Abstract
Sequence recognition through base pairing is essential for DNA repair and gene regulation but the basic rules governing this process remain elusive. In particular, the kinetics of annealing between two imperfectly matched strands is not well characterized despite its potential importance in nucleic acids-based biotechnologies and gene silencing. Here we use single molecule fluorescence to visualize the multiple annealing and melting reactions of two untethered strands inside a porous vesicle, allowing us to quantify precisely the annealing and melting rates. The data as a function of mismatch position suggest that seven contiguous base pairs are needed for rapid annealing of DNA and RNA. This phenomenological rule of seven may underlie the requirement of seven nucleotides complementarity to seed gene silencing by small non-coding RNA and may help guide performance improvement in DNA and RNA-based bio- and nano-technologies where off-target effects can be detrimental.
Introduction
Double helix formation1 of nucleic acids has been under investigation for over six decades. Thermodynamic parameters have been determined from compiled data of temperature-induced melting of DNA duplex and theoretical analysis2,3, allowing the prediction of melting temperatures Tm with 2°C accuracy. The equilibrium constant, Kd, defined as the ratio of the rates of melting, koff, and annealing, kon, can also be determined. However, the determinants of individual rates are still poorly understood due to the difficulty in directly observing the annealing and melting reactions.
kon could be deduced from changes in diffusion times4–6, from fluorescence resonance energy transfer (FRET) analysis in bulk7,8, through relaxation analysis following electric shock9 or temperature jumps10–12 and by nuclear magnetic resonance13. Recently, single molecule techniques have enabled the determination of opening and closing rates for DNA and RNA hairpins12,14,15 or oligonucleotides tethered inside membrane pore proteins16 but not the observation of freely diffusing intermolecular reaction. Moreover, single molecule mechanical studies gave extrapolated zero-force koff 100–10,000 folds different from the fluorescence-based estimate17. In none of the previous studies was the effect of base pair mismatches on the annealing and melting kinetics examined despite the likelihood that the kinetics may play an important role in a variety of cellular processes where two slightly mismatched oligonucleotides interact.
Here, we aimed to quantify precisely the effect of a single base pair mismatch on the annealing and melting rates between two untethered DNA or RNA. We developed an assay based on single molecule FRET18 that can directly observe multiple rounds of melting and annealing reactions of a pair of DNA or RNA strands freely diffusing inside a porous vesicle. Confinement by the vesicle 19,20,21,22 enables us to observe single molecule reactions even when the Kd is as high as 100 μM, which cannot be achieved by conventional methods. We observe that a single base pair mismatch can cause over 3 orders of magnitude change in Kd depending on the mismatch position. koff increased gradually as the mismatch was placed closer to the middle of the sequence whereas kon exhibited a response more like a step function to the mismatch position such that preventing 7 contiguous base pairs resulted in up to 100 times lower annealing rate. These results suggest that at least 7 contiguous Watson-Crick base pairs are necessary for rapid duplex formation.
Results
Kinetics of the full 9 bp DNA-DNA interaction
First, we designed two 9 nt long, complementary DNA strands (Fig. 1a) with Tm near room temperature2,3,23,24 and void of secondary structures and dinucleotide repeats. The two DNA strands are end-labeled fluorescently with Cy3 (donor) and Cy5 (acceptor) so that their proximity can be detected using FRET. We used a vesicle encapsulation method described earlier which allows exchange of ions while keeping the nucleic acid oligomers within21 (see Methods section). Only those vesicles with fluorescence intensities and subsequent (single step) photo-degradation consistent with one donor and one acceptor were included in the analysis.
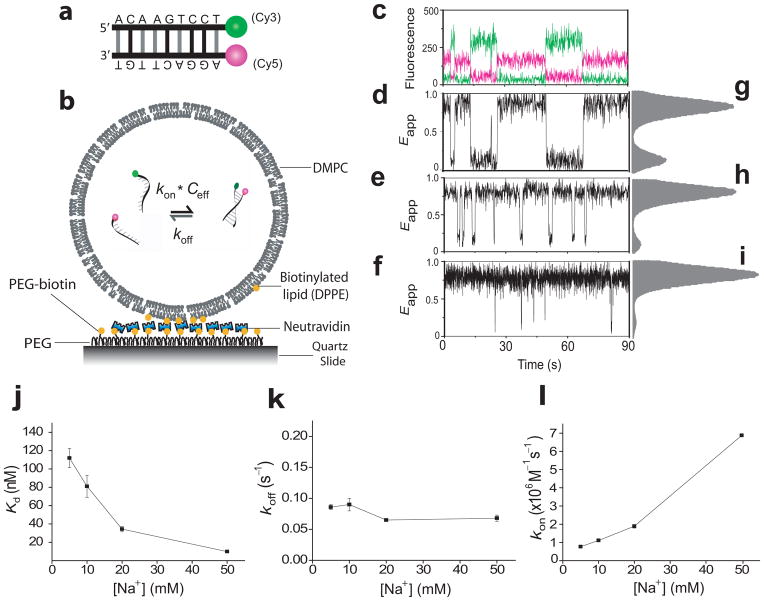
Figure 1.
Single-molecule porous vesicle encapsulation assay. (a) Schemes for the 9 bp DNA duplex and (b) DNA encapsulation; (c–l) Salt dependent dynamics for 9 bp DNA in 200 nm vesicles: single molecule time traces (c, d, e, f), and histograms from over 100 vesicles (g, h, i), at 5 mM (c, d, g), 20 mM (e, h), and 50 mM Na+ (f, i). Kd, koff, and kon vs [Na+] (j, k, l); 541 vesicles were used to calculate the rates in j, k and l. All error bars represent standard error from triplicate experiments at 23 °C. In c, green curve denotes the donor intensity and red the acceptor intensity.
Figure 1 shows single vesicle time traces and apparent FRET efficiency (Eapp) histograms obtained under various NaCl concentrations, each condition representing at least three different preparations of encapsulated samples. The lower salt conditions show two clear peaks centered around Eapp = 0.85 and 0.1 (Figs. 1g–i), corresponding to the annealed and melted states, respectively. Time trajectories of single molecule FRET (Figs. 1d–f) show repetitive transitions between two Eapp states due to multiple annealing and melting transitions. Via dwell time analysis, we determined the average dwell times of the high and low Eapp states, τhigh and τlow, respectively. koff is given by τhigh−1 whereas kon is obtained as (τlow ceff)−1. The effective concentration ceff is given by 6/NAπd3, where d is the vesicle diameter in decimeter estimated as the filter diameter used in the vesicle preparation (see Methods section) and NA is Avogadro’s number.
Kd≡ koff/kon decreases by an order of magnitude when [Na+] increases from 5 mM to 50 mM (Fig. 1j). Kd vs. [Na+] agrees well with a logarithmic function derived from the unified thermodynamic database for 10 mM to 50 mM salt concentrations19 (Supplementary Fig. 1a–b), and also for the lowest concentration, 5 mM, after accounting for the effects of buffer electrolyte (Supplementary Fig. 1c). This dependence of Kd on salt is therefore attributable to the well known trend2,10–12 of increased duplex stability with increasing ionic strength. The salt-dependence of kon and koff, measured independently in this study, reveal that the salt effect on Kd mostly originates from rapidly increasing kon (Fig. 1l) consistent with previous NMR studies13, and we see relatively little variation in koff (Fig. 1k). The large dependence of kon on salt concentration is attributable to effective charge screening of the electro-negative DNA backbone.
Potential effects of the fluorescent labels were tested by comparing with a construct with the labels on the opposite ends of the duplex. The cyanine dyes stack on the terminal base pair of the duplex25, with possible stabilization of the double stranded DNA. At 10 mM Na+, kon for the original 9 bp duplex with labels at the same side is 1.1 (± 0.1)×106 M−1s−1 (Fig. 1l) which is well within the range of previous estimates of 104 to 107 M−1s−1 for duplexes varying from 8 bp to 20 bp (ref. 4–6,13). koff under the same condition is 0.1 (±0.01) s−1 (Fig. 1k). When the dyes were positioned at opposite ends, we indeed observed a factor of 2 decrease in koff (0.05 (±0.01) s−1 vs. 0.10 (±0.01) s−1) while kon remained unaffected (1.0 (±0.1) ×106 M−1s−1 vs. 1.1 (±0.1) ×106 M−1s−1) (Supplementary Fig. 2). The effect of fluorescent labeling therefore must be negligible compared to the orders of magnitude variations we report below for mismatched sequences.
Effect of single base pair mismatches on DNA
We introduced a single base pair mismatch by changing the A-T base pair at one end of the 9 bp duplex into T-T mismatch, and observed a factor of ~ 2.5 increase in Kd (Fig. 2a). The decrease in stability is mostly due to a factor of 3.5 increase in koff (Fig. 2b). There is however an increase by a factor of 1.5 in kon (Fig. 2c), which we cannot explain.
Figure 2.
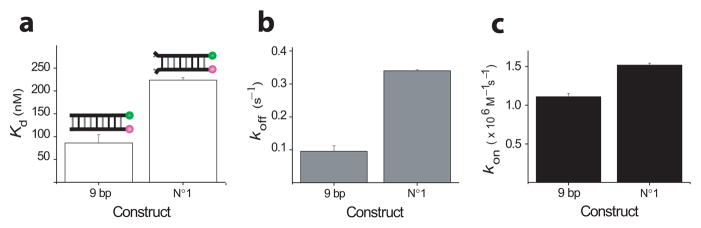
The effect of terminal base pair mismatch. (a) Kd, (b) koff, and (c) kon at 10 mM Na+; A total of 309 vesicles were used to calculate the rates in this figure.
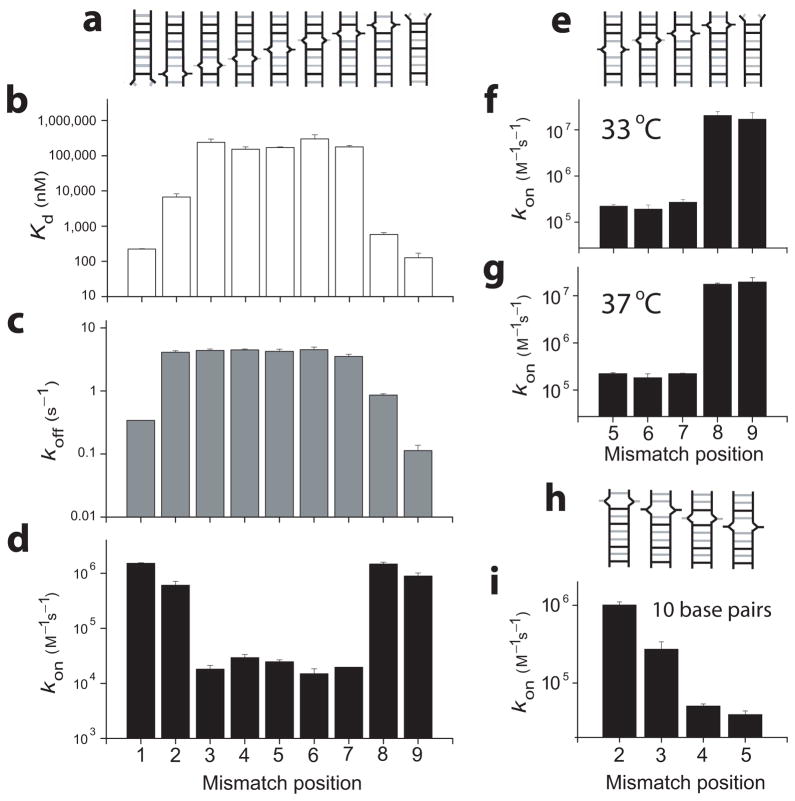
We then proceeded to characterize the effect of a mismatch at each of the nine possible positions. The constructs were designed similarly to the 1st bp mismatch above by changing a single base from the original 9 bp sequence; for example in the 2nd bp the original C-G was changed to G-G mismatch (Fig. 3a and Supplementary Table 1). For each construct, the experiments were first attempted in 200 nm diameter vesicles, but as the mismatch makes the DNA-DNA interactions weaker, some duplexes were not well captured at the lower concentrations required for capturing < 1 duplex per vesicle; in those cases, we increased the DNA concentrations and used vesicles of smaller diameters (100, 50, and 30 nm). The same DNA construct in different vesicle sizes shows similar τhigh (and therefore similar koff) values but very different τlow values. However, after adjusting for the different vesicle volumes, we obtained nearly identical kon values (Supplementary Fig. 3).
Figure 3.
The effect of DNA mismatch position. (a, e) Schemes of mismatched constructs; (b) Kd, (c) koff, and (d) kon were measured at 23 °C, 10 mM Na+; (e) For mismatch positions 5 through 9, kon was additionally measured at 33 °C, 150 mM Na+ and (f) at 37 °C, 200 mM Na+; A total of 723 vesicles were used for b, c, and d, 937 vesicles in f, 511 vesicles in g, and 697 vesicles in i.
Depending on the position of a single bp mismatch, there is a variation by over a factor of 3,000 in Kd with the mismatch having a bigger negative impact on the duplex stability when it is closer to the middle (Fig. 3b). The equilibrium constant Kd is higher than 100 μM for the middle mismatches, which would have made it difficult to perform kinetic analysis using conventional single-molecule methods. A relative high penalty cost of a middle mismatch compared to an end mismatch is consistent with observations from previous thermodynamic analyses26,27. We note that changing the middle construct (mismatch position 5) from a G-G to a C-C mismatch resulted in a duplex that we were unable to capture with adequate encapsulation yield even in our smallest vesicle size, likely because the middle C-C mismatch is more disruptive than the corresponding G-G mismatch as shown by previous thermodynamic analyses27,28.
The variation in koff was ~ 30 fold, and the general trend shows a gradual, bell-shaped variation, peaking with a mismatch toward the middle of the duplex (Fig. 3, Supplementary Fig. 4). The only outlier from the symmetric bell shape is the 2nd bp mismatch which showed a four times higher koff compared to the reciprocal mismatch on the 8th bp; this effect may arise from the fact that the 2nd bp is the only G-C pair in the first half of the duplex.
The kon vs mismatch position result shows the most striking effect (Fig. 3d). For mismatches on the 1st and 2nd bp, kon is of the same order of magnitude as with the full 9 bp construct (~ 106 M−1s−1), but moving the single mismatch to the 3rd bp reduced kon by a factor of ~ 100. The same observation is made comparing the mismatches on the other end of the duplex: for the mismatch on the 9th and 8th bp comparable kon values are measured, then a factor of ~ 100 decrease in kon for the mismatch on the 7th bp. The position of this transition remained the same at different conditions: room temperature with 10 mM Na+ (Fig. 3d), 33 °C with 150 mM Na+ (Fig. 3f), and 37 °C with 200 mM Na+ (Fig. 3g), and after testing for possibly missed transitions using a hidden Markov analysis on the FRET trajectories29 (Supplementary Fig. 4i–k).
Furthermore, there is little variation of kon for mismatches between the 3rd and the 7th bp. Overall, DNA constructs with mismatches on the 1st, 2nd, 8th, and 9th bp (with 7 or more contiguous base pairs) have up to two orders of magnitude higher kon than the constructs with a mismatch position between the 3rd and 7th bp (< 7 contiguous base pairs). These results suggest 7 bp cooperativity during duplex formation (but not during melting), whereby a single mismatch that prevents 7 contiguous base pairs results in the lowest annealing rate. The necessity for at least 7 contiguous base pairs remains valid over a range of biologically relevant temperature and salt conditions.
As a further test of the apparent requirement of 7 contiguous base pairs for rapid annealing, we designed 10 nt constructs of unrelated sequence. When a single mismatch reduces the maximal contiguity to 6 bp or 5 bp we obtained the lowest annealing rate kon ~104 M−1s−1 (Fig. 3i), and kon increased by about one order of magnitude with 7 bp contiguity (see Supplementary Figure 4 for koff and Kd). The lowest annealing rate was achieved when the mismatch is at least 4 nt away from the end in contrast to the 9 nt constructs which gave the lowest annealing rate when the mismatch is placed at the 3rd nt from the end, indicating that the rule of seven does not arise from an end effect. In the case of the 10 nt sequence, however, 8 bp contiguity further increased the annealing rate by a factor of ~ 3, suggesting that 7 contiguous base pairs are necessary but not always sufficient for the highest annealing rate.
Effect of single base pair mismatches on RNA
The rule of seven, suggested by our measurements on DNA, is reminiscent of the empirical observation that mammalian targets of micro RNA (miRNA) can be predicted by searching for conserved 7 nt matches 30–32 and that messenger RNAs are under selective evolutionary pressure to conserve (targets) or avoid (non-targets) 7 nt binding sites that match miRNAs seed sequence 33,34. In the canonical model for miRNA target recognition35,36 the target recognition is suggested to be driven by Watson-Crick base pair interactions of residues 2 to 8 of the 5′ portion of the miRNA, and consequently the protein in the silencing complex is assigned the role of preorganizing the geometry so as to favor the optimal presentation of this heptameric core for hybridization30. If the naked RNA molecules themselves require 7 contiguous base pairs for rapid annealing, we may deduce that the proteins in the silencing complex may have evolved to utilize such an intrinsic property of nucleic acids alone. We therefore tested if there exists a large difference in annealing rate between 7 and 6 contiguous base pairs in the case of RNA using a well-known miRNA as an example.
We synthesized the 8 nt sequence corresponding to miR125 seed in human, a homolog to Lin-4 in C. Elegans37, labeled with Cy5 at the 3′ end. Its complementary target sequence derived from human p53 gene38, with the 1st bp mismatched (U-U) and 7 contiguous bp, was labeled at the 5′ end with Cy3 (#1RNA_7cont.bp). It was compared with the second construct with a (C-U) mismatch in the 2nd bp (#2RNA_6cont.bp, see Supplementary Table 1).
Figure 4 shows the average rates obtained from the spontaneous melting and annealing of individual RNA duplexes at 5mM Na+ inside porous vesicles. In terms of Kd, the 7 contiguous bp RNA is a factor of ~ 450 times more stable than 6 contiguous bp RNA (Fig. 4a). koff values were 0.014 (±0.004) s−1 for 7 bp RNA and 0.14 (±0.03) s−1 for 6 bp RNA, exhibiting difference of a factor of 10 (Fig. 4b). The predominant difference in stability between the two RNA constructs, however, comes from a difference of a factor of 45 in kon, with 3.4 (±0.8)×106 M−1s−1 for 7 bp RNA, and 7.6 (± 1.2)×104 M−1s−1 for 6 bp RNA (Fig. 4c). These results support further the requirement of 7 contiguous Watson-Crick base pairs for rapid annealing of two oligonucleotides.
Figure 4.
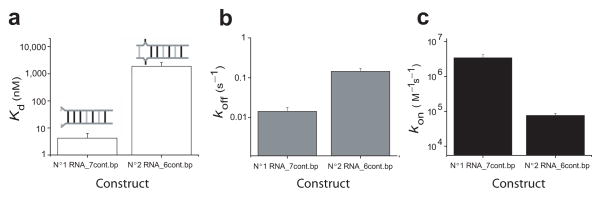
The effect of mismatch position on RNA. (a) Kd, (b) koff, and (c) kon for 8 bp RNA constructs with 7 and 6 contiguous base pairs at 23 °C, 5 mM Na+; A total of 228 vesicles were used to calculate the rates in this figure.
Discussion
In the ‘zipper-up’ model39, DNA annealing is said to proceed first through a slow nucleation step followed by microseconds time scale zipping. One implication of the model, the so-called ‘all or none’ aspect requiring that the strands should be either fully annealed or fully unzipped within our 30 ms time resolution, is consistent with the 2-states nature of the single molecule FRET trajectories observed. However, thermodynamic estimation suggests that 2 to 3 base pairs should be enough for this nucleation step in the zipper-up model39. Therefore, we do not believe that this model is directly applicable to the 7 bp cooperativity observed in this study.
A heptamer core was previously postulated as the optimal compromise in miRNA target recognition: a larger core would impose topological difficulties for the silencing complex whereas a smaller core would result in a drop in the initial Watson-Crick base pairing interaction. Recent crystal structures of Argonaute yielded a picture consistent with this model, with the protein prearranging the core residues in A-form helix40. In vivo investigation has shown that 6 mer seed match was not sufficient for miRNA regulation even when inserted in multiple copies in the 3′ UTR of messenger RNAs, whereas a single insertion of a 7 mer was sufficient for gene silencing41. However, previous studies could not test if such a difference was intrinsically due to a systematic drop in the formation rate of miRNA-mRNA helix or due to the effect of the proteins in the silencing complex. Our results suggest that specificity in the microRNA target recognition may arise from the intrinsic properties of oligonucleotides themselves that require 7 contiguous base pairs for rapid annealing.
Many questions remain to be addressed, particularly regarding the origin of the 7 bp cooperativity and the limits of its validity. Different approaches providing more atomistic details, for example using molecular dynamics simulation, may prove useful in elucidating the molecular mechanism. A key advantage of our porous vesicle encapsulation assay is that the oligonucleotides are untethered, and therefore free to adopt any preferred geometry during the double helix formation. However, the method is laborious compared to measurements using tethered oligonucleotides. Although we have tested three unrelated sequences in DNA and RNA, these represent a tiny fraction of the sequence space and we do not know yet whether similar rules will hold for oligonucleotides longer than 10 nt. New technical developments to improve the co-encapsulation efficiency or to parallelize the experiments may improve the data throughput, allowing us to sample a much bigger sequence space and to test the validity and generality of the rule more thoroughly.
It is possible that other instances of 7 bp cooperativity occur in other cellular processes, for example during transcription initiation and termination. Moreover, understanding the rules governing the rate of duplex formation in the presence of a mismatch should also be useful for hybridization based applications in biotechnology where off-target effects can be detrimental. Examples include improvement on target specificity of small interference RNA (siRNA), DNA-based computing, DNA-based nanotechnologies and microarrays. In many cases, the overall hybridization efficiency would be governed by the annealing kinetics (kon) instead of the thermodynamic stability (e.g. Tm, as most commonly considered) because even in the presence of mismatches, melting can be slower than the relevant time scale.
Methods
The encapsulation and single molecule detection methods were as described previously21 with the following optimizations.
Vesicle Encapsulation
Lipid films were prepared by mixing biotinyl cap phosphoethanolamine with dimyristoyl phosphatidylcholine (DMPC) dissolved in chloroform (1:100 molar ratio) then dried in vacuum for ~ 1 h. The lipids were hydrated with solution containing DNA or RNA according to the following specifications:
Solution condition for DNA encapsulations: 500 mM NaCl, 10 mM MgCl2, and 25 mM Tris pH 8.0
Solution condition for RNA encapsulations: 500 mM NaCl, and 25 mM Tris pH 8.0 (MgCl2 absence intentional, and special care was taken to minimize potential RNase contamination during sample preparation).
-
Oligo concentrations:
For the full 9 bp, 9 bp with dyes on separate ends, #1, #9 DNA constructs: successful encapsulations were conducted at 400 nM oligo concentrations (for extrusion with 200 nm filter pores) or 3.2 μM oligo concentrations (for extrusion with100 nm filter pores).
For N°2 and #8 DNA constructs: successful encapsulations were conducted at 3.2 μM oligo concentrations (for extrusion100 nm filter pores).
For N°3, N°4, N°5, N°6, N°7 DNA constructs: successful encapsulations were conducted with 6.4 μM oligo concentrations (for extrusion with either 50 nm or 30 nm filter pores)
For N°1RNA_7cont.bp construct: successful encapsulations were conducted with 100 nM oligo concentration (for extrusion with 200 nm filter pores)
For N°2RNA_6cont.bp construct: successful encapsulations were conducted with 1 μM oligo concentration (for extrusion with 50 nm filter pores)
After hydration, the mixture of lipid and nucleic acids was frozen in liquid nitrogen and thawed about 7 times to create large unilamellar vesicles. The solution was then processed through an extrusion set with a filter with small pores to create the small unilamellar vesicles of the desired size (see specifications above for the filter pore sizes used for each construct). Dynamic light scattering measurements gave vesicle size estimates that are in line with the filter pore diameter used (Supplementary Fig. 3). The effective co-encapsulation yield (defined as the fraction of vesicles detected with a pair of donor and acceptor among all vesicles with any signal) was ~ 20% for 200nm and 100 nm vesicles. The co-encapsulation yield for 50 nm and 30 nm depended on the stability of the construct used and was more variable; only those preparations with 10% or better yield were included in the analysis. Higher co-encapsulation yield may be obtained if stock-level concentrations of labeled DNA could be used to match the expected local concentrations after encapsulation: for example ~ 25 μM for 50 nm vesicles, and ~ 117 μM for 30 nm vesicles. However, in practice, such high concentrations of labeled DNA and RNA were unreasonable due to high cost.
All lipids, extrusion sets and vesicle size filters were obtained from Avanti Polar Lipids, Inc (Alabaster, AL 35007). All oligonucleotides (DNA and RNA) were custom-designed and purchased from Integrated DNA Technology (Coralville, IA 52241). Additional information on DNA and RNA design is available online as Supplementary Table 1.
Single Molecule detection and analysis
We used total internal reflection fluorescence microscopy for imaging as described previously21. Imaging solution contains 1 mg/ml glucose oxidase, 0.04 mg/ml catalase, 0.8% dextrose, and saturated trolox (~ 3 mM) in 50 mM Tris. Because the transition temperature of DMPC is at room temperature, coexistence of liquid and gel phases of the lipids would result in membranes with pores wide enough for exchanging ions and small molecules but small enough to keep larger biomolecules like DNA oliomers inside the vesicles21. All solution exchanges were made at room temperature. Data acquisition was performed under various salt and temperature conditions as indicated in the text and figure captions.
We calculated the apparent FRET efficiency by Eapp= IA/(ID+IA) where ID and IA are the emission intensity of the donor and acceptor, respectively. Since multiple molecules may be confined in the same vesicle, the criterion set to determine the rates of bi-molecular interactions was that only vesicles with fluorescence consistent with one donor and one acceptor were selected; any additional criteria (e.g. encapsulation yield for successful experiment) were as described in the manuscript. To obtain the transition rates from repetitive reactions inside vesicles, we selected the region where both donor and acceptor were photoactive in the individual traces. Eapp of the selected region was then regarded as a two-state trajectory with a midpoint cutoff at Eapp ~ 0.5, and the individual dwell times were extracted for each low Eapp (< 0.5) and high Eapp (> 0.5) residence. The kinetic rates for each condition were calculated from the average dwell times, obtained from multiple vesicles. Alternatively, a previously described hidden Markov model29 has been adopted to test for events which might be missed because of short dwell times. For all rate calculations, the vesicle diameter is estimated to be the pore size of filter selected for vesicle extrusion (200 nm, 100 nm, 50 nm, or 30 nm).
Supplementary Material
Acknowledgments
We thank B. Okumus, R. Clegg, and Z. Bryant for critical suggestions. We acknowledge J. Chen, C. Joo, and members of Narry Kim Group for discussion on microRNA. We thank current and past members of the Ha Group for various suggestions. The project was supported by NIH Grants GM074526 and GM065367 and NSF grant 0822613 to T.H.; H.K was supported in part by grant (KRF-2006-352-C00019) of the Korean Research Foundation (Seoul, S.Korea).
Footnotes
Author contributions: IC and TH designed the initial experiments. IC and HK performed the experiments and analyzed the data. IC, HK, and TH wrote the manuscript.
The authors declare no conflict of interests.
References
- 1.Watson JD, Crick FHC. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004;33:415–40. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 3.Owczarzy R, et al. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004;43:3537–54. doi: 10.1021/bi034621r. [DOI] [PubMed] [Google Scholar]
- 4.Kinjo M, Rigler R. Ultrasensitive hybridization analysis using fluorescence correlation spectroscopy. Nucleic Acids Res. 1995;23:1795–9. doi: 10.1093/nar/23.10.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wetmur JG, Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968;31:349–70. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- 6.Wetmur JG. DNA probes: applications of the principles of nucleic acid hybridization. Crit Rev Biochem Mol Biol. 1991;26:227–59. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- 7.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1988;85:8790–4. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkhurst KM, Parkhurst LJ. Kinetic studies by fluorescence resonance energy transfer employing a double-labeled oligonucleotide: hybridization to the oligonucleotide complement and to single-stranded DNA. Biochemistry. 1995;34:285–92. doi: 10.1021/bi00001a035. [DOI] [PubMed] [Google Scholar]
- 9.Porschke D. Short electric-field pulses convert DNA from “condensed” to “free” conformation. Biopolymers. 1985;24:1981–93. doi: 10.1002/bip.360241010. [DOI] [PubMed] [Google Scholar]
- 10.Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–40. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y, Montrichok A, Zocchi G. Length and statistical weight of bubbles in DNA melting. Phys Rev Lett. 2003;91:148101. doi: 10.1103/PhysRevLett.91.148101. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci U S A. 1998;95:8602–6. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braunlin WH, Bloomfield VA. 1H NMR study of the base-pairing reactions of d(GGAATTCC): salt effects on the equilibria and kinetics of strand association. Biochemistry. 1991;30:754–8. doi: 10.1021/bi00217a026. [DOI] [PubMed] [Google Scholar]
- 14.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–4. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–7. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 16.Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc Natl Acad Sci U S A. 2001;98:12996–3001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodside MT, et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci U S A. 2006;103:6190–5. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of the National Academy of Sciences. 1996;93:6264. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boukobza E, Sonnenfeld A, Haran G. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. The Journal of Physical Chemistry B. 2001;105:12165–12170. [Google Scholar]
- 20.Okumus B, Wilson TJ, Lilley DMJ, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophysical journal. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cisse I, Okumus B, Joo C, Ha T. Fueling protein–DNA interactions inside porous nanocontainers. Proceedings of the National Academy of Sciences. 2007;104:12646. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benitez JJ, et al. Probing transient copper chaperone-Wilson disease protein interactions at the single-molecule level with nanovesicle trapping. Journal of the American Chemical Society. 2008;130:2446–2447. doi: 10.1021/ja7107867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–94. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 24.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 1998;95:1460–5. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iqbal A, et al. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci U S A. 2008;105:11176–81. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petruska J, et al. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proceedings of the National Academy of Sciences. 1988;85:6252. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aboul-ela F, Koh D, Tinoco I, Jr, Martin FH. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T) Nucleic Acids Res. 1985;13:4811–24. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–77. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 29.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical journal. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 32.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farh KKH, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 34.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3,ÄA≤ UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 35.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 36.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nature Reviews Molecular Cell Biology. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 37.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 38.Le MT, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porschke D, Eigen M. Co-operative non-enzymatic base recognition III. Kinetics of the helix-coil transition of the oligoribouridylic - oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. Journal of Molecular Biology. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biology. 2005;3:0404–0418. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.