20 °C. Thermal denaturation of samples was conducted between 4 °C and 97 °C while measuring the CD signals at 208 and 222 nm.
Chemical denaturation by guanidine hydrochloride (GdnHCl) was induced by mixing 15 μM designed protein in 0M GdnHCl with 15 μM designed protein in 7.8 M GdnHCl. Great care was taken to ensure the concentration of designed protein in each sample was the same. The protein calculation was calculated using predicted extinction coefficients. The GdnHCl concentration was monitored by the change in refractive index. Thermodynamic parameters were calculated assuming and observing that the unfolding of the designed protein was a reversible two-state process by fitting both the thermal and chemical denaturations to the Gibbs-Helmholtz equation (Kuhlman and Raleigh, 1998).
Nuclear Magnetic Resonance Spectroscopy
The NMR samples of U-13C, 15N-DRNN and 5% 13C, U-15N-DRNN were prepared at concentrations of ~1.0 mM in 90% H2O/10% D2O solution containing 20 mM sodium phosphate (pH 6.5). An isotropic overall rotational correlation time of about 5 ns was inferred from averaged 15N spin relaxation times, indicating that DRNN is monomeric in solution.
The following spectra were recorded for U-13C, 15N-DRNN at 25 °C on a Varian INOVA 750 spectrometer (total measurement time: 6.5 days) equipped with a conventional 1H[13C, 15N] probe: 2D [15N, 1H]-HSQC, aliphatic and aromatic 2D constant-time [13C, 1H]-HSQC, 3D HNCO, HNCACB, CBCA(CO)NH, HBHA(CO)NH, HN(CA)CO, aliphatic (H)CCH, (H)CCH-TOCSY (Cady et al., 2007; Feher et al., 1997), and simultaneous 3D 15N/13Caliphatic/13Caromatic-resolved [1H, 1H]-NOESY (mixing time 70 ms)(Shen et al., 2005). For 5% 13C, U-15N-DRNN, aliphatic 2D constant-time [13C, 1H]-HSQC spectra were acquired as described(Penhoat et al., 2005) at 25 °C on a Varian INOVA 600 spectrometer (total measurement time: 12 hours) equipped with a conventional 1H[13C, 15N] probe in order to obtain stereo-specific assignments for Val and Leu isopropyl groups (Neri et al., 1989).
All NMR spectra were processed using PROSA (Guntert et al., 1992) and analyzed using CARA(Keller, 2004). Sequence-specific backbone (HN, N, Cα, Hα, and CO) and Hβ/Cβ resonance assignments were obtained by using the program AutoAssign (Moseley et al., 2001; Zimmerman et al., 1997). Resonance assignment of side-chains was accomplished using 3D (H)CCH, 3D (H)CCH-TOCSY, and 3D 15N/13Caliphatic/13Caromatic-resolved [1H, 1H]-NOESY. Overall, for residues 1-113 sequence-specific resonance assignments were obtained for 95.2% of backbone and 95.7% of side chain resonances assignable with the NMR experiments listed above (Supplemental Table 3). Chemical shifts were deposited in the BioMagResBank (BMRB ID: 17612). 1H-1H upper distance limit constraints for structure calculation were obtained from 3D 15N/13Caliphatic/13Caromatic-resolved [1H, 1H]-NOESY, and backbone dihedral angle constraints for residues located in well-defined regular secondary structure elements were derived from chemical shifts using the program TALOS+ (Cornilescu et al., 1999).
Automated NOE assignment was performed iteratively with CYANA (Guntert et al., 1997a, b; Herrmann et al., 2002), and the results were verified by interactive spectral analysis. Stereospecific assignments of methylene protons were performed with the GLOMSA module of CYANA, and the final structure calculation was performed with CYANA followed by refinement of selected conformers in an ‘explicit water bath’ (Linge et al., 2003) using the program CNS (Brunger et al., 1998). Validation of the 20 refined conformers was performed with the Protein Structure Validation Software (PSVS) server (Bhattacharya et al., 2007). The NMR structure was deposited in the PDB (PDB ID: 2LCH).
Protein Crystallization and X-ray Crystallography
Crystallization of the designed protein was performed using the hanging-drop vapor-diffusion method at 20 °C. Crystals formed in a drop consisting of 0.5 μl of protein (20 mg/ml in 100 mM ammonium acetate) and 0.5 μl of well solution (0.2 M magnesium acetate and 20% (w/v) PEG 3350. Prior to data collection, crystals were cryo-protected by transferring them into well solution supplemented with 15% (v/v) ethylene glycol before plunging into liquid nitrogen. Crystals diffracted X-rays to a resolution of better than 1.8Å, exhibited the symmetry of space group P1 with cell parameters of a=25.6 Å, b=43.9 Å, c=47.7 Å, α=63.89°, β=80.02°, γ=87.00°, and contained two molecules in the asymmetric unit (solvent content = 36%). Diffraction data were collected at 100 K at the Advanced Proton Source GM/CA CAT 23IDB beamline. The diffraction data were processed using HKL2000 (Otwinowski and Minor, 1997). The crystal suffered from directional diffraction anisotropy. This was corrected using an automated webserver (Strong et al., 2006).
The structure was determined by molecular replacement using the program Phaser (McCoy et al., 2007); the computationally designed model was used as a search model. To test for model bias, side-chain atoms were not included in the search model. After molecular replacement and an initial round of refinement the designed side-chain positions were clearly visible in Fo-Fc and 2Fo-Fc electron density maps. Iterative rounds of refinement were conducted with Refmac5 (Vagin et al., 2004) from the CCP4 suite (Winn et al.) interspersed with manual adjustments to the model using the program COOT (Emsley et al.). The final model contains two molecules in the asymmetric unit with all residues defined in the electron density, except for residue 1 in chain A and residues 1-3 in chain B. Ramachandran statistics for the final DRNN structure model show that the backbone dihedral angles of all residues are in the favored region (Supplemental Table 2). The structure was deposited in the protein data bank as pdb code 3U3B.
Supplementary Material
Highlights.
-
-
Flexible backbone design has been used to mutate every position in a protein core
-
-
The redesign is hyperthermostable (melting temperature > 140°C).
-
-
An NMR structure and an X-Ray structure closely match the design model.
-
-
Designed backbone perturbations are accurately recapitulated in the experimentally determined structures.
Acknowledgments
This work was supported by National Institutes of Health Grant RO1GM073960 and an award from the W.M. Keck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apgar JR, Hahn S, Grigoryan G, Keating AE. Cluster expansion models for flexible-backbone protein energetics. J Comput Chem. 2009;30:2402–2413. doi: 10.1002/jcc.21249. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Borgo B, Havranek JJ. Automated selection of stabilizing mutations in designed and natural proteins. Proc Natl Acad Sci U S A. 2012;109:1494–1499. doi: 10.1073/pnas.1115172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cady SD, Goodman C, Tatko CD, DeGrado WF, Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: a 2H, 13C, AND 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Correia BE, Ban YE, Friend DJ, Ellingson K, Xu H, Boni E, Bradley-Hewitt T, Bruhn-Johannsen JF, Stamatatos L, Strong RK, et al. Computational protein design using flexible backbone remodeling and resurfacing: case studies in structure-based antigen design. J Mol Biol. 405:284–297. doi: 10.1016/j.jmb.2010.09.061. [DOI] [PubMed] [Google Scholar]
- Dahiyat BI, Mayo SL. Probing the role of packing specificity in protein design. Proc Natl Acad Sci U S A. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas G, Corrent C, Reichow SL, Havranek JJ, Eletr ZM, Isern NG, Kuhlman B, Varani G, Merritt EA, Baker D. High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J Mol Biol. 2007;366:1209–1221. doi: 10.1016/j.jmb.2006.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J Mol Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Raha K, Head MS, Baker D. Blind docking of pharmaceutically relevant compounds using RosettaLigand. Protein Sci. 2009;18:1998–2002. doi: 10.1002/pro.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrado WF, Nilsson BO. Engineering and design Screening, selection and design: standing at the crossroads in three dimensions. Curr Opin Struct Biol. 1997;7:455–456. doi: 10.1016/s0959-440x(97)80106-6. [DOI] [PubMed] [Google Scholar]
- Desjarlais JR, Handel TM. Side-chain and backbone flexibility in protein core design. J Mol Biol. 1999;290:305–318. doi: 10.1006/jmbi.1999.2866. [DOI] [PubMed] [Google Scholar]
- Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL., Jr. Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feher VA, Zapf JW, Hoch JA, Whiteley JM, McIntosh LP, Rance M, Skelton NJ, Dahlquist FW, Cavanagh J. High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition. Biochemistry. 1997;36:10015–10025. doi: 10.1021/bi970816l. [DOI] [PubMed] [Google Scholar]
- Friedland GD, Linares AJ, Smith CA, Kortemme T. A simple model of backbone flexibility improves modeling of side-chain conformational variability. J Mol Biol. 2008;380:757–774. doi: 10.1016/j.jmb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung HK, Floudas CA, Taylor MS, Zhang L, Morikis D. Toward full-sequence de novo protein design with flexible templates for human beta-defensin-2. Biophys J. 2008;94:584–599. doi: 10.1529/biophysj.107.110627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev I, Donald BR. Dead-end elimination with backbone flexibility. Bioinformatics. 2007;23:i185–194. doi: 10.1093/bioinformatics/btm197. [DOI] [PubMed] [Google Scholar]
- Gordon DB, Marshall SA, Mayo SL. Energy functions for protein design. Curr Opin Struct Biol. 1999;9:509–513. doi: 10.1016/s0959-440x(99)80072-4. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Degrado WF. Probing designability via a generalized model of helical bundle geometry. J Mol Biol. 405:1079–1100. doi: 10.1016/j.jmb.2010.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Ochoa A, Keating AE. Computing van der Waals energies in the context of the rotamer approximation. Proteins. 2007;68:863–878. doi: 10.1002/prot.21470. [DOI] [PubMed] [Google Scholar]
- Guntert P, Dotsch V, Wider G, Wuthrich K. Processing of multidimensional NMR data with the new software PROSA. J Biomol NMR. 1992:619–629. [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Automated NOE assignment was performed iteratively with CYANA Methods in Molecular Biology. 1997a;278 [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997b;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- Harbury PB, Tidor B, Kim PS. Repacking protein cores with backbone freedom: structure prediction for coiled coils. Proc Natl Acad Sci U S A. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek JJ, Baker D. Motif-directed flexible backbone design of functional interactions. Protein Sci. 2009;18:1293–1305. doi: 10.1002/pro.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht MH, Richardson JS, Richardson DC, Ogden RC. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990;249:884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Hill RB, DeGrado WF. A polar, solvent-exposed residue can be essential for native protein structure. Structure. 2000;8:471–479. doi: 10.1016/s0969-2126(00)00130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wang H, Ke H, Kuhlman B. High-resolution design of a protein loop. Proc Natl Acad Sci U S A. 2007;104:17668–17673. doi: 10.1073/pnas.0707977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. How good are predictions of protein secondary structure? FEBS Lett. 1983;155:179–182. doi: 10.1016/0014-5793(82)80597-8. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- Keller R. The computer aided resonance assignment tutorial. Cantina Verlag. 2004 [Google Scholar]
- Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, O’Neill JW, Kim DE, Zhang KY, Baker D. Accurate computer-based design of a new backbone conformation in the second turn of protein L. J Mol Biol. 2002;315:471–477. doi: 10.1006/jmbi.2001.5229. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Raleigh DP. Global analysis of the thermal and chemical denaturation of the N-terminal domain of the ribosomal protein L9 in H2O and D2O. Determination of the thermodynamic parameters, deltaH(o), deltaS(o), and deltaC(o)p and evaluation of solvent isotope effects. Protein Sci. 1998;7:2405–2412. doi: 10.1002/pro.5560071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Hodel A, Sauer RT, Richards FM. The crystal structure of a mutant protein with altered but improved hydrophobic core packing. Proc Natl Acad Sci U S A. 1994;91:423–427. doi: 10.1073/pnas.91.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Lovejoy B, Choe S, Cascio D, McRorie DK, DeGrado WF, Eisenberg D. Crystal structure of a synthetic triple-stranded alpha-helical bundle. Science. 1993;259:1288–1293. doi: 10.1126/science.8446897. [DOI] [PubMed] [Google Scholar]
- Malakauskas SM, Mayo SL. Design, structure and stability of a hyperthermophilic protein variant. Nat Struct Biol. 1998;5:470–475. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- Mandell DJ, Kortemme T. Backbone flexibility in computational protein design. Curr Opin Biotechnol. 2009;20:420–428. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL, Jr., Kim PS. Measurement of the beta-sheet-forming propensities of amino acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- Munson M, Balasubramanian S, Fleming KG, Nagi AD, O’Brien R, Sturtevant JM, Regan L. What makes a protein a protein? Hydrophobic core designs that specify stability and structural properties. Protein Sci. 1996;5:1584–1593. doi: 10.1002/pro.5560050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy PM, Bolduc JM, Gallaher JL, Stoddard BL, Baker D. Alteration of enzyme specificity by computational loop remodeling and design. Proc Natl Acad Sci U S A. 2009;106:9215–9220. doi: 10.1073/pnas.0811070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography. 1997;276(Pt A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Penhoat CH, Li Z, Atreya HS, Kim S, Yee A, Xiao R, Murray D, Arrowsmith CH, Szyperski T. NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure. J Struct Funct Genomics. 2005;6:51–62. doi: 10.1007/s10969-005-5277-z. [DOI] [PubMed] [Google Scholar]
- Pokala N, Handel TM. Review: protein design--where we were, where we are, where we’re going. J Struct Biol. 2001;134:269–281. doi: 10.1006/jsbi.2001.4349. [DOI] [PubMed] [Google Scholar]
- Sammond DW, Bosch DE, Butterfoss GL, Purbeck C, Machius M, Siderovski DP, Kuhlman B. Computational design of the sequence and structure of a protein-binding peptide. J Am Chem Soc. 133:4190–4192. doi: 10.1021/ja110296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiker KL, Makhatadze GI. Protein stabilization by the rational design of surface charge-charge interactions. Methods Mol Biol. 2009;490:261–283. doi: 10.1007/978-1-59745-367-7_11. [DOI] [PubMed] [Google Scholar]
- Shapovalov MV, Dunbrack RL., Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Atreya HS, Liu G, Szyperski T. G-matrix Fourier transform NOESY-based protocol for high-quality protein structure determination. J Am Chem Soc. 2005;127:9085–9099. doi: 10.1021/ja0501870. [DOI] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A, Mayo SL. Coupling backbone flexibility and amino acid sequence selection in protein design. Protein Sci. 1997;6:1701–1707. doi: 10.1002/pro.5560060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Walsh ST, Sukharev VI, Betz SF, Vekshin NL, DeGrado WF. Hydrophobic core malleability of a de novo designed three-helix bundle protein. J Mol Biol. 2001;305:361–373. doi: 10.1006/jmbi.2000.4184. [DOI] [PubMed] [Google Scholar]
- Willis MA, Bishop B, Regan L, Brunger AT. Dramatic structural and thermodynamic consequences of repacking a protein’s hydrophobic core. Structure. 2000;8:1319–1328. doi: 10.1016/s0969-2126(00)00544-x. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Zimmerman DE, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, Chien C, Powers R, Montelione GT. Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol. 1997;269:592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.