Abstract
The Podospora anserina PaMpk1 MAP kinase (MAPK) signaling pathway can generate a cytoplasmic and infectious element resembling prions. When present in the cells, this C element causes the crippled growth (CG) cell degeneration. CG results from the inappropriate autocatalytic activation of the PaMpk1 MAPK pathway during growth, whereas this cascade normally signals stationary phase. Little is known about the control of such prion-like hereditary units involved in regulatory inheritance. Here, we show that another MAPK pathway, PaMpk2, is crucial at every stage of the fungus life cycle, in particular those controlled by PaMpk1 during stationary phase, which includes the generation of C. Inactivation of the third P. anserina MAPK pathway, PaMpk3, has no effect on the development of the fungus. Mutants of MAPK, MAPK kinase, and MAPK kinase kinase of the PaMpk2 pathway are unable to present CG. This inability likely relies upon an incorrect activation of PaMpk1, although this MAPK is normally phosphorylated in the mutants. In PaMpk2 null mutants, hyphae are abnormal and PaMpk1 is mislocalized. Correspondingly, stationary phase differentiations controlled by PaMpk1 are defective in the mutants of the PaMpk2 cascade. Constitutive activation of the PaMpk2 pathway mimics in many ways its inactivation, including an effect on PaMpk1 localization. Analysis of double and triple mutants inactivated for two or all three MAPK genes undercover new growth and differentiation phenotypes, suggesting overlapping roles. Our data underscore the complex regulation of a prion-like element in a model organism.
Keywords: regulatory inheritance, prion-like element, MAP kinase, filamentous fungi, Podospora anserina
MAP kinase (MAPK) modules are signaling devices used by all eukaryotes to signal various developmental programs or to respond to environmental stresses (Widmann et al. 1999). They are composed of three kinases that sequentially phosphorylate their targets: a MAPKKK that phosphorylates one or several MAPKK(s), which in turn phosphorylate(s) one or several MAPK(s). These latter kinases phosphorylate various cellular components, especially transcription factors, resulting in gene expression modification (Garrington and Johnson 1999). These MAPK modules are embedded in more complex regulatory networks, including other MAPK pathways (Saito 2010), whose complexity is known to generate emergent properties that rule cellular behavior (Neves and Iyengar 2002; Ferrell et al. 2009). This includes bistability, memory, hysteresis, and threshold responses. In some instances, the alternate states adopted by the cells are so stable that they can be transmitted through cell divisions, as exemplified by the crippled growth (CG) degeneration of the filamentous fungus Podospora anserina (reviewed in Lalucque et al. 2010). This kind of regulatory inheritance is now suspected of playing an important role in various physiological processes, including tumor formation, cell differentiation, and degeneration (Blagosklonny 2005; Lalucque et al. 2010), yet it is still poorly studied.
In filamentous fungi, MAPKs are mostly studied for their involvement in the infectious process of plant pathogenic species (reviewed in Zhao et al. 2007) and in model saprobes such as Aspergillus nidulans (Bussink and Osmani 1999; Kawasaki et al. 2002) and Neurospora crassa (Maerz et al. 2008; Fleissner et al. 2009) or in the human pathogen A. fumigatus (May et al. 2005). Through complete genome sequence analyses, three MAPK pathways have been characterized in most filamentous ascomycetes (Zhao et al. 2007; Rispail et al. 2009). One is orthologous to the MPK1 pathway involved in the control of the cell wall integrity in Saccharomyces cerevisiae, the other to the FUS3 pathway involved in the control of cell fusion during sexual reproduction, and the third one to the HOG1 pathway regulating the response to high osmolarity. Unlike yeasts, in which many connections between the MAPK pathways have been described, little is known about the entire MAPK network and potential cross-talks in filamentous fungi. Indeed, comparative analyses of the MAPK pathways have been carried out only in Cochliobolus heterostrophus (Igbaria et al. 2008) and N. crassa (Maerz et al. 2008), although inactivation of the three pathways has been performed in more fungi. Only in N. crassa was the complete inactivation set for the nine kinase genes reported; as in C. heterostrophus, only inactivation of the MAPK gene was achieved (Igbaria et al. 2008). Moreover, a double mutant inactivated for two MAPKs, the MPK1-like and HOG1-like MAPK of C. heterostrophus, has been described (Igbaria et al. 2008).
In P. anserina, we previously identified a MAPK pathway composed of the PaASK1 MAPKKK, the PaMKK1 MAPKK, and the PaMpk1 MAPK, which appears to be able to generate C, a hereditary unit that has properties exhibited by prions. Especially, C spreads in the cytoplasm in an infectious manner (Silar et al. 1999; Kicka and Silar 2004; Kicka et al. 2006). When present in dividing cells, C triggers the CG degenerative process, characterized by slower mycelium growth, higher accumulation of pigment, and female sterility. However, unlike classical prions based on alternate conformations of proteins, the C hereditary unit seems to rely on the “ON” state of the PaMpk1 cascade. As described for the JNK cascade of Xenopus oocytes (Bagowski and Ferrell 2001), the PaMpk1 cascade would present a positive regulatory loop whereby a component downstream of the cascade upregulates an upstream component in trans (Kicka et al. 2006). Therefore, once one molecule of the pathway is activated, the activation could spread to the other nonactive molecules and lock them in the ON state. In this system, the hereditary unit would not be a protein with a particular conformation, but the activation status of the MAPK cascade, implying that any active component of the entire cascade could propagate the infectious activation process. Accordingly, the genetic control of this element is highly complex and depends on numerous genes (Haedens et al. 2005). The PaMpk1 cascade normally signals stationary phase, because mutants of the cascade are unable to differentiate aerial hyphae, to accumulate pigments and to undergo sexual reproduction, which are three hallmarks of the P. anserina stationary phase (Kicka and Silar 2004). Activation of the cascade and thus the production of C is a normal part of P. anserina development during stationary phase. As a consequence, CG cultures can be easily recovered by incubating hyphae into stationary phase and replicating them onto fresh medium (Silar et al. 1999). Sustained activation of the cascade in the hyphae renewing growth results in the CG altered pattern of growth. Interestingly, other genes controlling CG appeared to be involved in the correct nuclear localization in the mycelium of PaMpk1 rather than its phosphorylation (Kicka et al. 2006; Jamet-Vierny et al. 2007). To date, in no other fungus was a process akin to CG detected. However, because CG of P. anserina is highly sensitive to nutrient conditions (Haedens et al. 2005), it may remain undetected in other fungi.
Involvement of the PaMpk1 MAPK pathway was discovered through a traditional genetics approach, i.e., we identified mutants unable to develop CG (IDC mutants with Impaired Development of CG) and found that two of the mutated genes encode the MAPKKK and MAPKK of the cascade (Kicka and Silar 2004; Kicka et al. 2006). Reverse genetics showed that the PaMpk1 MAPK gene also controls CG (Kicka et al. 2006). Here, we present a thorough analysis of the MAP kinases of P. anserina and investigate the roles of these kinases in the physiology of this fungus, especially with regard to CG. We show that another MAP kinase cascade, composed of PaTLK2 MAPKKK, PaMKK2 MAPKK, and PaMpk2 MAPK is crucial at all stages of the P. anserina life cycle and for the development of CG, while the third cascade composed of PaHOK3, PaMKK3, and PaMpk3 is solely dedicated to signaling osmotolerance. Part of the phenotypes of PaMpk2 is possibly due to an indirect action on the PaMpk1 pathway. With the aim of uncovering a redundant role of the MAPK, we constructed all combination of mutants lacking two or all three P. anserina MAPK kinases. The data showed an overlap in the activity of the kinases restricted to female gamete differentiation, branching, and direction of apical growth. In addition to being unable to present CG, strains devoid of all three MAPKs had a very limited repertoire of differentiation processes, although they normally mated as males and aged as wild-type strains.
Materials and Methods
Strains and growth conditions
All the strains used in this study derived from the “S” (uppercase S) wild-type strain that was used for sequencing (Espagne et al. 2008). The genome sequence and EST derived from the S strain are available at http://podospora.igmors.u-psud.fr. The ΔPaMpk1, IDC404 PaMKK1, and IDC118 PaASK1 mutants have been previously described and differ from wild type by a single mutation (Kicka and Silar 2004; Kicka et al. 2006) as do the pks1-193 mutants (Coppin and Silar 2007). Construction of the Δmus51::su8-1 strain lacking the mus-51 subunit of the complex involved in end joining of broken DNA fragments was described previously (Lambou et al. 2008). DNA integration in this strain proceeds almost exclusively by homologous recombination. Standard culture conditions, media, and genetic methods for P. anserina have been described (Rizet and Engelmann 1949) and the most recent protocols can be accessed at http://podospora.igmors.u-psud.fr/more.php. The M2 minimal medium is a medium in which carbon is supplied as dextrin and nitrogen as urea. Ascospores do not germinate on M2, thus germination is assayed with a specific G medium containing ammonium acetate. The methods used for nucleic acid extraction and manipulation have been described (Ausubel et al. 1987; Lecellier and Silar 1994). Transformation of P. anserina protoplasts was carried out as described previously (Brygoo and Debuchy 1985).
Detection of MAPK genes in the P. anserina genome
The full complement of MAP kinases of P. anserina, including MAPK, MAPKK, and MAPKKK, was determined by searching with BLAST (Altschul et al. 1990) the complete set of predicted P. anserina coding sequences (CDS) available at http://podospora.igmors.u-psud.fr with the S. cerevisiae MAPK (Slt2p/Mpk1p, Kss1p, and Hog1p), MAPKK (Mkk1p, Mkk2p, ste7p, and Pbs2p) and MAPKKK (Bck1p, Ste11p, Ssk2p, and ssk22p) as queries. Nine CDSs resembling these queries with significant score were retrieved. Phylogenetic trees constructed with the S. cerevisiae proteins as well as that of MAPK of other filamentous fungi available in GenBank showed that three complete MAPK cascades with one MAPK, one MAPKK, and one MAPKKK in each pathway, are present in P. anserina (Supporting Information, Table S1). One cascade composed of PaASK1, PaMKK1, and PaMpk1, is orthologous to the S. cerevisiae cell integrity pathway (Mpk1(Slt2)-like). The cascade composed of PaTLK2, PaMKK2, and PaMpk2, is orthologous to the pheromone/filamentation pathway (KSS1/FUS3-like) and the last one, composed of PaHOK3, PaMKK3, and PaMpk3, is orthologous to the high osmolarity pathway (Hog1-like). As observed in the other MAPK pathways described (Rispail et al. 2009), the MAPKKKs are larger than the MAPKKs and MAPKs, probably because cis-regulatory domains are present in the polypeptides, as demonstrated for the PaASK1 MAPKKK (Kicka and Silar 2004). Expressed sequence tags (ESTs) are found that derive from all the corresponding genes except for PaASK1, which has been shown to be weakly expressed (Kicka and Silar 2004).
Deletions of the MAPK, MAPKK, and MAPKKK genes
PaMpk2 was inactivated by replacing the PaMpk2 CDS with a hygromycin-resistance marker. The flanking regions of the PaMpk2 gene were amplified by PCR, using the primers fus3gauche and fus3G-sphI for the upstream sequence and fus3D-NotI and fus3droit for the downstream sequence (Table S2). Two primers contain sites for restriction enzymes to help in cloning. The PCR products were digested with SphI/HindIII for the upstream region and with HindIII/NotI for the downstream region and cloned into the pMO-CosX vector (Orbach 1994) digested with SphI and NotI. The plasmid recovered was then linearized with HindIII and introduced into P. anserina by transformation. Numerous transformants were obtained, ∼25% of which developed unpigmented thalli. Southern blot analysis confirmed that for three unpigmented transformants, the PaMpk2 gene was correctly deleted. One such mutant was selected for further phenotypic analyses.
PaMKK2 was inactivated by inserting through a single crossing-over event the entire pBC-hygro vector (Silar 1995) into the DNA sequence corresponding to the catalytic domain of PaMKK2. To this end, 500 bp of the catalytic domain of PaMKK2 were amplified with the primers MKK2A and MKK2B (Table S2). The resulting amplified product was cloned into pBC-hygro at the SmaI site to yield plasmid pDMKK2. This plasmid was introduced by transformation into the mus51::su8-1 strain. Numerous hygromycin-resistant transformants displaying the same phenotypes as the ΔPaMpk2 mutants were obtained. One, ΔPaMKK2, was selected for further analyses and crossed with the wild-type strain. Progeny analyses showed cosegregation of resistance and the germination and mycelium phenotypes. This mutant was used in subsequent analyses.
To construct the deletion cassette of the entire coding sequence of PaTLK2, a nourseothricin resistance marker was fused by PCR with the upstream or the downstream sequence of the PaTLK2 gene. Two 1-kb regions, located upstream and downstream of the TLK2 coding sequence, were amplified from the wild-type genomic DNA with primers TLK-1F and Mk_TLK2-2R for the 5′ region and Mk-TLK2-3F and TLK2-4R for the 3′ region (Table S2). The primers contained additional bases allowing fusion with the nourseothricin resistance marker (in lowercase in Table S2). This marker (nourseo) was amplified using the TLK2_Mk-2F and TLK2_Mk-3R primers from pBC-Nourseo, a plasmid containing the nourseothricin resistance cassette inserted at the XmnI site of pBluescript. In a second step, two amplification reactions were performed using the distal primers to obtain the fusion of two PCR products, the 5′ region/nourseo and nourseo/3′ region. The mixture of these two PCR products was used to transform a mus51::su8.1 strain. Numerous nourseothricin-resistant transformants were obtained thanks to three crossing-over events between the transformed PCR products and the P. anserina genome. Three transformants with a phenotype similar to ΔPaMpk2 were selected for further analyses. They were crossed with wild-type, and homokaryotic nourseothricin-resistant progeny were analyzed by Southern blotting to confirm the deletion of PaTLK2 in the resistant progeny of the three candidates.
To construct the deletion cassette of the entire coding sequence of PaMpk3, a 1804-bp-long PaMpk3 5′ noncoding fragment was PCR amplified using primers HOG1AML and HOG1AMR (Table S2), along with a 1827-bp-long PaMpk3 3′ noncoding fragment using primers HOG1VML and HOG1VMR (Table S2). Both fragments were cloned into the pGEMT vector (Promega). Because a SalI site and an ApaI site were present in HOG1AML and HOG1AMR, respectively, the corresponding 5′ noncoding fragment was released from the pGEMT vector by performing a SalI/ApaI double digestion. A NotI site in HOG1AML and a SalI one in HOG1VMR allowed the release from the pGEMT vector of the corresponding 3′ noncoding fragment. These two fragments were ligated with the pBC-hygro plasmid (Silar 1995) previously hydrolyzed with the ApaI and NotI enzymes. This generated the pΔPaNox3 plasmid. P. anserina transformation was performed using the pΔPaNox3 plasmid previously linearized at the unique SalI site to generate homologous recombination ends. A total of 79 hygromycin-B resistant transformants were obtained and 3 of them showing the same growth defect in osmotic-rich medium were selected for further analyses. After purification of the primary transformants by appropriate crossing, their genomic DNA was extracted and deletion of PaMpk3 was confirmed by PCR and Southern blot analyses.
To delete PaMKK3, a 530-bp-long PaMKK3 5′ noncoding fragment was PCR amplified using primers M3K2GF and M3K2GR (Table S2), as well as a 516-bp-long PaMKK3 3′ noncoding fragment using primers M3K2DF and M3K2DR (Table S2). Both fragments were cloned into the pGEMT vector. Because an EcoRV site and a XbaI site were present in M3K2GF and M3K2GR, respectively, the corresponding 5′ noncoding fragment was released from the pGEMT vector by performing a EcoRV/XbaI double digestion and cloned into the pBC-hygro plasmid at the corresponding sites. A XhoI site in M3K2DF and an EcoRV one in M3K2DR allowed the release from the pGEMT vector of the corresponding 3′ noncoding fragment, which was then cloned at the XhoI/EcoRV sites into the plasmid harboring the PaMkk3 5′ noncoding fragment, generating the pΔPaMkk3 plasmid. Transformation of the P. anserina Δmus51::su8-1, pks1–193 strain (Lambou et al. 2008) was performed using pΔPaMkk3 plasmid previously linearized at the unique EcoRV site to generate homologous recombination ends. Three hygromycine B-resistant transformants were selected for further analyses. All three transformants showed a similar phenotype as the PaMpk3 mutants. After appropriate crosses, both the Δmus51::su8-1 and the pks1-193 mutant alleles segregated from the ΔPaMkk3 allele. DNA was extracted from these three purified transformants to confirm the deletion of the PaMkk3 gene using PCR and Southern blot analyses.
To delete PaHOK3 a 492-bp-long PaHok3 5′ noncoding fragment was PCR amplified using primers HOK3GF and HOK3GR (Table S2), as well as a 648-bp-long PaHOK3 3′ noncoding fragment using primers HOK3DF and HOK3DR (Table S2). Both fragments were cloned into pGEMT vector. Because an EcoRV site and a XbaI site were present in HOK3GF and HOK3GR, respectively, the corresponding 5′ noncoding fragment was released from the pGEMT vector by performing an EcoRV/XbaI double digestion and cloned into the pBC-hygro plasmid at the corresponding sites. A XhoI site in HOK3DF and an EcoRV site in HOK3DR allowed the release from the pGEMT vector of the corresponding 3′ noncoding fragment, which was then cloned at the XhoI/EcoRV sites in the plasmid harboring the PaHOK3 5′ noncoding fragment, generating the pΔPaHok3 plasmid. Transformation of the P. anserina Δmus51::su8-1, pks1-193 strain (Lambou et al. 2008) was performed using the pΔPaHok3 plasmid previously linearized at the unique EcoRV site to generate homologous recombination ends. Three hygromycine B-resistant transformants were selected for further analyses. All three transformants showed a phenotype similar to the PaMpk3 mutants. After appropriate crosses, both the Δmus51::su8-1 and the pks1-193 mutant alleles segregated away from the ΔPaHOK3 allele. DNA was extracted from these three purified transformants to confirm the deletion of the PaHOK3 gene using PCR and Southern blot analyses.
Complementation of ΔPaMpk2 and creation of constitutive mutants
The PaMpk2 gene including its own promoter was amplified by PCR using the primers Mpk2_for and Mpk2_rev (Table S2) and cloned into the pGEMT vector. The resulting plasmid was introduced with the pBC-phleo vector by cotransformation into a ΔPaMpk2 mutant strain. The phenotype of 36 phleomycin-resistant transformants was tested; 18 of them presented aerial hyphae and pigmentation as did the wild type. Four were crossed with wild type. Progeny analyses revealed that the transgene had integrated in an ectopic locus and complemented all the defects of the ΔPaMpk2 mutant strain in the four transformants.
Directed mutagenesis was used to change the S210IADT214 (SXXXT) motif in MKK2 into a phosphomimetic DXXXD motif. To this end, a PCR reaction was performed using primers MKK2_GF_CH1_for and MKK2_GF_CH1_rev and plasmid pBC-phleo (Silar 1995) containing the PaMKK2 gene as template. After DpnI digestion to eliminate the original plasmid, bacteria were transformed with the reaction product. Candidate plasmids carrying the mutant MKK2 allele were recovered and confirmed by sequencing. The plasmid was then introduced by transformation into a ΔPaMKK2 mutant strain and several phleomycin-resistant transformants were obtained. Candidates were analyzed by crossing with wild type. Partial or complete complementation of the germination defect of the ΔPaMKK2 mutant was observed for the two transformants used in the present study, indicating that the transgenes were expressed. As control, a wild-type PaMKK2 allele was introduced by transformation into the ΔPaMKK2 mutants.
Construction of P. anserina mutants devoid of three MAP kinase genes
The ΔPaMpk3 mutant was first crossed as female with the ΔPaMpk1 and ΔPaMpk2 mutants. In the progeny of the crosses, double ΔPaMpk1 ΔPaMpk3 and ΔPaMpk2 ΔPaMpk3 mutants were recovered as having the mycelium phenotype of ΔPaMpk1 and ΔPaMpk2, respectively, and the osmosensitivity of ΔPaMpk3. To construct the double ΔPaMpk1 ΔPaMpk2 mutants, advantage was taken of the fact that heterokaryotic Δmat/ΔPaMpk1 strains are fertile as females (Jamet-Vierny et al. 2007), unlike the ΔPaMpk1 and ΔPaMpk2 mutants. Because the Δmat nuclei are unable to engage in fertilization, crossing Δmat/ΔPaMpk1 as female with ΔPaMpk2 yielded a progeny with double mutants. To circumvent the fact that ΔPaMpk2 naturally pigmented ascospores do not germinate, the cross was set up on plates containing tricyclazole, an inhibitor of melanin biosynthesis (Coppin and Silar 2007). Unpigmented ascospores germinated efficiently and the double ΔPaMpk1 ΔPaMpk2 mutants were identified by crossing candidates with wild type and observing the segregation of the single mutants in the progeny. The triple ΔPaMpk1 ΔPaMpk2 ΔPaMpk3 mutants were constructed using the same scheme as above but with heterokaryotic Δmat/ΔPaMpk1 ΔPaMpk3 strains as female and ΔPaMpk2 ΔPaMpk3 as males. Because it was the first recovery of a eukaryote lacking all its MAPK, the genotype of the triple ΔPaMpk1 ΔPaMpk2 ΔPaMpk3 mutants was also verified by backcrossing the mutants with wild type and observing segregation of the three mutations in the progeny.
Phenotypic analysis
CG was assayed both as described (Silar et al. 1999) and according to the following protocol. The strains to be tested were inoculated at the edge of M2 Petri plates. After 6 days of growth, the resulting cultures were sliced from the growing edge to the inoculation point every millimeter. Slices were then transferred onto fresh plates containing either M2 or M2 supplemented with 5 g/liter of yeast extract. Slices were deposited in such a way that the aerial and submerged parts of the culture could give birth to new hyphae. Because C, the hereditary unit responsible for CG, is induced in stationary phase, the area of the slices corresponding to the inoculation point regenerated a CG mycelium, while the part corresponding to the growing edge regenerated a normal growing (NG) mycelium, yielding a mosaic culture (Figure 1). CG developed on M2 + yeast extract and not on M2 as described (Haedens et al. 2005).
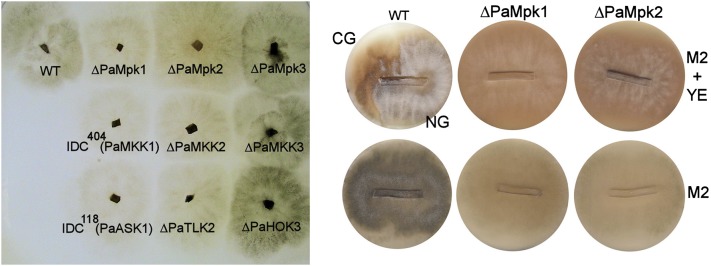
Figure 1 .
(Left) Mycelium phenotypes of the indicated strains grown on M2 medium. (Right) CG test of WT and the ΔPaMpk1 and ΔPaMpk2 mutants. Slices of the indicated strains were inoculated on M2 medium (M2) and M2 medium supplemented with yeast extract (M2 + YE). Parts of the inoculated slices corresponding to the growing edge are on the right; parts corresponding to the stationary phase area are on the left. The top of the slice corresponding to the aerial mycelium is oriented toward the top of the plate. Wild-type develops CG on M2 + YE (visible on the left of the culture as the pigmented area that lacks aerial hyphae) but not on M2 as previously described (Haedens et al. 2005), while the MAPK mutants never develop CG on either medium. Note the similar mycelium phenotypes of the ΔPaMpk1 and ΔPaMpk2 mutants.
Anastomosis was assayed as described (Kicka and Silar 2004). Briefly, strains carrying the investigated allele and either leu1-1 or lys2-1 were constructed and the ability of these strains to form a prototrophic mycelium was assayed on M2 minimal medium. Branching was observed as described (Arnaise et al. 2001) and differentiation of appressorium-like structures was observed as reported (Brun et al. 2009). Efficiency of cellulose degradation was measured as in Brun et al. (2009), longevity as in Silar and Picard (1994), and fertility as in Rizet and Engelmann (1949). Hyphal interference including peroxide production and cell death was assayed as described (Silar 2005) and DAB (diaminobenzidine) and NBT (nitroblue tetrazolium) staining to detect production of superoxides and peroxides, respectively, as in Malagnac et al. (2004).
Western blot analysis
Phosphorylation of PaMpk1 and PaMpk2 was evaluated as described (Kicka et al. 2006). For mycelium assays, the mycelium was recovered after 48 hr of growth on M2 Petri dishes covered with a cellophane layer. At this time, the mycelium formed a disk of ∼3 cm in diameter. The mycelium was totally collected or as three concentric samples corresponding to 1, 2, and 3 days of growth. Proteins were then extracted as indicated previously (Kicka et al. 2006). Ascospores were collected overnight on cellophane overlaying a medium containing agar, NaCl (10 g/liter), and sterile water. Noninduced spores were stored in liquid nitrogen, while spores to be induced were transferred along with the cellophane on G medium for 2 hr. Perithecia were obtained at the confrontation lines between mat+ and mat− strains. Three days after inoculation, fertilization proceeded at the confrontation lines. Developing perithecia were collected 2 and 4 days later by scraping them from the cross-plates. To extract proteins from ascospores and perithecia, the samples were frozen with a steal bead in liquid nitrogen and crushed by shaking in a Mikro-Dismembrator (Sartorius) at 2600 rpm for 90 sec. After adding loading buffer, the samples were incubated at 100° for 5 min, centrifuged, and the resulting supernatants were stored at −20°.
Microscopy
Mycelia were grown directly on a slide covered with a thin M2 medium layer. Mounting was performed in water supplemented with 0.5 μg/ml of DAPI. Pictures were taken with a Leica DMIRE 2 microscope coupled with a 10-MHz Cool SNAPHQ charge-coupled device camera (Roper Instruments). They were analyzed with ImageJ. The GFP filter was the GFP-3035B from Semrock (exciter, 472 nm/30; dichroïc, 495 nm; and emiter, 520 nm/35).
Results
The PaMpk2 pathway is the major MAPK cascade controlling development in P. anserina
The entire set of MAPK, MAPKK, and MAPKKK was searched in the P. anserina genome sequence (Table S1). In addition to the PaMpk1 pathway, two additional, complete, and expressed MAPK pathways were found. One is related to the S. cerevisiae STE11–Ste7–FUS3 cascade and is composed of PaTLK2 (MAPKKK), PaMKK2 (MAPKK), and PaMpk2 (MAPK). The other is related to the SSK2/22–Pbs2–Hog1 cascade and is formed by PaHOK3 (MAPKKK), PaMKK3 (MAPKK), and PaMpk3 (MAPK). To investigate the role of these cascades, the six genes were inactivated (see Materials and Methods) and the phenotypes of the mutants were examined (see Figure S1 for the phenotypes that may be studied in P. anserina and Table 1 for a summary of the phenotypes observed).
Table 1 . Phenotypes associated with inactivation of the PaMpk1, PaMpk2, and PaMpk3 MAPK modules.
| WT | ΔPaMpk1 | PaMKK1 (IDC404) | PaASK1 (IDC118) | ΔPaMpk2 | ΔPaMKK2 | ΔPaTLK2 | ΔPaMpk3 | ΔPaMKK3 | ΔPaHOK3 | Δ1 Δ2 | Δ1 Δ3 | Δ2 Δ3 | Δ1 Δ2 Δ3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ascospore | ||||||||||||||
| Germination (%) | 100 | 100 | 100 | 100 | <0.01 | <0.01 | <0.01 | 100 | 100 | 100 | <0.01 | 100 | <0.01 | <0.01 |
| Mycelium | ||||||||||||||
| Apical growth speed (mm/day) | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 5.7 + 0.1 | 6.3 + 0.1 | 6.3 + 0.1 | 5.0 + 0.1 |
| Branching | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | Altered | WT | WT | Altered |
| Anastomosis | +++ | ++ | NT | NT | − | NT | NT | +++ | NT | NT | NT | NT | NT | NT |
| Vegetative incompatibility with lowercase s | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Appressorium-like differentiation | + | + | + | + | − | − | − | + | + | + | − | + | − | − |
| Aerial hyphae | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Pigments | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Longevity (cm) | 10.5 ± 1.0 | 11.5 ± 1.5 | 10.5 ± 1.5 | 11.5 ± 1.5 | 10.5 ± 1.0 | 10.0 ± 1.0 | 9.5 ± 1.5 | 11.0 ± 1.5 | 11.0 ± 2.0 | 10.5 ± 1.5 | 10.5 ± 1.0 | 10.5 ± 1.5 | 9.5 ± 0.5 | 11.0 ± 0.5 |
| Hyphal interference | + | − | − | − | +/− | +/− | +/− | + | + | + | − | − | +/− | − |
| CG on M2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| CG on M2 + YE | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Gamete | ||||||||||||||
| Microconidia (male) | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ascogonia (female) | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| Protoperithecia (female) | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Sexual development postfertilization | ||||||||||||||
| Envelope development | + | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Dikaryon formation | + | + | + | + | + | NT | NT | + | + | + | NT | + | NT | NT |
| Meiosis | + | + | + | + | + | + | + | + | + | + | NT | + | NT | NT |
| Ascosporogenesis | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
NT, not tested; WT, similar to wild type.
First, as for the PaMpk1 pathway, the MAPK, MAPKK, and MAPKKK mutants exhibited the same phenotypes, indicating that the PaMpk2 and PaMpk3 pathways are not branched, at least in the conditions investigated here (Figure 1, Table 1). Second, the PaMpk3 pathway had no role in any developmental process of P. anserina in laboratory conditions. Its role appeared restricted to osmotolerance and resistance to antifungal compounds (Table 1, Table S3), hence the name of the PaHOK3 MAPKKK (hyperosmolarity kinase). Third, the PaMpk2 pathway appeared to be the major regulator of P. anserina development, since ascospores, mycelia, and fruiting bodies were affected in the mutants of this pathway (Table 1). We thus named the MAPKKK PaTLK2 (throughout the lifecycle kinase). Since the ΔPaMpk2, ΔPaMKK2, and ΔPaTLK2 mutants displayed the same phenotypes, only experiments with ΔPaMpk2 are described below for the sake of clarity. Complementing ΔPaMpk2 by introducing by transformation a wild-type allele restores a wild-type phenotype showing that all phenotypes are due to inactivation of PaMpk2.
Ascospore germination is impaired in ΔPaMpk2 mutants
ΔPaMpk2 ascospores looked like wild-type ascospores. However, in the progeny of wild type × ΔPaMpk2 crosses, ΔPaMpk2 ascospores germinated with very low efficiency, while wild-type ascospores germinated with ∼100% efficiency. Evaluation of the germination frequency in ΔPaMpk2 × ΔPaMpk2 crosses showed that ∼1 out of 10,000 ΔPaMpk2 ascospores germinated. Impairment of PaMpk2 germination occurred at an early stage since no germination peg was produced. This ascospore-autonomous germination phenotype was identical to the one exhibited by the PaNox2 and PaPls1 null mutants (Malagnac et al. 2004; Lambou et al. 2008). PaNox2 encodes a NADPH oxidase and PaPls1, a tetraspanin. The germination defect of the PaNox2 and PaPls1 mutants can be relieved by removing melanin from ascospore. The same was true for the germination defect of the ΔPaMpk2 mutant since addition of tricyclazole, an inhibitor of melanin synthesis, in the cross-plates resulted in efficient germination of ΔPaMpk2 ascospores. Similarly, the nonpigmented pks1-193 ΔPaMpk2 ascospores obtained after crossing ΔPaMpk2 with pks1-193, a mutant that lacks melanin at all stage of its life cycle (Coppin and Silar 2007), germinated efficiently. This provided a simple method to recover ΔPaMpk2 mycelia and indicated that PaMpk2 and Pls1/Nox2 could act in the same pathway.
Mycelium growth and differentiation in the MAPK mutants
Vegetative growth of ΔPaMpk2 mycelia proceeded at the same speed as wild-type mycelia and branching at the growing edge was normal. However, several features occurring beyond the growing edge were altered. First, hyphal fusion (anastomosis) was impaired in ΔPaMpk2. This was detected by the inability of ΔPaMpk2 to form prototrophic heterokaryotic mycelia from auxotrophic homokaryotic mycelia. Anastomosis was completely abolished in homozygous assays of ΔPaMpk2 mutants and occurred very infrequently in heterozygous assays with wild type. This is different from what occurs in the PaMpk1 mutants, as these latter mutants normally engage in anastomosis in heterozygous confrontations and are only partially impaired in homozygous confrontations (Kicka and Silar 2004). As expected of mutants unable to engage anastomoses, the ΔPaMpk2 mutants did not accumulate dead cells at the contact point with an incompatible strain [the “s” (lowercase s) strain was used as tester; Rizet 1952]. Interestingly, the ΔPaMpk1 mutants also did not accumulate dead cells at the contact with the s strain. Since they were able to engage in anastomosis, later stages of the incompatibility reaction were impaired in these mutants.
Unlike the PaMpk1 and PaMpk3 mutants, the PaMpk2 mutants were unable to correctly differentiate the appressorium-like structures that enable P. anserina to penetrate cellophane (Brun et al. 2009). As seen in Figure S2, the mycelium of ΔPaMpk2 could reorient its growth toward cellophane and to produce swellings, but failed to produce the needle-like hyphae that penetrate the substrate. This was accompanied by a strong decrease in the capacity of the mutant to degrade cellulose, as the mutants degraded half the cellulose degraded by the wild type in 7 days (Table S4). Interestingly, while able to differentiate appressorium-like structures, the ΔPaMpk1, but not the ΔPaMpk3 mutants, also presented a diminished ability to degrade cellulose (Table S4).
Senescence and longevity were not affected in the ΔPaMpk2 mutants (as in the case of the other MAPK mutants; Table 1). Additionally, the ΔPaMpk2 mutants had an abnormal pattern of staining with DAB and NBT, which are supposed to measure accumulation of peroxides and superoxides, respectively (Figure S3) (Munkres 1990). This shows that mycelium redox activities are modified in the ΔPaMpk2 mutants as in the ΔPaMpk1 mutants (Malagnac et al. 2004) and is in line with their impaired ability to degrade cellulose as redox activities are known to be involved in the degradation of this polysaccharide (Morel et al. 2009). Unlike the ΔPaMpk1 mutants, the ΔPaMpk2 mutants were slightly affected in hyphal interference, a defense mechanism exerted by P. anserina when it encounters another filamentous fungus (Silar 2005). This process is associated with an oxidative burst at the confrontation with the contestant. Moreover, when P. anserina encounters Penicillium chrysogenum, it kills its hyphae upon contact. While hyphal inteference is completely abolished in the PaMpk1 mutants (Figure S4; Silar 2005), it is only partially impaired in the PaMpk2 mutants and not at all in the PaMpk3 mutants (Figure S4, Table 1).
Because MAPK are known to signal not only development but also stress responses, resistance to various stresses and drugs was assayed (Table S3). If the Mpk3 pathway is clearly devoted to osmotic stress response in P. anserina, neither the Mpk1 nor the Mpk2 cascades have stress responses as their primary role. Most noticeable was the resistance of the ΔPaMpk2 mutants to cell wall stresses. This outcome indicates that the cell walls of these mutants may have original properties, which remain to be characterized.
Finally, the same four hallmarks of stationary phase in P. anserina also affected in the ΔPaMpk1 mutants were altered in the ΔPaMpk2 mutants (Figure 1): they accumulated much less pigment than the wild type, differentiated few aerial hyphae, and were unable to differentiate fruiting bodies and to develop CG.
Fruiting body development in the ΔPaMpk2 mutants
The ΔPaMpk2 mutants could differentiate male (spermatia) and female (ascogonia) gametes, but, like ΔPaMpk1 (Kicka and Silar 2004), the mutants did not develop protoperithecia and perithecia. To check at which stage perithecium development was impaired, mosaics were first constructed as previously described (Jamet-Vierny et al. 2007). Mosaics of the ΔPaMpk2 and pks1-193 mutants of opposite mating types produced pigmented and nonpigmented perithecia, indicating that expression of PaMpk2 was not required in the envelope of the developing fructification, similarly to PaMpk1 (Jamet-Vierny et al. 2007). However, ΔPaMpk2 mat+/ΔPaMpk2 mat−/Δmat mosaics were nearly sterile, as only few abnormal-looking perithecia were produced. This is unlike the ΔPaMpk1 mat+/ΔPaMpk1 mat−/Δmat mosaics that produce numerous mature fruiting bodies (Jamet-Vierny et al. 2007). However, because true heterokaryotic mosaic needs anastomosis to form and because the ΔPaMpk2 mutants were unable to undergo anastomosis, these mosaics may not be completely informative. We thus constructed a true ΔPaMpk2 mat−/Δmat heterokaryon following a protocol described for N. crassa (Pandey et al. 2004). In such a heterokaryon, only PaMpk2 mat− nuclei can engage in fertilization, permitting to test whether PaMpk2 is required for fertilization and for the subsequent steps of sexual reproduction. To this end, the method of heterokaryon formation designed for N. crassa was adapted to P. anserina (Pandey et al. 2004). ΔPaMpk2 leu1-1 mat− and lys2-1 Δmat were mixed together in the presence of limiting amounts of lysine and leucine. True heterokaryons carrying nuclei from both strains were formed a few days later, likely through very rare cell fusion events different from anastomoses, and could be detected by their prolific growth on minimal medium. These were used as female partners in crosses with mat+ wild-type or the ΔPaMpk2 mat+ mutants as males. In both cases, a few perithecia that matured and ejected ascospores were obtained (Figure S5). This indicates that PaMpk2 is not required for fertilization, the dikaryotic stage, meiosis, and ascospore differentiation. However, in addition to normal-looking perithecia, numerous abnormally shaped perithecia were observed (Figure S5), suggesting that development was impaired even in the heterokaryon. Hence the defect of the ΔPaMpk2 mutants was most likely a combination of defects associated with the lack of PaMpk1 activity and the absence of anastomosis. This later defect could account for the lack of perithecium production in mosaics with Δmat and the partial rescue of fruiting body production in true heterokaryons. Anastomosis impairment of the mycelium is supported by the fact that grafted wild-type fruiting bodies onto ΔPaMpk2 mycelia did not mature, unlike what was observed when they were grafted onto wild-type mycelia (Silar 2011).
Lack of CG in the ΔPaMpk2 mutants
As seen in Figure 1, CG was not induced after passage into the stationary phase in mutants of the PaMpk2 pathway like those of the PaMpk1 cascade (Kicka and Silar 2004; Kicka et al. 2006). Since the ΔPaMpk2 hyphae were unable to efficiently fuse with other hyphae, even wild-type hyphae, we could not directly test whether the lack of CG in the ΔPaMpk2 mutants was due to the inability of the mutants to make C, to propagate C, or whether it was merely due to a masking of its effects on cell physiology. Indeed, these possibilities must be assayed by transfer of cytoplasm from donor to recipient thallus and hence require cell fusion (Kicka and Silar 2004; Kicka et al. 2006). The effect of the ΔPaMpk2 mutation on the functioning of the PaMpk1 pathway was thus measured by immunoblotting with specific antibodies.
Phosphorylation of PaMpk1 and PaMpk2 are independent
To investigate whether PaMpk2 is necessary for the phosphorylation of PaMpK1, the phosphorylation of PaMpk1 in the ΔPaMpk2 mutant was measured. Both MAPK were recognized by the same anti-p44/p42 and antiphospho-p44/p42 antibodies (Figure 2). As a first step, the phosphorylation of both PaMpk1 and PaMpk2 was evaluated throughout the life cycle in the wild-type strain. Both MAPKs were present and phosphorylated in 1-, 2-, and 3-day-old mycelia (Figure 2). They were also present and phosphorylated in developing fruiting bodies (perithecia). PaMpk1 and PaMpk2 were detected in ascospores. Neither MAPK was phosphorylated in noninduced ascospores. However, intense phosphorylation of PaMpk2 was detected in ascospores triggered to germinate, in agreement with its role during germination (Figure 2). To address the possibility that PaMpk2 acts downstream of PaNox2 and PaPls1, phosphorylation of PaMpk2 was measured in the ΔPaNox2 and ΔPaPls1 mutant ascospores (Figure S6). PaMpk2 was phosphorylated in both mutants to the same extent as in wild type, showing that PaNox2 and PaPls1 do not act upstream of PaMpk2 during germination.
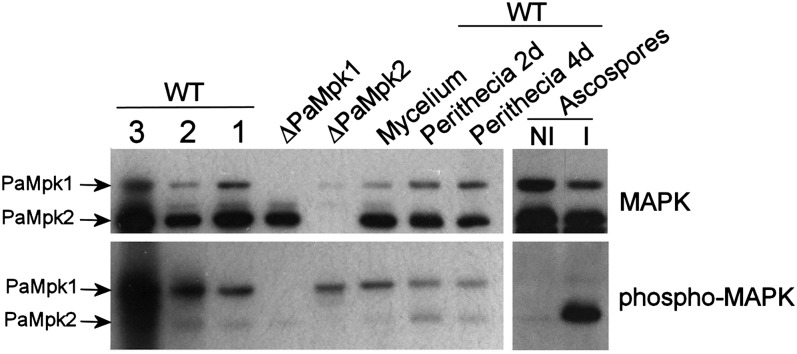
Figure 2 .
Immunoblot analysis of amounts of PaMpk1 and PaMpk2 (MAPK) and phosphorylation (phospho-MAPK) in wild type throughout the life cycle. Extracts from ΔPaMpk1 and ΔPaMpk2 mycelia were loaded as controls. Mycelium of 3-day-old WT cultures was separated into three zones corresponding to 1 day of growth (1), 2 days (2), and 3 days (3) or extracted as a whole (mycelium). Gel loading with extract corresponding to 3 days contained more proteins. PaMpk1 and PaMpk2 are present and phosphorylated in all three zones in roughly equal amounts. They are also present and phosphorylated in maturing 2-day- (perithecia 2 d) and mature 4-day-(perithecia 4 d)-old fruiting bodies. They are present in an unphosphorylated form in ascospores not induced for germination (NI) and PaMpk2 is phosphorylated upon induction of germination (I).
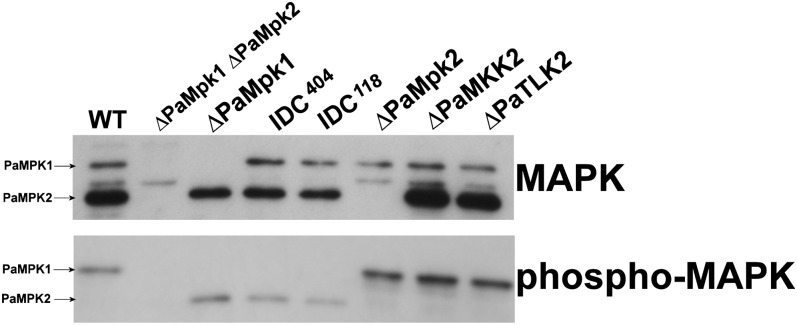
As seen in Figure 2, PaMpk1 was phosphorylated in the ΔPaMpk2 mutants and PaMpk2 was phosphorylated in the ΔPaMpk1 mutants, indicating that each MAPK was required for the phosphorylation of its own module but not for the other module. This was confirmed in further experiments where phosphorylation was measured in the MAPKK and MAPKKK mutants. Indeed, as described for the PaMKK1 and PaASK1 mutants (Kicka et al. 2006; Figure 3) in which phosphorylation of PaMpk1 is abolished, phosphorylation of PaMpk2 did not occur in the PaMKK2 and PaTLK2 mutants, confirming that PaMKK2 and PaTLK2 act upstream of PaMpk2. Moreover, MAPK phosphorylation was conserved in the MAPK, MAPKK, and MAPKKK mutants of the other pathway. Hence, the PaMpk2 pathway was not required for PaMpk1 phosphorylation and vice versa.
Figure 3 .
Amounts and phosphorylation of PaMpk1 and PaMpk2 were measured in the indicated strains in 3-day-old mycelia. Legend as in Figure 2.
Constitutive mutants show that PaMpk2 is not part of the C hereditary unit
To determine whether the PaMpk2 pathway is an integral part of C or merely controls its production, mutants expressing a constitutive allele of PaMKK2, PaMKK2c were constructed (Pages et al. 1994; Shiozaki et al. 1998). To this end, the S210IADT214 site of PaMKK2 was mutated in vitro to DIADD, and the mutant allele was introduced into ΔPaMKK2 by transformation with a phleomycin resistance gene as marker (see Materials and Methods). When crossed with wild-type strains of opposite mating type, several phleomycin-resistant transformants produced ascospores capable of germinating on noninducing medium, such as M2 (Figure 4A). This phenotype cosegregated with resistance to phleomycin, which was expected if the PaMpk2 pathway was constitutively activated by the PaMKK2c allele. To confirm this assertion, PaMpk2 and PaMpk1 phosphorylation was monitored in ascospores. As seen in Figure 4B, PaMpk2 was indeed phosphorylated in the noninduced ascospores carrying PaMKK2c, but not in wild-type ascospores. PaMpk1 remained nonphosphorylated, confirming that its phosphorylation does not rely on PaMpk2.
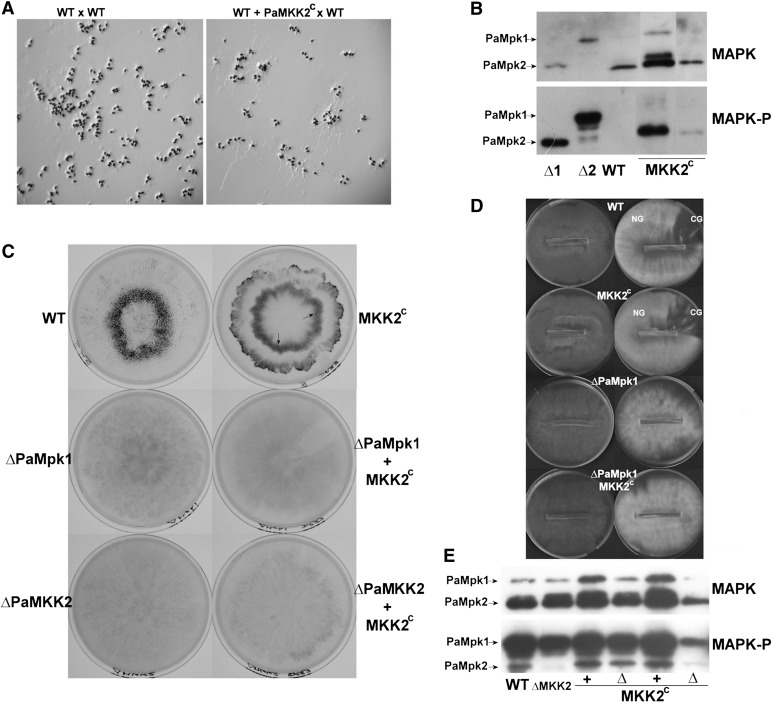
Figure 4 .
(A) Ascospores carrying the PaMKK2c mutation issued from a wild-type (WT) × PaMKK2c mutant cross germinate on a noninducing medium, while wild-type ascospores (issued from a wild-type × wild-type cross) do not. Note that the structures that decorate the ascospores on the WT × WT panel are appendages and not germinating hyphae. (B) Phosphorylation of PaMpk1 and PaMpk2 was measured on noninduced ascospores in WT and in two strains carrying constitutive PaMKK2c alleles (MKK2c). Extract from 3-day-old mycelia from ΔPaMpk1 (Δ1) and ΔPaMpk2 (Δ2) mutants were loaded to identify both MAPKs. (C) Phenotypes of strains carrying PaMKK2c constitutive allele. Fruiting bodies are visible as small black dots. They are numerous in WT and present in low amounts in strains carrying a PaMKK2c constitutive allele. Arrows point toward matured fructifications. Inactivation of PaMpk1 and PaMKK2 abolished fertility in strains with or without PaMKK2c. (D) CG is slightly diminished in strains carrying PaMKK2c. The presence of PaMKK2c does not rescue the CG defect of the ΔPaMpk1 mutants. (E) Phosphorylation of PaMpk1 and PaMpk2 is not modified in 3-day-old mycelia of strains carrying PaMKK2c in association with either a wild-type PaMKK2 allele (+) or ΔPaMKK2 (Δ). Fewer proteins were loaded in the well located on the far right.
Two transformants expressing constitutive PaMKK2 were selected for further studies. After crossing both with wild type, strains containing PaMKK2c in association with a wild-type PaMKK2+ allele were recovered in the progeny. They exhibited phenotypes similar to the ΔPaMKK2 PaMKK2c mutants. First, in addition to having ascospores germinating without induction, they possessed a mycelium with very few aerial hyphae. Their fertility was severely impaired (Figure 4C). However, heterokaryon tests showed that anastomosis formation was not affected in these strains. Moreover, they normally differentiated appressorium-like structures. Unlike what is expected if the PaMpk2 cascade were part of C, PaMKK2c strains did not exhibit constitutive CG, but on the contrary were slightly impaired in their ability to develop CG on M2 supplemented with yeast extract (Figure 4D). Intriguingly, phosphorylation of PaMpk2 (and PaMpk1) was not increased in mycelia of PaMKK2c strains, suggesting that the overall PaMpk2 phosphorylation level was tightly regulated in mycelia, possibly by unidentified phosphatases (Figure 4E).
Strains carrying the ΔPaMpk1 and PaMKK2c alleles were obtained after crossing PaMKK2c and ΔPaMpk1 mutants. Like the ΔPaMpk1 mutants, ΔPaMpk1 PaMKK2c were completely sterile (Figure 4C) and did not develop CG, showing that ΔPaMpk1 was epistatic over PaMKK2c. Overall, these data show that constitutively activating the PaMpk2 cascade did not result in constitutive CG, as expected if the cascade were part of the hereditary unit. On the contrary, it led to a defect resembling in part the inactivation of PaMpk1 and PaMpk2. Therefore, correct activation of PaMpk2 appears required for proper functioning of PaMpk1.
Finally, constitutive germination of the PaMKK2c mutant permitted to confirm that PaNox2 and PaPls1 did not act upstream of PaMpk2. Indeed, crossing the constitutive PaMKK2c mutant with ΔPaNox2 and ΔPaPls1 yielded progeny in which only PaMKK2c ascospores, and not PaMKK2c ΔPaNox2 and PaMKK2c ΔPaPls1 ascospores, germinated on M2.
PaMpk2 inactivation causes abnormal cell morphology in stationary phase and impairs PaMpk1 nuclear localization
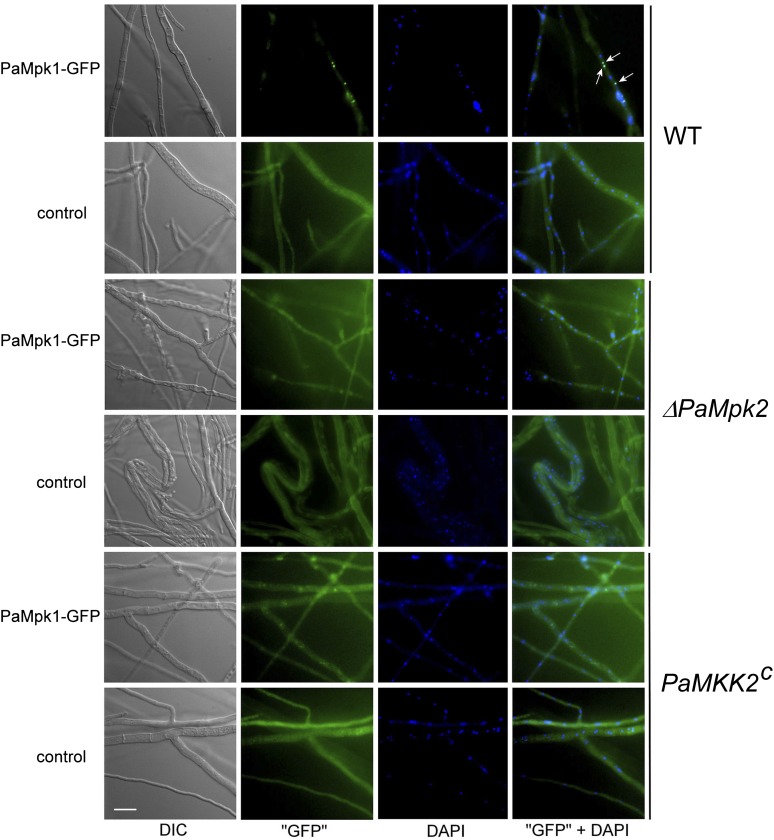
In previous studies (Kicka et al. 2006; Jamet-Vierny et al. 2007), we observed that correct nuclear localization of PaMpk1 is important for the activity of the cascade and for CG. ΔPaMpk2 and PaMKK2c strains carrying a transgene expressing PaMpk1–GFP were obtained by crossing the ΔPaMpk2 and PaMKK2c mutants with a strain carrying a PaMpk1–GFP transgene, which was functional because it could complement the defects of the ΔPaMpk1 mutants (Kicka et al. 2006). The apical hyphae of these strains appeared normal, and PaMpk1–GFP exhibited the same diffuse localization as the wild-type strain (Figure S7). As previously reported (Kicka et al. 2006), 10–20% of the wild-type nuclei accumulated PaMpk1–GFP during the stationary phase (Figure 5). Natural fluorescence in wild-type hyphae was not an issue because focalization of GFP in nuclei generated a signal clearly above the background level. On the contrary, we could not be entirely sure of the localization of PaMpk1 in the mutants because GFP fluorescence was masked by the natural fluorescence of the mutant hyphae. However, nuclear accumulation in the ΔPaMpk2 and the PaMKK2c mutants was never observed. Moreover, many stationary phase hyphae of ΔPaMpk2 presented profound morphological alterations (Figure 5), suggesting that the physiology of the ΔPaMpk2 mutants was severely altered.
Figure 5 .
Localization of PaMpk1–GFP in stationary phase hyphae of wild type, ΔPaMpk2, and PaMKK2c mutants. In WT, PaMpk1–GFP accumulates in 10 to 20% of the nuclei (arrows). In the ΔPaMpk2 and the PaMKK2c mutants, PaMpk1–GFP never accumulates in nuclei (n > 500 nuclei). In these strains, fluorescence of the hyphae carrying the PaMpk1–GFP transgene is similar to autofluorescence of hyphae with no transgene (control). Hyphae have an abnormal morphology in the ΔPaMpk2 mutant, since they look crooked and bulging. “GFP” are hyphae observed with a GFP–3035B filter from Semrock (Ex, 472 nm/30; dichroïc, 495 nm; and Em, 520 nm/35); “GFP” + DAPI are overlays of PaMpk1–GFP (in green) and DAPI-stained nuclei (in blue).
Life without MAPK
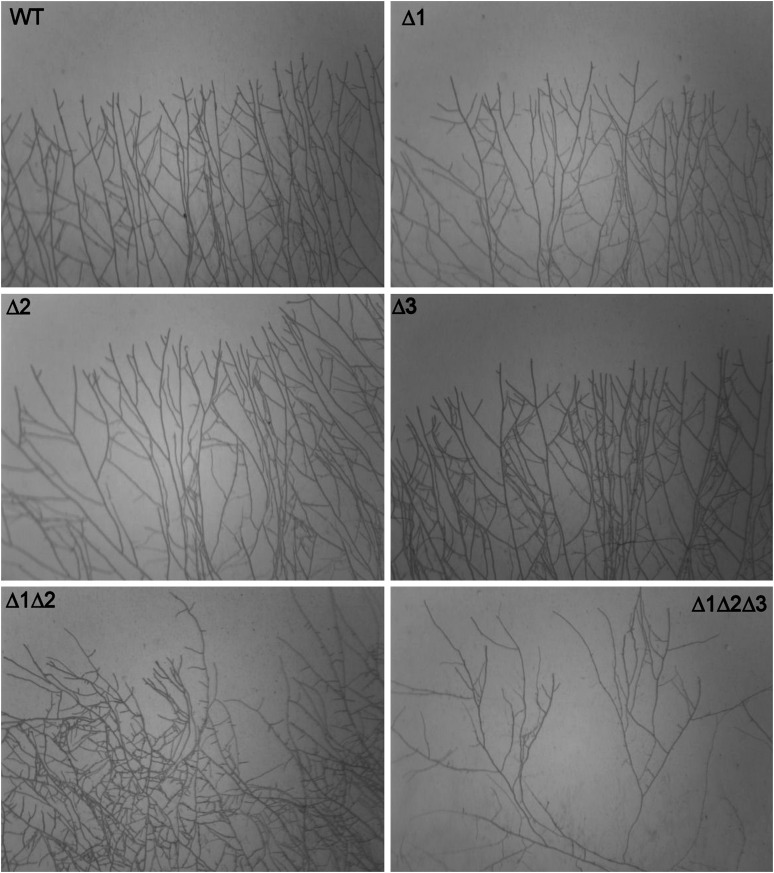
Because P. anserina crosses were easily performed, the availability of the single MAPK mutants served to construct the double and triple MAPK mutants (see Materials and Methods; Table 1). The double and triple mutants were viable and presented a combination of the phenotypes of the single mutants (Table 1). Surprisingly, despite severe physiological alteration, senescence (Marcou 1961) was not modified even in the triple mutant. However, the ΔPaMpk1 ΔPaMpk2 and the ΔPaMpk1 ΔPaMpk2 ΔPaMpk3 mutants grew slightly more slowly than wild type and did not differentiate ascogonia. Slow growth was correlated with a defect in branching and direction of apical growth (Figure 6). Indeed, while the single MAPK mutants had a regular pattern of outward-growing hyphae as wild type, the ΔPaMpk1 ΔPaMpk2 double and ΔPaMpk1 ΔPaMpk2 ΔPaMpk3 triple mutants presented irregular branching and some hyphae were curved and did not grow outwardly, especially in the triple mutant. Interestingly, the triple mutant was able to differentiate male gametes and could thus be crossed with female–fertile strains with success.
Figure 6 .
Growing edge showing branching and outward growth of the wild type and MAPK mutants. Δ1, ΔPaMpk1; Δ2, ΔPaMpk2; and Δ3, ΔPaMpk3.
Dicussion
Because of its interesting prion-like properties, we investigated the genetic determinants that lead to the formation of the MAPK-based nonconventional hereditary unit C of P. anserina (Silar et al. 1999). Through a forward genetic screen, the PaMpk1 MAP kinase cascade was previously identified as a key player in C formation (Kicka and Silar 2004; Kicka et al. 2006). Here, using a reverse genetic approach, we investigated the roles of the two other MAPK pathways present in this fungus in the formation of C, but also in the other developmental processes, including sexual reproduction, since the formation of C is tightly correlated with the ability to complete sexual reproduction (Kicka and Silar 2004). The data obtained in N. crassa and C. heterostrophus suggest that connections between the MAPK cascades may exist in filamentous fungi (Igbaria et al. 2008; Maerz et al. 2008). Our results show that (1) as in N. crassa (Maerz et al. 2008), the MAPK pathways are not branched in P. anserina, since mutants of the MAPK, MAPKK, and MAPKKK genes exhibit the same phenotype; (2) the PaMpk2 cascade has major developmental roles; (3) the PaMpk3 pathway function is restricted to osmotolerance. This is unlike many other filamentous fungi, in which this cascade often plays major roles not only in osmoresistance and resistance of antifungal compounds, but also during development (Zhao et al. 2007). Such a preeminent role of the orthologous mak-2 cascade has also been demonstrated in N. crassa (Pandey et al. 2004; Maerz et al. 2008). However, mutants of the N. crassa os-2 MAPK gene orthologous to PaMpk3 are sterile (Maerz et al. 2008).
It was observed that the N. crassa cascades orthologous to the PaMpk1 and PaMpk2 pathways are linked since they share similar phenotypes (Maerz et al. 2008). Similarly, in C. heterostrophus these two pathways control the same developmental processes and regulate the expression of the same genes (Igbaria et al. 2008). Here, we provide further evidence of such a link between the two pathways. Indeed, lack of CG and the other defect observed during stationary phase in the mutants of the PaMpk2 cascade are probably due to improper activation of PaMpk1, which is not caused by a defect in phosphorylation. That phosphorylation of PaMpk1 occurs independently of PaMpk2 and reciprocally, was also observed in C. heterostrophus (Igbaria et al. 2008). On the basis of cytological observations, it appears that hyphae lacking or harboring a constitutive PaMpk2 do not differentiate properly, resulting in a defect in PaMpk1 activation, as seen by its mislocalization. We thus propose that this is a typical case of developmental epistasis, in which correct activity of PaMpk2 prior to that of PaMpk1 is necessary for proper hyphal differentiation and subsequent functioning of PaMpk1. This epistasis may stem from several causes, including the inability to correctly express the machinery required to translocate PaMpk1 into the nucleus or an incorrect trafficking inside the hyphal network due to the lack of anastomosis.
Intriguingly, while the expected effect of PaMKK2 constitutive activation during germination was visible, i.e., constitutive ascospore germination on noninducing media correlated with constitutive phosphorylation of PaMpk2 in ascospores, no increased PaMpk2 phosphorylation was detected in mycelia. Thus the overall phosphorylation level of PaMpk2 is presumably tightly regulated in hyphae, possibly by unidentified phosphatases. A factor regulating PaMpk1 activity was also identified on the basis of mutant analyses (Silar et al. 1999; Haedens et al. 2005). The data also showed that the constitutive mutants exhibited a phenotype resembling inactivation of PaMpk1. Indeed, while appressorium-like differentiation and anastomoses are normal, the constitutive mutants lack aerial hyphae, pigments and are female sterile, which are the phenotypes shared by PaMpk1 and PaMpk2 mutants. This confirms our proposal that correct activity of PaMpk2 is required for PaMpk1 function.
The double mps1 hog1 MAPK mutant of C. heterostrophus displayed the hypersensitivity to hyperosmotic stress of the hog1 mutant as well as the pigmentation and autolysis defect of the mps1 mutant (Igbaria et al. 2008). In P. anserina, the corresponding double mutant ΔPaMpk1 ΔPaMpk3 also exhibited the osmotic defect of ΔPaMpk3 and the developmental defects of ΔPaMpk1. Analyses of the other double mutants showed that they also combine the defect of the corresponding single mutants. However, additional defects during female gamete differentiation, branching, and direction of apical growth are seen in the double ΔPaMpk1 ΔPaMpk2 and the triple ΔPaMpk1 ΔPaMpk2 ΔPaMpk3 mutants, indicating redundancy in MAPK functions. Therefore, the three MAPKs regulate an overlapping set of factors and only inactivation of the three MAPK genes revealed the full set of developmental processes they control. P. anserina devoid of its MAPK is thus a mere assembly of cells unable to differentiate most of the structures that enable them to adapt to their surroundings and reproduce efficiently.
In P. anserina, the life cycle may now be described according to the roles of the MAP kinases and in connection with the other known factors involved. During ascospore germination, only PaMpk2 is required, possibly in association with the PaNox2 NADPH oxidase and PaPls1 tetraspanin (Malagnac et al. 2004; Lambou et al. 2008). Indeed, mutants of the three genes exhibit the same germination defect, which is alleviated by removal of melanin. Phosphorylation of PaMpk2 is not abolished in the ΔPaNox2 and ΔPaPls1 mutants, suggesting that PaNox2 and PaPls1 act downstream of PaMpk2 or in a parallel pathway. This hypothesis is confirmed by the fact that PaNox2 and PaPls1 inactivation is epistatic over the constitutive activation of PaMKK2. Thereafter, apical growth and branching is controlled by the three MAPKs, having somewhat redundant roles in P. anserina. Interestingly, in other fungi, inactivation of a single MAPK gene may result in slow growth (see for example Bussink and Osmani 1999). Hyphae then undergo several developmental processes, including anastomosis and formation of appressorium-like structures. In P. anserina, both stages are mostly controlled by PaMpk2, but apparently not by PaMpk1. Moreover, appressorium-like differentiation also requires PaNox2 and PaPls1, while anastomosis does not. On the contrary, anastomosis requires the IDC1 factor (Silar 2011). This argues for a change of upstream activators and/or downstream targets for PaMpk2 during these two different processes. Proper maturation of hyphae also requires PaMpk2 as these are abnormally shaped in the ΔPaMpk2 mutants. The next stages of mycelium development, i.e., aerial hyphae and pigment formation, are clearly under the control of PaMpk1. However, at this stage a direct role for PaMpk2 independent of its action on PaMpk1 cannot be excluded. Finally, either one of the MAPK is required for female gamete formation, while male gamete formation is independent of the MAPK pathways. The reproductive stages from fertilization to the production of mature ascospores appear to require PaMpk1 and PaMpk2, but only in the mycelium and not in the fruiting bodies per se. Indeed, through a combination of mosaic, heterokaryon, and grafting experiments, we showed that, as for PaMpk1, PaMpk2 is required in neither the perithecium wall nor the sexual tissue. Defect of PaMpk2 mutants are more pronounced than those of PaMpk1, likely because lack of anastomoses in the mosaics impairs proper transfer of nutrients to the maturing fruiting bodies.
Despite severe defects, senescence was not affected in the MAPK mutants, even in the strain lacking all its MAPK. Because senescence is caused by a cytoplasmic and infectious factor (Marcou 1961), it is surprising that lack of anastomosis had no effect in the ΔPaMpk2 mutants. This suggests that the senescence factor does not spread through anastomoses that occur in the inner region of the culture. It was previously demonstrated that this factor amplifies solely at the growing edge in accordance with the present data (Marcou 1961). Lack of anastomoses also prevented direct testing of whether C is never produced in the ΔPaMpk2 mutants, is poorly amplified or whether only its effects are masked. Indeed, although cell fusion occurs in these mutants (likely by a mechanism different from typical anastomoses), their frequency was too small to perform contamination experiments as previously described (Silar et al. 1999; Kicka and Silar 2004; Kicka et al. 2006). Importantly, constitutive activation of the PaMpk2 pathways does not result in a constitutive CG, but rather in a diminished ability to present CG, demonstrating that this pathway is not part of the regulatory loop triggering C. Moreover, growth renewal after incubation into the stationary phase of ΔPaMpk2 mutant mycelia never results in CG. Since stationary phase induction of C should be independent of the occurrence of anastomoses, this suggests that lack of CG in the ΔPaMpk2 mutants is not solely due to the inability for C to propagate from cell to cell, but rather corresponded to an innate inability of its formation or expression of its effects. We proposed a model in which the PaMpk1 pathway is positively self-regulated (Kicka et al. 2006). According to this model, C would be the active state of the cascade and spreading would be due to the trans-activation of nonactive MAPK modules by active modules. We thus favor the first hypothesis of impairment of C formation, since in our model of a developmental epistasis, inactivation or constitutive activation of PaMpk2 should abolish C induction, because C is the active state of the PaMpk1 pathway. The effect of the constitutive PaMKK2c mutants can be explained by the same model. These mutants may be less affected than ΔPaMpk2, permitting some function of PaMpk1 allowing anastomoses, a weak CG, and residual sexual reproduction. Such a similar effect of excess and lack of activity of PaMpk2 on C is reminiscent of what has been observed for HSP104 in the control of yeast prions, for which too much or too little HSP104 inhibits propagation or maintenance (Chernoff et al. 1995). These data confirm the complex genetic determinism of C formation that was previously uncovered through a forward genetic analysis (Haedens et al. 2005). It remains to be studied whether the genes previously identified in the genetic analysis correspond to those encoding PaMpk2, PaMKK2, or PaTLK2 or whether they correspond to additional new factors.
Supplementary Material
Acknowledgments
We thank Sylvie François for expert technical assistance, Anne-Lise Haenni for reading the manuscript, and all members of the Génétique and Epigénétique des Champignons laboratory for discussions. This work was supported by grant ANR-05-Blan-0385.
Footnotes
Communicating editor: E. U. Selker
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arnaise S., Zickler D., Poisier C., Debuchy R., 2001. pah1: A homeobox gene involved in hyphal morphology and microconidiogenesis in the filamentous ascomycete Podospora anserina. Mol. Microbiol. 39: 54–64 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al., (Editors), 1987. Current Protocols in Molecular Biology, Wiley Interscience, New York [Google Scholar]
- Bagowski C. P., Ferrell J. E., Jr, 2001. Bistability in the JNK cascade. Curr. Biol. 11: 1176–1182 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M. V., 2005. Molecular theory of cancer. Cancer Biol. Ther. 4: 621–627 [DOI] [PubMed] [Google Scholar]
- Brun S., Malagnac F., Bidard F., Lalucque H., Silar P., 2009. Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Mol. Microbiol. 74: 480–496 [DOI] [PubMed] [Google Scholar]
- Brygoo Y., Debuchy R., 1985. Transformation by integration in Podospora anserina. I. Methodology and phenomenology. Mol. Gen. Genet. 200: 128–131 [Google Scholar]
- Bussink H. J., Osmani S. A., 1999. A mitogen-activated protein kinase (MPKA) is involved in polarized growth in the filamentous fungus, Aspergillus nidulans. FEMS Microbiol. Lett. 173: 117–125 [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W., 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- Coppin E., Silar P., 2007. Identification of PaPKS1, a polyketide synthase involved in melanin formation and its utilization as a genetic tool in Podospora anserina. Mycol. Res. 111: 901–908 [DOI] [PubMed] [Google Scholar]
- Espagne E., Lespinet O., Malagnac F., Da Silva C., Jaillon O., et al. , 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E., Jr, Pomerening J. R., Kim S. Y., Trunnell N. B., Xiong W., et al. , 2009. Simple, realistic models of complex biological processes: positive feedback and bistability in a cell fate switch and a cell cycle oscillator. FEBS Lett. 583: 3999–4005 [DOI] [PubMed] [Google Scholar]
- Fleissner A., Leeder A. C., Roca M. G., Read N. D., Glass N. L., 2009. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. USA 106: 19387–19392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrington T. P., Johnson G. L., 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11: 211–218 [DOI] [PubMed] [Google Scholar]
- Haedens V., Malagnac F., Silar P., 2005. Genetic control of an epigenetic cell degeneration syndrome in Podospora anserina. Fungal Genet. Biol. 42: 564–577 [DOI] [PubMed] [Google Scholar]
- Igbaria A., Lev S., Rose M. S., Lee B. N., Hadar R., et al. , 2008. Distinct and combined roles of the MAP kinases of Cochliobolus heterostrophus in virulence and stress responses. Mol. Plant Microbe Interact. 21: 769–780 [DOI] [PubMed] [Google Scholar]
- Jamet-Vierny C., Prigent M., Silar P., 2007. IDC1, a Pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet. Biol. 44: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Kawasaki L., Sanchez O., Shiozaki K., Aguirre J., 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Kicka S., Silar P., 2004. PaASK1, a mitogen-activated protein kinase kinase kinase that controls cell degeneration and cell differentiation in Podospora anserina. Genetics 166: 1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kicka S., Bonnet C., Sobering A. K., Ganesan L. P., Silar P., 2006. A mitotically inheritable unit containing a MAP kinase module. Proc. Natl. Acad. Sci. USA 36: 13445–13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalucque H., Malagnac F., Silar P., 2010 Prions and prion-like phenomena in epigenetic inheritance, pp. 63–76 in Handbook of Epigenetics, edited by T. Tollefsbol. Academic Press, San Diego
- Lambou K., Malagnac F., Barbisan C., Tharreau D., Lebrun M. H., et al. , 2008. The crucial role during ascospore germination of the Pls1 tetraspanin in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot. Cell 7: 1809–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier G., Silar P., 1994. Rapid methods for nucleic acids extraction from Petri dish grown mycelia. Curr. Genet. 25: 122–123 [DOI] [PubMed] [Google Scholar]
- Maerz S., Ziv C., Vogt N., Helmstaedt K., Cohen N., et al. , 2008. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics 179: 1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F., Lalucque H., Lepere G., Silar P., 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41: 982–997 [DOI] [PubMed] [Google Scholar]
- Marcou D., 1961. Concept of longevity and the cytoplasmic basis of the senecence determinant in a few fungi. Ann. Sci. Natur. Bot. Paris Sér. 12 2: 653–764 (in French) [Google Scholar]
- May G. S., Xue T., Kontoyiannis D. P., Gustin M. C., 2005. Mitogen activated protein kinases of Aspergillus fumigatus. Med. Mycol. 43(Suppl 1): S83–S86 [DOI] [PubMed] [Google Scholar]
- Morel M., Ngadina A. A., Jacquota J. P., Gelhayea E., 2009. Reactive oxygen species in Phanerochaete chrysosporium relationship between extracellular oxidative and intracellular antioxidant systems. Adv. Bot. Res. 52: 153–186 [Google Scholar]
- Munkres K. D., 1990. Histochemical detection of superoxide radicals and hydrogen peroxide by Age-1 mutants of Neurospora. Fungal Genet. Newsl. 37: 24–25 [Google Scholar]
- Neves S. R., Iyengar R., 2002. Modeling of signaling networks. Bioessays 24: 1110–1117 [DOI] [PubMed] [Google Scholar]
- Orbach M. J., 1994. A cosmid with a HyR marker for fungal library construction and screening. Gene 150: 159–162 [DOI] [PubMed] [Google Scholar]
- Pages G., Brunet A., L ’Allemain G., Pouyssegur J., 1994. Constitutive mutant and putative regulatory serine phosphorylation site of mammalian MAP kinase kinase (MEK1). EMBO J. 13: 3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Roca M. G., Read N. D., Glass N. L., 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3: 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispail N., Soanes D. M., Ant C., Czajkowski R., Grunler A., et al. , 2009. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46: 287–298 [DOI] [PubMed] [Google Scholar]
- Rizet G., 1952. The “barrage” phenomena in Podospora anserina. I Genetic analysis of “barrage” between strains S and s. Rev. Cytol. Biol. Veg. 13: 51–92 (in French) [Google Scholar]
- Rizet G., Engelmann C., 1949. Contribution to the genetical study of a four-spored Ascomycete: Podospora anserina (Ces.) Rehm. Rev. Cytol. Biol. Veg. 11: 201–304 (in French) [Google Scholar]
- Saito H., 2010. Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 13: 677–683 [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Shiozaki M., Russell P., 1998. Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol. Biol. Cell 9: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silar P., 1995. Two new easy-to-use vectors for transformations. Fungal Genet. Newsl. 42: 73 [Google Scholar]
- Silar P., 2005. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol. Res. 109: 137–149 [DOI] [PubMed] [Google Scholar]
- Silar P., 2011. Grafting as a method for studying development in the filamentous fungus Podospora anserina. Fungal Biol 115: 793–802 [DOI] [PubMed] [Google Scholar]
- Silar P., Picard M., 1994. Increased longevity of EF-1 alpha high-fidelity mutants in Podospora anserina. J. Mol. Biol. 235: 231–236 [DOI] [PubMed] [Google Scholar]
- Silar P., Haedens V., Rossignol M., Lalucque H., 1999. Propagation of a novel cytoplasmic, infectious and deleterious determinant is controlled by translational accuracy in Podospora anserina. Genetics 151: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C., Gibson S., Jarpe M. B., Johnson G. L., 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79: 143–180 [DOI] [PubMed] [Google Scholar]
- Zhao X., Mehrabi R., Xu J. R., 2007. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6: 1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.