Abstract
Type II hyperprolinemia is an autosomal recessive disorder caused by a deficiency in Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH, aka ALDH4A1), the aldehyde dehydrogenase that catalyzes the oxidation of glutamate semialdehyde to glutamate. Here we report the first structure of human P5CDH and investigate the impact of the hyperprolinemia-associated mutation of Ser352 to Leu on the structure and catalytic properties of the enzyme. The 2.5 Å resolution crystal structure of human P5CDH was determined using experimental phasing. Structures of the mutant enzymes S352A (2.4 Å) and S352L (2.85 Å) were determined to elucidate the structural consequences of altering Ser352. Structures of the 93%-identical mouse P5CDH complexed with sulfate ion (1.3 Å resolution), glutamate (1.5 Å), and NAD+ (1.5 Å) were determined to obtain high resolution views of the active site. Together, the structures show that Ser352 occupies a hydrophilic pocket and is connected via water-mediated hydrogen bonds to catalytic Cys348. Mutation of Ser352 to Leu is shown to abolish catalytic activity and eliminate NAD+ binding. Analysis of the S352A mutant shows that these functional defects are caused by the introduction of the nonpolar Leu352 side chain rather than the removal of the Ser352 hydroxyl. The S352L structure shows that the mutation induces a dramatic 8-Å rearrangement of the catalytic loop. Because of this conformational change, Ser349 is not positioned to interact with the aldehyde substrate, conserved Glu447 is no longer poised to bind NAD+, and Cys348 faces the wrong direction for nucleophilic attack. These structural alterations render the enzyme inactive.
Keywords: X-ray crystallography, aldehyde dehydrogenase, ALDH4A1, proline catabolism, isothermal titration calorimetry, metabolic disorders
Introduction
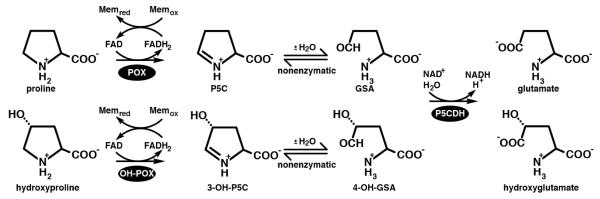
The enzyme Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (P5CDH, EC 1.5.1.12) is a nuclear-encoded mitochondrial matrix protein that catalyzes the final step of proline catabolism (Fig. 1, upper reactions).1 Despite its name, P5CDH is an aldehyde dehydrogenase (ALDH); it catalyzes the NAD+-dependent oxidation of L-glutamate-γ-semialdehyde (GSA) to L-glutamate. GSA is the hydrolysis product of Δ1-pyrroline-5-carboxylate (P5C), which is generated from proline by proline dehydrogenase (PRODH). In humans, PRODH is called proline oxidase (POX) in recognition of its function as a superoxide-generating tumor suppressor protein.2 GSA is also produced from ornithine by ornithine aminotransferase.
Fig. 1.
The reactions of proline (upper) and hydroxyproline (lower) catabolism in humans.
P5CDH is also the second enzyme of hydroxyproline catabolism in humans (Fig. 1, lower reactions).3 Analogous to proline, catabolism of hydroxyproline begins with the oxidation of hydroxyproline (trans-4-hydroxy-L-proline) to Δ1-pyrroline-3-hydroxy-5-carboxylate (3OH-P5C) by hydroxyproline oxidase (OH-POX, 45 % identical to POX). The nonenzymatic hydrolysis of 3OH-P5C produces 4-hydroxyglutamate semialdehyde (OH-GSA), which is oxidized to 4-erythro-hydroxy-L-glutamate (OH-Glu) by P5CDH. The structural basis for the dual substrate specificity of human P5CDH (HsP5CDH) is unknown.
P5CDH belongs to the ALDH superfamily, a vast group of divergently evolved enzymes that share a common three-dimensional fold and catalytic strategy for oxidizing aldehydes to carboxylic acids. As of a 2002 census,4 the superfamily comprises several hundred enzymes from all three kingdoms of life, which have been classified into 20 families based on sequence identity. P5CDH belongs to the ALDH4 family and is encoded by the ALDH4A1 gene. The generally accepted catalytic mechanism of ALDHs begins with nucleophilic attack by a universally conserved Cys residue at the C atom of the aldehyde group of the substrate to form a hemithioacetal. Hydride transfer to NAD(P)+ yields a thioacyl intermediate and NAD(P)H. Lastly, hydrolysis of the thioacyl generates the carboxylic acid product. Crystal structures of P5CDH from Thermus thermophilus suggest that this mechanism holds for P5CDH.6
Certain mutations in the ALDH4A1 gene cause type II hyperprolinemia (HPII), an inherited autosomal recessive disorder characterized by a deficiency in P5CDH activity.7-11 Individuals with HPII have elevated levels of P5C and proline in plasma, urine, and cerebrospinal fluid.12 Elevation of P5C is consistent with the block in the conversion of P5C to glutamate. The increase in proline reflects the conversion of some of the accumulated P5C pool to proline by P5C reductase.11
HPII is causally linked to neurologic manifestations and is associated with an increased incidence of seizures and possibly mental retardation.13 Hyperprolinemia is also a risk factor for schizophrenia. Precisely how a deficiency of P5CDH contributes to these disorders is uncertain but may reflect the role of proline as a neurotransmitter.16-19 Animal studies have established a connection between high proline levels and neurological dysfunction and suggest that excessive proline disrupts energy metabolism in the brain, leading to mitochondrial dysfunction and oxidative stress.12 Recent studies of a fly model of HPII show that loss of P5CDH activity results in swollen mitochondria and suggest that proper P5CDH activity is necessary for normal mitochondrial homeostasis.20
Human genetics studies have provided insight into the molecular basis of the HPII. Several nonsynonymous single-nucleotide polymorphisms (SNPs) in the P5CDH gene are associated with the disease.9 Known pathological SNPs include the missense mutation of Ser352 to Leu (S352L) and frameshift mutations that cause premature termination of translation.
Here we provide the first structural analysis of HsP5CDH and investigate the impact of the S352L mutation on the structure and catalytic properties of HsP5CDH. Several crystal structures of HsP5CDH and the 93%-identical mouse P5CDH (MmP5CDH) are reported, including complexes of the latter enzyme with NAD+ and glutamate. The effect of mutating Ser352 to Leu or Ala is probed using steady-state kinetics assays, NAD+ binding measurements, and X-ray crystallography. The data provide insight into three-dimensional structural basis of HPII as well as the dual substrate recognition of GSA and OH-GSA.
Results
Overall fold
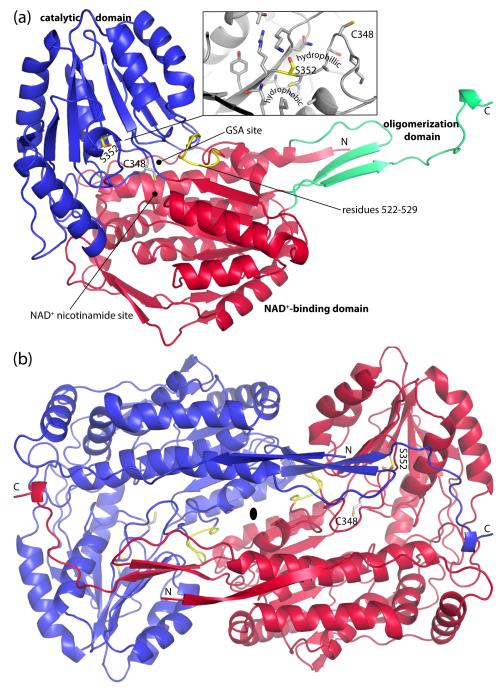
The structure of HsP5CDH was determined at 2.5 Å resolution using single-wavelength anomalous diffraction (SAD) phasing (Table 1). HsP5CDH exhibits the classic ALDH fold (Fig. 2a), which consists of an N-terminal NAD+-binding domain (residues 23-182, 199-315, 525-542), C-terminal catalytic domain (residues 316-524), and oligomerization domain (residues 183-198, 543-562). At the core of the NAD+-binding domain is an open α/β substructure that resembles the Rossmann dinucleotide-binding fold (residues 203 - 314). As first described for ALDH3, the Rossmann fold of ALDH differs from the classic one in that the final helix and strand of the classic Rossmann fold are absent in ALDHs.21 The catalytic domain also exhibits an open α/β fold and features a twisted 7-stranded β-sheet with all but one strand in parallel. This domain furnishes the essential cysteine nucleophile (Cys348) and several residues that bind GSA, including Ser349. The oligomerization domain is a bipartite structure consisting of a β-hairpin protruding from the NAD+-binding domain and the C-terminal ~20 residues of the polypeptide chain. The latter part forms a β-strand followed by an extended section. The hairpin and C-terminal strand combine to form a two-stranded antiparallel β-sheet that resembles a flap.
Table 1.
Data collection and refinement statistics for HsP5CDHa
| Se-Met-1 | Se-Met-2 | S352A | S352L | |
|---|---|---|---|---|
| Space group | P65 | P65 | P65 | P65 |
| Unit cell parameters (Å) | a = 150.7 | a = 150.7 | a = 149.1 | a = 150.4 |
| c = 191.6 | c = 192.0 | c = 190.7 | c = 192.5 | |
| Resolution (Å) | 50.0 - 2.85 | 50.0 - 2.50 | 48.8 - 2.40 | 48.8 - 2.85 |
| (2.95 - 2.85) | (2.59 - 2.50) | (2.53 - 2.40) | (3.00 - 2.85) | |
| Total observations | 661829 | 321938 | 342969 | 177351 |
| Unique reflections | 113595 | 84960 | 93350 | 55880 |
| Multiplicity | 5.8 (5.8) | 3.8 (3.8) | 3.7 (3.2) | 3.2 (3.1) |
| Rmergeb | 0.100 (0.524) | 0.075 (0.539) | 0.072 (0.450) | 0.101 (0.626) |
| Rmeasb | 0.083 (0.539) | 0.119 (0.742) | ||
| Rpimb | 0.041 (0.291) | 0.062 (0.390) | ||
| <I /σ(I)> | 22.9 (4.4) | 17.0 (2.5) | 10.7 (2.3) | 8.6 (2.2) |
| Completeness (%) | 100.0 (100.0) | 99.7 (100.0) | 99.7 (99.5) | 97.5 (99.0) |
| Rwork / Rfreec | 0.199 / 0.237 | 0.210 / 0.251 | 0.209 / 0.264 | |
| Number of atoms | 16032 | 15844 | 13924 | |
| Protein residues | 2162 | 2151 | 1982 | |
| Water molecules | 0 | 167 | 0 | |
| RMSD bond lengths (Å) | 0.009 | 0.008 | 0.011 | |
| RMSD bond angles (°) | 1.18 | 1.16 | 1.23 | |
| Ramachandran plotd | ||||
| Favored (residues) | 2099 | 2059 | 1846 | |
| Allowed (residues) | 55 | 73 | 72 | |
| Outliers (residues) | 0 | 1 | 6 | |
| Average B-factors | ||||
| Protein (Å2) | 62 | 49 | 56 | |
| Water (Å2) | 34 | |||
| Coordinate error (Å)e | 0.70 | 0.72 | 0.90 | |
| PDB code | 3V9G | 3V9H | 3V9I |
Values for the outer resolution shell of data are given in parenthesis.
Definitions of Rmerge, Rmeas, and Rpim can be found in Weiss.48
A common test set of reflections was used for all refinements (2.4 %).
The Ramachandran plot was generated with RAMPAGE.49
Maximum - likelihood based coordinate error from PHENIX.
Fig. 2.
Protomer and dimer structure of HsP5CDH. (a) Ribbon drawing of the protomer. The NAD+-binding, catalytic, and oligomerization domains are colored red, blue, and green, respectively. Ser352 and residues 522-529 are colored yellow. The side chain of catalytic Cys348 is shown. Inset: close-up view of the environment around Ser352. (b) The HsP5CDH dimer. The two chains are colored red and blue. This figure and others were created with PyMOL.50
Oligomeric state and quaternary structure
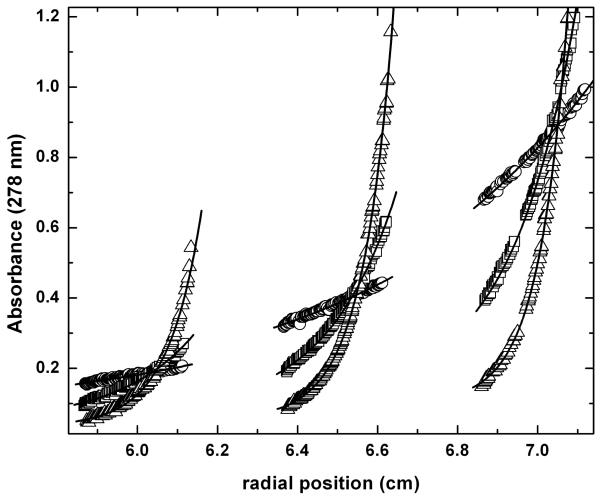
The oligomeric state of HsP5CDH in solution was determined using equilibrium analytical ultracentrifugation (Fig. 3). The data from nine sedimentation experiments corresponding to three loading concentrations and three rotor speeds were fitted globally to a single-species model. The molar mass is estimated to be 122 kDa (120 – 125 kDa), which is within 2 % of the value predicted for the dimer (124 kDa).
Fig. 3.
Analytical ultracentrifugation data for HsP5CDH. Data were acquired at 4000 (circles), 8000 (squares), and 12000 (triangles) rpm at three nominal loading concentrations. The curves represent the best least-squares fit to an ideal single-species model. The molar mass obtained from global fitting is 122 kDa (120 – 125 kDa), which is within 2 % of the value predicted for the dimer (124 kDa).
Inspection of the crystal lattice revealed a two-body assembly that represents the dimer formed in solution (Fig. 2b). This dimeric assembly is also found in the crystal lattices of MmP5CDH and other ALDHs.21-23 The dimer is a domain-swapped assembly in which the oligomerization domain of one protomer engages the other protomer. The domain swapping results in the formation of an intermolecular β-sheet involving the C-terminal strand of the oligomerization domain of one protomer and the final strand of the catalytic domain of the other protomer. Also, the extended region at the C-terminus of one protomer wraps over the catalytic domain of the other protomer forming several hydrogen bonds. Another major part of the dimer is located on the face opposite to that of the intermolecular β-sheet and consists of extensive nonpolar and electrostatic interactions involving the 290s helices of the two protomers (residues 288-304). In total, the dimer interface buries 4300 Å2 of surface area.
Binding of ligands to the active site
MmP5CDH was used for structure determination of enzyme-ligand complexes because of the exceptional crystallographic resolution (Table 2). The overall conformations of MmP5CDH and HsP5CDH are essentially identical except for a deviation at the N-terminus, which is due to different crystal lattice interactions (Fig. S1). The root mean square deviation between the two structures is 0.3 Å for 533 residues. The sequence identity is 100 % for active site residues. Moreover, the conformations of active site residues in the two enzymes are essentially identical. Thus MmP5CDH is an excellent surrogate for HsP5CDH.
Table 2.
Data collection and refinement statistics for MmP5CDHa
| Sulfate | Glutamate | NAD+ | |
|---|---|---|---|
| Space group | P212121 | P212121 | P212121 |
| Unit cell parameters (Å) | a = 85.2 | a = 84.8 | a = 84.9 |
| b = 94.0 | b = 93.9 | b = 94.0 | |
| c = 132.4 | c = 132.2 | c = 132.4 | |
| Resolution (Å) | 50.0 - 1.30 | 47.0 - 1.50 | 47.0 - 1.50 |
| (1.32 - 1.30) | (1.58 - 1.50) | (1.58 - 1.50) | |
| Total observations | 914888 | 1222700 | 1221816 |
| Unique reflections | 247337 | 168017 | 168338 |
| Multiplicity | 3.7 (3.6) | 7.3 (6.7) | 7.3 (6.8) |
| Rmergeb | 0.055 (0.502) | 0.066 (0.418) | 0.061 (0.399) |
| Rmeasb | 0.071 (0.454) | 0.065 (0.432) | |
| Rpimb | 0.026 (0.174) | 0.024 (0.164) | |
| <I /σ(I)> | 26.6 (2.4) | 25.3 (4.8) | 25.3 (4.9) |
| Completeness (%) | 94.9 (86.0) | 99.5 (96.5) | 99.6 (97.6) |
| Rwork / Rfreec | 0.132 / 0.160 | 0.151 / 0.169 | 0.158 / 0.176 |
| Number of atoms | 9479 | 9233 | 9246 |
| Protein residues | 1091 | 1081 | 1081 |
| Water molecules | 1035 | 896 | 872 |
| Active site ligand atoms | 10 | 20 | 46 |
| RMSD bond lengths (Å) | 0.005 | 0.006 | 0.006 |
| RMSD bond angles (°) | 1.05 | 1.09 | 1.11 |
| Ramachandran plotd | |||
| Favored (%) | 98.3 | 98.2 | 98.0 |
| Allowed (%) | 1.7 | 1.8 | 2.0 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
| Average B-factors | |||
| Protein (Å2) | 12 | 10 | 11 |
| Water (Å2) | 25 | 20 | 21 |
| Active site ligand (Å2) | 17 | 11 | 14 |
| Coordinate error (Å)e | 0.30 | 0.29 | 0.31 |
| PDB code | 3V9J | 3V9K | 3V9L |
Values for the outer resolution shell of data are given in parenthesis.
Definitions of Rmerge, Rmeas, and Rpim can be found in Weiss.48
A common test set of reflections was used for all refinements (5.0 %).
The Ramachandran plot was generated with RAMPAGE.49
Maximum - likelihood based coordinate error from PHENIX.
The 1.3 Å resolution structure of MmP5CDH was determined from crystals grown in the presence of 0.2 M Li2SO4. Electron density maps clearly indicate a sulfate ion bound in the active site (Fig. S2). The ion interacts with Gly512, Ser513, and Ser349. We note that the sulfate site corresponds to the binding site for the phosphate group of glyceraldehyde-3-phosphate of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase, which also belongs to the ALDH superfamily.24
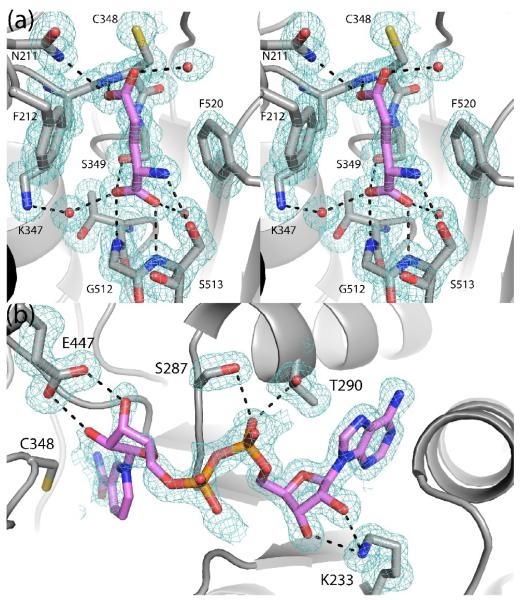
The structure of MmP5CDH complexed with the product glutamate was solved at 1.5 Å resolution using data from a crystal soaked in ~300 mM glutamate. Electron density maps clearly showed that glutamate had displaced the bound sulfate ion (Fig. 4a). Glutamate binds with its α-carboxylate in the sulfate site and its side chain extending toward catalytic Cys348. The amino and α-carboxylate groups of glutamate form hydrogen bonds with Ser349, Gly512, and Ser513. The aliphatic part of the glutamate side chain packs between Phe212 and Phe520. One of the oxygen atoms of the γ-carboxylate interacts with Asn211 and the main chain of Cys348. This atom likely represents the O atom of the aldehyde group of GSA. The other O atom of the carboxylate points into a solvent-filled cavity and represents the O atom derived from the attack of water on the thioacyl intermediate. This mode of product binding is similar to that observed for Thermus thermophilus (TtP5CDH, 30 % identical to HsP5CDH).6
Fig. 4.
Electron density and interactions for (a) glutamate (stereographic view) and (b) NAD+ bound to MmP5CDH. The cages represent simulated annealing σA-weighted Fo - Fc omit maps contoured at 3.0 σ. The NMN part of NAD+ in panel b was modeled based on other ALDH structures and has zero occupancy in the structure deposited in the PDB.
A 1.5 Å resolution structure of MmP5CDH complexed with NAD+ was determined. Electron density for the AMP moiety is strong, while the density for NMN is weaker (Fig. 4b). Nevertheless, the weak density is consistent with the hydride transfer NAD+ conformation that is seen in other ALDHs, including TtP5CDH. We note that conformational disorder is common for ALDHs. Based on these observations, the NMN and AMP moieties were modeled with occupancy values of 0.0 and 1.0, respectively. The adenine ring is wedged between the last two helices of the Rossmann dinucleotide-binding fold. Lys233 engages the adenine ribose, while Ser287 and Thr290 interact with the pyrophosphate. The interactions involving Ser287 and Lys233 are also seen in TtP5CDH. The NMN ribose is predicted to form hydrogen bonds with Glu447. This interaction is universally conserved in ALDHs and is presumably essential for binding NAD+.
The structural context of G521fs(+1)
The frameshift mutation G521fs(+1) consists of an insertion of T following nucleotide 1563 in exon 16 of the ALDH4A1 gene.9 This mutation alters the sequence of residues 522-529 (yellow in Fig. 2a) from GARASGTN to WGPSLWNQ and introduces a stop codon that truncates translation after residue 529. Residue 529 is located in the inter-domain linker that connects the end of the catalytic domain to the strand of the oligomerization domain. The truncated protein thus lacks the final β-strand and C-terminal extension. These elements form a major part of the dimer interface, suggesting that they are essential for proper dimer formation. Since there are no examples of functional monomeric ALDHs, the truncated protein is probably nonfunctional. The lack of proper dimer formation most likely explains why P5CDH-deficient yeast expressing G521fs(+1) failed to grow on proline and had no detectable P5CDH activity.9
The structural context of S352L
Ser352 is located at one end of the catalytic loop (Fig. 2a inset). This loop contains catalytic Cys348 and Ser349, a residue that interacts with the aldehyde substrate (Fig. 4a). The MmP5CDH structure provides a high resolution view of the three-dimensional context of Ser352 (Fig. 5). We note that the conformations of the catalytic loops in HsP5CDH and MmP5CDH are almost identical (Fig. S3a).
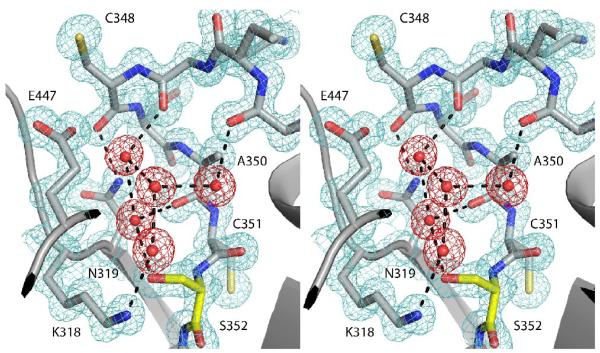
Fig. 5.
High resolution (1.3 Å) view of the catalytic loop of MmP5CDH (stereographic view). The cage represents a 2Fo - Fc map contoured at 1.5 σ. Protein density is colored aquamarine. Water density is colored red. Ser352 is colored yellow.
Ser352 sits at the bottom of a water-filled pocket (Fig. 5). The adjacent residue, Cys351, is turned away from this hydrophilic space, and its side chain is tucked into a hydrophobic region. Catalytic Cys348 and Glu447 sit at the top of the water-filled pocket where it flows into the crevice between the catalytic and NAD+-binding domains. Ser352, Lys318, Asn319 and backbone carbonyls of the catalytic loop project into the pocket and form hydrogen bonds with water molecules, thus creating an electrostatic network that appears to stabilize the conformation of the catalytic loop (Fig. 5). This region appears to be conserved among ALDHs. For example, the constellation of water molecules in the pocket is also found in human mitochondrial aldehyde dehydrogenase (PDB code 1O04).5
Characterization of the S352L and S352A mutants of HsP5CDH
The mutation of Ser352 to Leu in HsP5CDH abolishes catalytic activity. The mutant enzyme was inactive under all conditions tested. For example, the activity of S352L was below detection in an assay containing 100 μg/mL of enzyme, 355 μM NAD+, and 200 μM P5C (Fig. S4). For reference, an identical assay performed with HsP5CDH present at 10-fold lower concentration exhibited substrate depletion within 1 minute (Fig. S4). The absence of measurable catalytic activity for recombinant S352L is in agreement with previous studies showing that P5CDH-deficient yeast expressing S352L failed to grow on proline and had no P5CDH activity.9
The functional consequences of mutating Ser352 was probed further using mutagenesis, kinetics, and isothermal titration calorimetry. Mutation of Ser to Leu has the dual effect of removing a hydroxyl group and adding a branched aliphatic side chain. The mutant enzyme S352A was created to examine the contribution of the former effect.
In contrast to S352L, the mutant enzyme S352A exhibits catalytic activity that is commensurate with that of HsP5CDH. The kinetic properties of HsP5CDH and S352A were thus more closely compared. Competitive type inhibition by L-P5C at concentrations > 50 μM was observed for HsP5CDH and S352A in assays varying NAD+ as previously reported but not characterized25 (Fig. S5). In order to obtain kinetic constants and a substrate inhibition constant for L-P5C (KI), reaction progress curves at various NAD+ concentrations using different fixed P5C concentrations were globally fitted to a Theorell-Chance mechanism (Fig. S6). Global fitting using KinTek Global Kinetic Explorer26 yielded kcat values of 10 and 5 s−1 for HsP5CDH and S352A, respectively (Table S1). The kcat/K for HsP5CDH and S352A are 98.7 and 102 mM−1s−1 m , respectively. The Km values for NAD+ and L-P5C are thus 100 and 32 μM for HsP5CDH, and 49 and 51 μM for S352A respectively. The similar kinetic parameters for HsP5CDH and S352A suggest that the catalytic defect of S352L is due primarily to the addition of the nonpolar Leu side chain rather than the removal of the hydroxyl group of Ser352.
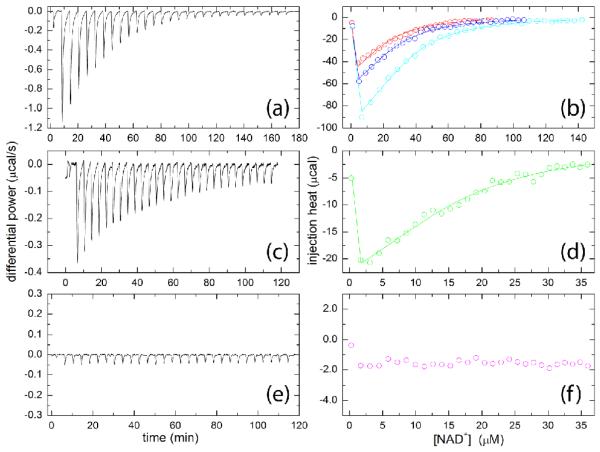
Isothermal titration calorimetry (ITC) was used to study the binding of NAD+ to HsP5CDH, S352A, and S352L (Fig. 6). The association of NAD+ with HsP5CDH is exothermic (Fig. 6a). Global fitting of data from three replicate titrations (Fig. 6b) yielded an apparent enthalpy change of −18 ± 1 kcal/mol and dissociation constant (Kd) of 15 ± 1 μM (Table 3). The binding of NAD+ to S352A is likewise exothermic (Fig. 6c), and the binding parameters are similar to those of HsP5CDH (ΔH = −18 ± 2 kcal/mol, Kd, = 10 ± 2 μM). In contrast, the injection heats for S352L are small and nearly constant during the titration (Fig. 6e, 6f), implying that S352L has negligible affinity for NAD+. This result implies that the catalytic defect of S352L is due, in part, to a lack of NAD+ binding. Furthermore, the ITC results show that the defect in NAD+ binding of S352L results primarily from the introduction of the nonpolar Leu side chain rather than removal of the hydroxyl group of Ser352.
Fig. 6.
ITC analysis of NAD+ binding to (a-b) HsP5CDH, (c-d) S352A, and (e-f) S352L. (a) Raw data for the titration of 47 μM HsP5CDH with 0.77 mM NAD+. (b) Integrated data for titrations of 30, 34, and 47 μM HsP5CDH with NAD+. The solid lines represent the optimal global fit of the three data sets to a single-site binding model. (c) Raw data for the titration of 18 μM S352A with 0.20 mM NAD+. (d) Integrated data for the experiment displayed in panel c, along with the optimum least-squares fit to a single-site model. (e) Raw data for titration of 18 μM S352L with 0.20 mM NAD+. (f) Integrated data for the experiment displayed in panel e.
Table 3.
ITC data for NAD+ binding to HsP5CDH and S352A
| ΔH (kcal/mol) | K (M−1) | Kd (μM) | |
|---|---|---|---|
| HsP5CDH | −18. ± 1. | 6.6 ± 0.5 × 104 | 15. ± 1. |
| S352A | −18. ± 2. | 10. ± 2. × 104 | 10. ± 2. |
The structure of S352A was determined at 2.4 Å resolution (Table 1). The structure is essentially identical to that of HsP5CDH. In particular, the catalytic loop and constellation of residues that bind GSA and NAD+ have the same conformations as in the native enzyme (Fig. S3b). The S352A structure, in agreement with the ITC and kinetic data, shows that the catalytic defect of S352L is due primarily to the introduction of the nonpolar Leu side chain.
Crystal structure of the S352L mutant of HsP5CDH
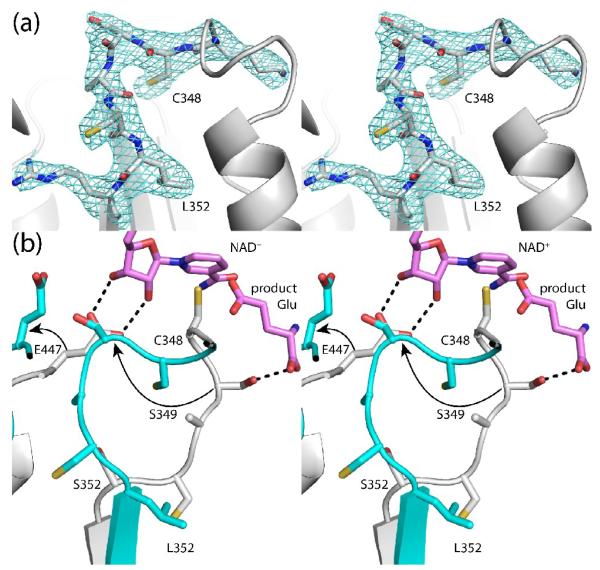
The structure of S352L was determined at 2.85 Å resolution (Table 1). The mutant enzyme exhibits the same overall fold and dimeric structure as HsP5CDH. The active site of S352L, however, is highly perturbed. In particular, the catalytic loop adopts a non-native conformation (Fig. 7a).
Fig. 7.
The active site of S352L. (a) Electron density map for the catalytic loop of S352L (stereographic view). The cage represents a simulated annealing σA-weighted Fo - Fc omit map contoured at 2.5 σ. (b) Superposition of HsP5CDH (white) and S352L (cyan) (stereographic view). The arrows denote the movements of Ser349 and Glu447 that are induced by the mutation of Ser352 to Leu. For reference, NAD+ and the product glutamate from the MmP5CDH structures are shown in pink. The dashed lines indicated hydrogen bonds to NAD+ and the product glutamate in the MmP5CDH structures.
Mutation of Ser352 to Leu profoundly changes the structure of the catalytic loop (Fig. 7b). Leu352 packs into a hydrophobic pocket that is occupied by Cys351 in the native enzyme. Consequently, Cys351 and Ala350 have moved into the hydrophilic pocket of the native enzyme, forcing Lys318 out of the pocket. Ser349 moves by 8Å into the space that is reserved for Glu447 in the native enzyme, ejecting Glu447 from the active site (see arrows in Fig. 7b). These changes are significant because Ser349 and Glu447 interact with the aldehyde substrate and the nicotinamide ribose of NAD+, respectively (Fig. 4). The conformational change of Glu447 may be particularly significant, since mutagenesis of this residue in ALDH2 (Glu399) substantially lowers kcat and increases K for NAD+.27 m Finally, catalytic Cys348 shifts by 3 Å, and its side chain is turned into the enzyme rather than pointing into the open space between the catalytic and NAD+-binding domains.
Discussion
Human and bacterial P5CDHs share a common fold. Structures of P5CDH from Thermus thermophilus, Bacillus licheniformis (PDB code 3RJL), and Bacillus halodurans (3QAN) have been determined. The root mean square deviation between HsP5CDH and the bacterial enzymes is 1.5 Å over about 500 residues, indicating a high level of structural similarity at the fold level (Fig. S7). A notable difference is that the C-terminal extension is not found in bacterial P5CDHs. The extension is involved in dimerization in HsP5CDH. It remains to be determined whether bacterial and mammalian P5CDHs share a common oligomeric state in solution.
The MmP5CDH structures provide insight into the dual substrate specificity of HsP5CDH for GSA and OH-GSA. A model of the ternary enzyme-OH-GSA-NAD+ complex was created by superimposing the MmP5CDH-Glu and MmP5CDH-NAD+ structures and replacing the glutamate ligand with a model of OH-GSA (Fig. S8). The hydroxyl group occupies a solvent-filled cavity and is bounded by Phe520 and the nicotinamide of NAD+. The shortest contact distances with the O atom of the OH-GSA hydroxyl are 3.2 Å with the phenyl group of Phe520 and 3.1 Å with the nicotinamide carbonyl. Thus, there are no severe steric clashes in the model. The absence of a protein group that clashes with hydroxyl of OH-GSA and the potential for hydrogen bonding with water molecules and the carboxamide of NAD+ provide an explanation for the dual recognition of GSA and OH-GSA.
The S352L mutant is reminiscent of the E487K mutant of ALDH2. As with S352L, E487K is associated with a human health disorder, impaired ethanol metabolism. In both cases the mutated residue does not directly contact the substrate or NAD+, yet the mutant enzymes are catalytically deficient. Mutation of Glu487 to Lys in ALDH2 results in a 200-fold increase in Km for NAD+,29 and structural studies have shown that NAD+ induces partial reordering of the E487K active site.30 In contrast, we see no evidence for association of NAD+ with S352L and hence no reordering of the active site.
Finally, our data provide a satisfying explanation of the role of the S352L mutation in HPII. It was previously suggested, based on crystal structures and molecular dynamics simulations, that the main consequence of this mutation is to disrupt a water network that connects Ser352 to the catalytic Cys. This idea was based, in part, on a static model derived from a TtP5CDH structure and assumed that the Leu side chain projects into the solvent-filled pocket that connects Ser352 to Cys348.6 In fact, the S352L structure shows that Leu352 avoids the solvent-filled pocket and instead seeks a hydrophobic pocket (Fig. 7b). The structure further shows that the S352L mutation dramatically alters the constellation of residues responsible for aldehyde binding, NAD+ binding, and covalent bond formation. In particular, Ser349 is not positioned properly to interact with the carboxylate of the aldehyde substrate. Conserved Glu447 is no longer poised to bind the nicotinamide ribose of NAD+, an interaction that is common to all ALDHs. And Cys348 is facing the wrong direction for nucleophilic attack. These structural features are consistent with the observed absence of catalytic activity and NAD+ binding in S352L. In conclusion, the mutation of Ser352 to Leu disrupts three major aspects of catalysis - aldehyde recognition, NAD+ binding, and nucleophilic attack - rendering the enzyme nonfunctional.
Materials and Methods
Subcloning
The genes encoding HsP5CDH (NCBI RefSeq NP_003739) and MmP5CDH (NCBI RefSeq NP_780647.3) were obtained from ATCC. The coding sequences for HsP5CDH residues 18 - 563 and MmP5CDH residues 21 - 562, which omit the mitochondrial leader sequences, were subcloned into pET28a between NdeI and XhoI restriction sites. The site-directed mutants S352L and S352A of HsP5CDH were created with the QuikChange XL II mutagenesis kit (Agilent Technologies).
Expression and purification of HsP5CDH
HsP5CDH and site-directed mutants of HsP5CDH were expressed in Bl21(DE3)pLysS as follows. Two liters of LB media, supplemented with 50 μg/L kanamycin, were inoculated with 1 % of a starter culture and grown at 37 °C with constant aeration at 250 rpm until the OD600 reached 0.6. IPTG was added and protein expression proceeded for about 20 hours at 22 °C and 200 rpm. The cells were harvested by centrifugation at 3500 rpm for 30 minutes in a Sorvall SLC 6000 rotor at 4 °C. The cells were resuspended into 20 mM HEPES, 60 mM NaCl, and 5 % glycerol at pH 8.0, quick frozen in liquid nitrogen, and stored at −80 °C until further purification.
Frozen cells were thawed and broken by sonication. Cell debris and unbroken cells were separated by centrifugation at 16500 rpm for 1 hour in a SS34 rotor. The supernatant was applied to a HisTrap HP column that had been charged with NiCl2 and equilibrated with buffer A (20 mM HEPES, 300 mM NaCl, 5 % glycerol, pH 8.0). The column was washed with buffer A followed by a second wash with buffer A supplemented with 30 mM imidazole. The protein was eluted with 300 mM imidazole in buffer A. The sample was dialyzed into 50 mM Tris, 0.5 mM EDTA, 0.5 mM THP, and 5 % glycerol at pH 8.0 in preparation for further purification using anion exchange chromatography (HiTrap Q). His-tagged HsP5CDH was collected in the flow-through, while contaminating proteins were retained in the column. The purified protein was dialyzed into pre-crystallization buffer (50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM THP, 5 % glycerol, pH 8.0). The His-tag was not removed.
Expression and purification of MmP5CDH
MmP5CDH was expressed as described above for HsP5CDH ([IPTG] = 0.5 mM, overnight induction at 22 °C and 200 rpm). The protein was purified using Ni2+-affinity and ion exchange chromatography. Unlike His-tagged HsP5CDH, His-tagged MmP5CDH bound to the anion exchange column when loaded in a buffer of 50 mM Tris, 0.5 mM EDTA, 0.5 mM THP, and 5 % glycerol at pH 7.5, and was eluted with a linear 0 – 1.0 M NaCl gradient. The purified protein was dialyzed into 50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM THP and 5 % glycerol at pH 7.5 in preparation for crystallization trials. The His-tag was not removed.
Preparation of Se-Met HsP5CDH
The Se-Met derivative of HsP5CDH was prepared using the method of metabolic inhibition.32 An overnight starter culture was grown in LB media at 37 °C. Cells from the starter culture were pelleted and resuspended in minimal media supplemented with 2 mM MgSO4, 0.1 mM CaCl2 and 0.4 % glucose as the carbon source. Cells were grown until the OD600 reached 0.5, at which time, the Met biosynthetic pathway was inhibited by adding 1 mg each of lysine, threonine, and phenylalanine, and 0.5 mg each of leucine, isoleucine, valine, and L-selenomethionine per liter of media. The temperature was lowered to 22 °C and the rotation rate was set to 200 rpm. After 30 minutes, IPTG was added (50 μM). The cells were harvested after ~20 hours. Se-Met HsP5CDH was purified as describe above for the native protein.
Crystallization
Se-Met HsP5CDH was crystallized at 4 °C in sitting drops using reservoirs containing 20 - 25 % (w/v) PEG 3350, 0.2 M (NH4)2SO4, and 0.1 M HEPES at pH 7.0 - 8.0. The drops were formed by mixing equal volumes of the reservoir and the protein stock solution (6.0 mg/mL protein in 50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM THP, 5 % glycerol, pH 8.0). The crystals were cryoprotected in 25 % PEG 3350, 0.2 M (NH4)2SO4, 0.1 M HEPES, 25 % glycerol at pH 7.5. The space group is P65 with unit cell parameters of a = 150.7 Å and c = 192.0 Å. The asymmetric unit includes two dimers, which implies 53 % solvent and VM of 2.6 Å3/Da.33
Crystals of S352A and S352L were grown at 20 °C using the recipe described above for HsP5CDH except that microseeding was used. Sitting drops were formed by mixing equal volumes of the protein stock (7.0 mg/ml for S352A, 2.0 mg/ml for S352L) and reservoir solutions, and the drops were streak seeded 12 hours later using a horse hair. The seed stock was prepared by diluting crushed S352A crystals by a factor of 5000-10000 with the reservoir. Crystallization of S352L was unsuccessful without microseeding.
MmP5CDH was crystallized in sitting drops (1 μl of protein, 1 μl of reservoir) at 20 °C using reservoir solutions containing 20 - 25 % (w/v) PEG 3350, 0.2 M Li2SO4, 0.1 M Bis-Tris, and pH of 6.0 - 7.0. The protein stock solution contained 6 mg/mL His-tagged MmP5CDH in 50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM THP and 5 % glycerol at pH 7.5. The crystals were cryoprotected with 25 % PEG 3350, 0.2 M Li2SO4, 0.1 M Bis-Tris, 25 % glycerol, and pH 6.25. The space group is P212121 with unit cell lengths of a = 85.2 Å, b = 94.0 Å, and c = 132.4 Å. The asymmetric unit contains one dimer, implying 44 % solvent and VM of 2.2 Å3/Da. The glutamate complex was formed by soaking crystals for 6 – 8 hours in the cryobuffer without Li2SO4 and supplemented with 300 mM sodium glutamate. The NAD+ complex was obtained similarly using 20 mM NAD+.
X-ray diffraction data collection
Crystals of Se-Met HsP5CDH were analyzed at NE-CAT beamline 24-ID-C at the Advanced Photon Source using a Quantum 315 detector (Table 1). The data set that was used for single-wavelength anomalous diffraction (SAD) phasing was collected at the energy corresponding to the experimentally-determined peak of f” (λ = 0.979181 Å). This data set consisted of 180 frames collected with an oscillation width of 1.0° per image, detector distance of 375 mm, exposure time of 1.0 s/image, and transmission of 2.5 %. The data were processed to 2.85 Å resolution using HKL2000,34 with I+ and I− treated as nonequivalent reflections during scaling. The crystal was translated, and a second data set was collected at higher transmission for purpose of phase extension and refinement. Denoted as Se-Met-2 in Table 1, this data set consisted of 60 frames collected with an oscillation width of 1.0° per image, detector distance of 350 mm, exposure time of 1.0 s/image, and transmission of 10.0 %. The data were processed using HKL2000 to 2.5 Å resolution, with I+ and I-treated as equivalent reflections during scaling.
Data from crystals of S352A and S352L were collected at 24-ID-C using continuous vector scanning, in which the crystal is translated after each image to reduce radiation damage (Table 1). The S352A data set consisted of 68 frames with an oscillation width of 1.0° per image, detector distance of 275 mm, exposure time of 1.0 s/image, and a transmission of 50%. The S352L data set comprised 50 frames with an oscillation width of 1.0° per image, detector distance of 320 mm, exposure time of 1.0 s/image, and a transmission of 100.0 %. The data were integrated with XDS35 and scaled with SCALA36 via CCP4i.37
A 1.3 Å resolution data set for the MmP5CDH-sulfate complex was collected at 24-ID-C (Table 2). The data set consisted of 120 frames collected with an oscillation width of 1° and detector distance of 150 mm. The data were processed with HKL2000.
Data for the MmP5CDH-Glu and MmP5CDH-NAD+ complexes were collected at beamline 4.2.2 of the Advanced Light Source (Table 2). Each data set consisted of 360 frames with an oscillation width of 0.5°, detector distance of 100 mm, and exposure time of 1 s. These data sets were processed with XDS and SCALA.
Phasing and refinement
The structure of HsP5CDH was determined using SAD phasing based on data collected from crystals of Se-Met HsP5CDH. The phasing potential of each data set was analyzed with the HKL2MAP38 interface to the SHELXC/D/E programs.39-41 Promising data sets were input to PHENIX AutoSol for SAD phasing, density modification, and automated building calculations.42 For the best data set, 30 of the expected 36 Se sites (i.e., four HsP5CDH molecules per asymmetric unit) were identified, which resulted in a figure of merit of 0.39 for reflections to 2.85 Å resolution. Density modification, which included 4-fold non-crystallographic symmetry (NCS) averaging, increased the figure of merit to 0.69. The model from automated building included 1790 residues (10194 atoms) and had an R-factor of 0.48 and map-model correlation coefficient of 0.59. PHENIX AutoBuild42 was used for phase extension to 2.5 Å resolution and additional model building. The resulting model included 1751 protein residues and 505 water molecules, with Rcryst of 0.32, Rfree of 0.37, and map-model correlation coefficient of 0.72.
The protein part of the model from automated building was used as the starting point for several rounds of manual building in COOT43 and simulated annealing refinement against the 2.5 Å resolution data set in PHENIX.44 The B-factor model used during refinement consisted of an isotropic B-factor for each non-hydrogen atom plus one TLS group per chain. Four-fold NCS restraints were used during refinement. The final structure served as the starting point for NCS-restrained refinement of the S352A and S352L structures. Refinement statistics are listed in Table 1
The 1.3-Å structure of MmP5CDH complexed with sulfate was determined using molecular replacement as implemented in the programs MOLREP45 and PHASER46 with the HsP5CDH dimer serving as the search model. Several rounds of model building in COOT and refinement in PHENIX were performed. Initially, the B-factor model consisted of an isotropic B-factor for each non-hydrogen atom and one TLS group per protein chain, but anisotropic B-factors were used during the final few rounds of refinement. The refined MmP5CDH-sulfate structure was the starting point for refinements of the 1.5 Å resolution glutamate and NAD+ complexes. For these two structures, the B-factor model consisted of an isotropic B-factor for each non-hydrogen atom and one TLS group per protein chain. Refinement statistics are listed in Table 2.
Analytical ultracentrifugation
The quaternary structure of HsP5CDH in solution was analyzed using equilibrium analytical ultracentrifugation. Data were acquired at 20 °C using a Beckman XL-I Optima analytical ultracentrifuge equipped with an An50Ti rotor. Prior to centrifugation, a protein sample having concentration of 0.8 mg/mL was dialyzed into a buffer containing 50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM THP, and 5 % glycerol at pH 8.0. Absorbance data (λ = 278 nm) were collected at three protein concentrations (0.2 mg/mL, 0.4 mg/mL, 0.8 mg/mL) and three rotor speeds (4000 rpm, 8000 rpm, 12000 rpm). The nine sets of data were fit globally to a single-species model (Eq. 9 of Lebowitz et al.47). A solvent density of 1.02 g/cm3 and partial specific volume of 0.74 cm3/g were used in the calculations. The 68 % confidence interval for the molar mass was estimated by analyzing the χ2 surface using the F-statistic.
Isothermal titration calorimetry
The binding of NAD+ to HsP5CDH, S352A, and S352L was studied at 25°C using a VP-ITC calorimeter (MicroCal, LLC). Prior to the titration, the protein sample was dialyzed in 50 mM Tris, 50 mM NaCl, 0.5 mM THP, 0.5 mM EDTA, and 5 % glycerol at pH 8.0. The protein concentration was estimated using absorbance with a theoretical extinction coefficient of 1.1 A /mg. NAD+ 280 was dissolved in the dialysate, and the pH was adjusted to that of the dialyzed protein sample. The concentration of NAD+ was estimated by absorbance using an extinction coefficient of 16.9 A259/mM. The protein and NAD+ solutions were loaded into the sample cell and buret, respectively. Following thermal equilibration, 20 μl aliquots of the titrant were injected into the sample cell at 240-s intervals with a stirring speed of 250 rpm. A pre-injection of 4 μl was used and discarded during fitting. Fitting of the data to a single-site binding model was performed using Origin software.
Kinetic characterization
P5CDH activity was measured at 20 °C by monitoring the production of NADH at 340 nm. The assay buffer contained 0.1 M sodium phosphate (pH 7.0) and 1 mM EDTA. The enzyme concentration was 11 μg/ml (0.16 μM). Specific activity is expressed as μmoles of NADH produced per minute per mg of enzyme. Initial velocity data were collected using NAD+ concentrations from 1 to 1500 μM at different fixed concentrations of a 50/50 DL-P5C mixture with L-P5C concentrations from 10 to 500 μM. Data were globally fitted to a kinetic scheme for the Theorell-Chance mechanism as shown in Supplemental Information. The fitted rate constants and kinetic parameters for HsP5CDH are reported in Table S1.
Tryptophan fluorescence quenching
An estimate of NAD+ binding to HsP5CDH was performed to better constrain the k−1/k1 ratio in the global fitting analysis shown in Fig. S6. HsP5CDH (1 μM) in 0.1 M phosphate buffer (pH 7.0, 1 mM EDTA) was excited at 295 nm and the fluorescence emission maximum at 330 nm was recorded. Quenching of tryptophan fluorescence by NAD+ (0 - 30 μM) was monitored at 330 nm and the emission was normalized to represent fractional quenching. Increasing amounts of NAD+ were added to the HsP5CDH solution allowing for a 2 min equilibration time after each addition of NAD+. The tryptophan fluorescence quenching data from the NAD+ titration was then simulated with Kinetic Global explorer software and fitted globally along with the kinetic data to better constrain the K for NAD+ binding d to HsP5CDH (Fig. S6).
PDB accession numbers
Coordinates and structure factor amplitudes have been deposited under the following accession codes: HsP5CDH, 3V9G; S352A, 3V9H; S352L, 3V9I; MmP5CDH-sulfate, 3V9J; MmP5CDH-Glu, 3V9K; MmP5CDH-NAD+, 3V9L. The 3V9G deposition includes the anomalous data that was used for SAD phasing.
Supplementary Material
Highlights.
The structures of human and mouse P5CDH are determined using Se-Met SAD phasing.
These are the first structures of any eukaryotic P5CDH.
The impact of the HPII-associated S352L mutation is examined.
S352L is inactive and fails to bind NAD+.
The structure of S352L is determined.
Mutation of Ser352 to Leu profoundly alters the active site of human P5CDH.
Acknowledgements
This research was supported by the NIH grant GM065546 and the National Center for Research Resources Grant P20 RR-017675. We thank Dr. Jay Nix and Dr. Jonathan Schuermann for help with X-ray diffraction data collection and processing. Part of this research was performed at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Abbreviations used
- 3OH-P5C
Δ1-pyrroline-3-hydroxy-5-carboxylate
- ALDH
aldehyde dehydrogenase
- GSA
L-glutamate-γ-semialdehyde
- HPII
type II hyperprolinemia
- HsP5CDH
human Δ1-pyrroline-5-carboxylate dehydrogenase
- ITC
isothermal titration calorimetry
- MmP5CDH
mouse Δ1-pyrroline-5-carboxylate dehydrogenase
- OH-Glu
4-erythro-hydroxy-L-glutamate
- OH-GSA
4-hydroxyglutamate semialdehyde
- OH-POX
hydroxyproline oxidase
- P5C
Δ1-pyrroline-5-carboxylate
- P5CDH
Δ1-pyrroline-5-carboxylate dehydrogenase
- POX
proline oxidase
- PRODH
proline dehydrogenase
- S352A
human Δ1-pyrroline-5-carboxylate dehydrogenase with Ser352 mutated to Ala
- S352L
human Δ1-pyrroline-5-carboxylate dehydrogenase with Ser352 mutated to Leu
- SAD
single-wavelength anomalous diffraction
- TtP5CDH
Thermus thermophilus Δ1-pyrroline-5-carboxylate dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu. Rev. Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Borchert GL, Donald SP, Diwan BA, Anver M, Phang JM. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res. 2009;69:6414–22. doi: 10.1158/0008-5472.CAN-09-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valle D, Goodman SI, Harris SC, Phang JM. Genetic evidence for a common enzyme catalyzing the second step in the degradation of proline and hydroxyproline. J. Clin. Invest. 1979;64:1365–70. doi: 10.1172/JCI109593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem. Biol. Interact. 2003;143-144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42:7100–9. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- 6.Inagaki E, Ohshima N, Takahashi H, Kuroishi C, Yokoyama S, Tahirov TH. Crystal structure of Thermus thermophilus Delta1-pyrroline-5-carboxylate dehydrogenase. J. Mol. Biol. 2006;362:490–501. doi: 10.1016/j.jmb.2006.07.048. [DOI] [PubMed] [Google Scholar]
- 7.Efron ML. Familial Hyperprolinemia. Report Of A Second Case, Associated With Congenital Renal Malformations, Hereditary Hematuria And Mild Mental Retardation, With Demonstration Of An Enzyme Defect. N. Engl. J. Med. 1965;272:1243–54. doi: 10.1056/NEJM196506172722401. [DOI] [PubMed] [Google Scholar]
- 8.Baumgartner MR, Rabier D, Nassogne MC, Dufier JL, Padovani JP, Kamoun P, Valle D, Saudubray JM. Delta1-pyrroline-5-carboxylate synthase deficiency: neurodegeneration, cataracts and connective tissue manifestations combined with hyperammonaemia and reduced ornithine, citrulline, arginine and proline. Eur. J. Pediatr. 2005;164:31–6. doi: 10.1007/s00431-004-1545-3. [DOI] [PubMed] [Google Scholar]
- 9.Geraghty MT, Vaughn D, Nicholson AJ, Lin WW, Jimenez-Sanchez G, Obie C, Flynn MP, Valle D, Hu CA. Mutations in the Delta1-pyrroline 5-carboxylate dehydrogenase gene cause type II hyperprolinemia. Hum. Mol. Genet. 1998;7:1411–5. doi: 10.1093/hmg/7.9.1411. [DOI] [PubMed] [Google Scholar]
- 10.Scriver CR, Sly WS, Childs B, Beaudet AL, Valle D, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. 8th edit McGraw-Hill; New York: 2001. [Google Scholar]
- 11.Valle D, Goodman SI, Applegarth DA, Shih VE, Phang JM. Type II hyperprolinemia. Delta1-pyrroline-5-carboxylic acid dehydrogenase deficiency in cultured skin fibroblasts and circulating lymphocytes. J. Clin. Invest. 1976;58:598–603. doi: 10.1172/JCI108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyse AT, Netto CA. Behavioral and neurochemical effects of proline. Metab. Brain Dis. 2011;26:159–72. doi: 10.1007/s11011-011-9246-x. [DOI] [PubMed] [Google Scholar]
- 13.Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Metabolic and molecular basis of inherited disease. McGraw Hill; New York: 2001. pp. 1821–1838. [Google Scholar]
- 14.Jacquet H, Demily C, Houy E, Hecketsweiler B, Bou J, Raux G, Lerond J, Allio G, Haouzir S, Tillaux A, Bellegou C, Fouldrin G, Delamillieure P, Menard JF, Dollfus S, D’Amato T, Petit M, Thibaut F, Frebourg T, Campion D. Hyperprolinemia is a risk factor for schizoaffective disorder. Mol. Psychiatry. 2005;10:479–85. doi: 10.1038/sj.mp.4001597. [DOI] [PubMed] [Google Scholar]
- 15.Clelland CL, Read LL, Baraldi AN, Bart CP, Pappas CA, Panek LJ, Nadrich RH, Clelland JD. Evidence for association of hyperprolinemia with schizophrenia and a measure of clinical outcome. Schizophr. Res. 2011;131:139–45. doi: 10.1016/j.schres.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gogos JA, Santha M, Takacs Z, Beck KD, Luine V, Lucas LR, Nadler JV, Karayiorgou M. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat. Genet. 1999;21:434–9. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- 17.Felix D, Kunzle H. The role of proline in nervous transmission. Adv. Biochem. Psychopharmacol. 1976;15:165–73. [PubMed] [Google Scholar]
- 18.Takemoto Y, Semba R. Immunohistochemical evidence for the localization of neurons containing the putative transmitter L-proline in rat brain. Brain Res. 2006;1073-1074:311–5. doi: 10.1016/j.brainres.2005.12.064. [DOI] [PubMed] [Google Scholar]
- 19.Fremeau RT, Jr., Caron MG, Blakely RD. Molecular cloning and expression of a high affinity L-proline transporter expressed in putative glutamatergic pathways of rat brain. Neuron. 1992;8:915–26. doi: 10.1016/0896-6273(92)90206-s. [DOI] [PubMed] [Google Scholar]
- 20.He F, DiMario PJ. Drosophila delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDh) is required for proline breakdown and mitochondrial integrity-Establishing a fly model for human type II hyperprolinemia. Mitochondrion. 2011;11:397–404. doi: 10.1016/j.mito.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 1997;4:317–26. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 22.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–11. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 23.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–51. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 24.Lorentzen E, Hensel R, Knura T, Ahmed H, Pohl E. Structural Basis of allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde 3-Phosphate dehydrogenase from Thermoproteus tenax. J. Mol. Biol. 2004;341:815–28. doi: 10.1016/j.jmb.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Forte-McRobbie C, Pietruszko R. Human glutamic-gamma-semialdehyde dehydrogenase. Kinetic mechanism. Biochem. J. 1989;261:935–43. doi: 10.1042/bj2610935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009;387:20–9. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Ni L, Sheikh S, Weiner H. Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde dehydrogenase. J. Biol. Chem. 1997;272:18823–6. doi: 10.1074/jbc.272.30.18823. [DOI] [PubMed] [Google Scholar]
- 28.Inagaki E, Ohshima N, Sakamoto K, Babayeva ND, Kato H, Yokoyama S, Tahirov TH. New insights into the binding mode of coenzymes: structure of Thermus thermophilus [Delta]1-pyrroline-5-carboxylate dehydrogenase complexed with NADP+ Acta Cryst. 2007;F63:462–465. doi: 10.1107/S1744309107021422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farres J, Wang X, Takahashi K, Cunningham SJ, Wang TT, Weiner H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J. Biol. Chem. 1994;269:13854–60. [PubMed] [Google Scholar]
- 30.Larson HN, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J. Biol. Chem. 2007;282:12940–50. doi: 10.1074/jbc.M607959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hempel J, Kraut A, Wymore T. Gamma glutamyl semialdehyde dehydrogenase: simulations on native and mutant forms support the importance of outer shell lysines. Chem. Biol. Interact. 2009;178:75–8. doi: 10.1016/j.cbi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Doublie S. Preparation of selenomethionyl proteins for phase determinations. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 33.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 37.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 38.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. [Google Scholar]
- 39.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 40.Sheldrick GM. Macromolecular phasing with SHELXE. Z. Kristallogr. 2002;217:644–650. [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Rad. 2004;11:53–5. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 45.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 46.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–79. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss M. Global indicators of X-ray data quality. J. Appl. Cryst. 2001;34:130–135. [Google Scholar]
- 49.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. The PyMOL User’s Manual. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.