Abstract
DNA barcode is a new tool for taxon recognition and classification of biological organisms based on sequence of a fragment of mitochondrial gene, cytochrome c oxidase I (COI). In view of the growing importance of the fish DNA barcoding for species identification, molecular taxonomy and fish diversity conservation, we developed a Fish Barcode Information System (FBIS) for Indian fishes, which will serve as a regional DNA barcode archival and analysis system. The database presently contains 2334 sequence records of COI gene for 472 aquatic species belonging to 39 orders and 136 families, collected from available published data sources. Additionally, it contains information on phenotype, distribution and IUCN Red List status of fishes. The web version of FBIS was designed using MySQL, Perl and PHP under Linux operating platform to (a) store and manage the acquisition (b) analyze and explore DNA barcode records (c) identify species and estimate genetic divergence. FBIS has also been integrated with appropriate tools for retrieving and viewing information about the database statistics and taxonomy. It is expected that FBIS would be useful as a potent information system in fish molecular taxonomy, phylogeny and genomics.
Availability
The database is available for free at http://mail.nbfgr.res.in/fbis/
Keywords: Genetic Divergence, Fish, LAMP, Phylogeny, Phylogeography, Taxonomy
Background
The limitations inherent in traditional taxonomy and morphology based identification lead to emergence of genomic approaches to taxon diagnosis that exploit diversity among DNA sequences to identify organisms [1, 2]. DNA barcoding technique is a new approach for taxon recognition and classification of biological organism based on sequence of a small fragment (650 base pairs) near the 5'-terminus of the cytochrome c oxidase I (COI) mitochondrial gene [3] and has been widely used for species-level identification across a wide range of both invertebrate [4] and vertebrate [5, 6] organisms. DNA barcoding can provide a ‘biological barcode’ to enable identification of any organism at the species level [7–9] and allows accurate and relatively simpler species identification. India has a very important place in globe for variety of fishes and recognized as one of the 12 mega biodiversity countries of world [10]. Out of 32,300 extant fish species, 2438 belong to Indian subcontinent [11]. The global importance of Indian fish species demands a well developed system for species identification, classification and divergence analysis. A reference database with collection of diverse assemblage of sequences and other information under the registry of accession number is essential for performing DNA-based identifications in unknown samples, because DNA barcodes cannot be a useful identification tool without a comprehensive and reliable reference database [12]. With the advent of the new intellectual property rights (IPR) regime and implementation of Biological Diversity Act 2002, it is imperative to record relevant information on valuable fish genetic resources of the country in order to avoid conflicts and protect rights on native species with geographical indications. The DNA barcoding exercise and development of automated system in the Indian context was perhaps weighed from two important angles, namely (a) meeting the taxonomic challenges and providing a robust identification of species and (b) securing IPRs for some of the country's important bio resources [13]. The present study was focused to collect the DNA barcode information about Indian fishes from published data sources with objective to develop an automated web version database integrated with search, browse, identification and analytical tools. For this, we developed Fish Barcode Information System (FBIS), a database on DNA barcodes for 472 Indian aquatic species along with 2334 sequences using MySQL database management system, Perl and PHP programming languages under Red Hat Linux Enterprises 5.2 environment and integrated it with tools viz. ‘identification’, ‘database statistics’, ‘divergence estimation’ and ‘keyword search’. Although ‘Barcode of Life Data System (BOLD)' has been developed as a repository of DNA barcodes for all living organism at international level [14], yet our system is specific for fishes and has multi-fold applications in identification of larvae/ invasive species/ cryptic species/ new species, illegal trade of protected species, stock management, biodiversity assessment, ecosystem monitoring, revisions of certain taxa, estimation of intra and inter-specific divergence, phylogenetic relationships, phylogeographic [15] and speciation patterns.
Methodology
Data source:
A majority of sequences and annotation data in FBIS were generated from the Project entitled ‘DNA Barcoding of Indian Fishes’ undertaken at ‘National Bureau of Fish Genetic Resources (NBFGR)’ following systematic DNA extraction, amplification of COI gene, sequencing and submission of COI sequences into NCBI GenBank. The DNA barcode information of Indian shellfish's viz. molluscs and shrimps generated elsewhere were also downloaded from the GenBank of NCBI. The entire data was downloaded in GenBank and FASTA format for annotation and sequence analysis respectively. A Perl parsing program was written to extract important features from the files and manage into the database. The phenotypic and other physical information on habitat, distribution, IUCN Red List status of the fishes and shellfishes was collected from FishBase (http://www.fishbase.org). The methodology of data collection and its integration has been depicted in a data flow diagram (Figure 1).
Figure 1.
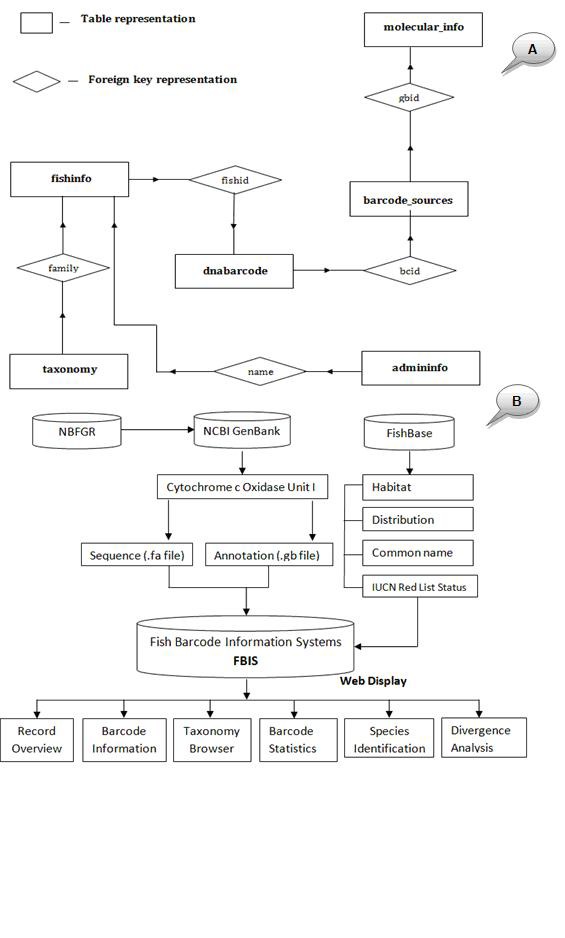
Schematic diagram of (A) the database architecture showing table's relationship and data flow; (B) data flow diagram showing data resources used in FBIS.
Database structure:
The FBIS database has been developed by using MySQL 5.2 relational database management system under Red Hat Linux 5.2 environment. To manage the DNA barcodes, phenotypic and physical information, the database was designed at two sublevel schemas for molecular information and phenotypic and physical information. The database contains six (6) main tables for each DNA barcode sequence entry viz.
fishinfo;
dnabarcode;
barcode_sources;
taxonomy;
molecular_info;
admin_info.
Table 1 (see supplementary materials) describes the details of different tables and its attributes, while (Figure 1) describes the FBIS structure and relationships among tables. The design of the database supported with query-based data integration preserves the inherent independence of data extracted from various parent databases.
Web interface and application:
FBIS has been created using open source technology i.e. LAMP (Linux, Apache, MySQL, PHP/Perl) to develop the database and web interfaces. MySQL, an object-relational database management system, works at the backend and provides commands to retrieve and store DNA barcode data into the database. PHP along with PERL, a server side scripting language provides interface and functions to analyze, fetch and displays data from the database. Besides, functions have been integrated in the interface to perform administration and management tasks with data submission facility. Perl DBI module was used to perform connectivity with MySQL database for accessing the data in the database. The graphical representation in the web interface was implemented using GD.pm (http://search.cpan.org/dist/GD/GD.pm). The whole FBIS run on Intel Server Class machine under Red Hat Enterprise Linux 5.2 environment using Apache httpd server.
Results
FBIS contains 2334 barcode records for 472 species belonging to 136 families, till February 2012 and regular update is being carried out using an automated program. Tools for search and browsing were integrated for better retrieval and analysis of the DNA barcode sequence data from the database. In addition, tools for species identification and sequence divergence estimation were also implemented to enhance the utility of FBIS. The details about the web tools integrated in FBIS are given below.
Keyword search:
This search option was designed to extract and analyse the relevant information. It uses a general keyword search which retrieves relevant records from the numerous tables of the database and provides facility for downloading and viewing the records. The design of search module has been divided in two parts: (1) program retrieves the fish name with relevant keyword; (2) once fish name is identified, the intelligent query system further process for retrieving all the information about the particular species.
Specimen record browsing:
The specimen record can be browsed based on alphabetic index on the first letter of family name of the aquatic species by navigating into the ‘Records overview’ menu item. We used the grid format to display records describing scientific name, family, specimen voucher, longitude-latitude, submission date and a cross link of NCBI Reference.
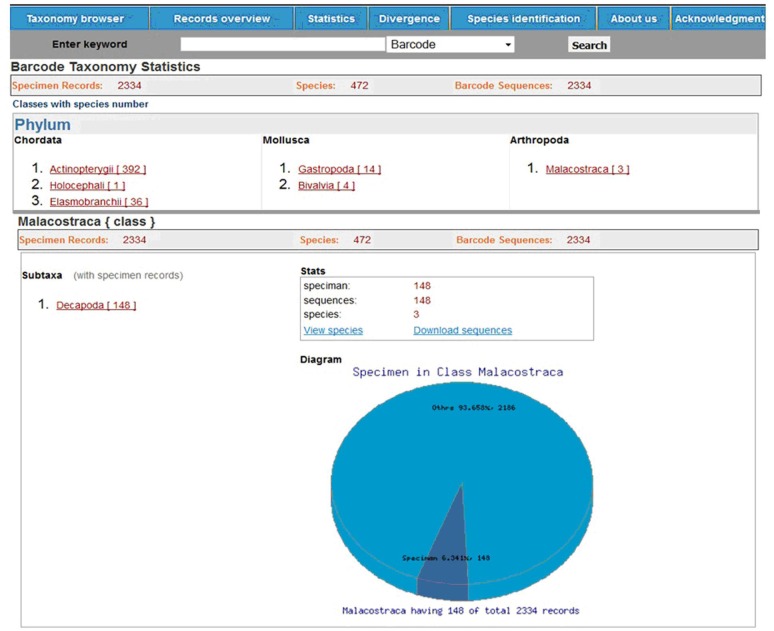
Taxonomy browser:
It retrieves taxonomy from phylum to species level and can be accessed by navigating into the menu bar of FBIS. The browser shows the hierarchal taxonomic representation of each Phylum, Class, Order, Family and Genus along with species it contains. For example, the Phylum ‘Arthropoda’ displays one Class ‘Malacostraca’ followed by [3] i.e. a number in square bracket to denote the total number of species available in that Class. The browser also generates pie diagram on data status and displays links for viewing the species and downloading all the corresponding sequences in FASTA format for the selected subtaxon (Figure 2).
Figure 2.
A screen view of FBIS taxonomy browser
Statistics of physical information:
The statistics application represents the dynamic numerical and graphical view of the current data statistics of number of COI sequences, families and species stored in the database. It also shows the statistics for habitat and conservation category (IUCN Red List Status) for all existing species of FBIS. The ‘Habitat Statistics’ describes total number of species distributed in freshwater, marine or other ecosystem. The ‘IUCN Red List Status’ category provides a link to invoke the physical and phenotypic information of species along with photograph of corresponding category.
Species identification:
This tool employs linear search to find out homologous sequence from the global alignment of all reference sequences. We used query-optimized search library to prepare the blast [16] compatible dataset FBISdb through ‘makeblastdb’ program maintained outside the main database with new records added at every occasions of database population. The homologous reference sequence identity of 100% or 99% with the query describes as accurately same species and the reference sequence identifier is used to retrieve same or similar species and other information from FBIS database through intelligent querying. With respect to query sequence, a table of top 30 similar and closely related records is displayed in the result.
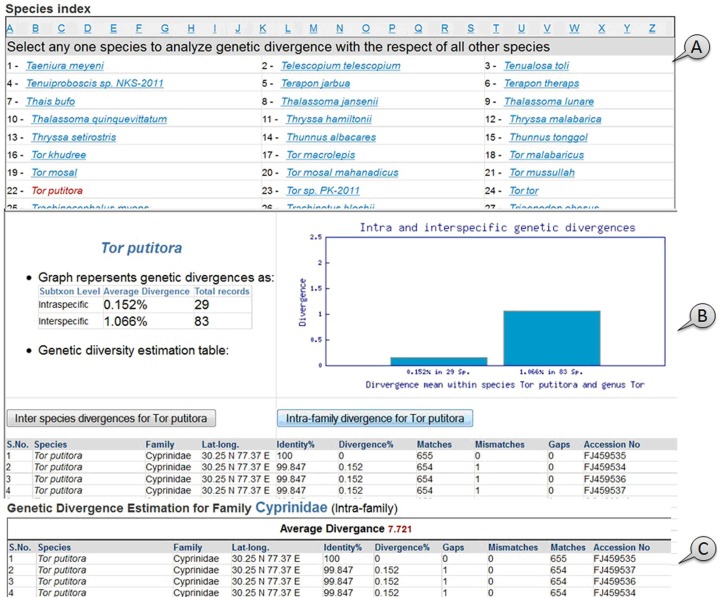
Divergence estimation:
In this application, species can be selected through index navigation from ‘Divergence’ menu item and a sequence divergence estimation table could be generated with the alignment of whole FBIS database. The lineage with the respect to genetic divergence was calculated on the basis of difference in nucleotide bases in same position during sequence comparison process and tabular estimation of sequence divergence among the closely related genera and species is generated. For example, a query sequence of Tor putitora was selected to align with FBIS sequence library through BLASTn program, 250 sequences having more than 85% identity among the aligned records were obtained. The analysis calculated 0.152% sequence divergence among individuals of T. putitora and 1.066% divergence for other species belonging to genus Tor, while it showed 7.721 % divergence with other genera of the same family ‘Cyprinidae’. The intraspecific and interspecific divergences can be visualized through dynamic bar diagram as shown in (Figure 3) along with other statistical details.
Figure 3.
A screen view of (A) species index; (B) divergence estimation; (C) intra-family divergence for Tor putitora
Data administration and management:
We integrated various administration, management and data submission operations in the web interface due to security reasons. The administrator of FBIS has full privileges for database administration and management of submission activities. Users may submit their data in FBIS by sending an email to the FBIS administrator, which will be verified before addition to the database.
Discussion
Analysis of 2334 barcode sequences belonging to 472 aquatic species under phylum Chordata, Mollusca and Arthropoda showed that FBIS is well suited to the identification of biological organisms at different taxonomic level. Further, this archival system has great potential to investigate DNA barcodes obtained from any fish tissue for taxonomic validation of species and determining genetic divergence between the query sequence and the reference sequences available in the database. The algorithm of FBIS delivers species identification if the query sequence shows less than 1% divergence to the reference sequence in FBIS and if the match shows more than 1% sequence divergence, it may assign the query sequence belonging to similar/closely related species. This is in agreement with the report of Avise (2000) who found genetic divergence to be less than 1% in majority of the records belonging to same species with the exception of few records that exhibited greater than 2% divergence for mitochondrial DNA [17]. Ratnasingham and Herbert (2007) also used similar criteria for identification of species through matching of query sequence with the reference barcode records [18]. Ward et al. (2005) reported a divergence level of 0.39% and 9.93% for individuals within species and species within genera for Australian fishes respectively [19], while Lakra et al. (2011) observed the average distances within species and genera as 0.30% and 6.60% respectively in Indian marine fishes [10]. Our system also provides a support for above demonstrations.
The ‘Divergence analysis’ module of our system enables the user to estimate intraspecific, interspecific and intra-family average divergence for a species. Further divergence analysis can be performed either through a query sequence or navigation through species name from 'Index Browser'. The present archival system offer several advantages over traditional taxonomy, image based recognition and digital taxonomy viz. providing a correct species identification tool for all life stages, complementing conventional taxonomic studies especially in securing IPRs for important taxa [13] and discriminate among species of a taxon. FBIS, the freely available open archival system, would be very useful to researchers in deciding and describing the new species. The online submission facility will be helpful for updating this database content with the additional curated and computed data. Keeping in view India's role in the aquatic animal species trade globally, FBIS would be useful in providing diagnostics for rapid and easy identification of species in case of adulterations and for drawing specific regulations to protect the national market.
Conclusion
The FIBS is a regional DNA barcode archival system for Indian fishes, and all the applications could be carried out through interactive user interfaces. The system enables fish species identification, taxonomic validation and genetic divergence estimation that may create great interest among the researchers and stakeholders. There is further scope to expand this archival system by incorporating novel data mining and visualization tools for increasing its analytical capabilities.
Supplementary material
Acknowledgments
Authors are grateful to National Agricultural Bioinformatics Grid (NABG), under National Agricultural Innovation Project (NAIP), ICAR, New Delhi for providing financial support and the Director, NBFGR, Lucknow for providing necessary facilities and guidance.
Footnotes
Citation:Nagpure et al, Bioinformation 8(10): 483-488 (2012)
References
- 1.CP Kurtzman. Yeast. 1994;10:1727. doi: 10.1002/yea.320101306. [DOI] [PubMed] [Google Scholar]
- 2.KH Wilson, et al. Clin Infect Dis. 1995;20:S117. doi: 10.1093/clinids/20.supplement_2.s117. [DOI] [PubMed] [Google Scholar]
- 3.PD Hebert, et al. Proc Biol Scie. 2003;270:313. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.FO Costa, et al. Science. 2007;64:272. [Google Scholar]
- 5.PD Hebert, et al. PLoS Biol. 2004;2:1657. [Google Scholar]
- 6.M Hajibabaei, et al. Genome. 2006;49:851. [Google Scholar]
- 7.PD Hebert, et al. Proc Biol Sci. 2003;270:S96. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M Hajibabaei, et al. Trends Genet. 2007;23:167. [Google Scholar]
- 9.E Marshall. Science. 2005;307:1037. doi: 10.1126/science.307.5712.1037. [DOI] [PubMed] [Google Scholar]
- 10.WS Lakra, et al. Mol Ecol Resour. 2011;11:60. doi: 10.1111/j.1755-0998.2010.02894.x. [DOI] [PubMed] [Google Scholar]
- 11. http://www.fishbase.org.
- 12.CP Meyer, G Paulay. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K Aravind, et al. Current Science. 2007;92:1213. [Google Scholar]
- 14.RD Ward, et al. J Fish Biol. 2009;74:329. doi: 10.1111/j.1095-8649.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 15.JC Avise, et al. Annual Review of Ecology and Systematics. 1987;18:489. [Google Scholar]
- 16.SF Altschul, et al. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.JC Avise, et al. Cambridge: Harvard University Press; 2000. [Google Scholar]
- 18.S Ratnasingham, PDN Hebert. Mol Ecol Notes. 2007;7:355. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.RD Ward, et al. Philos Trans R Soc Lond B Biol Sci. 2005;360:1847. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.