Abstract
Nemaline myopathy (NM), the most common non-dystrophic congenital myopathy, is a variably severe neuromuscular disorder for which no effective treatment is available. Although a number of genes have been identified in which mutations can cause NM, the pathogenetic mechanisms leading to the phenotypes are poorly understood. To address this question, we examined gene expression patterns in an NM mouse model carrying the human Met9Arg mutation of alpha-tropomyosin slow (Tpm3). We assessed five different skeletal muscles from affected mice, which are representative of muscles with differing fiber-type compositions, different physiological specializations and variable degrees of pathology. Although these same muscles in non-affected mice showed marked variation in patterns of gene expression, with diaphragm being the most dissimilar, the presence of the mutant protein in nemaline muscles resulted in a more similar pattern of gene expression among the muscles. This result suggests a common process or mechanism operating in nemaline muscles independent of the variable degrees of pathology. Transcriptional and protein expression data indicate the presence of a repair process and possibly delayed maturation in nemaline muscles. Markers indicative of satellite cell number, activated satellite cells and immature fibers including M-Cadherin, MyoD, desmin, Pax7 and Myf6 were elevated by western-blot analysis or immunohistochemistry. Evidence suggesting elevated focal repair was observed in nemaline muscle in electron micrographs. This analysis reveals that NM is characterized by a novel repair feature operating in multiple different muscles.
INTRODUCTION
In recent years, microarray studies have shed light on global gene expression patterns in a number of different neuromuscular disorders (1–6). Analysis of human clinical material, coupled with studies of murine models of various muscular dystrophies and myopathies, is leading to a new appreciation of the transcriptional basis for a variety of neuromuscular phenotypes. The ultimate goal remains the recognition of primary and secondary transcriptional and translational changes that occur during the course of the disease, and the determination of molecular pathways that will constitute promising therapeutic targets. Nemaline myopathy (NM) is one of the conditions starting to be explored at the transcriptional level. A slowly or non-progressive congenital myopathy, NM, is characterized by the presence of nemaline rods in affected muscle fibers, muscle weakness and absence of muscle regeneration. The clinical phenotype, muscle histology and gene mutations can be quite variable. Different forms of NM can range from severe neonatal lethal to mild adult onset, and histologically, there is a variation in the number, size and location of nemaline rods, as well as in the percentage of fast and slow fibers in affected muscles (7). Mutations have been described in five different genes that encode skeletal muscle thin filament proteins: alpha-skeletal muscle actin (ACTA1), nebulin (NEB), beta-tropomyosin (TPM2), alpha-tropomyosin slow (TPM3) and slow troponin T (TNNT1) (8). No absolute correlations exist between gene mutations and disease severity or any other phenotypic characteristic. However, gene expression analysis of a heterogeneous group of NM patients, in terms of disease severity, age and histology (none of whom had TPM3 mutations), revealed consistent changes in muscle energy metabolism (reduced), fibrosis (increased) and evidence for a partial regenerative response (5).
TPM3 mutations are a rare cause of NM in humans, counting for ~2–3% of cases (9–11). A mouse model of TPM3-related NM was recently generated by transgenic over-expression of a dominant negative mutation of this gene (Met9Arg) (10,12). Because most mouse muscles have a higher proportion of fast fibers (~90%) compared with humans (~33%), αTmslow(Met9Arg) was linked to the human skeletal actin (HSA) promoter that drives expression preferentially in 2B fibers of the mouse (13). The transgenic mice presented with all the main characteristics of NM as described for humans, including nemaline rods, increased slow/oxidative fiber content and muscle weakness, and therefore, to a large extent, they represent a phenocopy of the human condition. αTmslow(Met9Arg) mice exhibit a non-progressive increase in slow type 1 fibers and hypertrophy of fast type 2B fibers during the first few months, followed by the onset of muscle weakness at 7 months of age. Nemaline rods are located at the center and periphery of muscle fibers, and they commonly cluster around the Z-line in a similar fashion to the rods of patients with the TPM3(Met9Arg) mutation. The percentage of rod containing fibers increases between 2 and 12 months of age and varies considerably between different muscles. However, transgenic mice do not have an overt clinical phenotype, which is consistent with the mild form of the disease exhibited by patients carrying the TPM3(Met9Arg) mutation. In contrast to human muscles, cytoplasmic bodies and tubular aggregates were observed in the mouse superficial gastrocnemius, but not in any of the other mouse muscles examined.
The availability of a mouse model for NM provides a flexible and easily accessible system where multiple different skeletal muscles can be studied, sufficient amounts of tissue can be obtained and different therapeutic approaches can eventually be evaluated. To investigate the molecular pathways affected in the muscles of αTmslow(Met9Arg) mice, we analyzed the gene expression patterns of 12 488 genes and ESTs (U74Av2 Affymetrix GeneChips) in five different muscles: tibialis anterior (TA), gastrocnemius (GAS), plantaris (PLT), diaphragm (DIA) and extensor digitorum longus (EDL). Variation between the number of fibers with rods and the percent of slow/fast fibers in different muscles of the HSA-αTmslow(Met9Arg) mice warrants the study of a range of muscles (12). The specific muscles were chosen so as to include muscles presenting with varying levels of pathology and usage (e.g. DIA is a muscle in constant motion) while focusing on distal muscles, as TPM3 Met9Arg NM has a distal phenotype. Although, significant differences were seen in the transcriptional ‘response’ of different muscles to the mutation, the gene expression profiles of the different nemaline muscles were more similar to each other than between the same muscles in wild-type mice. Current findings are compared and contrasted with observations on human NM specimens. The data indicate an abrogation of normal muscle maturation and suggest the presence of focal muscle repair as key features of NM. Future studies will need to address whether focal repair is indeed a feature consistently present in the HSA-αTmslow(Met9Arg) mice and human patients.
RESULTS
To examine the transcriptional basis for the form of NM observed in αTmslowMet9Arg (‘NM’) mice, we conducted microarray analysis of three or four replicate total RNA pools from five different muscles (TA, PLT, GAS, DIA and EDL) each of NM and wild-type (‘WT’) control littermates. A total of 36 pools were analyzed, each of which contained RNA from three to five replicate littermates between 7 and 10 months of age. Labeled cRNA targets were hybridized to Affymetrix U74Av2 GeneChips, resulting in data for 12 488 annotated genes and ESTs.
Correlation coefficient analysis
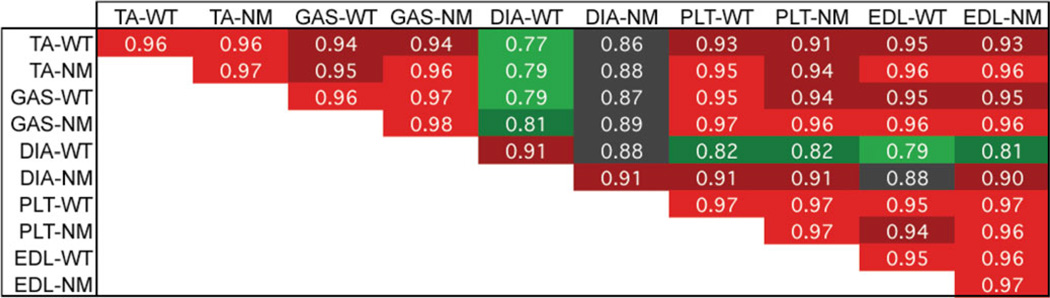
The correlation coefficients (r) for gene expression signals between all 36 mouse muscle pools (WT and NM) were calculated and the averages thereof were used to create a matrix of correlations between each of the 10 muscle/disease-state point (Fig. 1). The average r between all WT samples ranged from 0.77 (DIA-WT versus TA-WT) to 0.97 (different PLT-WT pools), whereas NM samples alone had an average r range from 0.88 (DIA-NM versus TA-NM) to 0.98 (different GAS-NM pools). The data sets from four of the five muscle types had high correlations with each other both in the WT and the NM state. DIA, however, was the exception with the lowest correlations to all other samples in its WT state. It is unlikely that this is associated with fiber-type differences between WT and NM DIA, as there were no significant expression changes in fast- or slow-fiber genes. Unexpectedly, NM DIA had higher correlations to all other WT and NM muscles than WT DIA.
Figure 1.
Correlation coefficients for all pairwise comparisons of data sets containing 12 488 probe set signals averaged for each muscle type/state. Heat map correlates with r-values in increments of 0.05. The DIA in WT (denoted ‘-WT’) mice is most dissimilar to the other skeletal muscles. NM transgenic (denoted ‘-NM’) DIA is more similar not only to other NM muscles but also to the other WT muscles.
The average r between WT and NM pools of the same muscle ranged from 0.88 (DIA) to 0.97 (GAS and PLT). In this instance, DIA again had the lowest correlation, suggesting that not only does it differ from other muscle types, but that it is also more profoundly affected by the expression of the mutant protein. Noise intrinsic to DIA may partly contribute but not fully explain these low inter-group correlations, as the average correlation coefficient within the same group of DIA specimens, be it WT or NM, was in both occasions higher (0.91). Taken together, our analysis indicates that the presence of the mutant alpha-tropomyosin slow protein reduces the dissimilarities in gene expression profile between muscles.
Significance analysis of microarrays
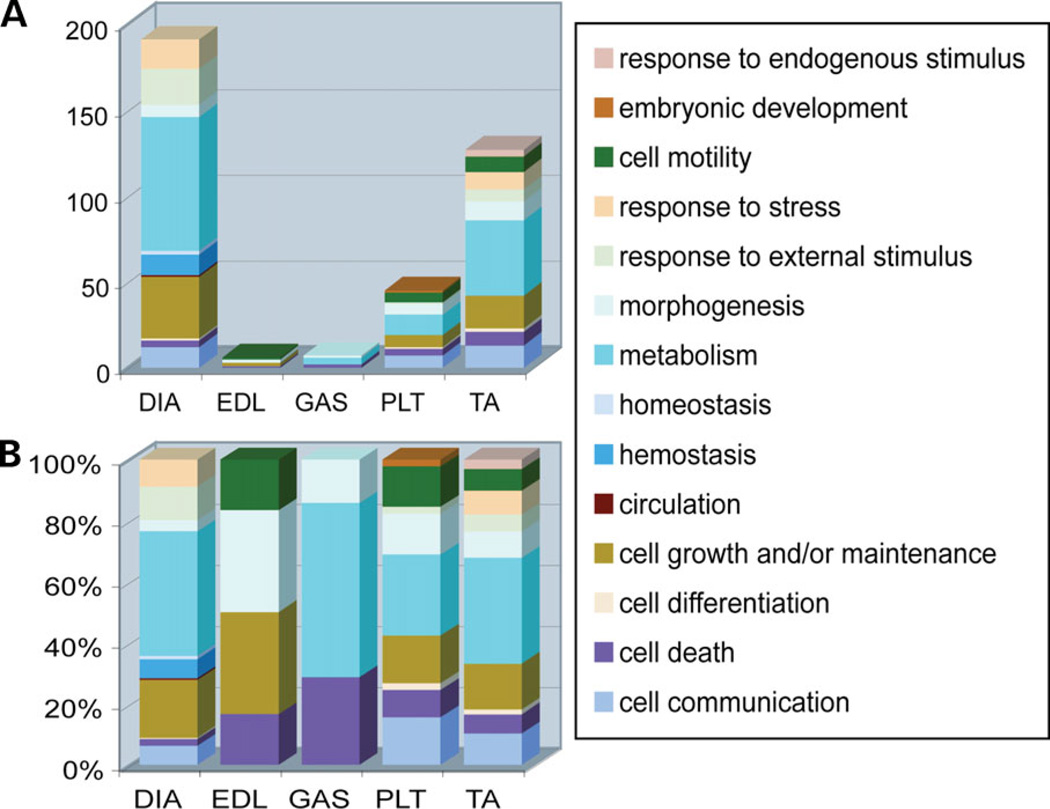
Significance analysis of microarrays (SAM) identified those probe sets with significantly different expression between NM and WT mice for each muscle type separately. The same fold cutoff (+ 1.5-fold change) and the same false discovery rate (FDR) cutoff (12%) were applied to all muscles, with the exception of EDL, where the lowest possible FDR was 24%. Probe sets with more than one-third absent calls, as determined by MAS5.0 (Affymetrix), were removed (Materials and Methods). The numbers of significantly changed probe sets, as well as their fold change, were highly variable between different muscle types: TA = 142 probe sets (fold-change range: 1.5–10.76), GAS = 8 (1.5– 2.73), DIA = 1386 (−7.74–6.68), PLT = 36 (1.5–3.67) and EDL = 4 (1.5–2.59) (Supplementary Material, Table S1). DIA was the most affected muscle with more than 10 times the number of significantly changed probe sets and a striking number of under-expressed probe sets (1380 out of 1386 were under-expressed in DIA as opposed to zero in other muscle types). The major molecular pathways represented by those probe sets are shown in Figure 2. It is also remarkable that despite the prominent pathological changes in NM EDL (12), only four probe sets met our statistical criteria for change, suggesting that the magnitude and number of gene expression changes in this muscle group was considerably smaller than for the other muscles studied.
Figure 2.
GO category analysis of genes considered significantly changed between WT and NM mouse muscles. All genes significantly changed in NM muscles were grouped in GO biological process categories on the basis of the function of their respective proteins. This graph shows the major GO categories implicated in each muscle studied when all significantly changed genes are considered. For DIA only, the top 200 significantly changed genes are tabulated. The results are presented as (A) the actual numbers of significantly changed probe sets in each muscle type and (B) the percent of significantly changed probe sets for each muscle type.
The lists of significantly changed probe sets for each muscle were compared, to identify probe sets commonly changed across all muscles and therefore likely to be highly related to the presence of the mutant protein in a muscle-independent manner. No probe sets were commonly changed across all five muscles but two probe sets (97786_at and 103084_at representing Ankrd2 and Csrp3, respectively) were common across four muscles (excluding EDL). Two more probe sets were common between TA, GAS and PLT (96167_at and 102213_at representing Bag3 and Xin, respectively) and one probe set was common between TA, DIA and PLT (103526_at, Padi2). Overall, the most similarities were seen between TA and PLT (19 probe sets), whereas at the other end of the spectrum, EDL had no probe sets significantly changed in common with GAS, DIA or PLT (Supplementary Material, Table S2).
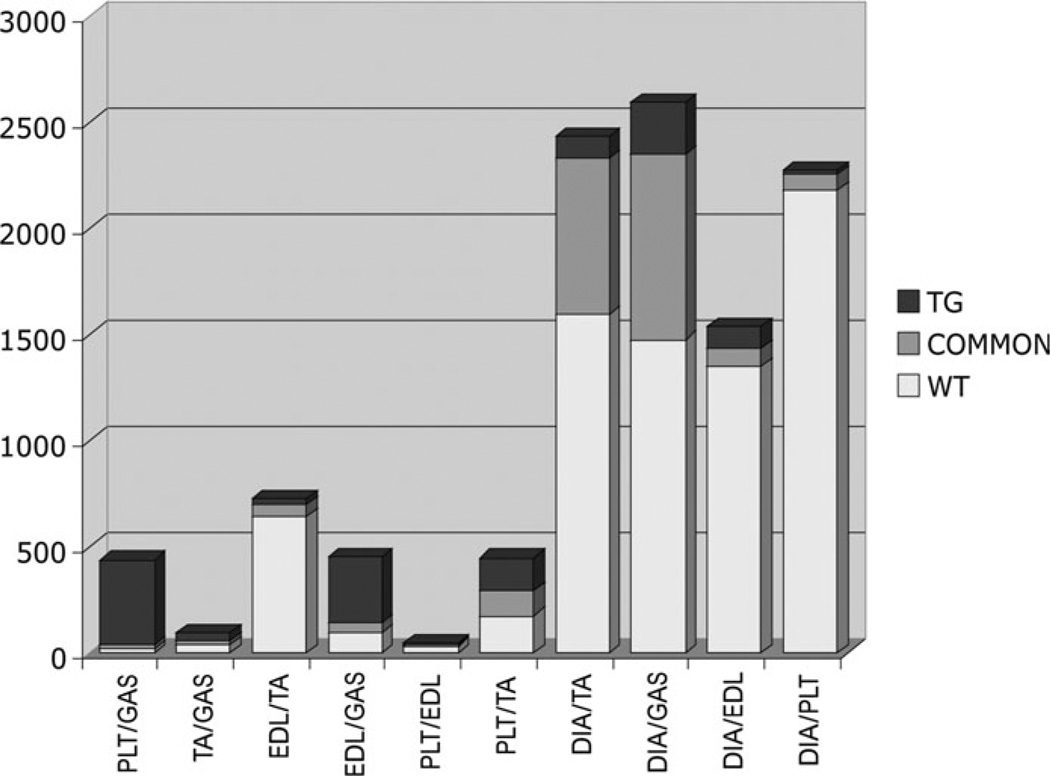
All WT muscle types were compared with each other: the largest number of differences was seen between DIA and all other muscles (between 1435 and 2350 probe sets were significantly changed), followed by TA versus EDL (696 probe sets) and TA versus PLT (294 probe sets). All NM muscles were also compared with each other: the largest number of differences was seen in DIA versus GAS (1118 probe sets), followed by DIA versus TA (845 probe sets) and PLT versus GAS (422 probe sets) (Fig. 3).
Figure 3.
Numbers of significantly changed probe sets in pairwise comparisons between the five different muscle types found (i) only in the WT versus WT comparisons (‘WT’), (ii) only in NM versus NM (‘TG’) and (iii) in common for both of these comparisons (‘COMMON’). Although WT DIA samples exhibited many differences from other muscle types, far fewer significant changes were seen in NM DIA comparisons with other muscle types.
The lists of WT and NM muscle type-related differences were compared and the significantly changed probe sets in common were determined. In general, probe sets significantly changed between WT muscles alone are likely to reflect muscle-specific differences suppressed by the mutant protein. Probe sets significantly changed between NM muscles alone likely reflect differences promoted by the mutant protein, i.e. muscle-specific responses to the mutant protein. Probe sets common in the two lists likely represent inter-muscle differences unaffected by the mutant protein. DIA presented with the most differences at the WT inter-muscle comparison level. However, NM DIA specimens had fewer differences compared with other NM specimens. The same pattern, although at a smaller scale, was true for all muscles studied, with WT samples having more significantly changed probe sets between them than the equivalent NM samples (Fig. 3). Exceptional were the comparisons of PLT versus GAS and EDL versus GAS, which showed the reverse pattern, suggesting that the mutant protein triggers similar molecular pathways across most, but not all muscles. Hence, the NM profiles were in general more similar than those for the WT muscles.
Evidence for increased satellite cell numbers and immature fibers in nemaline muscles
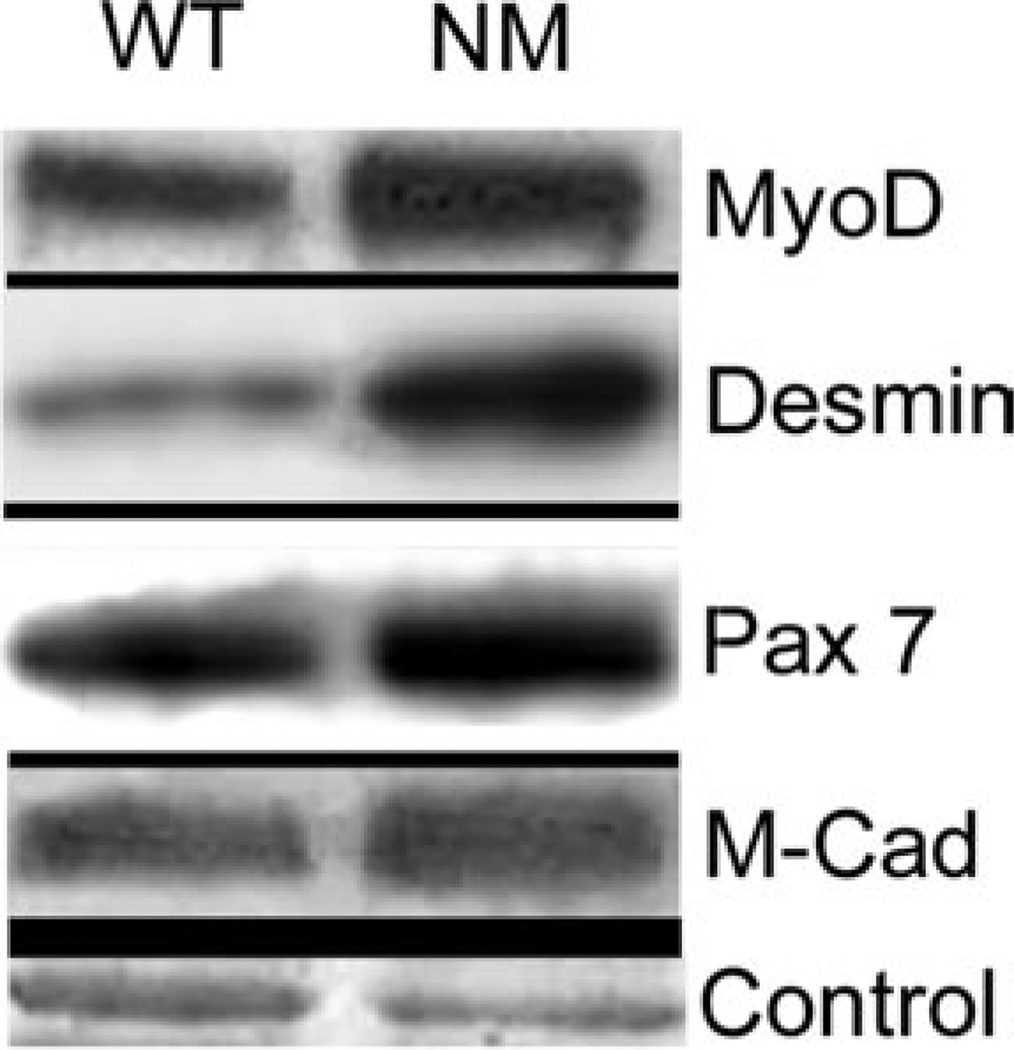
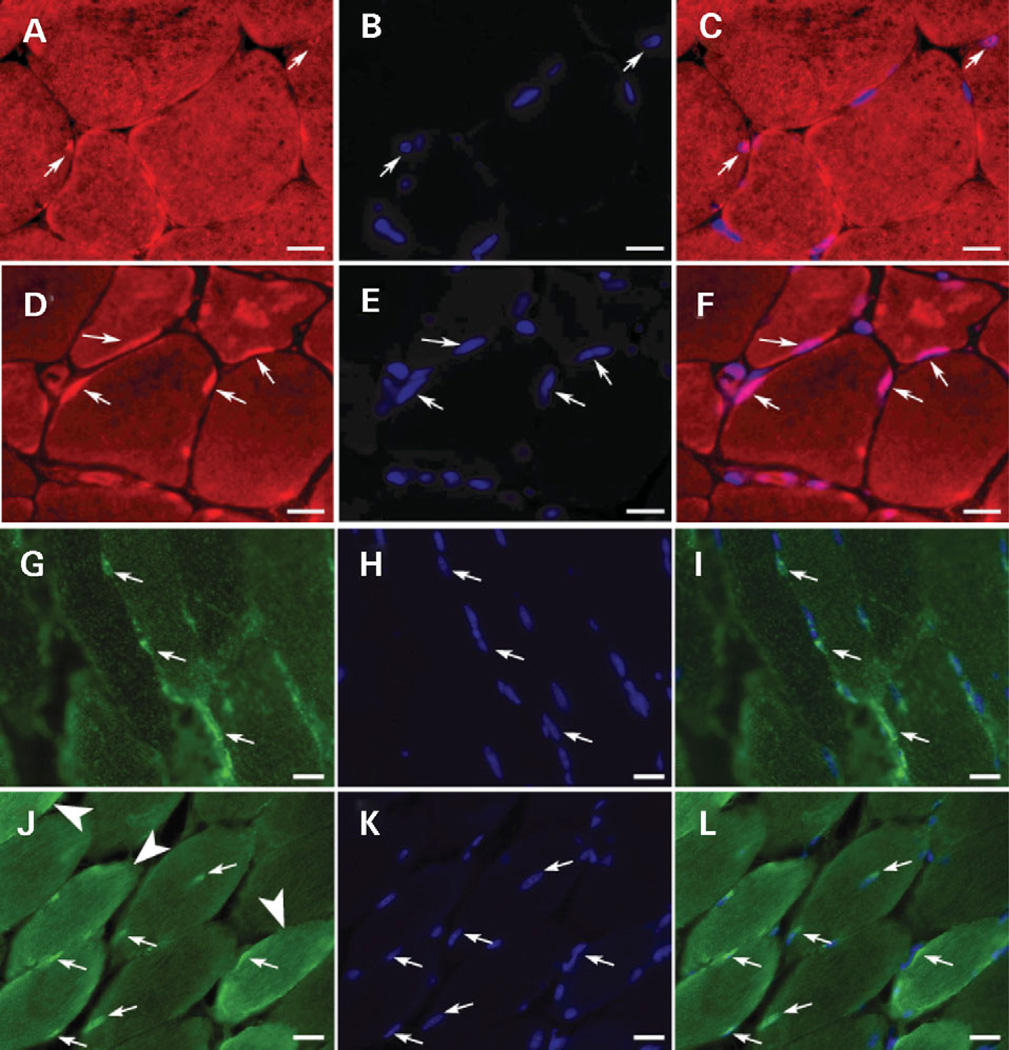
Transcripts for genes reflective of myogenesis, Six4, Pcaf, Myf6, Csrp3, Ankrd2, Tncc, Tnnt1, Tnni1, Mylpc and Lmna, were elevated in one or more of the five NM muscles analyzed. To determine whether this reflected an increase in the number of satellite cells present in nemaline muscles, we performed western-blot analysis of WT and NM TA muscles using antibodies for proteins present in quiescent and/or activated satellite cells: MyoD, desmin, Pax7 and M-Cadherin. Quantitative western-blot analysis showed that levels of these proteins were elevated within the range of 1.6- to 2.66-fold (Fig. 4, Table 1). A similar trend was also observed for the expression data of the respective genes (data not shown). This was, however, below our analysis thresholds for ‘significantly and highly’ changed. We also assessed the number of Myf6 expressing nuclei in WT and NM muscle to determine whether this indicator of immature muscle was elevated in NM muscle. Immunohistochemical analysis of the presence of My6 in myofiber nuclei as shown in Fig. 5A–F revealed a 1.8-fold increase (34.2–62.5%, P = 0.005 by ANOVA) in the number of Myf6-expressing myonuclei in NM TA muscle. Similarly, analysis of muscle LIM protein expression (MLP; encoded by Csrp3), a factor that promotes muscle differentiation, revealed marked differences in expression levels and intracellular localization between WT and NM TA muscles. Specifically, in WT TA muscle, 12% of the peripheral myofiber nuclei displayed prominent MLP expression, with a moderate cytoplasmic signal in all fiber types. In contrast, 50% of the peripheral nuclei in NM TA were MLP positive and 26% of the fast fibers showed a significant over-expression of MLP in the cytoplasm (Fig. 5G–L, a total of 400 nuclei and 250 fast fibers were analyzed). In addition, in NM TA muscle, 7% of the fibers had central nuclei and 90% of these were MLP positive. Taken together, these observations reflect ongoing fusion of satellite cells with existing myofibers and possibly the maintenance of transcriptional activity reminiscent of immature fibers.
Figure 4.
Western blot analysis of satellite cell markers, MyoD, desmin, Pax7 and M-Cadherin (M-Cad) (indicated at right) reveals elevations in αTmslow (Met9Arg) transgenic TA muscle from 7–8-month-old NM mice. Two to three replicate muscles from WT or NM mice were analyzed and a representative blot is shown. Control, Coomassie stained myosin light chain band.
Table 1.
Quantitation of satellite cell content in nemaline muscle
| Average density WT | Average density NM | Fold change | |
|---|---|---|---|
| MyoD | 226.5 ± 34.9 | 404.1 ± 33.6 | 1.8 |
| Desmin | 366.2 ± 92.2 | 973.2 ± 136.5 | 2.7 |
| Pax7 | 75.4 ± 7.7 | 179. 4 ± 41.9 | 2.4 |
| M-Cadherin | 27.6 ± 1.9 | 44.1 ± 1.5 | 1.6 |
TA muscles from 2–3 WT or NM mice, 7–8 months of age, were analyzed 2–4 times each by western blot. The optical density of each band was normalized to the corresponding Coomassie stained myosin light chain band and the mean optical density determined for each protein examined. Standard deviations were calculated and expressed as ± values of the mean density. Fold change was calculated from the mean optical densities.
Figure 5.
Immunohistochemical analysis of Myf6 and MLP reveals increased expression in nemaline muscle. Myf6 (A–F) and MLP (G–L) were detected immunohistochemically in TA muscles of 9- and 6-month-old, respectively, female WT (A–C, G–I) and NM (D–F, J–L) littermate mice. Small arrows indicate representative Myf6 containing nuclei for (A–F) and representative MLP positive nuclei for (G–L). The TA muscles from NM mice have twice as many nuclei expressing Myf6 (D–F) and four times as many MLP (J–L) containing nuclei compared with littermate controls (A–C and G–I, respectively). Furthermore, in NM mice, unlike littermate controls, increased MLP staining was observed in the cytoplasm of multiple muscle fibers [large arrow heads in (J)]. All nuclei were visualized using DAPI (stained blue in B, C, E, F, H, I, K and L). Scale bars = 16.7 µm.
Electron microscopic evidence for altered metabolism, myofibrillar degeneration and focal repair in nemaline muscles
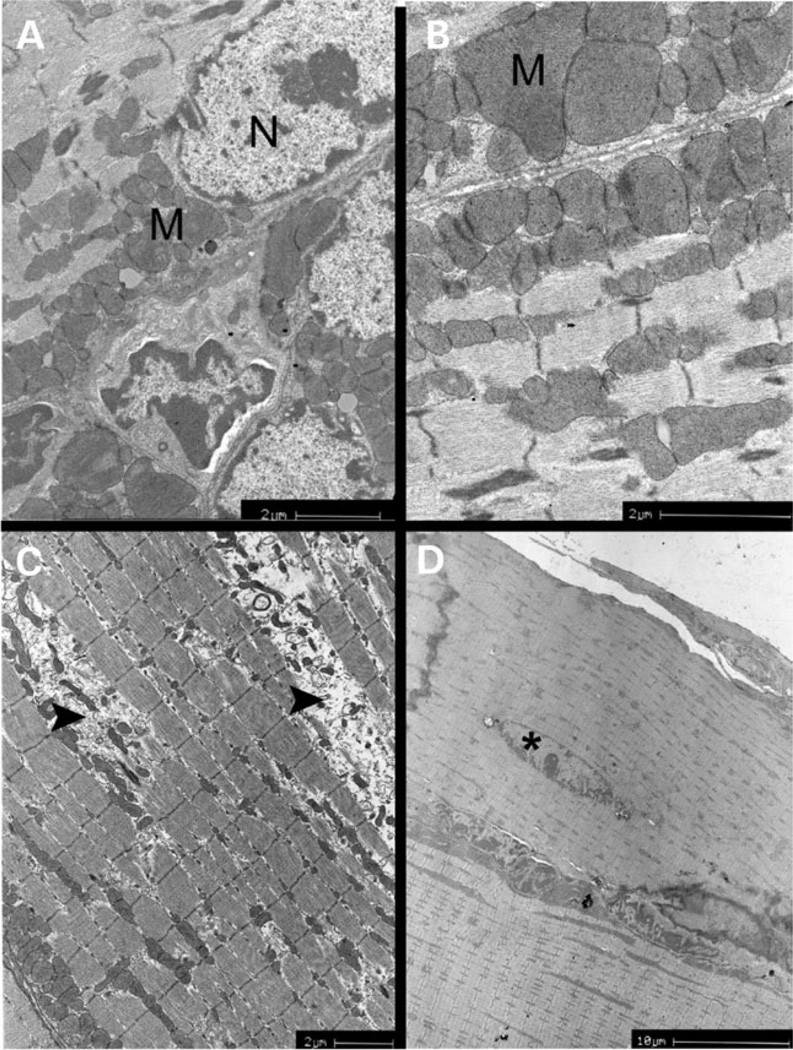
Tissue specimens from DIA and TA of WT and NM mice were used for electron microscopic (EM) analysis to identify pathological features correlating with gene expression data. Consistent with the gene expression profile, DIA demonstrated a marked variation in the size and structure of subsarcolemal mitochondria (Fig. 6A and B). Some mitochondria were significantly larger than others, and mitochondrial structure ranged from aberrant and variably shaped to partial lack of cristae. This lack of cristae was concomitant with the presence of vacuoles filled with material of medium electron density. EM analysis of NM TA muscles revealed segmental degeneration of myofibers highlighted by the absence of sarcomeres and the presence of mitochondria at different stages of degeneration (Fig. 6C). Finally, cytoplasmic lipofuscin inclusions, an indicator of oxidative stress, were encountered in both tissues (data not shown).
Figure 6.
EM analysis of αTmslow(Met9Arg) transgenic muscles reveals morphological evidence for metabolic disturbance, myofibrillar degeneration and repair in nemaline muscles. (A and B) Diaphragm of 8-month-old NM mouse: enlarged, abnormally shaped and increased numbers of mitochondria are present beneath the sarcolemma and in inter-fibrillar spaces. N, myonucleus; M, mitochondria; (C) Tibialis anterior muscle of 8-month-old NM mouse: myofibrillar degeneration is evident (arrowheads). Inter-fibrillar and subsarcolemmal mitochondria display abnormal shape, size and number. (D) Tibialis anterior muscle of 7-month-old NM mouse: localized repair is evident by the presence of an isolated, centrally localized nucleus (asterisk) within a myofiber.
Focal repair was evident in DIA and TA muscles as manifested by the presence of centrally located nuclei in regions or segments of myofibers (Fig. 6D). These myofibers did not display overt signs of necrosis or degeneration, consistent with a focal repair mechanism. Quantitation of fibers in NM DIA and TA that contained centrally located nuclei or nuclei not at the periphery of the fiber revealed that 12% of fibers in NM DIA and 7% of TA fibers exhibited focal repair (Table 2). In the fibers of WT mice, nuclei were only located at the periphery of the myofiber and membrane ligatures, seen in NM muscles, were not observed.
Table 2.
Numbers and percentage of fibers with central nuclei in diaphragm and tibialis anterior muscles of WT and αTmslow(Met9Arg) transgenic (NM) mice
| Diaphragm | Tibialis anterior | ||
|---|---|---|---|
| WT | NM | WT | NM |
| 0/95 (0%) | 11/92 (12%) | 0/90 (0%) | 7/94 (7%) |
The location of nuclei within myofibers was used as an indicator of local repair within the diaphragm and tibialis anterior muscles of wild-type (WT) and nemaline (NM) mice. Fibers scored as having central nuclei typically had only one nucleus within the field that was centrally located. For a given muscle, total fibers were scored in 10 EM images from each of three different muscles from mice of 7–8 months of age. Parentheses indicate percentage of fibers with central nuclei.
DISCUSSION
Correlation coefficient analysis revealed that different nemaline muscles had a considerably higher similarity between them in terms of gene expression patterns than did the corresponding wild-type muscles. This strengthens previous observations on the profound, naturally occurring differences in gene expression between wild-type muscles (6,14–16), yet more importantly, it indicates that the introduction of the αTmslow(Met9Arg) mutation triggers similar disease-related expression changes in the affected muscles. These results suggest that a common pathophysiologic mechanism or process is operating in nemaline muscles that have remarkably different pathologies. The DIA, TA and GAS muscles of the αTmslow(Met9Arg) mice have 50, 15 and 1% rod-containing fibers, respectively (12), indicating that the process common among nemaline muscles as revealed by gene expression analysis is not related to the pathological feature characteristic of the disease, i.e. nemaline rod formation.
In the present analysis of NM mouse muscles, gene expression data revealed that transcripts reflective of myogenesis, muscle development and myofiber immaturity, such as Six4, Pcaf, Myf6, Csrp3, Ankrd2, Tncc, Tnnt1, Tnni1, Mylpc and Lmna, were elevated in one or more of the five muscle types analyzed. The Six4 and Pcaf proteins regulate myogenesis through activation of myogenin and MyoD, respectively (17,18), whereas MLP (encoded by Csrp3) and Myf6 are essential and potent promoters of myogenesis (19,20). Ankrd2, Tncc, Tnnt1, Tnni1 and Mylpc encode sarcomeric proteins that participate in muscle development [Gene Ontology (GO) classification]. Western-blot analysis confirmed the elevated levels of M-Cadherin, MyoD, desmin and Pax7, classic markers of satellite cells, suggesting increased numbers of satellite cells present in the mouse muscles. Furthermore, increased numbers of Myf6-containing nuclei in myofibers suggested relative immaturity of the muscle. Further direct evidence of elevated focal repair in the DIA and TA muscles was found by EM analysis (Fig. 6). These data are further supported by our previous analysis of biopsies from human NM patients, which demonstrated an increase in the number of PAX7-positive muscle satellite cells, but no full blown repair process (5). Interestingly, a previous study of the response of these NM mouse muscles to chronic stretch revealed that regeneration occurred with relative lack of centralized nuclei, a hallmark indicator of repair (21). Taken together, these observations support the presence of ongoing active, but focal, repair processes in nemaline muscle that is not readily apparent by the classic indicator of muscle regeneration, extensive numbers of myofibers with centrally located myonuclei as seen in the mdx mouse (4).
The majority of over-expressed muscle contraction-related genes (such as Tncc, Tnnt1, Tnni1, Mylpc and Myh7), which overlap to a large extent with the muscle development genes mentioned earlier, coded for slow or cardiac muscle isoforms. Although WT mouse muscles are fast fiber predominant, the EDL muscle of αTmslow(Met9Arg) mice at 6 months of age was shown to contain almost three times the number of slow/fast oxidative fibers than their WT littermates (12). Furthermore, in the αTmslow(Met9Arg) mice, the relative fiber-type content remained static with age, unlike their WT littermates. Such an increase in numbers of slow fibers likely explains the over-expression of slow muscle gene isoforms in our gene expression data sets. The relatively static fiber-type composition in the Tpm3 transgenic muscles, together with the lack of slow/fast oxidative fiber number increase in a parallel analysis of HSA-αTmslow(WT) transgenic mice (without the Met9Arg mutation), supports earlier conclusions that the altered muscle fiber-type content in the HSA-αTmslow(Met9Arg) mice was due to the mutation and that the mechanism leading to this phenotype might indeed be the disruption of normal muscle maturation (12). The over-expression seen here for various cardiac muscle isoforms (e.g. Mylpc and Myh7), which can be expressed in skeletal muscle during the developmental stages, may be indicative of both delayed maturation and regeneration.
Ankrd2 and Csrp3 are the only two genes consistently over-expressed in four of the five muscles studied (fold-change range Ankrd2 = 2.7–8.6 and Csrp3 = 2.3–5.5; Fig. 5G–L), suggesting that this represents a key change in gene expression induced by the presence of the mutant protein. Although both Ankrd2 and Csrp3 are expressed predominantly in slow muscle fibers, the level and extent of their over-expression in all four out of five muscles in our study is beyond what could be justified by the altered fiber-type content of the transgenic mouse muscles. Interestingly, the proteins encoded by Ankrd2 and Csrp3 have a dual role by localizing both in the nucleus and the sarcomere, participating in cell-cycle arrest, initiation of muscle differentiation and/or sarcomere recruitment (22–25). Over-expression of MLP (encoded by Csrp3) has been associated with increased differentiation in C2C12 cells by enhancing the action of MyoD (26). Its localization seems to change from nuclear to cytoplasmic with muscle development, and changes in its localization can play a crucial role in disease pathophysiology (27). Furthermore, Ankrd2 and Csrp3 expression changes have recently been described in several muscular dystrophies and myopathies, although they did not pass the significance thresholds in our human NM analysis (5,28–32). Csrp3 has been shown to express significantly higher in WT TA (a representative of distal muscles) than quadriceps (a representative of proximal muscles) in WT mice (30). Despite potential gene expression differences between the five different WT muscles studied, significant Csrp3 expression changes were induced in 4 muscles, including three of four distal muscles and DIA.
Increased expression of neurogenesis-related genes (Cited2, Enah, Pten, Dctn1 and Wfs1) was also detected in the transgenic mice. The majority of these genes, such as Enah and Cited2 (33,34), are primarily involved in prenatal development. Their increased expression in the 7–10-month-old HSA-αTmslow (Met9Arg) mice further supports an ongoing repair process. Over-expression of myogenesis/muscle development genes was seen in all five muscles and neurogenesis in three (TA, PLT and EDL). However, the number of significantly changed genes and their fold change was higher in TA than in GAS, PLT, EDL and DIA (number of genes = TA: 9, all others: 2–6; fold change = TA: 1.5–10.7, all others: 1.5–6.7).
In contrast to our previous findings on human NM specimens, where the glycolytic energy production pathway was significantly down-regulated, the HSA-αTmslow(Met9Arg) mouse muscles TA, PLT, EDL and GAS exhibited no major changes in the expression of genes encoding the proteins involved in metabolic pathways. DIA, however, presented with a prominent down-regulation of over 25 inner mitochondrial membrane oxidoreductase genes (fold change −1.5 to −7.5). These are known to be involved in mitochondrial ATP production. Strongly supportive of these gene expression observations were EM findings of marked mictochondrial size and shape variation in DIA (Fig. 6A and B). Structural variation was also evident with some mitochondria lacking cristae but having vacuoles filled with material of medium electron density. It, therefore, seems likely that energy production defects (as seen at the gene expression level) are secondary changes associated with more severe stages of NM such as exhibited by DIA and the majority of human patients previously studied by microarrays (5,35).
Atp2a2 and Casq2, two proteins responsible for transporting and storing calcium ions, respectively, in the sarcoplasmic reticulum of cardiac and slow skeletal muscles, were up-regulated at the gene expression level in the transgenic TA (Atp2a2 and Casq2) and PLT (Atp2a2 only) muscles. In our previous study of NM patients, their human orthologs were also significantly changed (ATP2A1 = −4.1-fold; CASQ2 = 2.8-fold) together with several other calcium homeostasis-related genes (5). These were two of the few genes commonly changed in both species, suggesting a possible close link to the disease pathogenesis. The opposite directions of the ATP2A1 and Atp2a2 fold changes, together with the variability in fiber-type percentages of human specimens (six had slow-fiber predominance, three had fast-fiber predominance and four were normal), suggest that calcium homeostasis-related gene expression changes are likely to reflect the frequent slow-fiber predominance as seen in the case of the over-expressed slow-fiber isoforms Atp2a2, Casq2 and CASQ2 and the under-expressed ATP2A1. Consistent with this hypothesis, mechanical studies addressing the response of individual fibers to calcium ions did not find any changes in HSA-αTmslow(Met9Arg) mice (12).
Increased expression was also evident for a number of transcription, translation and protein modification genes such as Pdlim1, Atf5, D11Ertd619e, Eif4bp1 and Rpl22 (fold change 1.51–3.00). Indirectly, this evidence suggests an increase in overall transcription/translation/protein synthesis in the transgenic TA, PLT and GAS muscles. Apoptosis- and proteolysis-related genes (such as Faf1, Pdcd7, Dffa, Bcl2l11, Bag3 and Traf3) were also over-expressed in TA, PLT, GAS and EDL (fold change 1.5–2.1). It is difficult to speculate on the order of cellular events based on the present data. However, given the parallel increase in myo/neurogenesis, transcription, protein biosynthesis, apoptosis and proteolysis, we hypothesize that different cell populations are present in the transgenic muscles, with some cells going through a developmental phase and others undergoing proteolysis and apoptosis: consistent with a model involving some degree of regeneration. In support of this hypothesis, EM studies on TA revealed segmental degeneration of myofibers together with small areas of regenerating fibers, manifested by the presence of centrally located nuclei in small myofibers.
In all of the analyses performed, DIA consistently presented with the most pronounced transcriptional differences in comparison with those observed in the TA, PLT, GAS and EDL muscles. Not only was the number of significantly changed genes in the transgenic DIA 10–100-fold greater than that in the other muscles examined, but their functions and overall direction of change (99.6% probe sets were under-expressed in NM DIA) were markedly different (Supplementary Material, Table S1). Furthermore, the overall transcriptional profile of the NM DIA was more similar to the various WT limb muscles than to WT DIA (Fig. 1). Diaphragm has been shown to have a unique pattern of gene expression, relative to other skeletal muscles (6,16). Many of the 1386 significantly changed probe sets in the WT versus NM DIA comparison represent non-muscle transcripts that may reflect a relative absence or loss of increased vascularity and/or other functional specializations of the diaphragm (Supplementary Material, Table S1) (unpublished data). The functional categories of genes with highest expression differences between NM and WT DIA included metabolism, especially protein, lipid and organic acid metabolism, followed by cell growth. The latter consisted almost exclusively of transport-related genes. We hypothesize that these data reflect a relative lack of specialization of NM DIA muscles in HSA-αTmslow (Met9Arg) mice. Thus, the presence of the mutant protein appears to disrupt the skeletal muscle maturation process, with a more dramatic effect in the case of DIA.
The differential response of DIA compared with other skeletal muscles has also been described in the mdx mice (16,36). Similar to our findings, mdx DIA was more severely affected than other skeletal muscles, with a profound metabolic crisis reflected predominantly by the down-regulation of numerous mictochondrial transcripts. Unlike αTmslow(Met9Arg), mdx DIA showed a marked up-regulation of most differentially expressed transcripts. Furthermore, a number of other dystrophy related molecular pathways were more severely affected in mdx DIA, including regeneration, inflammation and fibrosis (16). Regardless of the disease, DIA appears to have pathophysiological mechanisms distinct from other skeletal muscles.
DIA data sets also had weaker correlations between them, both in the WT and the NM groups (average r = 0.91, as opposed to 0.95–0.97 seen for other muscles). Although the physical location of this muscle could give rise to concerns regarding the cellular purity of the samples used, our long experience in dissecting such muscles together with previous evidence showing the highly variable expression of DIA compared with other muscles points to the accuracy of these measurements (16,36). The increased difference between WT and NM DIA could be associated with the somewhat higher levels of αTmslow(Met9Arg) protein detected in this muscle, although sensitivity limitations in western-blot analysis preclude a definite association (12).
In conclusion, the lists of significantly changed genes between the different transgenic mouse muscle types varied considerably, yet there was a substantial overlap in the functional pathways affected by the mutant protein, and particularly muscle development. It is important to remember that some of these changes could be related to Tpm3 being overexpressed, and therefore, further studies comparing our findings with new NM mouse models are warranted. To a large extent, these molecular processes also overlap and extend our previous observations on human NM muscle specimens. The consistent finding of evidence for a repair process, characterized by some degree of degeneration and regeneration, in both species indicates that this is a basic aspect of NM pathophysiology. These findings reveal a novel feature of NM that has previously been considered uncharacteristic of congenital myopathies.
MATERIALS AND METHODS
Mouse samples and microarray experiments
Five different mouse skeletal muscles were studied from normal FVB/NJ and transgenic mice expressing an autosomal dominant mutant of alpha-tropomyosinslow under the HSA promoter (HSA-αTmslow(Met9Arg)) (12). All five muscles had similar levels of αTmslow protein expression but variable rod content (Supplementary Material, Table S3) (37,38). Three to four replicate total RNA pools were generated for each muscle type (TA, GAS, PLT, DIA and EDL). Each pool originated from muscle specimens of three to five mice. These specimens were obtained at the age of 7–10 months and were snap frozen.
RNA extraction (6–8 mg from each tissue sample), target preparation and hybridization to Affymetrix U74Av2 Gene-Chips were performed as previously described (5). The U74Av2 GeneChip contains 12 488 annotated genes and ESTs. The 36 raw data sets are publicly available at http://www.ncbi.nlm.nih.gov/geo/, under the series code GSE3384.
Data analysis
The 36 data sets (3–4 pools × five muscle types × normal and transgenic) were first processed by the Affymetrix MAS5.0 software, which computed the signal values (reflecting expression levels) and ‘present/absent’ calls (an Affymetrix computed measure representing confidence in gene expression presence) for each probe set. The signal values were adjusted to an overall target intensity of 1500 but were not normalized at this stage.
For correlation coefficient analysis, r-values for overall gene expression were calculated for all 36 data set pairwise comparisons. We then calculated the average of the r-values for the data sets of each specific muscle type (five muscles) and state (transgenic and wild-type). As a third step, we calculated the r of each muscle/state versus all others (Fig. 1).
SAM was used to identify significant fold changes between WT and NM specimens as described (39). Prior to this analysis, all data sets were normalized to a slope of 1, with the data set having the highest correlation to all others (PLTn2). For each muscle type, a two-class unpaired data analysis was performed comparing NM with WT specimens, using a fold threshold of 1.5 [where fold is calculated as: (average expression in NM specimens)/(average expression in WT specimens)] and a FDR cutoff of ≤12.5%. Because no genes were significantly changed for EDL samples at these thresholds, we present the four probe sets significantly changed at the lowest FDR available (24.9%). All significantly changed probe sets were assessed for present/absent calls and only those with ≥ 1/3 present calls were included in further analysis. SAM analysis was repeated to compare (i) the different WT muscle types and (ii) the different NM muscle types, using the same fold and FDR thresholds as earlier.
Immunohistochemistry
TA muscles from transgenic nemaline and littermate control mice were transversely sectioned at 5 µm thickness, placed onto polysine-coated microscope slides (Biolab Scientific) and fixed in 4% formaldehyde for 5 min. Slides were blocked and permeabilized in a mixture of 1×PBS, 0.2% fish gelatin and 0.1% Triton X for 1 h at RT. Sections were incubated in a rabbit polyclonal Myf6 primary antibody (Santa Cruz, SC301 C-19; 1:200 dilution) for 1 h at RT. The sections were washed thoroughly and incubated in an Alexa Fluor 555 goat anti-rabbit secondary antibody (Molecular Probes; 1:1000 dilution) for 1 h at RT. Nuclei were detected by co-staining sections with DAPI (Sigma; 0.4 mg/ml) for 5 min and a final 5 min wash in 1×PBS. Images were captured using a digital camera at 20× magnification and merged using Spot advanced program. Nuclei expressing Myf6 were scored using the Image-Pro software (Media Cybernetics, Silver Spring, MD, USA).
Immunohistochemistry for MLP (the Csrp3 gene product) was performed as previously described (5) using chicken polyclonal Csrp3 primary antibody (Abcam; 1:100 dilution) for 2.5 h at RT and Alexa Fluor 594 goat anti-chicken secondary antibody (Molecular Probes; 1:200) for 1 h.
Western blotting
Skeletal muscle tissue was homogenized in 20 volumes lysis buffer (25 mm Tris pH 7.8, 2 mm EDTA, 10% glycerol, 1% Triton-X 100, 1.5 mm MgCl2, 200 mm NaCl and 10 mm PMSF). Frozen samples were firstly crushed with a pestle and then resuspended in the lysis buffer in an Eppendorf tube. Samples were allowed to sit on ice for 1 h with constant resuspension, followed by centrifugation at maximum speed for 5 min at 4°C. The supernatant was saved. Samples were stored at −20°C. Equal volumes of protein were analyzed on 12.5% SDS–PAGE gels. Protein was transferred onto PVDF membranes (Millipore) for 1 h at 10 V using the Biorad semidry transblot. Blots were blocked overnight at 4°C in 5% skim milk in Tris-buffered saline containing 0.1% Tween-20 (TBST), washed in TBST and incubated with antibody overnight at 4°C (MyoD: Santa Cruz, M318; Desmin: NOVOcastra, NCL-L-DES-DERII; M-Cadherin: Santa Cruz, SC10734; Pax7: Developmental Studies Hybridoma Bank), followed by three washes with TBS 0.1% Tween-20. HRP-labeled secondary antibody was added at a 1:10 000 dilution for 1 h at RT. Excess antibody was removed with 4 × 20 min washes. Detection was performed using the western lightning chemiluminescence detection system (Perkin Elmer) on Fuji RX film for varying times (2–60 min).
Electron microscopy
Muscles were removed from anaesthetized mice and immediately cut into very thin slices while immersed in modified Karnovski’s fixative (2.5% glutaraldehyde/4% paraformaldehyde in 1 M cacodylate buffer, pH 7.4). Samples were further fixed overnight in the same fixative and post-fixed with 2% osmium tetroxide, dehydrated through an ascending series of ethanol and embedded in Spurr’s epoxy resin. Ultrathin sections (70 nm) were cut with a Reichert-Jung ULTRACUT ultramicrotome, double contrasted with uranyl acetate and lead citrate, viewed and photographed with a Philips CM120 BioTwin transmission EM.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Isaac S. Kohane for his invaluable advice on data analysis and Visalini Nair-Shalliker for her analysis of MyoD expression. Microarray experiments were conducted in the Children’s Hospital, Gene Expression Core Laboratory supported by NIH grant NS040828. This work was funded in part by NIH AR44345 and generous support from the Lee and Penny Anderson Family Foundation, the Joshua Frase Foundation and the Muscular Dystrophy Association of the USA to A.H.B. and by an NHMRC grant to E.C.H. ‘Funding to pay the Open Access publication charges for this article was provided by NIH AR44345’.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
Conflict of Interest statement. The authors declare that they have no conflicts of interest or competing financial interests to disclose.
REFERENCES
- 1.Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg SA, Sanoudou D, Haslett JN, Kohane IS, Kunkel LM, Beggs AH, Amato AA. Molecular profiles of inflammatory myopathies. Neurology. 2002;59:1170–1182. doi: 10.1212/wnl.59.8.1170. [DOI] [PubMed] [Google Scholar]
- 3.Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, Beggs AH, Kunkel LM. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc. Natl Acad. Sci. USA. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Guo W, Andrade FH. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2002;11:263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- 5.Sanoudou D, Haslett JN, Kho AT, Guo S, Gazda HT, Greenberg SA, Lidov HG, Kohane IS, Kunkel LM, Beggs AH. Expression profiling reveals altered satellite cell numbers and glycolytic enzyme transcription in nemaline myopathy muscle. Proc. Natl Acad. Sci. USA. 2003;100:4666–4671. doi: 10.1073/pnas.0330960100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haslett JN, Kang PB, Han M, Kho AT, Sanoudou D, Volinski JM, Beggs AH, Kohane IS, Kunkel LM. The influence of muscle type and dystrophin deficiency on murine expression profiles. Mamm. Genome. 2005;16:739–748. doi: 10.1007/s00335-005-0053-8. [DOI] [PubMed] [Google Scholar]
- 7.North KN, Laing NG, Wallgren-Pettersson C. Nemaline myopathy: current concepts. The ENMC International Consortium and Nemaline Myopathy. J. Med. Genet. 1997;34:705–713. doi: 10.1136/jmg.34.9.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanoudou D, Beggs AH. Clinical and genetic heterogeneity in nemaline myopathy—a disease of skeletal muscle thin filaments. Trends Mol. Med. 2001;7:362–368. doi: 10.1016/s1471-4914(01)02089-5. [DOI] [PubMed] [Google Scholar]
- 9.Durling HJ, Reilich P, Muller-Hocker J, Mendel B, Pongratz D, Wallgren-Pettersson C, Gunning P, Lochmuller H, Laing NG. De novo missense mutation in a constitutively expressed exon of the slow alpha-tropomyosin gene TPM3 associated with an atypical, sporadic case of nemaline myopathy. Neuromuscul. Disord. 2002;12:947–951. doi: 10.1016/s0960-8966(02)00182-7. [DOI] [PubMed] [Google Scholar]
- 10.Laing NG, Wilton SD, Akkari PA, Dorosz S, Boundy K, Kneebone C, Blumbergs P, White S, Watkins H, Love DR, et al. A mutation in the alpha tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy. Nat. Genet. 1995;9:75–79. doi: 10.1038/ng0195-75. [DOI] [PubMed] [Google Scholar]
- 11.Wattanasirichaigoon D, Swoboda KJ, Takada F, Tong HQ, Lip V, Iannaccone ST, Wallgren-Pettersson C, Laing NG, Beggs AH. Mutations of the slow muscle alpha-tropomyosin gene, TPM3, are a rare cause of nemaline myopathy. Neurology. 2002;59:613–617. doi: 10.1212/wnl.59.4.613. [DOI] [PubMed] [Google Scholar]
- 12.Corbett MA, Robinson CS, Dunglison GF, Yang N, Joya JE, Stewart AW, Schnell C, Gunning PW, North KN, Hardeman EC. A mutation in alpha-tropomyosin (slow) affects muscle strength, maturation and hypertrophy in a mouse model for nemaline myopathy. Hum. Mol. Genet. 2001;10:317–328. doi: 10.1093/hmg/10.4.317. [DOI] [PubMed] [Google Scholar]
- 13.Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- 14.Cheng G, Merriam AP, Gong B, Leahy P, Khanna S, Porter JD. Conserved and muscle-group-specific gene expression patterns shape postnatal development of the novel extraocular muscle phenotype. Physiol. Genomics. 2004;18:184–195. doi: 10.1152/physiolgenomics.00222.2003. [DOI] [PubMed] [Google Scholar]
- 15.Kang PB, Kho AT, Sanoudou D, Haslett JN, Dow CP, Han M, Blasko JM, Lidov HG, Beggs AH, Kunkel LM. Variations in gene expression among different types of human skeletal muscle. Muscle Nerve. 2005;32:483–491. doi: 10.1002/mus.20356. [DOI] [PubMed] [Google Scholar]
- 16.Porter JD, Merriam AP, Leahy P, Gong B, Feuerman J, Cheng G, Khanna S. Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle group-specific mechanisms in the pathogenesis of muscular dystrophy. Hum. Mol. Genet. 2004;13:257–269. doi: 10.1093/hmg/ddh033. [DOI] [PubMed] [Google Scholar]
- 17.Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl Acad. Sci. USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 19.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 20.Braun T, Bober E, Winter B, Rosenthal N, Arnold HH. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J. 1990;9:821–831. doi: 10.1002/j.1460-2075.1990.tb08179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joya JE, Kee AJ, Nair-Shalliker V, Ghoddusi M, Nguyen MA, Luther P, Hardeman EC. Muscle weakness in a mouse model of nemaline myopathy can be reversed with exercise and reveals a novel myofiber repair mechanism. Hum. Mol. Genet. 2004;13:2633–2645. doi: 10.1093/hmg/ddh285. [DOI] [PubMed] [Google Scholar]
- 22.Kojic S, Medeot E, Guccione E, Krmac H, Zara I, Martinelli V, Valle G, Faulkner G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J. Mol. Biol. 2004;339:313–325. doi: 10.1016/j.jmb.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 23.Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 24.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 25.Weiskirchen R, Gunther K. The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. Bioessays. 2003;25:152–162. doi: 10.1002/bies.10226. [DOI] [PubMed] [Google Scholar]
- 26.Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol. Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ecarnot-Laubriet A, De Luca K, Vandroux D, Moisant M, Bernard C, Assem M, Rochette L, Teyssier JR. Downregulation and nuclear relocation of MLP during the progression of right ventricular hypertrophy induced by chronic pressure overload. J. Mol. Cell. Cardiol. 2000;32:2385–2395. doi: 10.1006/jmcc.2000.1269. [DOI] [PubMed] [Google Scholar]
- 28.Nakada C, Tsukamoto Y, Oka A, Nonaka I, Sato K, Mori S, Ito H, Moriyama M. Altered expression of ARPP protein in skeletal muscles of patients with muscular dystrophy, congenital myopathy and spinal muscular atrophy. Pathobiology. 2004;71:43–51. doi: 10.1159/000072961. [DOI] [PubMed] [Google Scholar]
- 29.Pallavicini A, Kojic S, Bean C, Vainzof M, Salamon M, Ievolella C, Bortoletto G, Pacchioni B, Zatz M, Lanfranchi G, et al. Characterization of human skeletal muscle Ankrd2. Biochem. Biophys. Res. Commun. 2001;285:378–386. doi: 10.1006/bbrc.2001.5131. [DOI] [PubMed] [Google Scholar]
- 30.von der Hagen M, Laval SH, Cree LM, Haldane F, Pocock M, Wappler I, Peters H, Reitsamer HA, Hoger H, Wiedner M, et al. The differential gene expression profiles of proximal and distal muscle groups are altered in pre-pathological dysferlin-deficient mice. Neuromuscul. Disord. 2005;15:863–877. doi: 10.1016/j.nmd.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 32.Witt CC, Ono Y, Puschmann E, McNabb M, Wu Y, Gotthardt M, Witt SH, Haak M, Labeit D, Gregorio CC, et al. Induction and myofibrillar targeting of CARP, and suppression of the Nkx2.5 pathway in the MDM mouse with impaired titin-based signaling. J. Mol. Biol. 2004;336:145–154. doi: 10.1016/j.jmb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 33.Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 34.Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, Conlon RA, et al. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc. Natl Acad. Sci. USA. 2002;99:10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanoudou D, Frieden LA, Haslett JN, Kho AT, Greenberg SA, Kohane IS, Kunkel LM, Beggs AH. Molecular classification of nemaline myopathies: ‘nontyping’ specimens exhibit unique patterns of gene expression. Neurobiol. Dis. 2004;15:590–600. doi: 10.1016/j.nbd.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Rouger K, Le Cunff M, Steenman M, Potier MC, Gibelin N, Dechesne CA, Leger JJ. Global/temporal gene expression in diaphragm and hindlimb muscles of dystrophin-deficient (mdx) mice. Am. J. Physiol. Cell Physiol. 2002;283:C773–C784. doi: 10.1152/ajpcell.00112.2002. [DOI] [PubMed] [Google Scholar]
- 37.Corbett MA, Akkari PA, Domazetovska A, Cooper ST, North KN, Laing NG, Gunning PW, Hardeman EC. An alpha Tropomyosin mutation alters dimer preference in nemaline myopathy. Ann. Neurol. 2005;57:42–49. doi: 10.1002/ana.20305. [DOI] [PubMed] [Google Scholar]
- 38.Zardini DM, Parry DJ. Identification, distribution, and myosin subunit composition of type IIX fibers in mouse muscles. Muscle Nerve. 1994;17:1308–1316. doi: 10.1002/mus.880171110. [DOI] [PubMed] [Google Scholar]
- 39.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.