Abstract
Ypt/Rab are key regulators of intracellular trafficking in all eukaryotic cells. In yeast, Ypt1 is essential for endoplasmic reticulum (ER)-to-Golgi transport, whereas Ypt31/32 regulate Golgi-to-plasma membrane and endosome-to-Golgi transport. TRAPP is a multisubunit complex that acts as an activator of Ypt/Rab GTPases. Trs85 and Trs130 are two subunits specific for TRAPP III and TRAPP II, respectively. Whereas TRAPP III was shown to acts as a Ypt1 activator, it is still controversial whether TRAPP II acts as a Ypt1 or Ypt31/32 activator. Here, we use GFP-Snc1 as a tool to study transport in Ypt and TRAPP mutant cells. First, we show that expression of GFP-Snc1 in trs85Δ mutant cells results in temperature sensitivity. Second, we suggest that in ypt1ts and trs85Δ, but not in ypt31Δ/32ts and trs130ts mutant cells, GFP-Snc1 accumulates in the ER. Third, we show that overexpression of Ypt1, but not Ypt31/32, can suppress both the growth and GFP-Snc1 accumulation phenotypes of trs85Δ mutant cells. In contrast, overexpression of Ypt31, but not Ypt1, suppresses the growth and GFP-Snc1 transport phenotypes of trs130ts mutant cells. These results provide genetic support for functional grouping of Ypt1 with Trs85-containing TRAPP III and Ypt31/32 with Trs130-containing TRAPP II.
TRANSPORT of membranes and proteins through the endocytic and exocytic pathways connects membrane-bound intracellular compartments with the plasma membrane (PM) and the cell milieu. The endoplasmic reticulum (ER) is the compartment into which membrane and cargo proteins are translocated en route to all the other cellular compartments. From the ER, membrane and proteins are transported through the Golgi apparatus to the PM.
Ypt/Rab GTPases regulate trafficking between cellular compartments (Segev 2001a,b; Stenmark 2009). In yeast, Ypt1 is required for ER-to-Golgi transport (Segev et al. 1988; Jedd et al. 1995), whereas the Ypt31/32 functional pair plays a role at the trans-Golgi in both Golgi-to-PM and endosome-to-Golgi transport (Jedd et al. 1997; Chen et al. 2005). In addition, Ypt1 and Ypt31/32 play a role in autophagy (Segev and Botstein 1987; Geng et al. 2010; Lynch-Day et al. 2010). Autophagy is a cellular process induced by stress in which cytosolic and membrane proteins are engulfed by a double-membrane organelle termed autophagosome to be delivered to the lysosome for degradation (Nakatogawa et al. 2009; Yang and Klionsky 2009).
Ypt/Rabs are activated by guanine-nucleotide exchange factors (GEFs) and the GEF for Ypt1 and Ypt31/32 is the TRAPP complex. TRAPP is a multisubunit modular complex (Sacher et al. 2008), which exists in at least three forms. TRAPP I, which contains five essential subunits, acts as a GEF for Ypt1 (Jones et al. 2000; Wang et al. 2000), and is required for ER-to-Golgi transport (Sacher et al. 1998). Trs85, a TRAPP subunit, nonessential for cell viability, plays a role in autophagy (Meiling-Wesse et al. 2005; Nazarko et al. 2005). A Trs85-containing TRAPP complex, termed TRAPP III, also acts as a Ypt1 GEF (Lynch-Day et al. 2010). TRAPP II, which contains Trs120 and Trs130 in addition to TRAPP I subunits, functions at the trans-Golgi (Sacher et al. 2001). However, currently the Ypt GEF specificity of the TRAPP II complex is under dispute.
On the basis of biochemical and genetic evidence, we have proposed that TRAPP I activates Ypt1 in ER-to-Golgi transport, whereas Ypt31/32 act at the trans-Golgi in both Gogli-to-PM and endosome-to-Golgi transport, and TRAPP II acts as their GEF at least in the former step (Jones et al. 2000; Chen et al. 2005; Morozova et al. 2006). This model is currently being challenged claiming that Ypt1 functions in endosome-to-Golgi transport in addition to its role in ER-to-Golgi transport (Sclafani et al. 2010), and that TRAPP II acts as a GEF for Ypt1 in both steps (Cai et al. 2008; Barrowman et al. 2010; Yip et al. 2010). Here we study the physiological relationship between Ypt1 and Ypt31/32 and two TRAPP complexes, TRAPP III and TRAPP II. Data presented in this article provide support to the idea that TRAPP II acts with Ypt31/32, and not with Ypt1, in endosome-to-Golgi transport.
GFP-Snc1 has been extensively used as a PM recycling marker in yeast (Lewis et al. 2000; Chen et al. 2005; Montpetit and Conibear 2009; Sclafani et al. 2010). Snc1 is a v-SNARE that plays a role in fusion of trans-Golgi vesicles with the PM. For multiple rounds of vesicle fusion, Snc1 recycles back from the PM to the Golgi via endosomes (Lewis et al. 2000). We have previously used GFP-Snc1 to reveal a role for Ypt31/32 in endosome-to-Golgi transport (Chen et al. 2005). Recently, a role for Ypt1, together with TRAPP II, has been proposed in the recycling of GFP-Snc1 from the PM using YPT1 alleles defective specifically in Snc1-GFP trafficking (Sclafani et al. 2010). In addition, a role for Trs85 in endosome-to-Golgi transport was also suggested on the basis of intracellular accumulation of Snc1-GFP in trs85Δ mutant cells (Montpetit and Conibear 2009). In contrast, our results suggest that in ypt1ts and trs85Δts mutant cells, GFP-Snc1 accumulates in the ER, supporting a role for Ypt1 and Trs85 in exit of membrane proteins from the ER. Because accumulation of GFP-Snc1 in the ER of trs85Δts mutant cells is coupled with a cell-growth defect, we propose that accumulation of proteins in the ER can cause cell toxicity. Moreover, overexpression analyses support functional grouping of Ypt31/32 with the Trs130-containing TRAPP II complex in endosome-to-Golgi transport and of Ypt1 with the Trs85-containing TRAPP complex in ER exit of overexpressed membrane proteins. It is not clear whether this role of Trs85 is related to its established function in autophagy.
Results
Biochemical analysis has shown that TRAPP I acts as a Ypt1 GEF (Jones et al. 2000; Wang et al. 2000). However, the substrate for TRAPP II complex is currently controversial (Morozova et al. 2006; Cai et al. 2008). Recently, the Trs85-containing TRAPP III complex was suggested to act as a GEF for Ypt1 and not Ypt32 (Lynch-Day et al. 2010). However, in the published experiment there was no positive control for a Ypt32 GEF and does not include Ypt31. Here, we add biochemical support to the idea that TRAPP III acts as a GEF for Ypt1, but not Ypt31/32, by including the missing positive control. The Ypt GEF activity of the Trs85-containing TRAPP complex purified from yeast lysates was compared to that of the Bet5-containing TRAPP complexes, using a GDP-release assay. Because the Bet5 subunit is common to the three known TRAPP complexes, purified GST-Bet5 TRAPP contains all the TRAPP complexes including TRAPP II. Under our reaction conditions, in addition to acting as a Ypt1 GEF, GST-Bet5 purified TRAPP acts as a GEF for Ypt31 and Ypt32. In contrast, the GST-Trs85 purified complex acts as a GEF only for Ypt1 (Supporting Information, Figure S1). Thus, the Trs85-containing TRAPP, like TRAPP I, acts as a GEF for Ypt1, but not for Ypt31/32.
To study the physiological relationship between Ypt1 and Ypt31/32 and two TRAPP complexes, TRAPP II and TRAPP III, we followed the transport of GFP-Snc1 in mutants defective in Ypt and TRAPP function.
GFP-Snc1 accumulates in the ER of ypt1ts and trs85Δts mutant cells
The functional pair of Snc1 and Snc2 acts as a v-SNARE that cycles between the Golgi, PM, and endosomes (Protopopov et al. 1993). GFP-Snc1 has been used commonly as a PM recycling marker in yeast (Lewis et al. 2000). In wild-type (wt) cells, GFP-Snc1 localizes mostly to the PM, whereas in mutant cells defective in PM recycling, it accumulates in endosomes. Using this marker, we have previously shown that Ypt31/32 play a role in endosome-to-Golgi transport (Chen et al. 2005). A role for several TRAPP subunits in this step, including the two TRAPP II-specific subunits, Trs120 and Trs130 (Cai et al. 2005), and the three nonessential TRAPP subunit, Trs33, Trs65, and Trs85 (Montpetit and Conibear 2009), was also suggested on the basis of accumulation of intracellular GFP-Snc1. Recently, a role for Ypt1 in endosome-to-Golgi transport was suggested (Sclafani et al. 2010), and trs85Δ was identified in a screen for genes required for endosome-to-Golgi transport (Montpetit and Conibear 2009). Even though a role for Trs85 and Ypt1 in autophagy has been established (Segev and Botstein 1987; Lynch-Day et al. 2010), they could also play a role in endosome-to-Golgi transport, individually or together. For example, Ypt31/32 and Trs130 function both in exit from the trans-Golgi and in endosome-to-Golgi transport (Jedd et al. 1997; Sacher et al. 2001; Cai et al. 2005; Chen et al. 2005). Therefore, we reexamined whether Ypt1 and Trs85 function in the recycling of GFP-Snc1 from the PM.
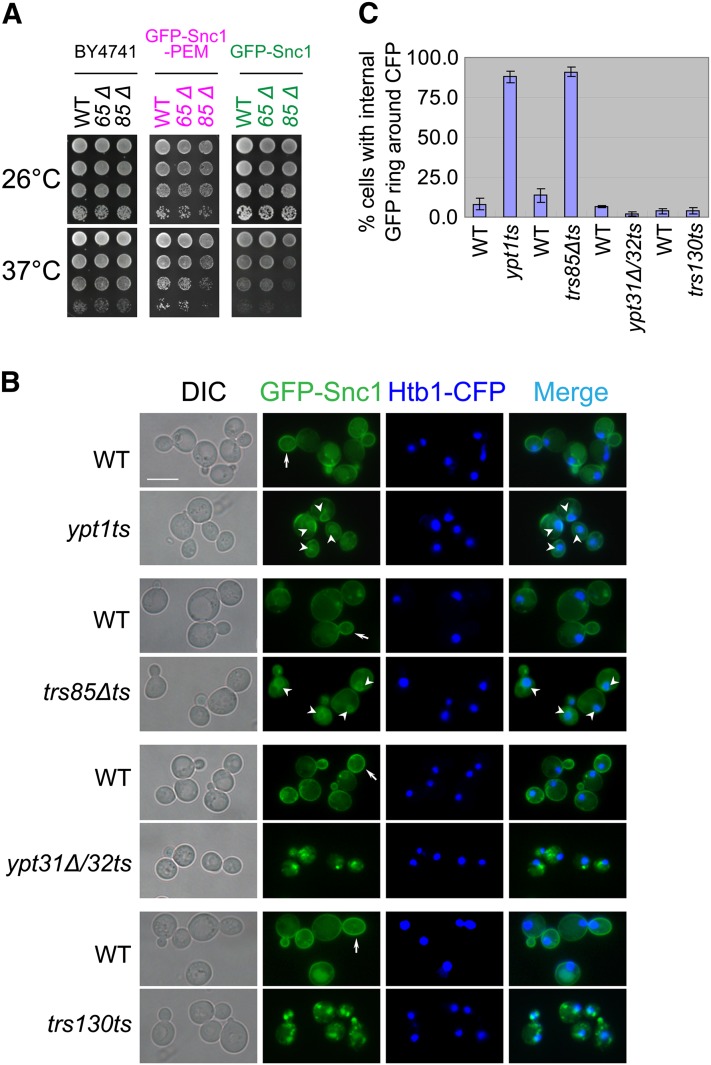
GFP-Snc1 was integrated into the URA3 locus in wt, ypt1ts, and trs85Δ mutant cells (these cells express both Snc1 and Snc2) (Lewis et al. 2000). GFP-Snc1–expressing trs85Δ, but not trs65Δ, mutant cells exhibit a temperature-sensitive growth phenotype (Figure 1A). The linkage of the temperature-sensitive growth phenotype with overexpression of GFP-Snc1 in trs85Δ mutant cells was verified by cosegregation of the temperature sensitivity with integrated GFP-Snc1 and trs85Δ, and by overexpression of GFP-Snc1 from a 2µ plasmid (Figure S2). Thus, overexpression of GFP-Snc1 in trs85Δ mutant cells confers a temperature-sensitive phenotype. We termed the mutant strain in which GFP-Snc1 is integrated and TRS85 is deleted trs85Δts. This mutant was used as a tool in microscopic analysis to study GFP-Snc1 transport phenotype, and in overexpression analysis, to determine genetic interactions between the Ypts and TRAPP subunits.
Figure 1 .
GFP-Snc1 accumulates in the rings around the nuclei of ypt1ts and trs85Δts mutant cells. (A) trs85Δ, but not trs65Δ mutant cells expressing GFP-Snc1 or GFP-Snc1-PEM exhibit a temperature-sensitive growth phenotype. The TRS85 or TRS65 gene was deleted in the following three strains: wild type (BY4741, left), chromosomally tagged GFP-Snc1-PEM (YLY1582, middle), and chromosomally tagged GFP-Snc1 (YLY130, right). Cells were grown on YPD plates at 26° (top) and 37° (bottom). Four 10-fold serial dilutions are shown from top to bottom. (B) GFP-Snc1 accumulates in the ER of ypt1ts and trs85Δts, but not of ypt31Δ/32ts and trs130ts mutant cells. Wild-type and mutant cells expressing chromosomally tagged GFP-Snc1 and Htb1-CFP, as a nuclear marker, were grown in YPD medium at 26° to midlog phase and then shifted to 37° for 1.5 hr. GFP-Snc1 and Htb1-CFP were visualized using live-cell fluorescence microscopy. In ypt1ts and trs85Δts, but not in ypt31Δ/32ts or trs130ts, GFP-Snc1 accumulates around nuclei, which is indicative of ER localization. Bar, 7 μm. Arrows point to GFP-Snc1 on the PM and arrowheads to perinuclear GFP-Snc1 circle. (C) Quantification of data presented in B: shown is percentage of cells with an internal GFP-Snc1 ring around the nucleus, which is marked with Htb1-CFP. At least 200 cells were counted in at least two fields for each strain. Error bars represent SD.
The accumulation of GFP-Snc1 in ypt1ts and trs85Δts mutant cells at their permissive (26°) and nonpermissive (37°) temperatures was determined using live-cell microscopy (Figure S3). In both ypt1ts and trs85Δts mutant cells, GFP-Snc1 accumulates internally at 37°. This internal GFP-Snc1 accumulation was compared to the accumulation in ypt31Δ/32ts and trs130ts mutant cells, in which GFP-Snc1 accumulates in endosomes already at the permissive temperature (under conditions that allow Golgi-to-PM transport) (Chen et al. 2005). However, whereas in ypt31Δ/32ts and trs130ts mutant cells, GFP-Snc1 accumulates in puncta, the internal accumulation in ypt1ts and trs85Δts mutant cells appears as rings. Because Ypt1 functions in ER-to-Golgi transport and in yeast ER appears as rings around the nucleus (Hampton et al. 1996; Huh et al. 2003), we wished to determine whether the GFP-Snc1 rings in ypt1ts and trs85Δts mutant cells are around the nucleus.
To determine the cellular compartment in which GFP-Snc1 accumulates in ypt1ts and trs85Δts mutant cells, it was colocalized with nuclear and endosomal markers. The nucleus was visualized by Htb1-CFP (histone H2B) (Michelsen et al. 2006). In both ypt1ts and trs85Δts mutant cells, GFP-Snc1 accumulates in rings around the nucleus labeled with Htb1-CFP. In contrast, in ypt31Δ/32ts and trs130ts mutant cells, the puncta are not associated with the nucleus (Figure 1, B and C).
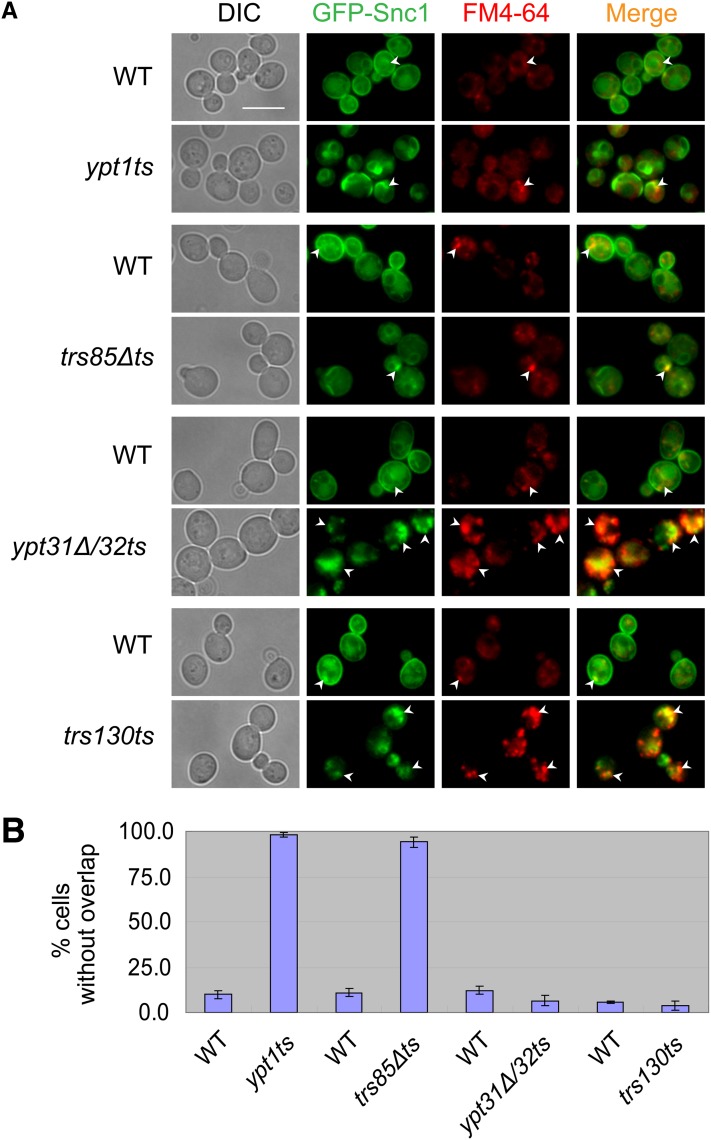
Localization of GFP-Snc1 to endosomes was determined by staining cells with the fluorescent dye FM4-64 for 5 min. As expected for a protein that recycles through endosomes, there is some localization of GFP-Snc1 to endosomes in wild-type cells. In ypt1ts and trs85Δts mutant cells the limited colocalization of GFP-Snc1 with the endosomal staining is similar to that seen in wild-type cells. In contrast, under the same conditions (as previously reported, Cai et al. 2005; Chen et al. 2005), in ypt31Δ/32ts and trs130ts mutant cells, intracellular GFP-Snc1 localizes mostly to endosomes (Figure 2).
Figure 2 .
GFP-Snc1 accumulates in endosomes of ypt31Δ/32ts and trs130ts, but not of ypt1ts and trs85Δts mutant cells. (A) Wild-type and mutant cells expressing chromosomally tagged GFP-Snc1 were stained with FM4-64 for 5 min to label early endosomes and were visualized using live-cell fluorescence microscopy. Cells grown in YPD medium at 26° to midlog phase were shifted to 37° for 85 min, stained with FM4-64 for 5 min, and kept on ice until visualization. Bar, 7 μm. Arrowheads point to GFP-Snc1 that localizes to early endosomes marked with FM4-64. (B) Quantification of data presented in A shown is percentage of cells with internal GFP-Snc1 that does not overlap with FM4-64. At least 200 cells were counted in at least three fields for each strain. Error bars represent SD.
Together, the colocalization analysis of GFP-Snc1 with nuclear and endosomal markers suggest that in ypt1ts and trs85Δts, but not in ypt31Δ/32ts and trs130ts, mutant cell Snc1 accumulates in the ER.
The nonrecycling marker, GFP-Snc1-PEM, accumulates in the ER of ypt1ts and trs85Δ mutant cells
To further characterize the GFP-Snc1 localization defect in ypt1ts and trs85Δts mutant cells, we used the GFP-Snc1-PEM protein. This hybrid protein contains the Sso1 trans-membrane domain and two point mutations, V40A and M43A, which interfere with endocytosis and cause accumulation on the PM (Lewis et al. 2000). Therefore, GFP-Snc1-PEM does not accumulate in endosomes in mutant cells defective in PM recycling (Galan et al. 2001; Lafourcade et al. 2004; Chen et al. 2005). Importantly, this protein can be used as a probe for defects in the biosynthetic transport of GFP-Snc1 because it is not endocytosed.
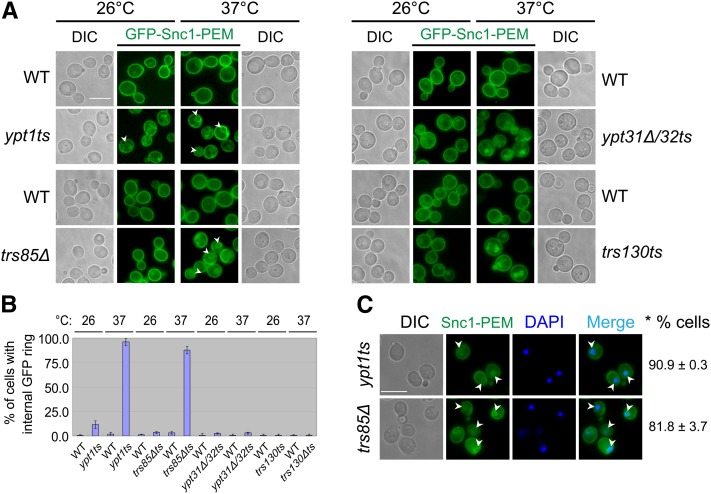
In wild-type cells, GFP-Snc1-PEM accumulates on the PM both at 26° and 37°. As expected, in ypt31Δ/32ts and trs130ts mutant cells at 26°, GFP-Snc1-PEM localizes mostly to the PM (Figure 3A). This localization supports the idea that in these mutant cells, intracellular accumulation of GFP-Snc1 is caused by a PM recycling defect. At 37°, GFP-Snc1-PEM accumulates in both ypt31Δ/32ts and trs130ts mutant cells (Figure 3A). This intracellular accumulation of GFP-Snc1-PEM indicates a block in transport of this protein to the PM. Because at 37°, ypt31Δ/32ts and trs130ts mutant cells are defective in Golgi-to-PM transport (Jedd et al. 1997; Sacher et al. 2000), GFP-Snc1-PEM probably accumulates in the Golgi.
Figure 3 .
GFP-Snc1-PEM accumulates in the rings around the nuclei of ypt1ts and trs85Δts mutant cells. (A) Like GFP-Snc1, GFP-Snc1-PEM accumulates in ring-like structures in ypt1ts and trs85Δ, but not in ypt31Δ/32ts or trs130ts mutant cells. Wild-type and mutant yeast cells expressing chromosomally tagged GFP-Snc1-PEM were used for live-cell fluorescence microscopy as described in Figure S3 legend. GFP-Snc1-PEM accumulates inside ypt1ts and trs85Δ mutant cells under the same conditions that cause intracellular accumulation of GFP-Snc1; namely 26° and 37° for ypt1ts and 37° for trs85Δ. In contrast, GFP-Snc1-PEM does not accumulate inside ypt31Δ/32ts or trs130ts mutant cells at 26°, under conditions that GFP-Snc1 does accumulate (see Figure S3). DIC images (on each side) show the contour of cells. Bar, 7 μm. Arrowheads point to internal GFP-Snc1-PEM. (B) Quantification of data presented in A: shown is percentage of cells with an internal GFP-Snc1-PEM ring. At least 200 cells were counted in at least three fields for each strain; error bars represent SD. (C) The GFP-Snc1-PEM rings in ypt1ts and trs85Δ mutant cells are around the nuclei. Ypt1ts and trs85Δ mutant cells were grown at 26° to midlog phase and shifted to 37° for 1.5 hr. The cells were fixed in ethanol, stained with DAPI to mark nuclei, and visualized by fluorescence microscopy. Arrowheads point to internalized GFP-Snc1-PEM. The GFP-Snc1-PEM ring-like structures accumulating in these mutant cells are around nuclei indicating ER staining. Bars, 7 μm. Right: *% cells indicates percentage of cells with an internal GFP-Snc1-PEM ring around the nucleus, which is stained with DAPI. ± represents SD.
In ypt1ts and trs85Δ mutant cells at 26°, GFP-Snc1-PEM localizes to the PM and, especially in ypt1ts mutant cells, it also accumulates inside the cells. At 37°, while there is GFP-Snc1-PEM on the PM, it also localizes to internal rings (Figure 3, A and B). Moreover, like the GFP-Snc1 rings that accumulate in these mutant cells at 37°, the GFP-Snc1-PEM rings also surround the nucleus (Figure 3C). Finally, like overexpression of GFP-Snc1, overexpression of GFP-Snc1-PEM in trs85Δ, but not trs65Δ mutant cells confers a cell growth defect (Figure 1A). Thus, we suggest that GFP-Snc1-PEM, like GFP-Snc1, accumulates in the ER of ypt1ts and trs85Δ mutant cells.
These results suggest that, whereas in ypt31Δ/32ts and trs130ts mutant cells GFP-Snc1 and GFP-Snc1-PEM accumulate in endosomes and Golgi, respectively, in ypt1ts and trs85Δ mutant cells both GFP-Snc1 and GFP-Snc1-PEM accumulate in the ER en route to the PM.
Overexpression of Ypts in TRAPP mutants
Recently, the observation that ypt1 mutant cells, like TRAPP II-specific subunit mutants, accumulate internalized GFP-Snc1 was used to support the claim that TRAPP II acts as a GEF for Ypt1 to regulate endosome-to-Golgi transport (Sclafani et al. 2010). To test this idea, we performed an overexpression analysis. The effect of overexpression of Ypt1 or Ypt31/32 on the following two phenotypes of trs85Δts and trs130ts mutant cells was determined: internal accumulation of GFP-Snc1 and cell growth.
Ypt1 and Ypt31 were overexpressed from 2μ plasmids. Overexpression of Ypt1 specifically suppresses the growth defect of ypt1ts, but not ypt31Δ/32ts mutant cells. Conversely, overexpression of Ypt31 suppresses the growth defect of ypt31Δ/32ts, but not ypt1ts mutant cells (Figure S4).
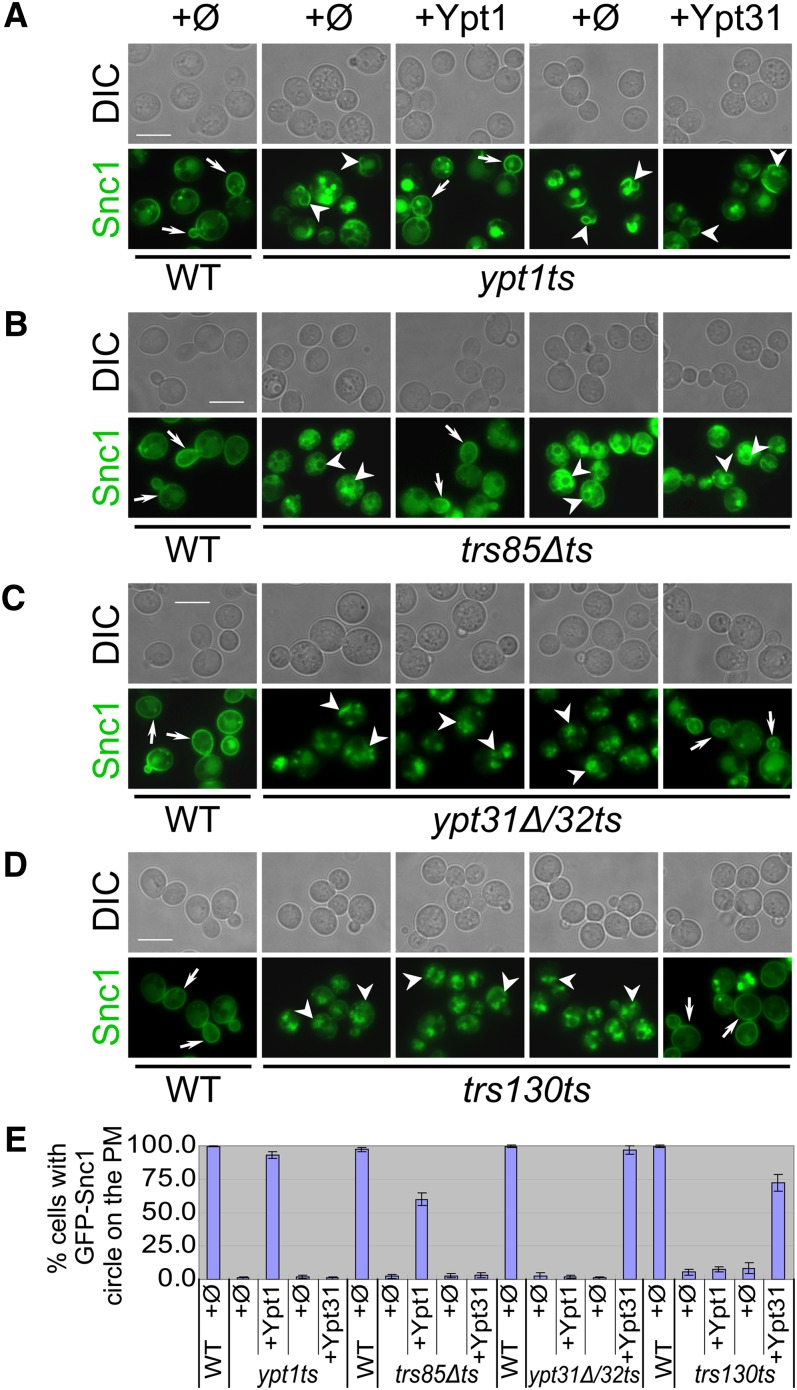
If Ypt1 plays a role in GFP-Snc1 recycling from the PM together with the Trs130-containing TRAPP II complex, it was expected that overexpression of Ypt1 would suppress the GFP-Snc1 recycling defect of trs130ts mutant cells. However, we found that Ypt31/32, but not Ypt1, can suppress this defect in ypt31Δ/32ts and trs130ts mutant cells (Figure 4, C–E and Figure S5, B and C). In contrast, Ypt1, but not Ypt31, can suppress the intracellular accumulation of GFP-Snc1 in ypt1ts and trs85Δts mutant cells (Figure 4, A, B, and E). Ypt32 also cannot suppress the intracellular accumulation of GFP-Snc1 in ypt1ts and trs85Δts mutant cells (data not shown). These results argue against the idea that Ypt1 plays a role with Trs130-containing TRAPP II in endosome-to-Golgi transport and further support the physiological relationship between Ypt31/32 and the Trs130-containing TRAPP II complex.
Figure 4 .
Ypt1 suppresses the GFP-Snc1 transport phenotype of trs85Δts, whereas Ypt31 suppresses the phenotype of trs130ts. The localization of chromosomally tagged GFP-Snc1 was determined by live-cell fluorescence microscopy in wild-type and mutant cells transformed with 2μ plasmids: Ypt1, and Ypt31, and their corresponding empty plasmids (ϕ, pRB684 and pRS425, respectively). Cells were grown in selective medium (to keep the plasmids) to midlog phase at 26° and then shifted to 37° for 1.5 hr. Plasmids are shown at the top of A. Bar, 7 μm. Wild-type cells transformed with the different plasmids show the same GFP-Snc1 PM localization; shown on the left are wild-type cells transformed with empty vector (ϕ, pRB684). All mutants accumulate internalized GFP-Snc1 (compare the left panels for wild type and mutant with empty plasmid (ϕ). Overexpression of Ypt1, but not Ypt31, suppresses the GFP-Snc1 phenotype of ypt1ts (A) and trs85Δts (B) mutant cells. In contrast, overexpression of Ypt31, but not Ypt1, suppresses the GFP-Snc1 phenotype of ypt31Δ/32ts (C) and trs130ts (D). In the GFP-Snc1 panels, arrows point to GFP-Snc1 on the PM in wild-type and suppressed mutant cells; arrowheads point to intracellular GFP-Snc1 seen in mutant cells. (E) Quantification of data presented in A–D. Shown is percentage of cells with GFP-Snc1 on the PM. At least 200 cells were counted in at least four fields for each strain. Error bars represent SD.
If the growth defect of the trs85Δts mutant cells is caused by ER accumulation of GFP-Snc1, we expected that suppression of the GFP-Snc1 ER accumulation by overexpression of Ypt1 should also suppress the temperature growth defect of these cells. Moreover, if Ypt31/32, and not Ypt1, function together with TRAPP II, we expect that overexpression of Ypt31/32, but not Ypt1, would suppress the growth defect of trs130ts mutant cell. Indeed, overexpression of Ypt1, but not Ypt31, can suppress the temperature-sensitive growth phenotype of ypt1ts and trs85Δts mutant cells (Figure 5A and Figure S4A). Overexpression of Ypt32 also cannot suppress the temperature-sensitive growth phenotype of ypt1ts and trs85Δts mutant cells (data not shown). In contrast, the temperature-sensitive growth phenotype of ypt31Δ/32ts and trs130ts mutant cells can be suppressed by overexpression of Ypt31 or Ypt32, but not Ypt1 (Figure 5B and Figure S4B; Figure S5A).
Figure 5 .
Ypt1 suppresses the growth defect of trs85Δts, whereas Ypt31 suppresses the growth defect of trs130ts. Mutant cells, trs85Δts (A), and trs130ts (B), and their corresponding wild-type cells, all expressing chromosomally tagged GFP-Snc1, were transformed with the following 2μ plasmids: Ypt1 and Ypt31, and their corresponding empty plasmids (ϕ, pRB684 and pRS425, respectively). Cell growth on plates was tested at 26° and 37°; four 10-fold serial dilutions are shown from top to bottom. Overexpression of Ypt1, but not Ypt31, suppresses the growth defect of trs85Δts (A). In contrast, overexpression of Ypt31, but not Ypt1, suppresses the growth defect of trs130ts (B). Overexpression of Ypt1 and Ypt31 in the transformants was verified by immunoblot analysis using anti-Ypt1 and anti-Ypt31 antibodies, respectively; G6PDH indicates equal loading (shown at the bottom of each panel). (C) Summary of the overexpression analysis described here: Specific suppression of trs85Δts and trs130ts mutant cells by Ypt1 and Ypt31/32, respectively, supports physiological grouping of Ypt1 with Trs85-containing TRAPP III and Ypt31/32 with Trs130-containing TRAPP II complex.
In summary, genetic analysis presented here reveals that while overexpression of Ypt1 can suppress the two GFP-Snc1–induced phenotypes of trs85Δts mutant cells, GFP-Snc1 accumulation in the ER and temperature sensitivity, it does not suppress those of trs130ts mutant cells. Importantly, the two GFP-Snc1–induced phenotypes of trs130ts mutant cells, internal accumulation of GFP-Snc1 and temperature sensitivity, can be suppressed by overexpression of Ypt31/32. Therefore, these results provide genetic support to grouping Ypt1 with Trs85-containing TRAPP III and Ypt31/32 with Trs130-containing TRAPP II (Figure 5C).
Discussion
Here, the v-SNARE GFP-Snc1 is used as a transport marker to support two ideas. First, GFP-Snc1 and GFP-Snc1-PEM accumulate in the ER of ypt1ts and trs85Δ mutant cells en route to the PM. Second, overexpression analysis supports physiological grouping of Ypt1 with the Trs85-containing TRAPP III complex and of Ypt31/32 with the Trs130-containing TRAPP II complex (Figure 5C). The importance of each of these conclusions is discussed below.
Using GFP-Snc1 as a cargo marker
Using traditional exocytic markers, like invertase and CPY, it has been established that ypt1ts mutant cells exhibit an ER transport block (Jedd et al. 1995), whereas ypt31Δ/32ts and trs130ts exhibit a Golgi block (Jedd et al. 1997; Sacher et al. 2001; Cai et al. 2005). Using CPY (Prc1) as a marker, it was recently concluded that trs85Δ mutant cells do not exhibit an ER block for this luminal vacuolar protease (even though a weak kinetic block is observed; Figure S3A) (Lynch-Day et al. 2010).
Here, we show that the membrane-associated GFP-Snc1 can also be used as an exocytic cargo marker. In wild-type cells, GFP-Snc1 is inserted into the ER membrane like all other proteins that contain a trans-membrane domain. From the ER, the v-SNARE Snc1 is transported to the Golgi where it plays a role in Golgi-to-PM transport. Snc1 can then be recycled back to the Golgi through endosomes for multiple rounds of vesicular transport (Lewis et al. 2000). GFP-Snc1 has been used as a PM-recycling marker to show that ypt31Δ/32ts and trs130ts mutant cells are defective in endosome-to-Golgi transport (Cai et al. 2005; Chen et al. 2005). Here, we show that GFP-Snc1 can also be used as an exocytic cargo marker in mutants defective in exit from the ER. Whereas in ypt31Δ/32ts and trs130ts it accumulates in endosomes or the Golgi, in ypt1ts and trs85ts it accumulates in the ER. The apparent discrepancy between transport of CPY (Lynch-Day et al. 2010) and GFP-Snc1 (here) in trs85Δ mutant cells might be due to the difference between the two markers: CPY is an endogenous luminal protein, whereas GFP-Snc1 is a tagged overexpressed membrane protein.
Ypt1 and Trs85 act in ER exit
Accumulation of internalized GFP-Snc1 in trs85Δ and ypt1 mutant strains has been previously reported. It was taken as an indication that Ypt1 and Trs85 play a role in endosome-to-Golgi transport (Montpetit and Conibear 2009; Sclafani et al. 2010). Here we show that in ypt1ts and trs85Δts mutant cells, GFP-Snc1 accumulates in the ER and not during PM recycling. This idea is supported by three observations: First, GFP-Snc1 accumulates in rings around the nucleus, which do not overlap with internalized FM4-64, suggesting ER localization. Second, similar accumulation of the nonrecycling GFP-Snc1-PEM protein, indicates that this accumulation is not caused by a PM recycling defect, but, rather by a transport defect. Third, the observation that overexpression of Ypt1 cannot suppress the endosomal accumulation of GFP-Snc1 in trs130ts mutant cells, but can suppress the ER accumulation of GFP-Snc1 in trs85Δts mutant cells, supports the idea that Ypt1 does not have a role in TRAPP II-mediated endosome-to-Golgi transport. Therefore, we can conclude that Trs85 does not play a role in endosome-to-Golgi transport. As for Ypt1, while ypt1ts mutant cells do not exhibit a block in this transport step, the idea that specific YPT1 alleles are defective in endosome-to-Golgi transport (Sclafani et al. 2010) is not addressed here, because these alleles were not used in this study. However, data presented here indicate that even if Ypt1 plays a role in endosome-to-Golgi transport, it does not act together with TRAPP II in that step (see below).
Why does GFP-Snc1 accumulate in the ER of ypt1ts and trs85Δts mutant cells? There are two possible explanations for this phenotype. Because Ypt1 is required for ER-to-Golgi transport and ypt1ts mutant cells exhibit a block in this step at their restrictive temperature (Jedd et al. 1995), it is possible that GFP-Snc1 is blocked in the ER of ypt1ts and trs85Δts mutant cells en route to the Golgi. In this scenario, at high temperatures, Trs85 plays a role in ER-to-Golgi transport. Alternatively, because both Ypt1 and Trs85 play a role in autophagy (Segev and Botstein 1987; Lynch-Day et al. 2010), it is possible that in ypt1ts and trs85Δts mutant cells excess GFP-Snc1 fails to be shuttled through the autophagy pathway for degradation in the lysosome/vacuole. Future studies should address this question.
Deletion of TRS85 results in temperature sensitivity only if GFP-Snc1 protein is overexpressed in this mutant strain. The correlation shown here between ER accumulation of GFP-Snc1 and GFP-Snc1-PEM at 37° and a temperature-sensitive growth phenotype of trs85Δ mutant cells expressing these tagged proteins, suggests that accumulation of proteins in the ER can cause cell toxicity. Such toxicity can be related to the role of Trs85 and Ypt1 in autophagy. Alternatively, it can be caused by an ER-to-Golgi transport block due to depletion of exocytic machinery by the ER-accumulated GFP-Snc1.
Grouping Ypt1 with Trs85 and Ypt31/32 with Trs130
We have previously shown that Ypt1 and Ypt31/32 are essential for Golgi entry and exit, respectively (Segev et al. 1988; Jedd et al. 1995, 1997). We have also shown that TRAPP I and TRAPP II act as GEF for Ypt1 and Ypt31/32, respectively (Jones et al. 2000; Morozova et al. 2006). This idea is supported by biochemical and genetic evidence (Morozova et al. 2006; Liang et al. 2007; Tokarev et al. 2009) and agrees with the assignment of TRAPP I and TRAPP II to the cis- and trans-Golgi, respectively, in the exocytic pathway (Sacher et al. 1998, 2001). However, this view has been challenged by negative biochemical results (Wang and Ferro-Novick 2002; Cai et al. 2008; Yip et al. 2010), and a role for Ypt1 with TRAPP II in endosome-to-Golgi transport was suggested (Sclafani et al. 2010). Data presented here further support a physiological role of Ypt31/32, and not Ypt1, with TRAPP II also in endosome-to-Golgi transport. While it is possible that GFP-Snc1 accumulates in endosomes in addition to its accumulation in the ER of ypt1 mutant cells not used in the current study, overexpression analysis presented here argues against the possibility that it acts with TRAPP II in this transport step. Thus, our genetic analysis further supports grouping of Ypt31/32, but not Ypt1, with TRAPP II in endosome-to-Golgi transport.
We propose that the modular TRAPP complexes act as GEFs for multiple Ypts, and not just of Ypt1. Currently, there is a dispute regarding the Ypt specificity of the different TRAPP complexes. Results presented here agree with previously published data that Trs85-containing TRAPP III acts as a GEF for Ypt1 (Lynch-Day et al. 2010). Importantly, we show here that TRAPP III does not act as a Ypt31/32 GEF under conditions in which GST-Bet5-purified TRAPP has such an activity (Figure S1). Our overexpression analysis provides genetic support to the idea that Ypt1 acts together with Trs85, whereas Ypt31/32 act together with Trs130 (Figure 5C).
In summary, studies presented here further support our view of a role for TRAPP I and Trs85-containing TRAPP III with Ypt1 in exit from the ER and a role for TRAPP II with Ypt31/32 in exit from the trans-Golgi and in the recycling of PM proteins to the Golgi.
Materials and Methods
Strains, plasmids, and reagents
Strains and plasmids used in this study are summarized in Table S1. For genetic interaction experiments, Ypt1, Ypt31, and Ypt32 in 2µ plasmids and their corresponding empty plasmids (ϕ, pRB684, pRS425, and yep351, respectively) were used. All yeast transformations were done using the lithium acetate method (Gietz et al. 1992). Escherichia coli transformation was done using electroporation.
For live-cell microscopy observation, GFP-Snc1 was integrated into NSY340 (ypt31Δ/32ts) and NSY128 (wild type) with pRS406 GS GFP-Snc1 (pNS568) (Lewis et al. 2000) to create YLY132 and YLY130, respectively. TRS85 was deleted from the wild-type strain (YLY130) with KANR cassette to create trs85Δts (YLY1347). YLY130 was used to mate with NSY1082 (ypt1ts), NSY991 (TRS130-HA), and NSY992 (trs130ts). The diploids were sporulated and tetrads were dissected to obtain the following GFP-Snc1–tagged mutants and wild-type strains: YLY1665 (ypt1ts) and YLY1664 (YPT1), YLY1771 (trs130ts) and YLY1770 (TRS130). GFP-Snc1-PEM was integrated into ypt3Δ/32ts, ypt1ts, trs130ts, and their corresponding wild-type strains with pRS406 GSSOM GFP-Snc1-PEM (pNS571) (Lewis et al. 2000) to create YLY1613 (ypt1ts-GFP-Snc1-PEM), YLY1583 (ypt31Δ/32ts-GFP-Snc1-PEM), YLY1585 (trs130ts-GFP-Snc-PEM), and their corresponding wild-type strains. TRS85 was deleted from YLY1582 to obtain YLY1651 (trs85Δ-GFP-Snc-PEM). Htb1-CFP was integrated into the above GFP-Snc1–tagged strains using pYL227 (a gift from B. Schwappach, Georg-August-Universität, Göttingen, Germany) for marking the nucleus.
Antibodies used in this study are: rabbit antiglucose-6-phosphate dehydrogenase (G6PDH; Sigma-Aldrich), affinity-purified rabbit anti-Ypt31 (Jedd et al. 1997), affinity-purified rabbit anti-Ypt1 (Segev et al. 1988), and horseradish peroxidase linked antirabbit and antimouse IgG (Amersham Biosciences, Little Chalfont, UK).
All chemical reagents were purchased from Amersco (Fair Lawn, NJ), unless otherwise noted. Other reagents used in this study: SynaptoRed, also known as FM4-64 (Molecular Probes, Eugene, OR), DAPI (Roche Diagnostics, Indianapolis, IN), Geneticin (Gibco Laboratories, Grand Island, NY), and restriction enzymes and buffers (Takara Biotechnology, Dalian, China).
Yeast culture conditions
For genetic interaction, cells were grown overnight at 26° in YPD or minimal (SC) media, normalized to the same density and spotted onto agar plates in 10-fold serial dilutions. Plates were incubated at different temperatures for genetic interaction assays. For live-cell fluorescence microscopy, yeast cultures were grown at permissive temperature (26°) in rich (YPD) or selective (when plasmid is used) media to log phase and switched to a restrictive temperature (37°) for 1.5 hr. Cells were either directly observed or subjected to DAPI staining or FM4-64 staining as described below.
Fluorescence microscopy
Direct fluorescence microscopy of temperature-sensitive yeast cells was performed as described in Chen et al. (2005) to localize Snc1, Snc1-PEM, and Htb1. DAPI staining was performed according to protocol suggested by the manufacturer. FM4-64 staining for endosomes (5 min) was done as previously described (Sclafani et al. 2010). Slides were visualized using Nikon inverted research microscope Eclipse Ti (Tokyo, Japan). More than five fields were collected for each sample. Each experiment was repeated at least twice.
Cell lysates and immunoblot analysis
For checking the expression level of Ypt1 or Ypt31 in cells used for the genetic interaction assay, five OD600 of overnight cell cultures were lysed in 100 µl of Laemmli buffer supplemented with equal volume of glass beads (BioSpec Products, Bartlesville, OK) and vortexed for 2 min. The supernatant of a 13,000 rpm spin (2 min) was subjected to immunoblot analysis with anti-Ypt1 or anti-Ypt31 antibodies, together with anti-G6PDH for serving as a loading control.
Supplementary Material
Acknowledgments
We thank B. Schwappach and M. Lewis for plasmids and S. Liebman, D. Taussig, and V. Mathur for critical reading of this manuscript. This work was supported by Nanjing Agricultural University (grant 680-804094-521 to Y. Liang), the Research Fund for the Doctoral Program of Higher Education of China (grant 20090097120039 to Y. Liang), the Fundamental Research Funds for the Central Universities (grant KYT201001 to Y. Liang), Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry ([2011]508 to Y. Liang), and by grant GM-45444 from the National Institutes of Health (to N.S.).
Footnotes
Communicating editor: M. D. Rose
Literature Cited
- Barrowman J., Bhandari D., Reinisch K., Ferro-Novick S., 2010. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11: 759–763 [DOI] [PubMed] [Google Scholar]
- Cai H., Zhang Y., Pypaert M., Walker L., Ferro-Novick S., 2005. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J. Cell Biol. 171: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chin H. F., Lazarova D., Menon S., Fu C., et al. , 2008. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 133: 1202–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Chen S., Tokarev A. A., Liu F., Jedd G., et al. , 2005. Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol. Biol. Cell 16: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Wiederkehr A., Seol J. H., Haguenauer-Tsapis R., Deshaies R. J., et al. , 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21: 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nair U., Yasumura-Yorimitsu K., Klionsky D. J., 2010. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H., 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R. Y., Koning A., Wright R., Rine J., 1996. In vivo examination of membrane protein localization and degradation with green fluorescent protein. Proc. Natl. Acad. Sci. USA 93: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Jedd G., Richardson C., Litt R., Segev N., 1995. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J. Cell Biol. 131: 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G., Mulholland J., Segev N., 1997. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J. Cell Biol. 137: 563–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Newman C., Liu F., Segev N., 2000. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell 11: 4403–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafourcade C., Galan J. M., Gloor Y., Haguenauer-Tsapis R., Peter M., 2004. The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p. Mol. Cell. Biol. 24: 3815–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R., 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11: 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Morozova N., Tokarev A. A., Mulholland J. W., Segev N., 2007. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol. Biol. Cell 18: 2533–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day M. A., Bhandari D., Menon S., Huang J., Cai H., et al. , 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107: 7811–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U. D., Krick R., Barth H., Appelles A., et al. , 2005. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280: 33669–33678 [DOI] [PubMed] [Google Scholar]
- Michelsen K., Mrowiec T., Duderstadt K. E., Frey S., Minor D. L., et al. , 2006. A multimeric membrane protein reveals 14–3-3 isoform specificity in forward transport in yeast. Traffic 7: 903–916 [DOI] [PubMed] [Google Scholar]
- Montpetit B., Conibear E., 2009. Identification of the novel TRAPP associated protein Tca17. Traffic 10: 713–723 [DOI] [PubMed] [Google Scholar]
- Morozova N., Liang Y., Tokarev A. A., Chen S. H., Cox R., et al. , 2006. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8: 1263–1269 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y., 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Nazarko T. Y., Huang J., Nicaud J. M., Klionsky D. J., Sibirny A. A., 2005. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy 1: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopov V., Govindan B., Novick P., Gerst J. E., 1993. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74: 855–861 [DOI] [PubMed] [Google Scholar]
- Sacher M., Jiang Y., Barrowman J., Scarpa A., Burston J. et al, 1998. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17: 2494–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Schieltz D., Yates J. R, 3rd, Ferro-Novick S., 2000. Identification and characterization of five new subunits of TRAPP. Eur. J. Cell Biol. 79: 71–80 [DOI] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Wang W., Horecka J., Zhang Y., et al. , 2001. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell 7: 433–442 [DOI] [PubMed] [Google Scholar]
- Sacher M., Kim Y. G., Lavie A., Oh B. H., Segev N., 2008. The TRAPP complex: insights into its architecture and function. Traffic 9: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani A., Chen S., Rivera-Molina F., Reinisch K., Novick P., et al. , 2010. Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic 11: 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., 2001a Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13: 500–511 [DOI] [PubMed] [Google Scholar]
- Segev N., 2001b Ypt/rab gtpases: regulators of protein trafficking. Sci. STKE 2001: re11. [DOI] [PubMed] [Google Scholar]
- Segev N., Botstein D., 1987. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol. Cell. Biol. 7: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., Mulholland J., Botstein D., 1988. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell 52: 915–924 [DOI] [PubMed] [Google Scholar]
- Stenmark H., 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Tokarev A. A., Taussig D., Sundaram G., Lipatova Z., Liang Y., et al. , 2009. TRAPP II complex assembly requires Trs33 or Trs65. Traffic 10: 1831–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ferro-Novick S., 2002. A Ypt32p exchange factor is a putative effector of Ypt1p. Mol. Biol. Cell 13: 3336–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Sacher M., Ferro-Novick S., 2000. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J., 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. K., Berscheminski J., Walz T., 2010. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat. Struct. Mol. Biol. 17: 1298–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.