ABSTRACT
The enterococci are Gram-positive lactic acid bacteria that inhabit the gastrointestinal tracts of diverse hosts. However, Enterococcus faecium and E. faecalis have emerged as leading causes of multidrug-resistant hospital-acquired infections. The mechanism by which a well-adapted commensal evolved into a hospital pathogen is poorly understood. In this study, we examined high-quality draft genome data for evidence of key events in the evolution of the leading causes of enterococcal infections, including E. faecalis, E. faecium, E. casseliflavus, and E. gallinarum. We characterized two clades within what is currently classified as E. faecium and identified traits characteristic of each, including variation in operons for cell wall carbohydrate and putative capsule biosynthesis. We examined the extent of recombination between the two E. faecium clades and identified two strains with mosaic genomes. We determined the underlying genetics for the defining characteristics of the motile enterococci E. casseliflavus and E. gallinarum. Further, we identified species-specific traits that could be used to advance the detection of medically relevant enterococci and their identification to the species level.
IMPORTANCE
The enterococci, in particular, vancomycin-resistant enterococci, have emerged as leading causes of multidrug-resistant hospital-acquired infections. In this study, we examined genome sequence data to define traits with the potential to influence host-microbe interactions and to identify sequences and biochemical functions that could form the basis for the rapid identification of enterococcal species or lineages of importance in clinical and environmental samples.
Introduction
The enterococci are a diverse group of Gram-positive gastrointestinal (GI) tract colonizers with lifestyles ranging from intestinal symbiont to environmental persister to multidrug-resistant nosocomial pathogen (1, 2, 3). Enterococci are used in food production, in probiotic products, and for tracking fecal contamination and thus also are of regulatory and industrial interest. Most enterococcal research has focused on the two species most associated with human GI tract colonization and infection, Enterococcus faecium and Enterococcus faecalis (2, 3). Certain lineages, defined by multilocus sequence typing (MLST), are associated with hospital-acquired infections (e.g., E. faecium sequence type 17 [ST17] ST18, ST78, and ST203 and E. faecalis ST6, ST9, and ST40) (4). Genome analysis has illuminated the extent of mobile content (5) and evolution of antibiotic resistance (6) in E. faecalis ST6 strain V583 and the mobile element content and metabolic capabilities of E. faecium (7). Using genomic data we recently developed for 28 enterococcal strains (8), we report and quantify divergence within what is currently classified as E. faecium and E. faecalis and identify the genetic bases for the defining characteristics of the motile enterococcal species Enterococcus gallinarum and Enterococcus casseliflavus. We identify loci homologous to those known to direct the synthesis of extracellular polymers that interact with host surfaces, including a putative E. faecium capsule locus. We additionally identify genetic sequences and biochemical functions that represent distinguishing features of potential value for the rapid identification of enterococci to the species level.
RESULTS AND DISCUSSION
Phylogenetic analysis of enterococci.
We recently announced the public release of genome sequence data for 28 enterococcal strains of diverse origin (8) (see Table S1 in the supplemental material). The 16 E. faecalis genomes sequenced represent the deepest nodes in the MLST phylogeny, providing the greatest diversity. The strains include those of clinical, animal, and insect origins and were isolated from 1926 to 2005 (9). These strains represent approximately 80 years of enterococcal evolution, spanning the periods prior to and during widespread antibiotic use. Additionally, the genomes of 6 E. faecium, 1 E. gallinarum, and 3 E. casseliflavus clinical isolates from 2001 to 2005 and 2 human fecal E. faecium strains were examined.
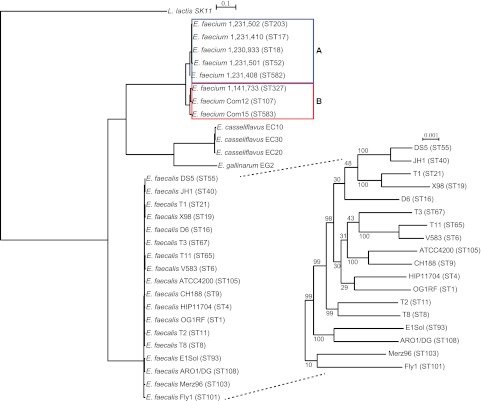
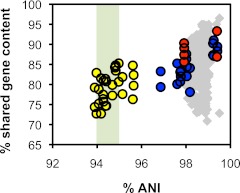
OrthoMCL (10) was used to identify ortholog groups in the 30 enterococcal genomes. Ortholog groups represented in all 31 genomes were considered core groups, which were further subdivided into single-copy (1 gene copy in each genome) and multicopy (>1 gene copy exists in at least 1 genome). Genes not clustered were considered orphans. A phylogenetic tree generated from the concatenated sequences of 847 single-copy core genes is shown in Fig. 1. Relationships among the 18 E. faecalis strains, despite their diverse origins, cannot be fully resolved by this analysis (based on lack of bootstrap support for branches within the E. faecalis branch; inset, Fig. 1). As expected, E. casseliflavus and E. gallinarum branch separately, supporting their designation as different species. Importantly, two clades were identified within the species E. faecium, as had been inferred by comparative genome hybridization, which suggested that hospital-associated isolates, including ST17 and ST18 isolates, may make up a distinct subspecies within E. faecium (11). The 3 vancomycin-resistant E. faecium strains in our collection are members of clade A, while the 2 human fecal isolates are members of clade B (Fig. 1). To quantify the relationships among these strains, we generated average nucleotide identity (ANI) plots (Fig. 2), which have been used to query and refine prokaryotic species definitions (12, 13).
FIG 1 .
Core gene tree. Concatenated sequences of 847 genes core to 30 enterococci and the outgroup species L. lactis were aligned, and a phylogenetic tree was generated using RAxML with bootstrapping. The bootstrap value for all nodes outside the E. faecalis clade is 100. E. faecium clades A (blue) and B (red) are indicated.
FIG 2 .
ANI plot. Each point represents a pairwise comparison of two genomes. Grey diamonds, E. faecalis-E. faecalis comparisons. Blue circles, clade A E. faecium-clade A E. faecium comparisons. Red circles, clade B E. faecium-clade B E. faecium comparisons. Yellow circles, clade A E. faecium-clade B E. faecium comparisons. A species threshold of 94 to 95% ANI is indicated by the green-shaded area.
E. faecium.
The E. faecium ANI analysis refines phylogenetic relationships among clade A and clade B strains (Fig. 2). Within clade A, ST17 strain 410 and double-locus variants (DLVs) 933 (ST18) and 502 (ST203) are closely related (99.2 to 99.4% ANI) whereas strains 501 (ST52) and 408 (ST582, an ST17 DLV) have lower ANI values with those strains, and each other (96.9 to 98.2% ANI). Similar ANI values were observed among clade B strains (97.9 to 99.4%). However, pairwise comparisons of clade A and clade B strains ranged from 93.9 to 95.6% ANI, overlapping an ANI species line of 94 to 95%. ANI values of 94 to 95% correlate with experimentally derived 70% DNA-DNA hybridization values, a commonly accepted threshold for species designation (12, 13, 14). Clade A and clade B may be endogenous to the GI tracts of different hosts and now coexist among human flora as a result of antibiotic elimination of competitors, or clade A and clade B may be diverging from each other as a result of antibiotic use and ecological isolation (less likely because of the short time involved).
For the 8 E. faecium strains in our collection, the two clades are recapitulated using the 7 housekeeping genes selected for E. faecium MLST (see Fig. S1 in the supplemental material). Between clade A and clade B strains, the nucleotide identities of concatenated MLST sequences range from 96.2 to 96.9% (compared to a 93.9 to 95.6% ANI range). To determine whether a single marker is representative of either E. faecium clade, we examined the distribution of individual MLST alleles among the E. faecium STs assigned to clade A or clade B (see Fig. S1 in the supplemental material). A “minority allelic population” composed of 5 divergent STs was reported in seminal E. faecium MLST work (15). The 5 divergent STs (ST39, ST40, ST60, ST61, and ST62) identified by that study belong to clade B (see Fig. S1 in the supplemental material). The genomes of 7 additional E. faecium strains were recently sequenced (7), and we used MLST to assign them to clade A or B (see Fig. S1 in the supplemental material). The assignment of one of these strains, E. faecium E980, to clade B is consistent with previous analyses demonstrating the phylogenetic distance of this strain from the other 6 (clade A) strains in that sequencing collection (7). In the first-pass analysis, the allele adk-6, which differs from the ST17 allele adk-1 at 3 synonymous sites, was observed to occur almost exclusively in clade B strains (see Fig. S1 in the supplemental material). To further explore the distribution of adk alleles among E. faecium isolates, we extracted sequences of all 617 available STs in the E. faecium MLST database and determined the extents of identity to an ST17 (clade A) reference. In the MLST database, adk-1, adk-5, and adk-6 are the most abundant adk alleles, representing 87% of the E. faecium STs. Of the 85 STs possessing adk-6, 66 (78%) share 96 to 96.9% nucleotide identity with ST17, comparable to that observed for clade B-ST17 comparisons. Conversely, adk-1 and adk-5 occur primarily in STs with ≥99% identity to ST17. These data suggest that adk allele exchange is restricted, perhaps resulting from a barrier to DNA uptake such as clustered regularly interspaced short palindromic repeats (CRISPR)-cas defense and/or from the proximity of adk to the replication origin (Fig. 3).
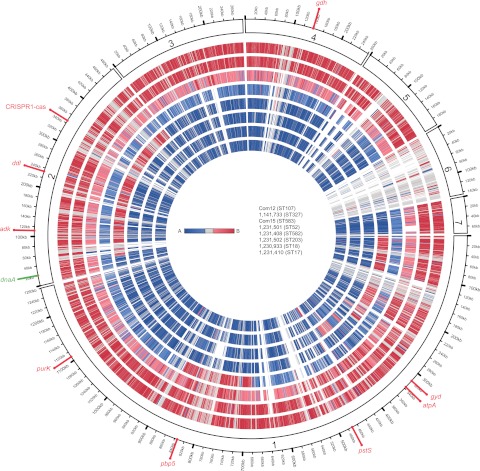
FIG 3 .
E. faecium genome mosaicism plot. The outermost ring shows E. faecium Com12 scaffolds, ordered by decreasing length clockwise from scaffold 1, with each gene represented as a radial position along the ring. Each of the remaining 7 E. faecium genomes is represented by the rings below Com12. Genes are colored by membership in clade A (blue) or clade B (red), as determined by individual gene trees built from ortholog groups. The strains shown, from the outermost to the innermost rings, are Com12, 733, Com15, 501, 408, 502, 933, and 410. The locations of dnaA, Com12 MLST alleles, pbp5, and the EfmCRISPR1-cas locus are shown.
E. faecium 408 is a DLV of ST17 that possesses adk-6 and ddl-13 (see Fig. S1 in the supplemental material). Because adk-6 occurs mostly among strains with lower relatedness to ST17, and ddl-13 is present in two clade B strains (see Fig. S1 in the supplemental material), we were curious about whether these alleles were acquired by recombination. Genome mosaicism is evident in E. faecium clade A strains 408 and 501 (Fig. 3). The occurrence of adk-6 and ddl-13 within a hybrid region in E. faecium 408 (Fig. 3; see data set S1 in the supplemental material) supports the acquisition of this region from a clade B strain. The putative genome defense system EfmCRISPR1-cas (16), present in 2 of 3 clade B strains and in E. faecium 408 (see Table S1 in the supplemental material), occurs within this region, suggesting that CRISPR-cas was acquired by E. faecium 408 from a clade B strain via recombination. The hybrid region in E. faecium 501 includes pbp5 (Fig. 3; see data set S1 in the supplemental material), which can confer ampicillin resistance. Our results indicate that pbp5-S was acquired by E. faecium 501 from a clade B strain. The hybrid region in 501 is flanked by a putative phage integrase (EFRG_00906) that is conserved among all of the E. faecium strains in our collection (see data set S1 in the supplemental material). We recently reported an Hfr-like mechanism for the transfer of chromosomal genes between E. faecalis strains (17), and it seems likely that a similar mechanism functions in E. faecium.
To determine whether specific traits define the two E. faecium clades, we searched for clade-specific ortholog groups present in and exclusive to all of the members of each clade. We then used representative gene sequences from each to search for similar sequences in 7 additional E. faecium genomes (7) assigned to clade A or B (see Fig. S1 in the supplemental material). Of the clade A-specific genes (see data set S2 and Table S2 in the supplemental material), 8 are associated with a locus that has high sequence identity with and almost the same gene content as the ycjMNOPQRSTUV locus of Escherichia coli, which is significantly enriched in enteric clades (18) and also occurs in Listeria (19). The organization of this locus is similar to that of a Lactobacillus acidophilus fructooligosaccharide (prebiotic) utilization locus (20). Of the genes unambiguously assigned to clade B (see data set S2 and Table S2 in the supplemental material), 5 encode putative transcriptional regulators with protein domain hits to Mga or Rgg, regulators of virulence, competence, and cell-cell signaling in streptococci (21, 22). Two of these putative regulators are divergently transcribed from genes that are also clade B specific, including a putative thioredoxin that could modulate the redox state of cellular targets in response to oxidative stress (23). A putative phospholipase C is also clade B specific. Finally, one clade B-specific gene (EFSG_01746) was useful in identifying a genomic insertion, composed of 17 genes, in E. faecium 733 (see Table S2 in the supplemental material). This region encodes a putative phosphotransferase system and a secreted hyaluronidase that could cleave the extracellular matrix of host cells. It is surprising that clade B (and not clade A, which contains all high-risk STs) strains encode a number of secreted factors that could interact with eukaryotic cell surfaces. This suggests that clade B strains may be more closely associated with host tissues in the GI tract than clade A strains are, which possibly contributes to their persistence in the GI tract, whereas clade A strains may be more transient and associated with the GI lumen, which contributes to their dissemination.
E. faecalis.
In contrast to E. faecium, little phylogenetic divergence was observed among E. faecalis strains (Fig. 2). Among 306 pairwise comparisons, ANI varies within a narrow range (97.8 to 99.5%). Instead, shared gene content among these strains varies (70.9 to 96.5%). For example, strain T11 shares 96.5% of its 2,511 genes with ST6 strain V583, while V583 shares only 72.8% of its 3,265 genes with T11; they possess 99.5% ANI in the genes that they share. The genome size of T11 is smaller than that of V583 (2.74 Mb versus 3.36 Mb) and is similar to that of the oral isolate OG1RF (24), likely representing the minimal E. faecalis genome. For all 18 E. faecalis strains, genome sizes vary between the extremes of T11 and V583 (see Table S1 in the supplemental material). We recently proposed that loss of CRISPR-cas in founders of modern E. faecalis high-risk MLST lineages facilitated the influx of acquired antibiotic resistance genes and other mobile traits into these lineages (16). Genome size distribution significantly differs between strains possessing or lacking CRISPR-cas (P = 0.026; one-tailed Wilcoxon rank-sum test), with a greater average genome size in strains lacking CRISPR-cas (3.1 Mbp versus 2.9 Mbp). The distribution of domain motifs associated with mobile elements is significantly different in strains with genomes >3 Mb in size (P < 0.05), including the plasmid mobilization MobC domain (PF05713; P = 0.001), the antirestriction protein ArdA (PF07275; P = 0.032), the replication initiation factor domain (PF02486; P = 0.001), a plasmid addiction toxin domain (TIGR02385; P = 0.032), and a transposase domain (PF01526; P = 0.021). This supports a model where increased genome size is the result of mobile element accretion, consistent with the proposition that compromised genome defense facilitated the accretion of mobile elements (16), resulting in larger genomes.
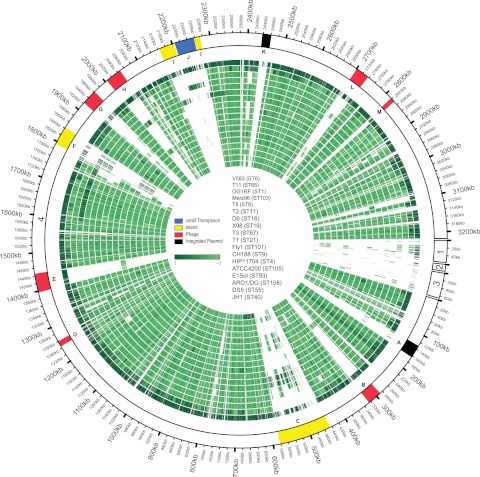
We analyzed the 18 E. faecalis genomes for mosaicism (Fig. 4). Thirteen variable regions were previously defined for E. faecalis genomes using comparative genome hybridization to a V583-based microarray (9). Regions of mosaicism were detected in strains Merz96, JH1, and T2 overlapping the E. faecalis pathogenicity island (25) and two putative genomic islands containing Tn916-like genes (5), respectively. These results are consistent with conjugative acquisition of these islands and surrounding sequence by Merz96, JH1, and T2 from strains closely related to V583 (17). Collectively, much of the diversity of E. faecalis can be attributed to the accretion of mobile genetic elements on a largely conserved genomic backbone, with those mobile elements facilitating recombinatorial exchange of chromosomally encoded traits.
FIG 4 .
E. faecalis genome mosaicism plot. The outermost ring shows E. faecalis V583 chromosomal (scaffold 4) and plasmid scaffolds (scaffold 1, pTEF2; scaffold 2, pTEF3; scaffold 3, pTEF1), with each gene represented as a radial position along the ring. Each of the remaining 17 E. faecalis genomes is represented by the rings below V583. Genes are colored by phylogenetic distance from E. faecalis V583 (from dark to light green with increasing phylogenetic distance), as determined by individual gene trees built from ortholog groups. The strains shown, from the outermost to the innermost rings, are V583, T11, OG1RF, Merz96, T8, T2, D6, X98, T3, T1, Fly1, CH188, HIP11704, ATCC 4200, E1Sol, AR01/DG, DS5, and JH1. The locations of E. faecalis variable regions are shown (9). A, integrated plasmid; B, prophage 1; C, E. faecalis pathogenicity island; D, prophage 2; E, prophage 3; F, putative island; G, prophage 4; H, prophage 5; I, putative island; J, vancomycin resistance (vanB) transposon; K, integrated plasmid; L, prophage 6; M, prophage 7.
The motile enterococci.
Very little is known about the genomes of E. casseliflavus and E. gallinarum. Once thought to be associated primarily with vegetation (E. casseliflavus [26]) and fowl (E. gallinarum [27]) and only rarely found in humans, these species appear to be increasingly implicated in infections and hospital outbreaks (28, 29). Motility is a defining characteristic of most strains of E. casseliflavus and E. gallinarum, while E. casseliflavus additionally produces a yellow pigment (30); however, there has been confusion because of phenotype variation (3). ANI analysis confirms that the E. casseliflavus and E. gallinarum strains in our collection, which possess ~74% ANI in shared genes, are members of two separate species (Table 1). Motile enterococci are reported to have ≤3 or 4 terminal or lateral flagella per cell (31). In the E. casseliflavus and E. gallinarum genomes, we identified conserved gene clusters encoding proteins predicted to synthesize, export, and power a flagellum, as well as a chemotactic response system (see data set S3 in the supplemental material). Most of the proteins predicted to be encoded by the representative E. casseliflavus EC10 motility gene cluster have best BLASTP hits to Lactobacillus ruminis proteins (see data set S3 in the supplemental material) (32).
TABLE 1 .
ANI and shared-gene analyses of E. casseliflavus and E. gallinarum
| Strain | % ANI, % shared gene contenta |
||||
|---|---|---|---|---|---|
| EC10 | EC20 | EC30 | EG2 |
E. faecalis and E. faeciumb |
|
| EC10 | 98, 85 | 100, 99 | 74, 72 | 65–66, 51–55 | |
| EC20 | 98, 88 | 98, 88 | 74, 74 | 65–66, 52–56 | |
| EC30 | 100, 99 | 98, 86 | 74, 72 | 65–66, 51–55 | |
| EG2 | 74, 78 | 74, 77 | 74, 78 | 65–67, 55–60 | |
The data shown are for genome 1 (left) compared to genome 2 (top). Values were rounded to the closest whole number.
Ranges of values are shown.
Bacterial motility is often regulated by the second messenger cyclic di-GMP (c-di-GMP), as are attachment to surfaces and production of extracellular polysaccharides (33). We identified putative diguanylate cyclases possessing GGDEF domains (for c-di-GMP synthesis) and phosphodiesterases possessing EAL domains (for c-di-GMP turnover) in all 3 E. casseliflavus strains (see data set S3 in the supplemental material) but not in E. gallinarum. Two GGDEF proteins and one EAL protein are encoded 5′ to a predicted protein possessing glycosyltransferase, cellulose synthase, and PilZ domains (see data set S3 in the supplemental material). This protein shares high identity with a Clostridium difficile protein thought to be regulated by c-di-GMP (CD2545 [34]) (see data set S3 in the supplemental material). PilZ domains bind c-di-GMP (33), and it is possible that this domain regulates cellulose synthesis or the production of another extracellular polymer in E. casseliflavus.
E. casseliflavus produces a cell-associated carotenoid pigment thought to facilitate its environmental persistence by protecting against photooxidation (35). Streptococcus aureus also produces a carotenoid pigment and virulence factor, staphyloxanthin, that protects it from host-induced oxidative damage and antimicrobial peptides, and inhibitors show promise as novel therapeutics (36). Compared to S. aureus CrtOPQMN, E. gallinarum and E. casseliflavus share CrtM and CrtN homologues that catalyze the first steps of staphyloxanthin biosynthesis (37). However, only E. casseliflavus possesses CrtO, CrtP, and CrtQ (see data set S3 in the supplemental material). Most ligand interaction sites (36) are conserved in the E. casseliflavus and E. gallinarum CrtM proteins (see Fig. S2 in the supplemental material), suggesting that CrtM inhibitors could be usefully applied to these bacteria.
E. faecalis and E. faecium extracellular polysaccharides.
Cell wall polymers produced by E. faecalis include a lipoteichoic acid (LTA) with a poly(glycerol-phosphate) backbone (38, 39); a putative wall teichoic acid (WTA) composed of glycerol, glucose, and phosphate (40); and a rhamnopolysaccharide (the enterococcal polysaccharide antigen or Epa) composed of rhamnose, N-acetylglucosamine, N-acetylgalactosamine, glucose, and galactose (40, 41, 42). The E. faecalis epa locus directs the synthesis of the Epa polymer (43), although the biochemical functions and essentiality of most Epa proteins are unknown. The production of an antiphagocytic capsule composed of galactose, glucose, and phosphate is strain variable in E. faecalis and dependent on the presence of the cps locus (9, 40). Other than the E. faecalis epa and cps loci and E. faecalis bgsA and bgsB, which are involved in LTA biosynthesis (44), the genetic bases of extracellular polymer biosynthesis in enterococci are largely uncharacterized, as is the genetic basis of variable phagocytosis resistance in the species E. faecium (45). We therefore examined the distributions of epa, cps, and a predicted LTA biosynthesis pathway (46) and searched for new loci potentially important for decoration of the enterococcal cell surface.
As expected based on previous work (42), the entire epa locus—encompassing epaA to epaR—is core to the species E. faecalis. An epa locus varying in organization and content from that of E. faecalis is also core to E. faecium. In E. faecium, the genes are ordered epaABCDEFGH-epaPQ-epaLM-[an E. faecium-specific gene]-epaOR. The intervening E. faecium-specific gene encodes a protein with N-terminal similarity to E. faecalis EpaN but was not identified as being orthologous to epaN by OrthoMCL. Both the E. faecalis EpaN and E. faecium-specific proteins have a predicted S-adenosylmethionine binding site in the N terminus but are divergent in C-terminal sequence. The E. faecium epa locus may direct the synthesis of a previously reported E. faecium tetraheteroglycan composed of galactose, rhamnose, N-acetylglucosamine, glucose, and phosphate (47). The conservation of most of the epa locus suggests that if Epa biosynthesis enzymes were targeted with novel antimicrobials, those antimicrobials could be effective against the enterococcal species of greatest concern to human health. Proteins predicted to be involved in E. faecalis V583 LTA biosynthesis (46) were additionally identified as being core to E. faecalis and E. faecium, as well as the other enterococcal species in our collection (see data set S4 in the supplemental material).
Potentially important variation in E. faecalis and E. faecium epa operons occurs between orthologs of epaR (EF2177 in V583) and EF2165 (see Fig. S3 in the supplemental material). Variation in this region was previously reported between E. faecalis strains V583 and OG1RF (42). The variable regions of the 26 E. faecalis and E. faecium strains, which consist of 37 ortholog groups and 11 orphans (excluding transposases), encode predicted glycosyltransferases and other proteins with likely roles in extracellular polysaccharide production (see data set S4 in the supplemental material). The 3 vancomycin-resistant E. faecium CC17 strains in our collection possess a unique epa locus configuration with putative sialic acid biosynthesis (neuABCD) genes within the variable region, and a divergently transcribed, predicted β-lactamase gene inserted in the core epa region between epaO and epaR (see Fig. S3 in the supplemental material). Sialic acid decoration by pathogenic bacteria is thought to be a form of molecular mimicry that interferes with detection by the host immune system (48). The neuABCD genes are not clade specific and are also present in clade B strain E980 and clade A strain U0317, suggesting that the epa region can be either lost or transferred between E. faecium clades. The potential for sialic acid decoration on high-risk, vancomycin-resistant strains has important implications for vaccine development. We additionally identified putative WTA biosynthesis genes (tagF/tarF and tagD/tarD [49]) in a subset of E. faecalis and E. faecium epa variable regions (see Fig. S3 and data set S4 in the supplemental material). The core epaA and epaLM genes encode proteins similar to Bacillus subtilis TagO and TagGH (43), which catalyze the initial step in WTA synthesis and the export of the assembled WTA polymer, respectively (49).
Phagocytosis resistance in E. faecalis is associated with capsule production (40), which is a variable trait of that species (9). We examined the distribution of the E. faecalis cps capsule locus and found that it occurs only in E. faecalis, with little variation (see data set S4 in the supplemental material). We identified a novel capsule-like region in E. faecium (Fig. 5; see data set S4 in the supplemental material) that includes a phosphoregulatory system conserved among all species except E. faecalis (see data set S4 in the supplemental material). The proteins encoded are similar to the YwqCDE proteins of B. subtilis and the CpsBCD proteins of Streptococcus pneumoniae (Table 2), protein-tyrosine kinase/dephosphorylase regulatory systems that regulate UDP-glucose dehydrogenase activity (50) and capsule production (51), respectively. This system is located 5′ to a variable cohort of putative extracellular polymer biosynthesis genes in E. faecium (Fig. 5; see data set S4 in the supplemental material). This genetic configuration is similar to that of S. pneumoniae cpsABCD, which is core to the capsule biosynthesis loci of 90 pneumococcal serotypes (52). In E. faecium, these genes are oriented cpsACDB (Table 3; see data set S4 in the supplemental material). Because cps nomenclature is already used for the unrelated E. faecalis capsule biosynthesis locus (40), we refer here to the E. faecium cpsACDB genes by the alternate S. pneumoniae gene names wzg, wzd, wze, and wzh, respectively (52).
FIG 5 .
Putative E. faecium capsule loci. The core wzg-wzd-wze-wzh genes and downstream variable region are shown for 8 E. faecium strains. Conserved anchor genes flanking the core and variable regions are indicated. Variable region genes are colored by BLASTP and Pfam conserved-domain hits shown in data set S4 in the supplemental material. Multiple Pfam domains were collapsed into categories (for example, glycosyltransferases). Only the most abundant Pfam categories are shown. Orphan genes not grouped by OrthoMCL are indicated. Contig gaps in scaffolds are indicated by black bars; the size of the black bar is proportional to the number of N’s inserted during genome assembly. In E. faecium 502, the nucleotide sequence of wze is conserved but is interrupted by a contig gap, and a scaffold gap occurs between wzh and the EFVG_00414 flanking gene homologue (indicated by vertical slashes). The drawing is to scale, and a scale bar is shown.
TABLE 2 .
BLAST and Pfam analyses of putative phosphoregulatory system present in E. faecium, E. casseliflavus, and E. gallinarum
| Representative locusa |
Pfam hit (E value)b |
BLASTP best hitc |
|
|---|---|---|---|
|
S. pneumoniae TIGR4 |
B. subtilis 168 |
||
| EFPG_02020 | None | None | None |
| EFPG_02021 | LytR_cpsA_psr (2.7e-49) | SP_1942, transcriptional regulator (9e-75, 65%, 95%)d |
Membrane-bound transcriptional regulator LytR (1e-83, 67%, 91%) |
| EFPG_02022 | Wzz (1.1e-19) | Capsular polysaccharide biosynthesis protein Cps4C (6e-15, 48%, 81%) |
YwqC, modulator of YwqD protein tyrosine kinase activity (3e-45, 60%, 84%) |
| EFPG_02023 | CbiA (1.8e-14) | Capsular polysaccharide biosynthesis protein Cps4D (9e-42, 56%, 87%) |
PtkA/YwqD, protein tyrosine kinase (7e-74, 71%, 93%) |
| EFPG_02024 | None | Capsular polysaccharide biosynthesis protein Cps4B (2e-25, 49%, 88%) |
YwqE, protein tyrosine phosphatase (5e-68; 61%, 100%) |
From E. faecium 933.
Pfam hits with E values of ≤10−5 are shown.
Values in parentheses are E value, % similarity, and % query coverage.
The second-best hit was capsular polysaccharide biosynthesis protein Cps4A (7e-31, 79%, 55%).
TABLE 3 .
Biolog carbon catabolic substrates with the strongest species-specific signatures
| Chemical | OD590 ratiosa |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
E. casseliflavus |
E. faecium |
E. faecalis |
||||||||||||||||
| EC10 | EC20 | EC30 | 408 | 410 | 933 | Com12 | Com15 | 733 | V583 | T8 | T1 | X98 | E1Sol | AR01/D.G. | Fly1 | T3 | ||
| α-Ketovalerate | 4.6, 3.5 | 3.8, 3.4 | 3.7, 4.5 | 3.8, 3.7 | 3.5, 2.7 | 5.4, 5.6 | 3.6, 3.6 | 3.8, 3.8 | ||||||||||
| γ-Cyclodextrin | 7.0, 7.2 | 7.4, 6.2 | 4.4, 3.4 | 7.5, 6.8 | 4.7, 5.3 | 6.7, 6.8 | 2.4, 2.6 | |||||||||||
| Inulin | 5.2, 2.8 | 3.6, 4.0 | 4.2, 4.6 | |||||||||||||||
Shown are ratios of the OD590 in a carbon-containing well to the OD590 of a no-carbon-added control well after 48 h incubation at 37°C. A ratio >2 was considered a positive result. Each strain was tested twice, and the data shown are for both trials. Ratios of <2 are not shown. All ratios for E. gallinarum were <2.
We subjected the variable region between wzh and EFVG_00414 in each E. faecium genome to BLASTP and conserved-domain analyses (see data set S4 in the supplemental material). In S. pneumoniae capsule production, a sugar transferase (WchA) initiates capsule biosynthesis at the membrane by transferring an initial sugar to an undecaprenyl-phosphate carrier; additional sugars are then transferred to the repeat unit by glycosyltransferases, the structure is flipped across the membrane by the Wzx flippase, and additional repeat units are added by the Wzy polymerase (52). WchA-like proteins were identified in each of the 8 E. faecium variable regions, as were many predicted glycosyltransferases and enzymes likely to generate activated sugar moieties for transfer (Fig. 5). Wzx and Wzy homologues were identified in some, but not all, E. faecium strains (Fig. 5). Phagocytosis resistance is variable among E. faecium isolates (45), and no mechanism has been reported for this clinically relevant phenotype. It is likely that the putative capsule locus and/or variable epa loci described here contribute. We additionally identified putative wzg, wzd, wze, and wzh sequences in the enterococcal species E. saccharolyticus and E. italicus (data not shown), suggesting that, at least among the sequenced enterococci, E. faecalis is the exception, lacking this capsule biosynthesis pathway.
Species-specific signatures.
We used a combination of data, including Biolog carbon substrate catabolism analysis of a subset of our strains (see Materials and Methods), ortholog groups, and the Comparative Metabolism tool within a computationally generated database of predicted metabolic pathways (EnteroCyc; http://enterocyc.broadinstitute.org), to identify species-specific biochemical traits and nucleotide sequences that could augment existing methodologies to classify enterococcal isolates. For Biolog analysis, we focused on carbon substrates having the strongest species-specific signatures (Table 3).
Inulin fermentation was reported to be a distinguishing characteristic of motile enterococci (31), and our Biolog analysis confirmed that inulin metabolism is restricted to E. casseliflavus. Additionally, genes for acetoin dehydrogenase (ECAG_02019 to ECAG_02022), which converts acetoin to acetaldehyde and acetyl coenzyme A, are unique to E. casseliflavus. Catabolism of α-ketovalerate is specific to E. faecalis, as are genes (bkdDABC; EF1661 to EF1658) encoding a previously characterized branched-chain α-keto acid dehydrogenase complex (53). The eutBC genes (EF1629 and EF1627, respectively) directing ethanolamine catabolism and the formate dehydrogenase gene fdhA (EF1390) are also E. faecalis specific. Catabolism of the cyclic oligosaccharide γ-cyclodextrin, an additive in pharmaceuticals and other products (54), is enriched in E. faecium, while a gene for glutaminase (EFTG_00235), which converts glutamine to glutamate and ammonia, is E. faecium specific. Probes targeting c-di-GMP signaling (see data set S3 in the supplemental material) and acetoin dehydrogenase genes (E. casseliflavus); the eutBC, fdhA, and bdkDABC genes (E. faecalis); and the glutaminase gene (E. faecium) could be used to discriminate these different enterococcal species. We did not detect E. faecium clade-specific metabolism using Biolog analysis or EnteroCyc predictions; however, clade-specific gene sequences (see Table S2 in the supplemental material) could be used as molecular probes.
Perspectives.
A comparative genomic approach was used to address gaps in our knowledge of Enterococcus, a bacterial genus of importance to human health. Our phylogenetic analysis of E. faecium reveals and quantifies the distance that separates two distinct phylogenetic clades between which gene exchange has occurred. E. faecium clade-specific genes (see data set S2 and Table S2 in the supplemental material) are suggestive of different niches for clade A and clade B E. faecium in the GI tract. Additionally, conserved and variable pathways that appear to be important for cell wall polymer biosynthesis were identified. In contrast to E. faecium, a multiclade structure was not observed in E. faecalis, for which the acquisition of mobile elements appears to be a major source of genome diversity. Antibiotic resistance and pathogenicity island traits have converged in E. faecalis lineages (9), represented by strains V583, T8, and CH188. Despite the convergence of similar traits in those lineages and similar genome sizes (>3 Mb), substantial differences in gene content exist. Ecotypes defined by specific mobile element cohorts may be identified within high-risk lineages or in lineages with variable CRISPR-cas status (e.g., ST40 and ST21 [16]). Finally, comparative genomics highlighted fundamental differences between E. casseliflavus and E. gallinarum. The importance of the occurrence of motility operons in both but of genes related to the formation and function of the c-di-GMP second messenger only in E. casseliflavus and the impact of motility on metabolism represent interesting areas for future exploration.
MATERIALS AND METHODS
Enterococcal strains and genome sequencing.
E. faecalis strains were selected for genome sequencing to represent the diversity of a collection of 106 isolates previously characterized (9). The E. faecalis V583 and OG1RF genome sequences were previously reported (5, 24). The E. casseliflavus, E. gallinarum, and 6 E. faecium strains were obtained from a repository of clinical isolates (Eurofins Medinet). E. faecium Com12 and Com15 were isolated from feces of healthy human volunteers under Schepens Eye Research Institute Institutional Review Board protocol 2006-02, Identification of Pathogenic Lineages of E. faecalis. E. faecium STs were previously determined (16, 55), and E. faecium MLST data were accessed at http://efaecium.mlst.net. The sequencing, assembly, annotation, and rapid public release of these genome sequences have been previously described (8).
Standard analyses, OrthoMCL, and EnteroCyc.
Orthologous gene groups were identified using OrthoMCL (10), with an all-versus-all BLAST cutoff of 1E−5. Lactococcus lactis subsp. cremoris SK11 plasmid (NC_008503 to NC_008507) and chromosomal (NC_008527) genes were included as the outgroup. Coding sequences were aligned using Muscle (56), and poorly conserved regions were trimmed using trimAI (57). All trimmed alignments were concatenated and used to estimate phylogeny using maximum likelihood and 1,000 bootstrap trials as implemented by RAxML (58) using the rapid-bootstrapping option and the GTRMIX model. Conserved protein domains were predicted using HMMER3 (59) to search the Pfam (release 24; http://pfam.janelia.org) (60) and TIGRfam (release 10) (61) databases. The statistical significance of differences in genome size and conserved protein domain distribution was assessed using the one-tailed Wilcoxon rank sum test. Membrane helix predictions were generated with transmembrane protein topology with a hidden Markov model (14). Protein subcellular localization predictions were generated using PsortB (62). Sequence alignments and phylogenetic trees in the figures in the supplemental material were generated with ClustalW in MacVector. Enzyme Commission (EC) numbers for the proteins in EnteroCyc (http://enterocyc.broadinstitute.org/) were predicted using gene coding sequences (CDS) and BLASTX to search the KEGG database (release 56) (63) and assigning EC numbers based on the KEGG annotation. Only significant hits with an E value of <1E−10 and 70% overlap were considered. Pathways, operons, transporters, and pathway holes were predicted using the Pathway Tools software suite (64, 65). Unless otherwise noted, BLASTP and nucleotide megaBLAST queries were executed against the NCBI nonredundant protein sequence, nucleotide collection, and whole-genome shotgun read databases using NCBI BLAST. Proteins encoded by the E. casseliflavus EC10 motility locus were compared to a B. subtilis 168 reference using BLASTP (see data set S3 in the supplemental material); the B. subtilis 168 flagellum is a reference Gram-positive flagellum in the KEGG database (http://www.genome.jp/kegg-bin/show_pathway?bsu02040).
ANI and shared-gene analyses.
OrthoMCL ortholog groups were used to determine shared gene contents in pairwise genome comparisons. For a genome pair (genome 1 and genome 2), the total number of genes in genome 1 was determined and the number of genes in genome 1 shared with genome 2 (based on shared ortholog group membership) was determined. Percent shared gene content was calculated by dividing the number of genome 1 genes shared with genome 2 by the number of genes in genome 1. Nucleotide alignments of shared genes were used to determine the numbers of identical and different nucleotide residues in shared genes. For comparisons within species, at least 2,113 gene sequences were utilized. Percent ANI was calculated by dividing the number of identical nucleotide residues in shared genes by the total number of nucleotide residues.
Recombination analysis.
See the Text S1 in the supplemental material for a description of the methods used for genome mosaicism analysis and plot generation.
Biolog analysis.
A subset of strains (8/18 E. faecalis, 6/8 E. faecium, 3/3 E. casseliflavus, and 1/1 E. gallinarum) representing the diversity of the collection were analyzed in duplicate by Biolog Phenotype microarrays in accordance with the manufacturer’s instructions. Optical density at 590 nm (OD590) was read using a synergy 2 microplate reader (Bio-Tek). The 48-h OD590 reading of each well containing a carbon source was divided by the OD590 value obtained for the negative-control well. A ratio which gave a reproducible value of 2× the background was considered to be a positive result.
SUPPLEMENTAL MATERIAL
Supplemental methods. Expanded methods for genome mosaicism analysis and plot generation. Download Text S1, DOCX file, 0.1 MB.
E. faecium 408 and E. faecium 501 mosaic genes. Download Data set S1, XLSX file, 0.1 MB.
E. faecium clade-specific ortholog group nucleotide BLAST analysis against 7 additional sequenced E. faecium isolates. Download Data set S2, XLSX file, 0.1 MB.
Motile enterococcus BLAST and Pfam analyses. Download Data set S3, XLSX file, 0.1 MB.
Extracellular polymer biosynthesis BLAST and Pfam analyses. Download Data set S4, XLSX file, 0.1 MB.
E. faecium MLST tree. Sequences were downloaded from the E. faecium MLST database and aligned using ClustalW in MacVector. A phylogenetic tree with bootstrapping (1,000 replications) was generated by the unweighted-pair group method using average linkages. For each ST, MLST allele profiles were extracted from the database and are shown on the right. adk-6 alleles, identified as being highly specific to clade B, with the exception of clade A strain 408, are in red. ST39, ST40, ST60, ST61, and ST62 are the minority allelic population identified in reference 15. Download Figure S1, PDF file, 0.1 MB.
Alignment of CrtM proteins from S. aureus Newman, E. gallinarum, and E. casseliflavus. Substrate interaction residues are indicated by dots. Two DxxxD motifs (red boxes) and Mg2+ interact with the diphosphates of the farnesyl-diphosphate molecules and with inhibitors. Identical residues are shaded, and similar residues are shaded lightly. Substrate interaction data are from reference 36. Download Figure S2, PDF file, 0.1 MB.
E. faecalis and E. faecium epa loci. The core epa genes and downstream variable regions are shown for 18 E. faecalis and 8 E. faecium strains. Conserved anchor genes flanking the epa core genes and the variable regions are indicated. Variable-region genes are colored by annotations and by BLASTP and Pfam conserved-domain hits as shown in data set S4. Multiple Pfam domains were collapsed into categories (for example, glycosyltransferases). Only the most abundant Pfam categories are shown. Orphan genes not grouped by OrthoMCL are indicated. Contig gaps in scaffolds are indicated by black bars; and the size of each black bar is proportional to the number of N’s inserted during genome assembly. In E. faecalis ATCC 4200, a scaffold gap occurs in the epa variable region (indicated by vertical slashes). The drawing is to scale, and a scale bar is shown. Download Figure S3, PDF file, 0.1 MB.
Bacterial strain information.
E. faecium clade-specific genes.
ACKNOWLEDGMENTS
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under grant AI072360, the Harvard-Wide Antibiotic Resistance Program (AI083214), and contract no. HHSN272200900018C. Additionally, K.L.P. was supported by NIH fellowships EY007145 and EY020734.
We gratefully thank Rob Willems, Marvin Whiteley, Nathan Shankar, Willem van Schaik, and Mark Huycke for helpful comments during manuscript preparation.
Footnotes
Citation Palmer KL, et al. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3(1):e00318-11. doi:10.1128/mBio.00318-11.
REFERENCES
- 1. Aarestrup FM, Butaye P, Witte W. 2002. Nonhuman reservoirs of enterococci, p 55–99 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 2. Malani PN, Kauffman CA, Zervos MJ. 2002. Enterococcal disease, epidemiology, and treatment, p 385–408 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 3. Tannock GW, Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p 101–132 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 4. Willems RJ, Hanage WP, Bessen DE, Feil EJ. 2011. Population biology of gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:872–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 6. Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55:3345–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Schaik W, et al. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palmer KL, et al. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192:2469–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Jr, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leavis HL, et al. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 3:e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goris J, et al. 2007. DNA-NDa hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57:81–91 [DOI] [PubMed] [Google Scholar]
- 14. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 15. Homan WL, et al. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo C, et al. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. U. S. A. 108:7200–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glaser P, Rusniok C, Buchrieser C. 2007. Listeria genomics, p 33–62 In Goldfine H, Shen H, Listeria monocytogenes: pathogenesis and host response. Springer Verlag, New York, NY. [Google Scholar]
- 20. Barrangou R, Altermann E, Hutkins R, Cano R, Klaenhammer TR. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. U. S. A. 100:8957–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fleuchot B, et al. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80:1102–1119 [DOI] [PubMed] [Google Scholar]
- 22. Kreikemeyer B, McIver KS, Podbielski A. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224–232 [DOI] [PubMed] [Google Scholar]
- 23. Aslund F, Beckwith J. 1999. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J. Bacteriol. 181:1375–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bourgogne A, et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shankar N, Baghdayan AS, Gilmore MS. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750 [DOI] [PubMed] [Google Scholar]
- 26. Mundt JO, Graham WF. 1968. Streptococcus faecium var. casselifavus, nov. var. J. Bacteriol. 95:2005–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Collins MD, Jones D, Farrow JA, Kilpper-Balz R, Schleifer KH. 1984. Enterococcus avium nom. rev., comb. nov.; E. casseliflavus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov.; and E. malodoratus sp. nov. Int. J. Syst. Bacteriol. 34:220–223 [Google Scholar]
- 28. Contreras GA, et al. 2008. Nosocomial outbreak of Enterococcus gallinarum: untaming of rare species of enterococci. J. Hosp. Infect. 70:346–352 [DOI] [PubMed] [Google Scholar]
- 29. Tan CK, et al. 2010. Bacteremia caused by non-faecalis and non-faecium Enterococcus species at a medical center in Taiwan, 2000 to 2008. J. Infect. 61:34–43 [DOI] [PubMed] [Google Scholar]
- 30. Facklam RR, Carvalho MD, Teixeira LM. 2002. History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci, p 1–54 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 31. Langston CW, Gutierrez J, Bouma C. 1960. Motile enterococci (Streptococcus faecium var. mobilis var. n. ) isolated from grass silage. J. Bacteriol. 80:714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Forde BM, et al. 2011. Genome sequences and comparative genomics of two Lactobacillus ruminis strains from the bovine and human intestinal tracts. Microb. Cell Factories 10(Suppl 1):S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mills E, Pultz IS, Kulasekara HD, Miller SI. 2011. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell. Microbiol. 13:1122–1129 [DOI] [PubMed] [Google Scholar]
- 34. Bordeleau E, Fortier LC, Malouin F, Burrus V. 2011. C-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7:e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor RF, Ikawa M, Chesbro W. 1971. Carotenoids in yellow-pigmented enterococci. J. Bacteriol. 105:676–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu CI, et al. 2008. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319:1391–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pelz A, et al. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280:32493–32498 [DOI] [PubMed] [Google Scholar]
- 38. Theilacker C, et al. 2006. Opsonic antibodies to Enterococcus faecalis strain 12030 are directed against lipoteichoic acid. Infect. Immun. 74:5703–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toon P, Brown PE, Baddiley J. 1972. The lipid-teichoic acid complex in the cytoplasmic membrane of Streptococcus faecalis N.C.I.B 8191. Biochem. J. 127:399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hancock LE, Gilmore MS. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. U. S. A. 99:1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hancock LE, Shepard BD, Gilmore MS. 2003. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J. Bacteriol. 185:4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. 2009. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect. Immun. 77:3759–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu Y, Murray BE, Weinstock GM. 1998. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 66:4313–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Theilacker C, et al. 2011. Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BMC Microbiol. 11:67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arduino RC, Jacques-Palaz K, Murray BE, Rakita RM. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 62:5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reichmann NT, Gründling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 319:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pazur JH, Anderson JS, Karakawa WW. 1971. Glycans from streptococcal cell walls. Immunological and chemical properties of a new diheteroglycan from Streptococcus faecalis. J. Biol. Chem. 246:1793–1798 [PubMed] [Google Scholar]
- 48. Vimr E, Lichtensteiger C. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10:254–257 [DOI] [PubMed] [Google Scholar]
- 49. Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mijakovic I, et al. 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 22:4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288–301 [DOI] [PubMed] [Google Scholar]
- 52. Bentley SD, et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ward DE, Ross RP, van der Weijden CC, Snoep JL, Claiborne A. 1999. Catabolism of branched-chain alpha-keto acids in Enterococcus faecalis: the bkd gene cluster, enzymes, and metabolic route. J. Bacteriol. 181:5433–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Z, et al. 2007. Gamma-cyclodextrin: a review on enzymatic production and applications. Appl. Microbiol. Biotechnol. 77:245–255 [DOI] [PubMed] [Google Scholar]
- 55. Galloway-Peña JR, Rice LB, Murray BE. 2011. Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob. Agents Chemother. 55:3272–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. TrimAI: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 59. Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Selengut JD, et al. 2007. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35:D260–D264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu NY, et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaneshisa M, et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Karp PD, et al. 2010. Pathway tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 11:40–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Romero PR, Karp PD. 2004. Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics 20:709–717 [DOI] [PubMed] [Google Scholar]
- 66. Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Price MN, Dehal PS, Arkin AP. 2010. Fast tree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Manson JM, Keis S, Smith JM, Cook GM. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 48:3743–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Expanded methods for genome mosaicism analysis and plot generation. Download Text S1, DOCX file, 0.1 MB.
E. faecium 408 and E. faecium 501 mosaic genes. Download Data set S1, XLSX file, 0.1 MB.
E. faecium clade-specific ortholog group nucleotide BLAST analysis against 7 additional sequenced E. faecium isolates. Download Data set S2, XLSX file, 0.1 MB.
Motile enterococcus BLAST and Pfam analyses. Download Data set S3, XLSX file, 0.1 MB.
Extracellular polymer biosynthesis BLAST and Pfam analyses. Download Data set S4, XLSX file, 0.1 MB.
E. faecium MLST tree. Sequences were downloaded from the E. faecium MLST database and aligned using ClustalW in MacVector. A phylogenetic tree with bootstrapping (1,000 replications) was generated by the unweighted-pair group method using average linkages. For each ST, MLST allele profiles were extracted from the database and are shown on the right. adk-6 alleles, identified as being highly specific to clade B, with the exception of clade A strain 408, are in red. ST39, ST40, ST60, ST61, and ST62 are the minority allelic population identified in reference 15. Download Figure S1, PDF file, 0.1 MB.
Alignment of CrtM proteins from S. aureus Newman, E. gallinarum, and E. casseliflavus. Substrate interaction residues are indicated by dots. Two DxxxD motifs (red boxes) and Mg2+ interact with the diphosphates of the farnesyl-diphosphate molecules and with inhibitors. Identical residues are shaded, and similar residues are shaded lightly. Substrate interaction data are from reference 36. Download Figure S2, PDF file, 0.1 MB.
E. faecalis and E. faecium epa loci. The core epa genes and downstream variable regions are shown for 18 E. faecalis and 8 E. faecium strains. Conserved anchor genes flanking the epa core genes and the variable regions are indicated. Variable-region genes are colored by annotations and by BLASTP and Pfam conserved-domain hits as shown in data set S4. Multiple Pfam domains were collapsed into categories (for example, glycosyltransferases). Only the most abundant Pfam categories are shown. Orphan genes not grouped by OrthoMCL are indicated. Contig gaps in scaffolds are indicated by black bars; and the size of each black bar is proportional to the number of N’s inserted during genome assembly. In E. faecalis ATCC 4200, a scaffold gap occurs in the epa variable region (indicated by vertical slashes). The drawing is to scale, and a scale bar is shown. Download Figure S3, PDF file, 0.1 MB.
Bacterial strain information.
E. faecium clade-specific genes.