Abstract
Polyploidization is an important mechanism for introducing diversity into a population and promoting evolutionary change. It is believed that most, if not all, angiosperms have undergone whole genome duplication events in their evolutionary history, which has led to changes in genome structure, gene regulation, and chromosome maintenance. Previous studies have shown that polyploidy can coincide with meiotic abnormalities and somatic cytogenetic mosaics in Arabidopsis allotetraploids, but it is unclear whether this phenomenon can contribute to novel diversity or act as a mechanism for speciation. In this study we tested the hypothesis that mosaic aneuploidy contributes to the formation of incipient diversity in neoallopolyploids. We generated a population of synthesized Arabidopsis allohexaploids and monitored karyotypic and phenotypic variation in this population over the first seven generations. We found evidence of sibling line-specific chromosome number variations and rapidly diverging phenotypes between lines, including flowering time, leaf shape, and pollen viability. Karyotypes varied between sibling lines and between cells within the same tissues. Cytotypic variation correlates with phenotypic novelty, and, unlike in allotetraploids, remains a major genomic destabilizing factor for at least the first seven generations. While it is still unclear whether new stable aneuploid lines will arise from these populations, our data are consistent with the notion that somatic aneuploidy, especially in higher level allopolyploids, can act as an evolutionary relevant mechanism to induce rapid variation not only during the initial allopolyploidization process but also for several subsequent generations. This process may lay the genetic foundation for multiple, rather than just a single, new species.
Keywords: somatic mosaicism, aneuploidy, allopolyploidy, polyploidy, incipient speciation
POLYPLOIDY is a major force in shaping angiosperm evolution and plant biodiversity (Ohno 1970; Stebbins 1971; Hegarty and Hiscock 2008; Leitch and Leitch 2008; Buggs et al. 2011). Polyploids are organisms with two or more complete sets of chromosomes. Polyploidy is frequent in nature and evidence shows that most, if not all, angiosperms have undergone at least one ancient genome doubling event in their evolutionary history (Bowers et al. 2003; Blanc and Wolfe 2004; Cui et al. 2006; Soltis and Soltis 2009; Jiao et al. 2011). Two major forms of polyploidy exist: autopolyploidy, which describes multiple genomes derived from a single species, and allopolyploidy, which refers to species in which genome doubling occurred concomitantly with the hybridization of two or more species.
Newly formed allopolyploids (neoallopolyploids) are subject to multiple changes from their progenitors in response to genome duplication, including structural chromosomal change, aneuploidy, genome rearrangement, epigenetic remodeling, and transcriptional change (Madlung et al. 2002; Henry et al. 2005; Huettel et al. 2008; Lim et al. 2008; Wright et al. 2009; Salmon et al. 2010; Chen 2010). Plants are generally quite plastic and can tolerate variation in their cytological composition (Leitch and Leitch 2008), including stable aneuploidies, supernumerary- and B chromosomes (Grant 1971). For example, in the eudicot Claytonia virginica, aneuploid cytotypes have been described with chromosome number variations between 12 and 191, depending on the geographic location of the population (Lewis et al. 1967). One hypothesis for why aneuploidy is tolerated in plants with higher ploidy levels is that the high degree of chromosomal duplication acts as a buffer to effects that would be more deleterious in diploids (Stebbins 1971). The allopolyploid Arabidopsis suecica, which is derived from A. thaliana and A. arenosa (O’Kane et al. 1996), is an ideal model for studying polyploidy. In resynthesized A. suecica genetic and genomic changes are frequently deleterious to the neoallopolyploids in their early generations. Allopolyploidization in these plants frequently results in moderate meiotic instabilities and in reduced fertility and fecundity (Comai et al. 2000; Madlung et al. 2005). In resynthesized A. suecica, allopolyploidization is also associated with stochastic changes in gene transcription that correlate, at least in part, with epigenetic modifications (Wang et al. 2004, 2006b; Madlung et al. 2005). These responses can result in reduced embryo viability and create an evolutionary bottleneck effect (Comai et al. 2000). In Tragopogon, resynthesized allopolyploid siblings vary in morphology, fertility (Tate et al. 2009), and tissue-specific transcriptional patterning (Buggs et al. 2010), possibly providing novel material upon which natural selection can act. Allopolyploidization can also lead to rapid advantages to the new species. Work by Ni et al. 2009 demonstrated that epigenetic changes in circadian-mediated pathway genes led to increases in photosynthetic output and overall hybrid vigor in the F7/F8 generation of a resynthesized A. suecica line as compared to the progenitors.
Mitotic chromosomal abnormalities, including aneuploidies, have been reported in both resynthesized and natural A. suecica, suggesting that these instabilities can arise during the early stages of allopolyploid formation and persist during the establishment of the species (Wright et al. 2009). Natural accessions of A. suecica display stable phenotypic variability despite low genetic diversity among accessions (Madlung et al. 2012). Work in polyploids of Tragopogon showed that chromosomal aberrations, such as intergenomic translocations, and mono- or trisomies, were variable between populations (Lim et al. 2008; Chester et al. 2012). Given the mostly stochastic nature of genomic changes during allopolyploidization that lead to genomic variability in allopolyploid offspring, we hypothesized that allopolyploidy not only leads to the formation of a single new species but to many potentially different variants, effectively promoting instant radiation. To test this hypothesis in a higher level allopolyploid, we produced populations of an Arabidopsis allohexaploid and analyzed cytogenetic stability in several distinct lines over eight generations. Here we report much greater cytological instability in early generation allohexaploids than previously reported in allotetraploids, somatic karyotypic variability within individuals, incipient establishment of various cytological groups among the different sibling lines tested, and correlative changes in phenotypes. Together, our data support the notion that higher allopolyploidization can result in cytologically variable sister lines of the new allopolyploid species, provide the raw material on which natural selection can act, and possibly lay the foundation for the subsequent evolution of distinct populations.
Materials and Methods
Plant material
Arabidopsis allohexaploids were synthesized by crossing diploid A. thaliana (2n = 2x = 10), accession Columbia (At) as the maternal parent, with allotetraploid A. suecica (As, Sue-1, TAIR accession CS22505, 2n = 4x = 26), as the paternal parent (Figure 1). One branch of the sterile triploid F1 hybrid spontaneously duplicated its genome and produced 37 viable hexaploid seeds, which were used as single-seed founders of individual, separate lines numbered 1–37. Lines were propagated, seeds harvested, and per F2 line one to several offspring were planted to produce the subsequent generation. Numbering followed the general scheme whereby the first number represents the original line. Additional numbers were used as needed to designate specific sibling lines. All offspring was self-fertile. All plants were grown in soilless peat mix. F1 and F2 plants were grown in a growth chamber at the University of Washington at 22° ± 3° under 16 h of light (TL80 fluorescent bulbs, Philips, Eindhoven, The Netherlands) and 8 h of darkness. F3–F6 and F8 plants were grown in a greenhouse at the University of Puget Sound under supplemental metal halide lamps (PL Light Systems, Beamsville, Ontario, 140–180 µmol m−2 s−1). The F7 generation was grown at the University of Missouri (Columbia, MO) first in a growth chamber (16/8 h light/dark cycle, 20°), and then in a greenhouse under natural daylight conditions (∼16/8 light/dark) and temperatures ranging between ∼20° and 30°. Seeds were not cold treated (vernalized) before germination. For FISH analysis both flower buds and root cells were collected. For the mitotic analysis, buds were harvested premeiotically. Later-stage flower buds were used for meiotic analysis. Root cell analysis was performed on root tips of 1-week-old seedlings grown on wet filter paper.
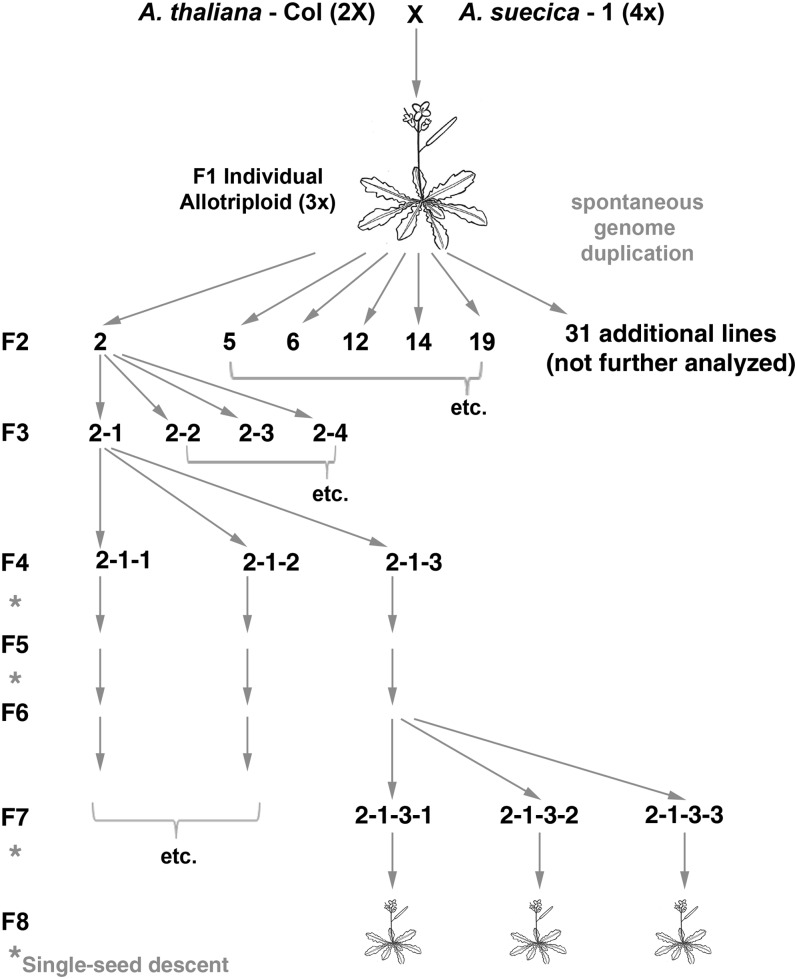
Figure 1 .
Pedigree and crossing scheme. Diploid Col A. thaliana was crossed with a tetraploid A. suecica to produce a triploid F1 generation individual, which spontaneously duplicated its genome, generating a set of allohexaploid siblings. Siblings were selfed to yield six distinct lines (2, 5, 6, 12, 14, and 19).
Flow cytometry
Nuclei were isolated from 4-week-old plants and stained with propidium iodide as described (Henry et al. 2005). The resuspended nuclei were spiked with 2.5 µl chicken erythrocyte nuclei (BioSure, Grass Valley, CA), which have a genome size of 1.05 billion bp (International Chicken Genome Sequencing Consortium 2004) per haploid genome. The nuclei were kept on ice in the dark for 2–4 hr and analyzed using a Becton-Dickinson FAC Scan (San Jose, CA). We collected 50,000 events per extract and the median fluorescence of each peak including the internal control was calculated using CellQuest software (Becton-Dickinson). Genome sizes were calculated and averaged for those preparations that were run in duplicate (N = 1–2).
Fluorescence in situ hybridization with CEN probes
Fluorescence in situ hybridization (FISH) was performed as described (Kato et al. 2004; Wright et al. 2009) with the following modifications: After fixing the tissue and before digestion with pectolyase and cellulase, premeiotic or meiotic floral bud tissue was washed twice with 100 μl of 1× citrate buffer (10 mM sodium citrate, 10 mM EDTA, pH 5.5). Root cells were digested using the same protocol, except that following enzymatic digestion, individual root tips were placed and directly squashed on the glass slide.
Probes for species-specific centromeric repeats were amplified from genomic DNA and labeled as described (Comai et al. 2003; Wright et al. 2009). Cells were randomly selected from FISH results that were interpretable with high confidence. In most cases, digestion had not fully removed the cytoplasm outlines of the cell, allowing us to exclude the possibility that chromosomes had been lost or gained from a cell spread during the slide preparation process producing artifacts. Cells were counted conservatively and cells with overlapping or uninterpretable signals were not counted.
From the original 37 F2 plants six sibling lines were randomly selected for analysis. FISH was used to determine the number of chromosomes from the two parental species in all analyzed offspring plants. Centromeric repeats from At were painted with fluorescein and from Aa with Texas Red. Thus, the diploid A. thaliana parent had 10 green chromosomes, and the tetraploid A. suecica parent had 10 green chromosomes from its A. thaliana and 16 red chromosomes from its A. arenosa progenitor. In an allohexaploid cell with fully additive genomes there should thus be 20 green At and 16 red Aa chromosomes (hereafter referred to as 20/16).
Generations F3–F5 were analyzed with a Zeiss fluorescence microscope. Digital images were acquired with a MicroPublisher 3.3 digital camera and QCapure software 2.71 (both from Quantitative Imaging, Surrey, British Columbia, Canada). Whole images were cropped and linearly adjusted for contrast, brightness, and color using Adobe Photoshop 7.0.1 (Adobe Systems, San Jose, CA). The F6 and F7 generations were analyzed using an Olympus BX41 and an Olympus BX61 equipped with F-View Soft Imaging system and Microsuite Basic Edition (Olympus) software, which was used to adjust whole images for contrast and brightness.
Chromosomes were counted on the basis of the number of centromeric signals. Eighteen plants were analyzed in the F3 and ∼10–21 cells were analyzed for each plant, (n = 204 cells, average = 11 cells/plant). Of the F6 generation, 18 plants were analyzed and 4–10 cells were counted in each plant (n = 171 cells, average = 5 cells/plant). Thirty-four plants were analyzed in the F7 generation and 4–40 cells were counted from each plant (n = 650 cells, average = 19 cells/plant). For root analysis a Nikon Eclipse Ti confocal microscope was used fitted with EZ-C1 3.9 acquisition software and NIS Elements BR 3.1 imaging software for analysis.
Determining and characterizing aneuploidy
Two measurements were collected to assess the degree of aneuploidy and genome stability. The first measurement was the number of cells that did not contain the expected additive number of chromosomes contributed from the two parents. Because a large proportion of cells displayed aneuploidy, a second method of analysis was developed on the basis of the assumption that some lines or individuals had established new adjusted chromosome numbers that were different from the additive value of the parents (the “adjusted euploid” vs. the “original euploid” value). These numbers, thus, reflect the fact that in some lines of later generations, euploidy, as defined by the expected additive genomes of the parent species, was no longer the predominant state. Means of chromosome counts for individuals or for sister plants from a common line were compared either within or between generations. Statistical significance of difference in chromosome numbers was assigned using either ANOVA or a two-tailed Student t-test.
Phenotypic analysis
Rosette leaf formation, fecundity, flower formation, and plant height were recorded for allohexaploids of the F7 and F8 generations. Pollen viability was estimated by live/dead staining of pollen from individual flowers with acetocarmine (45% acetic acid, 0.5% carmine w/v). The six anthers were extracted from each flower, placed in 20 µl acetocarmine, and squashed to release the pollen. Pollen was stained for 5 min and then viewed under a dissecting microscope. Ten separate fields of view of each slide were analyzed for live (stained red) or dead (not stained) pollen. Pollen stainability was statistically compared between the hybrid plants on the one hand and the combined parent species on the other hand, using an independent-samples Student t-test.
Transcriptional analysis
Reverse transcription PCR was performed as described previously (McCullough et al. 2010) with the exception that leaf RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA). Absence of genomic DNA in the c-DNA preparation was verified by using intron-spanning β-tubulin (Tub_P1: ATCCGTGAAGAGTACCCAGAT, Tub_P2: TCACCTTCTTCATCCGCAGTT) (Yu et al. 2002), and FLOWERING LOCUS C (FLC) primers (AtFLCF: GCTGATGATCTTAAAGCCTTGG, AtFLCR: ATCTGGCTAGCCAAAACCTGG). FLC primers amplified both copies of FLC from A. arenosa and the single copy from A. thaliana (Wang et al. 2006a). The amplicon also contained a ClaI restriction site for genotyping the parental origin of each transcript using CAPS analysis. This allowed the distinction between the single A. thaliana-, and the two A. arenosa-derived alleles. Treatment with ClaI resulted in two fragments of the A. thaliana allele, while the A. arenosa-derived A. suecica allele was not digested (Wang et al. 2006a). A total of eight F8 or F9 plants of equal age were sampled.
Statistical analysis
All statistical analyses were performed using SPSS versions 14 and 19, (SPSS, Chicago, IL), Microsoft Excel (Redmond, WA), and Graphpad (La Jolla, CA). Statistical significance was accepted at the P < 0.05 level.
Results
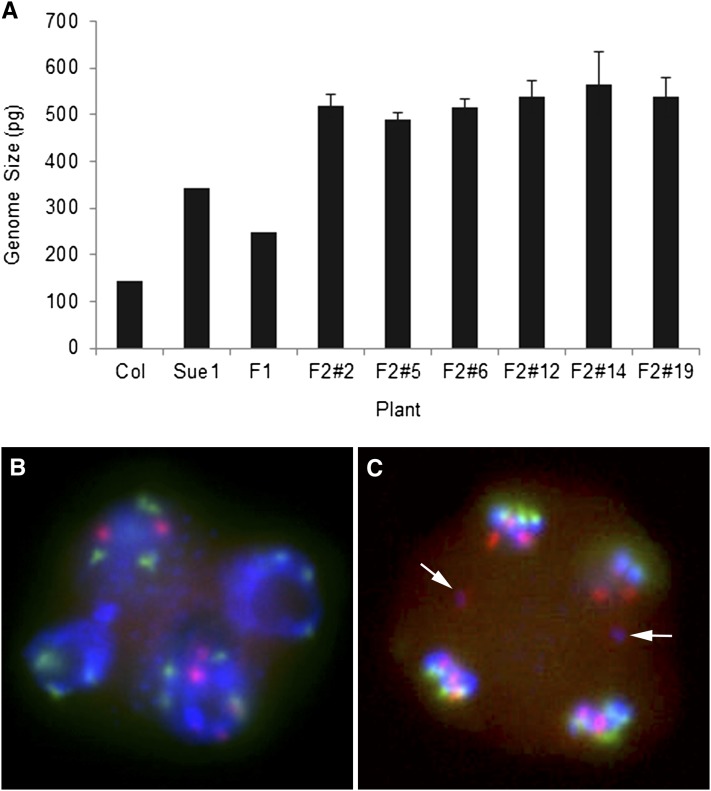
To verify ploidy levels in our experimental population, we measured the genome size of the triploid F1 and hexaploid F2 generations. Our genome size measurements of diploid A. thaliana (143 Mb) and 4x A. suecica (344 Mb) confirmed earlier measurements for A. thaliana (Davison et al. 2007) and A. suecica (Madlung et al. 2012). The F1 individual’s genome size (249 Mb) was as expected very close in size to the average of the two parental genomes (244 Mb), suggesting an additive triploid genome. Measurements for F2 plants (Figure 2A) showed slight variations between sibling lines with an average of 547 Mb (± SEM 4 Mb), which was ∼10% higher than would have been expected from a duplication of the F1 genome. Microsatellite analysis with species-specific markers showed additive signals of both parents in the hybrid (data not shown). F1 pollen FISH analysis of tetrads from the infertile branches of the F1 plant showed irregular numbers of chromosomes and chromosome laggards between developing pollen cells (Figure 2, B and C).
Figure 2 .
Genome size of parental species, triploid F1, and allohexaploid F2 individuals. (A) Genome sizes of 37 F2 plants (6 of them are shown) were measured using flow cytometry. Error bars represent SE, N = 1–2. (B) FISH of F1 pollen tetrad with irregular chromosome numbers. (C) Chromosome laggards (arrows) between developing tetrads of F1 triploid. Chromosomes were labeled with At- (green) or Aa (red)-specific centromeric probes and counterstained with DAPI.
Allohexaploids display frequent and sustained aneuploidy
To determine whether the genome of the neoallohexaploid was karyotypically uniform and stable, we analyzed cell spreads from the population for the occurrence of aneuploidy. Compared to the additive chromosome number (20/16), plants from the three generations analyzed displayed between 78 and 96% aneuploid cells per generation (Figures 3 and 4). Analysis of variance (ANOVA) suggested that the overall degree of aneuploidy between the F3, F6, and F7 generations was not significantly different (F = 2.722, P = 0.073, d.f. = 2). Each plant exhibited different levels of aneuploidy that ranged from 18 to 100% although the majority of the plants (57 of 68 plants) exhibited >80% aneuploid cells in their premeiotic tissues (Supporting Information, Table S1). Importantly, somatic cell chromosome numbers were generally not constant within a plant but instead formed mosaics within the premeiotic tissue. We observed abnormally dividing mitotic, and in older buds rare meiotic cells (Figure S1). In these cells bridges and laggards can be seen between daughter cells, suggesting a mechanism for the observed occurrence of aneuploidy.
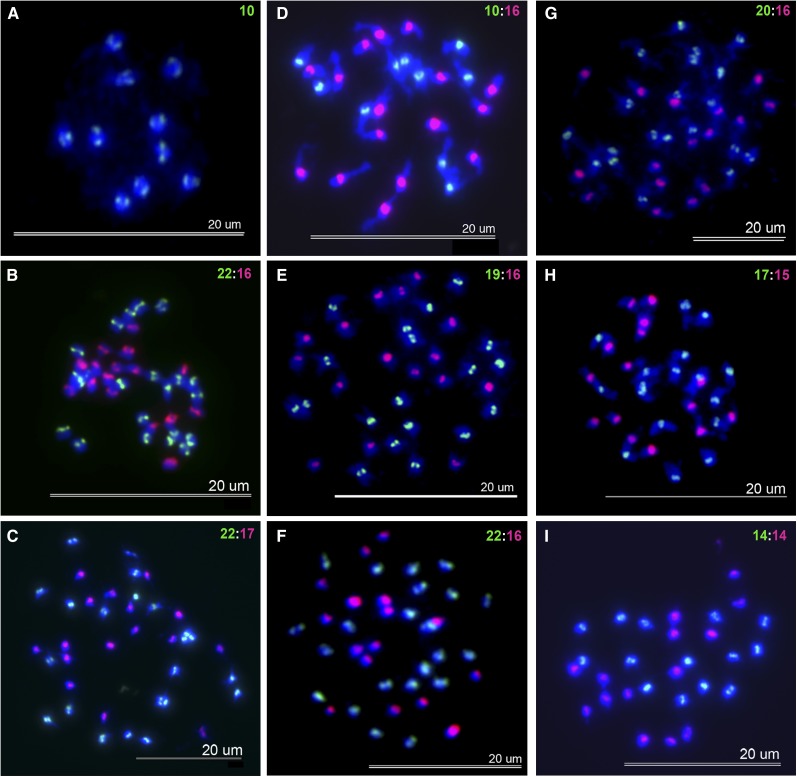
Figure 3 .
Mitotic chromosomes of somatic cells from parents and allohexaploids. Chromosomes were labeled with At- (green) or Aa (red)-specific centromeric probes and counterstained with DAPI (blue). (A) Maternal parent A. thaliana. (B) F7 6-1-2-3. (C) F7 6-1-2-3. (D) Paternal parent A. suecica. (E) F6 19-1-9. (F) F7 19-1-10-1. (G) F3 14-8 (example of euploid cell). (H) F6 14-4-4. (I) F7 14-4-4-1. Green and red numbers represent the chromosome count of that particular cell.
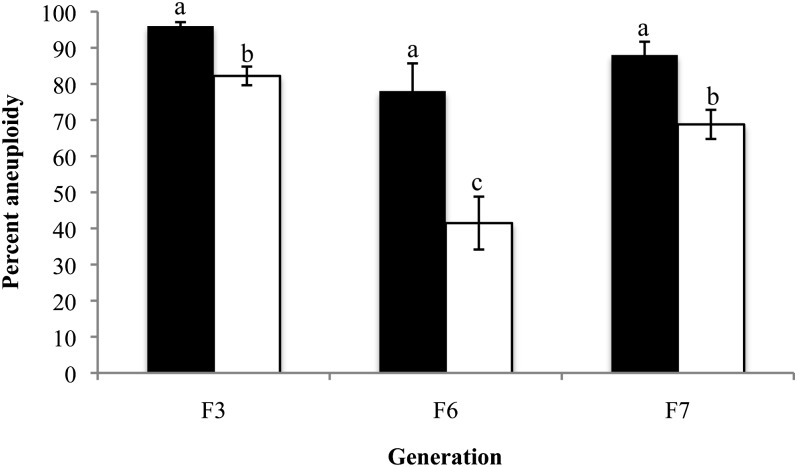
Figure 4 .
Somatic aneuploidy in individual cells of allohexaploids. The degree of aneuploidy was determined using FISH analysis (as in Figure 3) of plants from six sister lines over three generations. Aneuploidy was calculated on the basis of either divergence from the additive chromosome number (original euploid) of the two progenitors (solid bars) or adjusted modal (adjusted euploid) chromosome numbers (open bars) of each allohexaploid (see text for details). Error bars reflect the SEM. N = 204 cells (F3), N = 171 cells (F6), N = 626 cells (F7). Means between generations were compared within each type of analysis (solid or open bars) using ANOVA tests followed by Tukey post hoc analysis. Bars not connected by the same letter are statistically significantly different at P < 0.001 (adjusted euploid). (Original euploid comparisons (solid bars) P = 0.073, F = 2.72, d.f. = 2; adjusted euploid comparisons (open bars): P < 0.001, F = 14.74, d.f. = 2.
On the basis of the great amount of variation in chromosome numbers from cell to cell within individuals, one question was how the distribution of At and Aa chromosomes varied from plant to plant within sister lines, across lines, and across generations. To determine whether some lines lost chromosomes more frequently than others from generation to generation, and whether the chaotic chromosome constitution would stabilize over generations, we displayed our data as chromosome frequency charts for each individual analyzed (Figure S2), and determined from them the modal number of At and Aa chromosomes separately per cell per individual. This method of analysis assumed that each plant established a new adjusted chromosome number possibly distinct from the expected additive number (20/16), and we considered a cell as euploid (with respect to its new adjusted chromosome numbers) if it contained the modal numbers of both At and Aa chromosomes (Table S3; Figure S2). The modal chromosome numbers varied substantially between siblings of the same generation as well as between generations of the same line. For instance, some plants, especially in the F7 generation, had cells in which the number of At and Aa chromosomes were reduced (hypoploid) to 16 and 15 chromosomes, respectively (Table S3). Conversely, several plants had chromosome numbers above (hyperploid) the original euploid number (Figure 3, B, C, and F; Table S3). Other variants of the same generation exhibited chromosome numbers that ranged from 12 to 17 Aa chromosomes and from 13 to 23 At chromosomes in each cell (Figure 3 and data not shown). After determining modal chromosome numbers for each plant, there were 11 different cytotypes in the F3 and F6 generations, and 18 different cytotypes in the F7 generation (Table S3). Analysis of variance (ANOVA) based on the adjusted euploid values, suggested that the overall degree of aneuploidy was statistically significantly lower in the F6 generation compared to both the F3 and F7 generations, but the F3 and F7 did not show a significant difference between each other (F = 14.735, P < 0.001, d.f. = 2) (Figure 4).
To determine whether somatic mosaic aneuploidy is unique to immature floral tissue or is pervasive throughout the plant, we performed root squashes in four sibling lines of the F7 generation. All individuals tested showed mosaic aneuploidy, with chromosome numbers varying to a similar degree as seen in the premeiotic floral tissue (Figure S3).
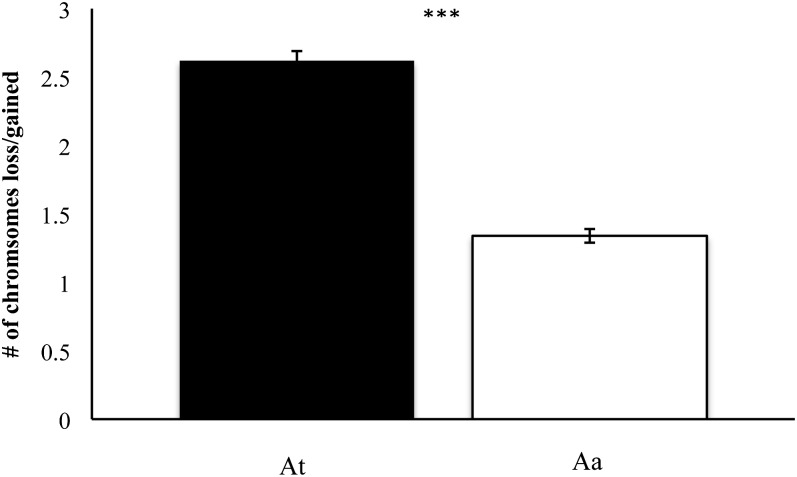
To test whether chromosomes from one progenitor were more or less likely to contribute to aneuploidy, we compared chromosome loss or gain between chromosomes inherited from At or Aa origin. A majority of cells exhibited the original euploid number of 16 Aa chromosomes; however, proper inheritance of the 20 At chromosomes was much less common (Figure 5; Table S1; Table S2). Of the 1035 cells assayed, an average of 2.62 ± 0.07 (SE) At chromosomes were lost or gained while Aa loss/gain on average occurred at a rate of only 1.34 ± 0.05 (SE) chromosomes per cell. Using an unpaired t-test, we tested whether aneuploidy was preferentially caused by loss/gain of At chromosomes on the one hand, or loss/gain of Aa chromosomes on the other hand (Figure 5). We observed a statistically significant difference indicating that At chromosomes were more frequently the cause of aneuploidy than Aa chromosomes (P < 0.001; t = 14.85; d.f. = 2068).
Figure 5 .
Most frequent origin of chromosomes lost or gained in aneuploid allohexaploids. The contribution to aneuploidy by At or Aa chromosomes was compared in the three investigated generations. Chromosomes from the At genome were more likely to be lost/gained than those from the Aa genome (error bars: SE; N = 1035 cells; P < 0.001, t = 14.85, d.f. = 2068). The difference was significant at P < 0.05.
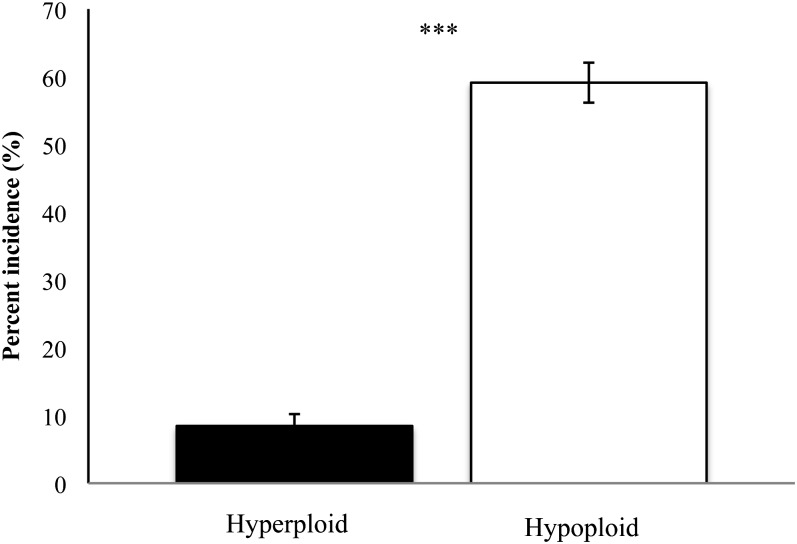
To test whether cells were more likely to lose or gain additional chromosomes, we classified all analyzed cells as either hyperploid (containing >36 chromosomes) or hypoploid (containing <36 chromosomes) and compared the frequency of the two states. Cases of hyperploidy (regardless of the parental origin of the extra or missing chromosomes) were statistically significantly less frequent than cases of hypoploidy, as compared by an unpaired t-test (Figure 6, for raw data see Table S1; P < 0.001, t = 14.8, d.f. = 270). Of the 136 plants surveyed, 8% of cells showed simultaneous hyperploidy of both parental types of chromosomes, and 59% of cells showed simultaneous hypoploidy of both At and Aa chromosomes (Table S1). Some sister lines were more prone to hyperploidy. For example, line 6 frequently displayed At chromosome numbers up to 23 (Table S1). The complete FISH data set can be found in Table S4.
Figure 6 .
Incidence of hyperploidy and hypoploidy among all allohexaploid cells analyzed. The frequency of cells displaying chromosome numbers above the original additive chromosome number of 20/16 was significantly lower than for cells with fewer than 20/16 chromosomes in cells of allohexaploid plants (P < 0.001, t = 14.8, d.f. = 270).
Cytogenetic stability of allohexaploids increases as generations progress
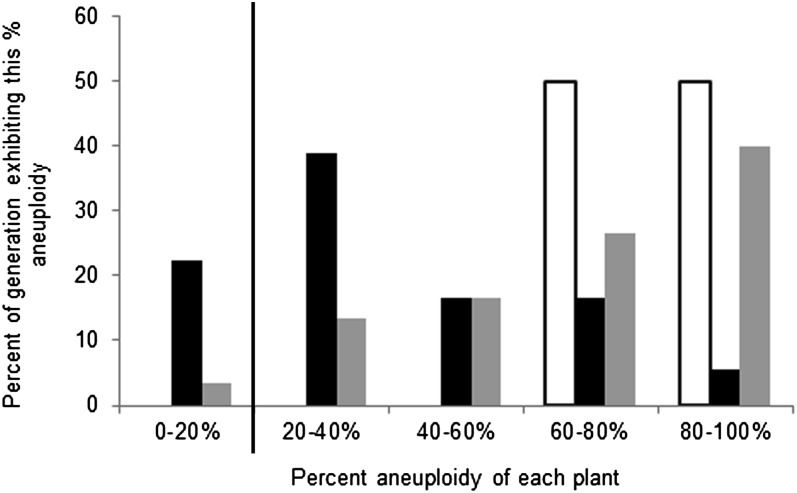
To assess the overall genetic stability of the allohexaploids in relation to their parent species, we compared the level of aneuploidy in each generation of the allohexaploids with that of its progenitors, A. thaliana and A. suecica. While diploid A. thaliana is very stable and does not display somatic aneuploidy, mosaic aneuploidy has been reported in premeiotic floral tissues for A. suecica at levels of 0–20%, depending on the assayed population (Wright et al. 2009). We compared the degree of aneuploidy (on the basis of adjusted euploid value) of the allohexaploids to the basal level of aneuploidy in these populations of A. suecica. As a conservative estimate, we assumed that any offspring of a cross between A. suecica and At could thus have an innate base level of aneuploidy of up to 20%. Any plant with >20% aneuploidy was considered to be more unstable than the progenitor species with respect to chromosome composition. While all of the F3 plants ranged in their level of aneuploidy from 60 to 100%, the F6 and F7 generations included many plants with levels of aneuploidy between 0 and 60% (Figure 7). The F6 generation exhibited the least amount of aneuploidy, making it the most genetically stable of the three generations.
Figure 7 .
Frequency of plants with high levels of aneuploidy compared by generation. Multiple cells from each individual plant were assessed for their level of aneuploidy and grouped into five bins on the basis of the frequency of aneuploid cells in their tissue. Frequencies of plants belonging to each bin are graphed separately by generation. Open bars, F3; solid bars, F6; shaded bars, F7. Previous results in natural A. suecica populations (Wright et al. 2009) display aneuploidy in 0–20% of cells. Therefore, as a conservative estimate of aneuploidy contributed by the paternal parent, bins with >20% of aneuploid cells are separated from the background noise aneuploidy by the vertical line.
Pollen stainability in allohexaploids is reduced
The fertility of 15 randomly selected F7 allohexaploid plants was assessed by pollen staining with acetocarmine and compared to that of the parent species (At and A. suecica) (Table 1; Figure S4). In general, pollen stainability of the allohexaploids was lower than in the parent species. There was no obvious correlation between the degree of aneuploidy and pollen stainability, suggesting that chromosomal aberrations were not directly linked to gamete viability. Interestingly, the allohexaploid with the greatest pollen stainability (F6 5-1-6-2) also had sustained the greatest overall loss of chromosomes from the original euploid number. The other hexaploids displayed lower pollen stainability than did the parents. The combined averages of the pollen stainability of the hexaploid plants (25 ± 5.6%, SE; N = 16) on the one hand, and the combined parent species (71 ± 2.5%, SE, N = 2) on the other hand, were statistically significantly different using an independent-samples Student t-test (P = 0.012, t = 2.84, d.f. = 16). Individual values for stainability are reported in Table 1.
Table 1 . Pollen viability of F6 and F7 allohexaploids and their parent species.
| Plant number | Viability (%) | No. of chromosomes | Total no. of chromosomes | |
|---|---|---|---|---|
| At | Aa | |||
| Col 2x | 68 | 10 | — | 10 |
| Sue 1 | 75 | 10 | 16 | 26 |
| F6 5-1-1-1 5pa | 34 | 17 | 16 | 33 |
| F6 5-1-1-1 4pa | 34 | 17 | 16 | 33 |
| F6 5-1-6-1 | 44 | 16 | 16 | 32 |
| F6 5-1-6-2 | 85 | 13 | 14 | 27 |
| F7 6-1-2-2 | 15 | 21 | 15 | 36 |
| F7 6-1-2-3 | 5 | 21 | 16 | 37 |
| F7 6-1-2-4 | 62 | 20 | 16 | 36 |
| F7 6-1-8-1 | 7 | 16 | 16 | 32 |
| F7 6-1-8-3 | 13 | 21 | 16 | 37 |
| F7 6-1-8-4 | 20 | 23 | 16 | 39 |
| F7 6-1-9-3 | 20 | 20 | 16 | 36 |
| F7 12-19-1-1 | 12 | 18 | 16 | 34 |
| F7 12-19-1-2 | 12 | 17 | 16 | 33 |
| F7 12-19-1-4 | 25 | 19 | 16 | 35 |
| F7 12-19-1-5 | 15 | 19 | 15 | 34 |
| F7 14-4-4-1 | 2 | 16 | 15 | 31 |
5p, five-petaled inflorescence; 4p, four-petaled inflorescence on the same plant.
Although we did not quantify fecundity of the allohexaploids, visual observation of the amount of collected seeds showed a steady decline in some but not all sibling lines over the seven generations. All six original lines used in this study were still represented by germinating seeds in the F8. However, when considering all 21 F7 sibling sublines within these main six original lines (see pedigree in Figure 1), only eight produced germinating seed in the F8 generation (data not shown).
Phenotypes of different allohexaploid lines vary substantially between sister lines
To assess whether there was any correlation between variation in the adjusted modal chromosome numbers and variation in novel phenotypes of the plants, we made visual inspections for plant height, flower size, leaf morphology, leaf size, leaf number, flowering time, and phenotype of the inflorescence. Flowering time varied widely between sister lines with some lines (lines 2 and 5) lagging behind a full generation. To determine whether the observed differences in flowering time between sibling allohexaploid lines were due to steady state levels of the floral inhibitor FLC, we performed RT–PCR analysis in eight individuals, of which six were flowering and two were not flowering at the time of analysis. FLC was expressed approximately equally in all tested plants, indicating that FLC expression was not likely to be the primary driving force behind the differences in flowering time (Figure S6). Digestion of the RT–PCR product with ClaI indicated that in all eight individuals, at least one thaliana- and one arenosa/suecica-derived allele was transcribed (data not shown).
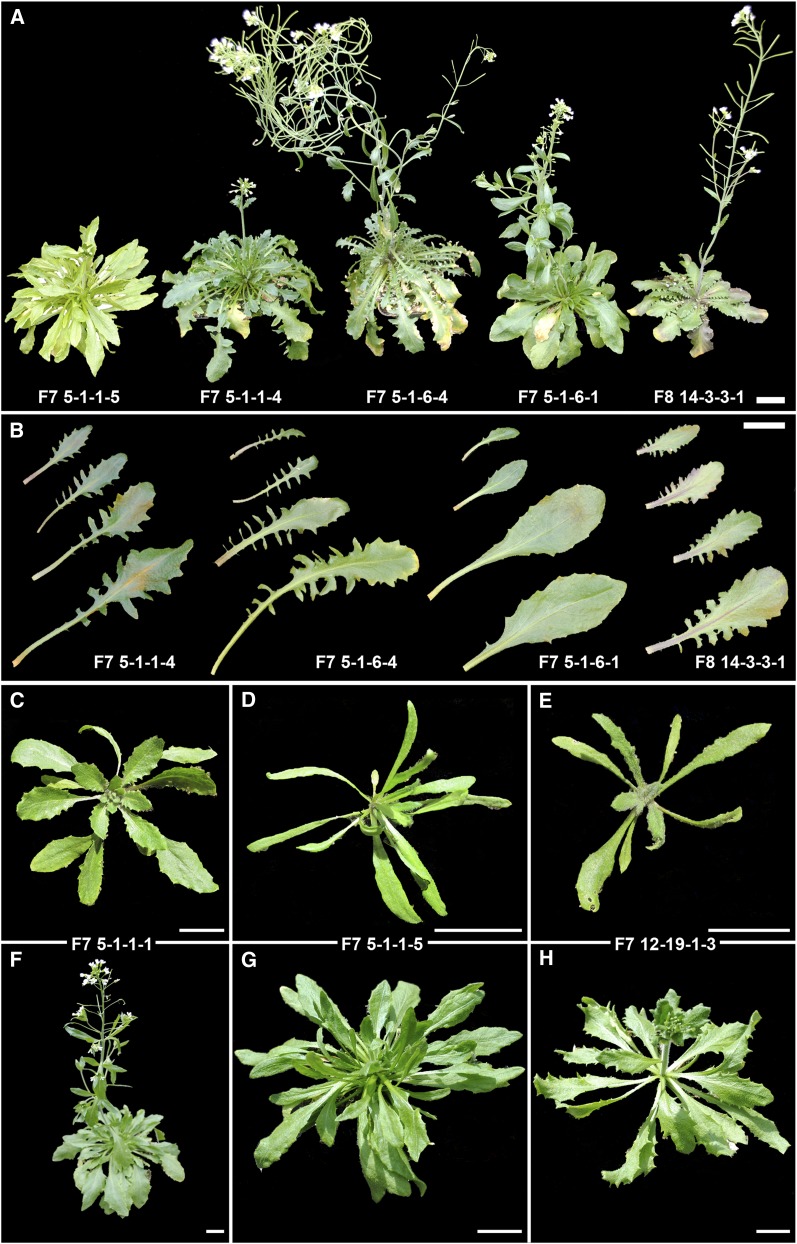
Variation in leaf morphology was particularly pronounced. Some lines displayed smaller, more rounded leaves that resembled those of A. thaliana, others showed long, slender, and highly serrated leaves, more similar to A. arenosa, while others had intermediate serration and a slightly wider leaf body, typical for A. suecica. To assess whether variation increased within sister lines derived from a single original 6x seed, we compared leaf shape and growth habit in four F7 sister lines of the original line 5 and one from line 14 (Figure 8). Flowering time varied between these lines by ∼4 weeks between the earliest and the latest flowering line (Figure 8A). Leaf coloration varied from light green (5-1-1-5) to dark green (5-1-1-4). Phenotypes varied from slightly spinose, not quite entire leaf margins (lines 5-1-6-1 and 5-1-1-5) to pinnatifid (pinnately lobed) leaves in line 5-1-6-4, to leaves that were lobed on the proximal half and serrated on the distal half of the leaf (5-1-1-4) (Figure 8B). Leaf length among the more serrated leaves varied from short (14-3-3-1) to long (5-1-6-4). Cauline leaves were serrated in 5-1-6-4 but smooth in 5-1-6-1 (Figure 8A). Several individuals from separate sister lines displayed very long, spindly leaves early on in development (Figure 8, D and E) compared to the usually rounded and only slightly serrated leaves commonly found in the paternal parent of the allohexaploids (Figure 8C). Some of the plants with spindly leaves died at the early rosette stage (data not shown), while others developed into fertile adult plants (Figure 8, G and H), albeit with a later flowering time than most of their same-age cohort siblings (Figure 8F).
Figure 8 .
Phenotypes of select F7 and F8 allohexaploids. (A) Flowering times varied between F7 siblings of the same line by up to 4 weeks. Note overall growth habit, light green foliage in individual 5-1-1-5, and variation in leaf morphology. For comparison with the most common leaf phenotype, an individual from line 14 is shown. (B) Variation in leaf serration between siblings of line 5 and an individual of line 14. (C–H) Developmental changes in three individuals between early rosette stage (∼3 weeks old) and onset of flowering (C/F; D/G; E/H). Note the slender leaves in D and E compared to the more common leaf shape in C. All plants were grown in the same conditions, were of the same age, and were photographed at the same times. Bars, 2 cm.
We correlated chromosome loss or gain with changes in leaf morphology. Table 2 shows that plants with a 20/16 cytotype have the most A. suecica-like phenotype with medium sized, oval, and slightly serrated leaves. An increase in At chromosomes seems to coincide with smaller, more rounded A. thaliana-like leaves, while a loss of chromosomes had several effects, including a variety of leaf shapes and sizes but no clear trend toward one specific phenotype. It must be noted that our analysis was not able to distinguish exactly which chromosomes were lost or gained and whether or not different lines with the same overall chromosome number had lost (or gained) the same or different chromosomes.
Table 2 . Rosette leaf morphology of select F6 and F7 plants and their modal chromosome numbers.
| Current modal chromosome nos. | Rosette leaf phenotype | |||||
|---|---|---|---|---|---|---|
| Plant no.a | At | Aa | Leaf size | Leaf edge | Additional | |
| F7 6-1-8-4 | 23 | 16 | Small, circular | Smooth | — | Smaller |
| F7 6-1-8-3 | 21 | 16 | Small, circular | Smooth | — | Circular |
| F7 6-1-2-3 | 21 | 16 | Medium, oval | Serrated | Round tips | Smooth |
| F7 6-1-6-4 | 21 | 16 | Medium, oval | Serrated | Curly stem | |
| F7 6-1-2-2 | 21 | 15 | Medium, oval | Slightly serrated | — | |
| F7 19-1-10-1 | 20 | 16 | Small, oval | Serrated | — | |
| F7 6-1-6-2 | 20 | 16 | Medium, oval | Slightly serrated | — | |
| F7 6-1-2-4 | 20 | 16 | Large, oval | Serrated | — | |
| F7 6-1-2-1 | 18 | 17 | Long, oval | Slightly serrated | — | |
| F7 6-1-8-2 | 18 | 11 | Small, oval | Serrated | Pointy tips | |
| F7 12-19-1-1 | 18 | 16 | Medium, thin | Serrated | Pointy tips | |
| F6 5-1-1-1 | 17 | 16 | Long, thin | Slightly serrated | Pointy tips | |
| F6 5-1-6-1 | 16 | 16 | Very small | — | — | |
| F7 14-4-4-1 | 16 | 15 | Long, very thin | Smooth | — | Longer |
| F7 19-1-10-2 | 14 | 12 | Short, oval | Serrated | — | Thinner |
| F6 5-1-6-2 | 13 | 14 | Very small | — | — | Serrated |
Plants are ordered from highest to lowest chromosome number.
Aside from leaf phenotypic variation, overall growth habit and development varied widely (Figure S5). One plant (F6 5-1-1-1) produced several flowers with five, rather than the normal four petals (Figure S5). We compared cells harvested from buds adjoining the flowers with the abnormal petal number to cells from a bud cluster on a separate side shoot of the same plant with normal petals. We found that buds neighboring the five-petaled flowers had an overall higher degree of aneuploidy (36%) than buds grown in proximity to the normal flowers (16%), but found no difference in their modal chromosome numbers (17 At/16 Aa; Table S2).
Discussion
Genetic diversity within a population allows natural selection, which promotes adaptation and evolution of a species (Seehausen 2004; Mallet 2007). Allopolyploidization leads to massive stochastic changes in gene activity and the genome landscape of the new allopolyploid species compared to its progenitors (Otto and Whitton 2000; Charlesworth and Wright 2001; Osborn et al. 2003; Adams and Wendel 2005; Comai 2005; Chen 2007, 2010; Soltis and Soltis 2009; Buggs et al. 2010, 2011). This sudden increase in novel variation lays the foundation for subsequent selection and evolutionary change. While molecular events shaping genomic change during and immediately after allopolyploidization have been studied intensely (Chen 2010), less experimental attention has been given to the question of whether allopolyploidization leads to the formation of a single new species or whether it can result in evolutionary radiation. In the example of the natural allotetraploid A. suecica genetic variation among populations is low (Lind-Hallden et al. 2002) and phenotypic differences between accessions are subtle (Madlung et al. 2012). By contrast, in resynthesized A. suecica genomic variation is frequent but newly formed allopolyploids also display poor fertility, fecundity, and viability (Comai et al. 2000), indicating that allopolyploidization presents a severe bottleneck. To our knowledge this is the first report using FISH in a controlled pedigree of resynthesized allopolyploids to address the question to what degree changes in karyotype concurrent with allopolyploidization provide the raw material for natural selection on other traits in a tractable, well-studied genetic system like Arabidopsis.
Somatic mosaicism and cytotypic variation is prevalent in early-generation allohexaploids
Synthetic allohexaploids of A. thaliana and A. suecica display extensive and lasting mosaic aneuploidy in somatic floral and root tissue. We observed frequent cytotypic variation between different lines, different siblings of the same line, and different cells of the same plant (Figure 3; Table S1). Variable numbers of chromosomes had previously been reported for several Finnish A. suecica accessions (Harmaja and Pellinen 1990) and somatic mosaics had been found in most natural and in one resynthesized A. suecica accessions (Wright et al. 2009). Chromosomal mosaics have been described in a number of polyploid plants such as synthetic hexaploid potatoes (Venkateswarlu and Krishna Rao 1969; Ramanna and Hermsen 1971), polyploids of various Triticinae (Sachs 1952; Pohlendt 1958), tobacco (Nuti Ronchi et al. 1981), and Rubus (Thompson 1962). The fact that somatic aneuploid mosaics have predominantly been reported in polyploids suggests that the additional homeologous chromosomes in polyploids provide the plant with added genomic flexibility. This increased genetic variation in the organism has been suggested as a mechanism for evolutionary change (Gill et al. 1995) but direct evidence for this hypothesis has been sparse. In human embryos, mosaic aneuploidy arising via nondisjunction of chromosomes during mitosis (Daphnis et al. 2005) or via premature centromere division (Furukawa et al. 2003) has mostly been associated with abnormal development.
When based on adjusted euploid chromosome numbers, the level of aneuploidy in our study fell statistically significantly from the F3 to the F6 generation, but that trend toward genomic stability was not continued in the F7 generation, and levels of aneuploidy remained overall relatively high (Figure 4). On the basis of original chromosome numbers, all three generations showed continually high levels of aneuploidy (Figure 4). To assess changes of karyotype stability throughout the generations more closely, we compared each plant’s adjusted aneuploidy level with the previously reported level of aneuploidy in the parent species A. suecica-1 (Wright et al. 2009). Any plant that exhibited aneuploidy levels higher than the highest observed levels in A. suecica accessions (20%) was considered genetically unstable. Under these analytical conditions, all plants of the F3 generation were unstable, whereas the F6 and F7 generations contained plants that showed both higher and lower levels of aneuploidy (Figure 7). Judging from this analysis we conclude that there is an increase in relative karyotype stability in the later generations compared to F3. However, we also note that stability was highest under this criterion in F6, and somewhat lower in the F7 generation. Our data are consistent with the notion that somatic aneuploidy and genome instability are greatest upon allopolyploidization, and this instability becomes less pronounced in subsequent generations. We note that the F7 generation was grown in a different location from where the other generations were grown and where slightly different greenhouse conditions might have had an influence on plant development. Should growth conditions play a role in genomic stability, the observations for the F7 generation might be viewed in this light. In any case, our experiments show that by the time these plants had reached the F7 generation, there was still a considerable amount of cytogenetic instability present.
Stable (nonmosaic) aneuploidy can lead to a variety of consequences. Blakeslee et al. (1920) described phenotypic variances in all possible trisomics of Datura noting that these aneuploids were less vigorous than diploids but viable. The loss or gain of a single gene on a mono- or trisomic chromosome can either lead to a gene dosage effect where affected gene products show changes in expression levels, or gene expression levels can be unaffected as a consequence of dosage compensation (Birchler 2010). Extrapolating from these findings, it appears possible that in somatic mosaics like in the population investigated here, each cell has somewhat different gene expression levels, either showing gene dosage effects or dosage compensation.
It is interesting to note that At chromosomes were more frequently lost than Aa chromosomes in the allohexaploids (Figure 5; Table S1; Table S2). In tetraploid A. suecica we had previously found Aa chromosomes to be less stably transmitted than At chromosomes (Wright et al. 2009). The karyotypic constitution of the allohexaploids used in this study contains the equivalent chromosome complement of autotetraploid A. thaliana and diploid A. arenosa. It is possible that the greater instability of autotetraploid meiosis compared to that of diploid meiosis in A. thaliana (Wright et al. 2009) is the reason for the higher transmission instability of At chromosomes.
Cases of chromosome loss were more prevalent than cases of chromosome gain (Figure 6). A possible mechanism for aneuploidy in mitotic cells is nondisjunction during mitosis (Daphnis et al. 2005), which would explain both chromosome losses and gains. Additionally, the formation of laggards, as observed in this material (Figure S1), can account for chromosome loss during cell division (Thompson 1962; Nuti Ronchi et al. 1981; Ford et al. 1988).
Phenotypic variation
The various allohexaploid lines showed widely varying phenotypes (Figure 8). Some of these phenotypes were loosely correlated with the degree of aneuploidy observed (Table 2). Sublines (Figure 1) that died out during the first seven generations sometimes displayed obvious developmental abnormalities, such as dwarfism or the inability to flower (data not shown). Other unsuccessful lines produced less or mostly inviable (nonstainable) pollen (Table 1; Figure S4) or did not set seed (data not shown). It was surprisingly difficult to obtain high-quality meiotic chromosome spreads from this material, which is consistent with the notion that pollen formation was reduced in all except one line. In the absence of sufficient meiotic data, it is difficult to speculate on chromosome pairing behavior in these allohexaploids.
The most conspicuous difference in phenotype between sister lines was the time needed until flowering. Previous microarray analysis work with resynthesized allotetraploid A. suecica had shown increased levels of the floral repressor FLC in late-flowering allopolyploid material (Wang et al. 2006a,b). FLC-mediated flower repression is alleviated by cold treatment (vernalization) (Sheldon et al. 2008; Amasino and Michaels 2010). Some A. thaliana ecotypes have low levels of FLC even without vernalization (Sheldon et al. 2000), and in nonvernalized A. thaliana FLC expression can be found in floral structures (Sheldon et al. 2008). The ability of a plant to flower is not solely controlled by FLC, as several other pathways are also involved in the control of flowering (Amasino and Michaels 2010). In allopolyploid Brassica species, flowering time variation between populations is correlated with chromosomal rearrangements involving FLC-carrying chromosome segments and changes in FLC transcription (Pires et al. 2004). To begin to address the molecular basis of the observed variability in flowering time between early and late flowering lines in this hexaploid population, we tested transcriptional activity of FLC but found no significant expression differences (Figure S6). CAPS analysis showed that alleles from both original parents were transcriptionally active in all tested lines (data not shown). Since the plants in our experiments were not vernalized, FLC expression differences may not have played a decisive role in flowering time (Sheldon et al. 2008). Given the observed flowering time differences and the lack of evidence that FLC is directly responsible for the phenotypes, it appears that flowering time variation is regulated in a more complex manner in these allohexaploids than solely via FLC gene dosage differences. In allopolyploids of Arabidopsis genomic rearrangements have been described at only a few loci (Madlung et al. 2005) and no evidence has been reported for bona fide homeologous exchanges.
Despite the current lack of data explaining the underlying molecular reasons for phenotypic variation in the allohexaploid sibling lines, our analysis brings up several important points. First, we have shown that allopolyploidization in a cross between A. thaliana and A. suecica does not produce a single homogeneous population but leads to an aneuploid swarm that displays cytogenetic heterogeneity, phenotypic variation, and variability in individuals’ fertility. In a relatively short period of time, lines have begun to separate from each other, displaying typical new chromosome numbers (Figures 4 and 7) and phenotypic characteristics (Figure 8; Figure S4; Figure S5). This novel variation incurred via allopolyploidy could thus represent the foundation for evolutionary radiation that may propel the new populations to produce many, rather than just a single new allopolyploid species. Second, our data show that neoallohexaploids, when compared to neoallotetraploids (Comai et al. 2000; Wright et al. 2009) produced from the same progenitor genomes, display a much greater degree of somatic aneuploid mosaicism and cytogenetic variability. This mosaicism is systemic and found both in root and shoot tissues (Figure 3; Figure S1; Figure S3). While phenotypic diversity is subtle in allotetraploid A. suecica (Comai et al. 2000; Madlung et al. 2012), it is more pronounced in allohexaploids, possibly due to the fact that the greater number of homeologs in the genome allows a greater degree of flexibility in genome reshuffling. Third, our study suggests that allopolyploidization may not be a singular bottleneck event incurred only during the first meiosis, in which the genome is rearranged, and which is followed by slow genomic recovery (Cifuentes et al. 2010). Instead, at least in allohexaploids of Arabidopsis, somatic aneuploidy appears to promote the reorganization of the genome in somatic cells for at least seven generations.
Despite their differences in cytotypic make up and physical appearance, most of these lines appear to be genomically and phenotypically still unstable. We cannot predict from the material at its current state if all or any of these lines will be able to survive and stabilize. Phenotypic variation, observed in tetraploid resynthesized A. suecica allopolyploids (Comai et al. 2000), was able to give rise to several stable, vigorous lines (Ni et al. 2009). On the other hand, previous work with 50 resynthesized allotetraploid Brassica napus lines showed over a period of 10 generations that these plants became less, rather than more stable. However, in this case the instability was likely due to homeologous transpositions (Gaeta et al. 2007; Gaeta and Pires 2010), and homeologous chromosome replacement (Xiong et al. 2011), but not to aneuploidy.
While we did not attempt in this study to assign specific chromosome losses or gains to corresponding phenotypes, it is intriguing to speculate on such relationships. We are currently testing BAC FISH markers that would allow us in the future not only to distinguish aneuploid cells, but to assign complete karyotypes to each cell. This might then allow the correlation of dosage effects of specific chromosomes with observed phenotypes.
Supplementary Material
Acknowledgments
We thank Michal Morrison, Susan Bennett, Paul Woodward, Johanna Morrow, Jen Holland, Mannie Liscum, and Zhiyong Xiong for technical and logistical help; Jerry Davison and Luca Comai for help with the flow cytometry; Jack Vincent, Cristina Walcher, and two anonymous reviewers for helpful suggestions to improve the manuscript; and the National Science Foundation (NSF) Functional Biology of Polyploids Consortium for stimulating discussions. This study was supported by an NSF Plant Genome grant (DBI-0501712) (A.M. and C.J.P.), a NSF Major Research Instrumentation grant (MRI-0619009) (A.M.), a summer undergraduate research fellowship from the American Society of Plant Biology (S.C.M.), and funds from the University of Puget Sound Enrichment Committee (S.C.M., G.M.T., and A.M.).
Footnotes
Communicating editor: T. Wu
Literature Cited
- Adams K. L., Wendel J. F., 2005. Novel patterns of gene expression in polyploid plants. Trends Genet. 21: 539–543 [DOI] [PubMed] [Google Scholar]
- Amasino R. M., Michaels S. D., 2010. The timing of flowering. Plant Physiol. 154: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., 2010. Reflections on studies of gene expression in aneuploids. Biochem. J. 426: 119–123 [DOI] [PubMed] [Google Scholar]
- Blakeslee A. F., Belling J., Farnham M. E., 1920. Chromosomal duplication and Mendelian phenomena in Datura mutants. Science 52: 388–390 [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K., 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J., Paterson A. H., 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Elliott N. M., Zhang L., Koh J., Viccini L. F., et al. , 2010. Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 186: 175–183 [DOI] [PubMed] [Google Scholar]
- Buggs R. J. A., Zhang L., Miles N., Tate J. A., Gao L., et al. , 2011. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr. Biol. 21: 551–556 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Wright S. I., 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11: 685–690 [DOI] [PubMed] [Google Scholar]
- Chen Z. J., 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., 2010. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M., Gallagher J. P., Symonds V. V., Cruz da Silva A. V., Mavrodiev E. V., et al. , 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M., Grandont L., Moore G., Chèvre A. M., Jenczewski E., 2010. Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 186: 29–36 [DOI] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A. P., Winter K., Holmes-Davis R., Reynolds S. H., et al. , 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tyagi A., Lysak M., 2003. FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res. 11: 217–226 [DOI] [PubMed] [Google Scholar]
- Cui L., Wall P., Leebens-Mack J., Lindsay B., Soltis D., et al. , 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daphnis D. D., Delhanty J. D., Jerkovic S., Geyer J., Craft I., et al. , 2005. Detailed FISH analysis of day 5 human embryos reveals the mechanisms leading to mosaic aneuploidy. Hum. Reprod. 20: 129–137 [DOI] [PubMed] [Google Scholar]
- Davison J., Tyagi A., Comai L., 2007. Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biol. 7: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J. H., Schultz C. J., Correll A. T., 1988. Chromosome elimination in micronuclei: a common cause of hypoploidy. Am. J. Hum. Genet. 43: 733–740 [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Azakami S., Kurosawa H., Ono Y., Ueda Y., et al. , 2003. Cystic partially differentiated nephroblastoma, embryonal rhabdomyosarcoma, and multiple congenital anomalies associated with variegated mosaic aneuploidy and premature centromere division: a case report. J. Pediatr. Hematol. Oncol. 25: 896–899 [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., 2010. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 186: 18–28 [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill D. E., Chao L., Perkins S. L., Wolf J. B., 1995. Genetic mosaicism in plants and clonal animals. Annu. Rev. Ecol. Syst. 26: 423–444 [Google Scholar]
- Grant V., 1971. Plant Speciation, Columbia University Press, New York [Google Scholar]
- Harmaja H., Pellinen K., 1990. Three different chromosome numbers from Finnish Arabidopsis suecica (Brassicaceae). Ann. Bot. Fenn. 27: 335–336 [Google Scholar]
- Hegarty M. J., Hiscock S. J., 2008. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 18: R435–R444 [DOI] [PubMed] [Google Scholar]
- Henry I. M., Dilkes B. P., Young K., Watson B., Wu H., et al. , 2005. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B., Kreil D. P., Matzke M., Matzke A. J. M., 2008. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 4: e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium, 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Kato A., Lamb J. C., Birchler J. A., 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A., Leitch I., 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483 [DOI] [PubMed] [Google Scholar]
- Lewis W. H., Oliver R. L., Suda Y., 1967. Cytogeography of Claytonia virginica and its allies. Ann. Mo. Bot. Gard. 54: 153–171 [Google Scholar]
- Lim K. Y., Soltis D. E., Soltis P. S., Tate J., Matyasek R., et al. , 2008. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS ONE 3: e3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind-Hallden C., Hallden C., Säll T., 2002. Genetic variation in Arabidopsis suecica and its parental species A. arenosa and A. thaliana. Hereditas 136: 45–50 [DOI] [PubMed] [Google Scholar]
- Madlung A., Masuelli R. W., Watson B., Reynolds S. H., Davison J., et al. , 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129: 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Tyagi A. P., Watson B., Jiang H., Kagochi T., et al. , 2005. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41: 221–230 [DOI] [PubMed] [Google Scholar]
- Madlung A., Henkhaus N., Jurevic L., Kahsai E. A., Bernhard J., 2012. Natural variation and persistent developmental instabilities in geographically diverse accessions of the allopolyploid Arabidopsis suecica. Physiol. Plant. 144: 123–133 [DOI] [PubMed] [Google Scholar]
- Mallet J., 2007. Hybrid speciation. Nature 446: 279–283 [DOI] [PubMed] [Google Scholar]
- McCullough E., Wright K. M., Alvarez A., Clark C. P., Rickoll W. L., et al. , 2010. Photoperiod-dependent floral reversion in the natural allopolyploid Arabidopsis suecica. New Phytol. 186: 239–250 [DOI] [PubMed] [Google Scholar]
- Ni Z., Kim E. D., Ha M., Lackey E., Liu J., et al. , 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuti Ronchi V., Nozzolini M., Avanzi L., 1981. Chromosomal variation in plants regenerated from two Nicotiana spp. Protoplasma 109: 433–444 [Google Scholar]
- O’Kane S. R. J., Schaal B. A., Al-Shehbaz I. A., 1996. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 21: 559–566 [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication, Springer Verlag, New York [Google Scholar]
- Osborn T., Pires J., Birchler J., Auger D., Chen Z., et al. , 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147 [DOI] [PubMed] [Google Scholar]
- Otto S. P., Whitton J., 2000. Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Pires J. C., Zhao J. W., Schranz M. E., Leon E., Quijada P. A., et al. , 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. Lond. 82: 675–688 [Google Scholar]
- Pohlendt G., 1958. Variabilty of chromosome counts and other nuclear pathologies in Aegilops triuncialis x Triticum aestivum hybrids. Z. Vererbungsl. 89: 170–188 (in German) [Google Scholar]
- Ramanna M. S., Hermsen J. G. T., 1971. Somatic chromosome elimination and meiotic chromosome pairing in the triple hybrid 6x-(Solanum acaule x S. bulbocastanum x 2x-S. phureja. Euphytica 20: 470–481 [Google Scholar]
- Sachs L., 1952. Chromsome mosaics in experimental amphiploids in the Triticinae. Heredity 6: 157–170 [Google Scholar]
- Salmon A., Flagel L., Ying B., Udall J. A., Wendel J. F., 2010. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 186: 123–134 [DOI] [PubMed] [Google Scholar]
- Seehausen O., 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19: 198–207 [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Rouse D. T., Finnegan E. J., Peacock W. J., Dennis E. S., 2000. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Hills M. J., Lister C., Dean C., Dennis E. S., et al. , 2008. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. USA 105: 2214–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P., Soltis D., 2009. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Stebbins G. L., 1971. Chromosomal Evolution in Higher Plants, Addison-Wesley, Reading, MA [Google Scholar]
- Tate J. A., Symonds V. V., Doust A. N., Buggs R. J. A., Mavrodiev E., et al. , 2009. Synthetic polyploids of Tragopogon miscellus and T. mirus (Asteraceae): 60 years after Ownbey’s discovery. Am. J. Bot. 96: 979–988 [DOI] [PubMed] [Google Scholar]
- Thompson M., 1962. Cytogenetics of Rubus. III. Meiotic instability in some higher polyploids. Am. J. Bot. 49: 575–582 [Google Scholar]
- Venkateswarlu J., Krishna Rao M., 1969. Chromosome numerical mosaicism in some hybrids of the Solanum nigrum complex. Genetica 40: 400–406 [Google Scholar]
- Wang J., Tian L., Madlung A., Lee H. S., Chen M., et al. , 2004. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Chen Z. J., 2006a Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics 173: 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Wei N. E., Jiang H., et al. , 2006b Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. M., Pires J. C., Madlung A., 2009. Mitotic instability in resynthesized and natural polyploids of the genus Arabidopsis (Brassicaceae). Am. J. Bot. 96: 1656–1664 [DOI] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R. T., Pires J. C., 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Xu Y., Tan E. L., Kumar P. P., 2002. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 99: 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.