Abstract
Prenatal environmental exposures play a critical role in determining late-life chronic disease susceptibility. However, the mechanisms linking the in utero environment and disease development in the offspring are poorly understood. Recent investigations have confirmed a central pathogenic role of T cell chemokine receptors, particularly C-C chemokine receptor (CCR) 2 and CCR5, in chronic inflammatory conditions. This study was designed to determine the effect of a synthetic prenatal micronutrient supplementation (MS) diet rich in methionine pathway metabolites on the T cell chemokine system in F1 C57Bl/6 mice. Female mice were fed either an MS or control diet 3 wk prior to mating, during pregnancy, and lactation. At 4 wk of age, F1 mice were killed for experiments or were fed the standard NIH-31 diet and allowed to age. Food consumption, maternal weight gain, and litter size were similar in dams fed the control and MS diets. However, the F1 offspring of dams fed the MS diet were smaller in size (P < 0.001). T cells from the MS F1 offspring had global hypermethylation compared with control F1 offspring (P < 0.005), corresponding to lower T cell chemokine receptor expression [CCR2 (P < 0.001), CCR5 (P < 0.001), and C-x-C chemokine receptor 3 (P < 0.01)] and cytokine expression [TNFα (P < 0.05), IL-2 (P < 0.001), and IL-4 (P < 0.01)]. Reduced T cell chemokine receptor gene expression in MS F1 mice was associated with decreased chemotaxis in vitro to C-C chemokine ligand (CCL) 2 and C-X-C chemokine ligand 10 (P < 0.01) and in vivo to CCL2 (P < 0.01). Taken together, the results suggest that epigenetic alteration through prenatal diet manipulation reduces the response to proinflammatory signals in mice.
Introduction
A tantalizing link between diet during oocyte/sperm development, epigenetics, and late-life disease in humans has recently been proposed. Previous studies report the association between ancestral food supply and longevity, cardiovascular disease, and diabetic mortality (1–3). These works suggest that these associations most likely are not related to classical fetal programming through maternal malnutrition but are the results of transgenerational epigenetic inheritance. However, experimentation during human pregnancy is difficult and the underlying mechanisms for the observations have not been defined.
Epigenetics refers to heritable changes in gene expression by mechanisms other than those resulting from alteration in DNA sequence (4). A number of epigenetic processes have been described, including DNA methylation, chromatin remodeling, and RNA splice variants. The data suggest that dietary supplementation with micronutrients from the methionine and folate cycles can induce hypermethylation of DNA in specific genomic regions. Several studies have examined the effect of high dietary methionine intake on rodents (5–7).
A prime example of the effect of maternal diet on F1 phenotype is provided by the viable yellow agouti mouse. In genetically identical mice, the agouti gene is differentially expressed due to epigenetic modifications established during early development, resulting in variations in coat color (8–11) and differing susceptibility to obesity, cancer risk, and life span. Maternal dietary supplementation shifts the offspring toward the healthier phenotype via methylation of the agouti locus. The prenatal diet-modifying effect on the germ-line continues into later generations. Importantly, all previous prenatal dietary studies have exclusively used commercial diets that do not control for micronutrient contents, raising the possibility that the observed changes may be in part related to differences in micronutrients present in different batches of commercial diets.
Chemokines are a superfamily of small, 8- to 10-kDa, proinflammatory cytokines (12). Chemokines and their receptors are important regulators of leukocyte recruitment to sites of inflammation. Although C-C chemokine receptor (CCR)7 2 and CCR5 are perhaps best known for their role in atherogenesis, another important role for these chemokine receptors also play important roles in other chronic inflammatory diseases (13). Based on these studies, we hypothesize that the prenatal maternal micronutrient environment can influence the offspring’s long-term immune function.
Materials and Methods
Designing a synthetic, prenatal, micronutrient supplementation diet.
To avoid the pitfall of uncertain micronutrient content in standard commercial diets, we designed a custom amino acid–defined control and micronutrient supplementation (MS) diet based on TD.99366 (Harlan Teklad), a synthetic version of the NIH31 diet (Supplemental Table 1). The MS diet was based on the maternal diet that has been shown to alter DNA methylation in the offspring (9) and was designed to affect DNA methylation through the methylation/methionine/folate cycle (Supplemental Fig. 1). Additionally, the diet had protein content sufficient for reproduction. Importantly, the overall energy and fat content of the control and MS diets were the same.
Mice.
Young adult (8 wk) C57Bl/6 mice were purchased from Harlan Laboratories. Female mice were fed 1 of 2 synthetic diets, control or MS for 3 wk prior to mating. A cohort of female mice from each diet was left unmated and their rate of diet consumption and weight gain was observed weekly. Mated female mice were fed the diet through pregnancy and lactation. Given that pups of MS-fed dams were significantly smaller than their control counterparts, mice were weaned at 28 d at the recommendation of animal husbandry technicians. Weaned pups were killed at 28 d by CO2 asphyxiation or were fed the standard NIH-31 diet and allowed to age, as determined by experimental needs. For experiments involving young and old mice, young mice (3 mo) were purchased from Harlan and old mice (18–20 mo) were purchased from the National Institute of Aging. All mice were maintained in a pathogen-free environment on a 12-h-light cycle and consumed feed ad libitum. Unless otherwise reported, all data presented are from 4-wk-old F1 mice. Male and female samples were prepared separately in pools of 2–5 mice each, depending on availability and experimental requirements. However, the analysis presented is gender neutral [except biometric data (Table 1; Supplemental Table 3)], because biochemical assay results were similar between male and female pups from the same diet group. Procedures involving the animals and their care were approved by the University of Michigan Committee on the Use and Care of Animals.
TABLE 1.
Long-term effect of prenatal MS diet on the body weight of F1 mice in 3 independent aging cohorts1
| F1 mice | 4 wk |
13 wk |
52 wk |
|||
| g | n | g | n | g | n | |
| Control male | 14 ± 0.39 | 40 | 25 ± 0.50 | 24 | 38 ± 2.2 | 11 |
| MS male | 10 ± 0.11* | 26 | 23 ± 0.22 | 28 | 34 ± 2.1 | 9 |
| Control female | 13 ± 0.27 | 37 | 21 ± 0.38 | 21 | 28 ± 1.1 | 7 |
| MS female | 11 ± 0.72 | 25 | 20 ± 0.25* | 16 | 34 ± 0.55 | 8 |
Values are mean ± SEM. *Different from control, < 0.005. MS, micronutrient-supplemented diet.
NMR-based body composition analyses.
Male and female F1 mice were placed in the measuring tube of a NMR Minispec LF90 II (Bruker Optics) and the percentages (by weight) of fat, muscle, and fluid content were measured.
T cell isolation.
Cluster of differentiation (CD)3+ or CD4+ T cells were isolated by the MACS microbeads technology from F1 spleens by negative selection according to the manufacturer’s instructions (Miltenyi Biotec). The purity of the isolated cells was confirmed by flow cytometric analysis by staining with fluorescein isothiocyanate–conjugated anti-CD3 or anti-CD4, or isotype IgG2b Abs (BD) and was consistently >90%.
Stimulation and 5-azacytidine treatment.
5-Azacytidine is a cytosine analogue that is incorporated into newly synthesized DNA, preventing maintenance of DNA methylation patterns and leading to hypomethylation. Freshly isolated murine T cells from wild-type C57Bl/6 mice were stimulated with 1 mg/L phytohemagglutinin (PHA) (Sigma) in the presence of 10 μg/L IL-2 (R&D Systems) in T cell media (RPMI 1640 plus 10% fetal calf serum, 105 U/L penicillin/streptomycin, 1× nonessential amino acids, 1× sodium pyruvate, 50 μmol/L β-mercaptoethanol, 25 mmol/L 4–2-hydroxyethyl-1-piperazineethanesulfonic acid). After 24 h of incubation, the cells were treated with 1 μmol/L 5-azacytidine or left untreated and cultured for an additional 48 h. Cells were then harvested and used as positive controls with the appropriate methylation detection assays (see below).
Nucleic acid isolation.
Total RNA was isolated from resting T cells obtained from F1 mice of each diet group at 4 wk of age using RNeasy Mini or Micro kits according to the manufacturer’s instructions (Qiagen). Carryover DNA contaminants were removed using the RNase-Free DNase Set (Qiagen). Genomic DNA was also isolated from F1 mice using the DNeasy Blood and Tissue Kit (Qiagen). λ phage and Sss1 methylase were purchased from New England Biolabs. In vitro methylation was performed following the manufacturer’s instructions.
Microarray analyses.
Gene expression in splenic T cells of control and MS 4-wk-old F1 mice was initially screened using the AffyMetrix GeneChip mouse-430v2 microarray gene expression system (AffyMetrix). We used 3 groups per diet per gender, with each group consisting of 5 μg RNA pooled from 5–10 mice. Arrays were processed according to the manufacturer’s protocol. Bioconductor software (http://www.bioconductor.org/packages/release/bioc/html/affy.html) was used in the analysis. Expression values were calculated using robust multi-array average (14). The software uses a prior estimate of the variance of the probe sets to adjust the denominator of each t-statistic. This allowed us to “weigh” the arrays, so unreliable arrays will not have an undue effect on the results (15, 16). We selected probe sets based on an unadjusted P value of 0.01. We removed all probe sets that had a variance <0.02 overall, then fit the same model as before, but this time using an unadjusted P value of 0.001. The analyses were conducted in collaboration with the University of Michigan Cancer Center Microarray Core facility.
Real-time qRT-PCR.
Real-time qRT-PCR was performed using a QuantiTect SYBR Green RT-PCR kit according to the manufacturer’s instructions (Qiagen). PCR reactions were carried out using a Rotor-Gene 6000 rotary analyzer (Corbett Research). Data were analyzed using Rotor-Gene 6000 software, version 1.7. The expression levels of genes of interest were normalized to β-actin expression. Primer sequences are listed in Supplemental Table 2A.
Bisulfite treatment and pyrosequencing.
Sodium bisulfite treatment of 1 μg genomic DNA was performed with the EZ DNA Methylation Gold kit (Zymo Research). PCR of the B1 element was performed using an assay designed by EpigenDx (Worcester). An assay for the intracisternal A Particle (Iap) repetitive element was designed using PSQ Assay Design Software (Biotage). Pyrosequencing was performed as previously described (17). Primer sequences are listed in Supplemental Table 2B.
Luminometric methylation assay.
T cell genomic DNA (300 ng) underwent luminometric methylation assay (LUMA) analysis as previously described (18). The sequence to analyze was modified to eliminate degradation products from analysis and was as follows: G/TGTGTCACACACATGTG. The dispensation order was GTGTGTCACACACATGTGTG, using dispensations 15 and 16 as the quantifying peaks.
Global methylation analysis by flow cytometry.
Freshly harvested T cells (2.5 × 105) were stained with fluorescein isothiocyanate–conjugated anti-CD3 Abs (BD) and then fixed in Cytofix/Cytoperm (BD) and permeabilized using PBS supplemented with 0.1% saponin, 1% FBS, and 0.1% sodium azide. Cells were treated with RNase A to eliminate the potential for detection of 5-methylcytidine in tRNA. Cells were exposed to primary antibody for 30 min at 37°C (Acris Antibodies), then incubated with secondary antibody for 30 min at 37°C (anti-mouse IgG1-PE or isotype rat-IgG1κ). Flow cytometry was performed using a FACScaliber machine. Results were analyzed with FCS Express software.
Methyl-cytosine ELISA.
DNA methylation was measured using the Methylamp Global DNA Methylation Quantification kit (Epigentek). DNA was prepared using the GenElute Mammalian Genomic DNA Miniprep kit (Sigma). Each DNA sample was pooled from 2–5 mice/diet and each assay performed in triplicate.
Western-blot analysis.
Total protein was isolated from CD3+ splenocytes of 4-wk-old F1 control and MS diet mice. Antibodies against CCR2 (Abcam), CCR5 (Abgent), C-x-C chemokine receptor (CXCR) 3 (Santa Cruz Biotech), and actin (Sigma) were used to monitor expression of the respective protein antigens and were detected using the Dura chemiluminescent HRP system (Pierce) as previously described (19). For quantification of Western-blot data, the integrated intensity of the protein band of interest was measured using ImageJ software and normalized to the integrated intensity of the actin band in the same sample.
In vitro chemotaxis assay.
Dual-chamber chemotactic assays were performed in triplicate to compare T cell C-C chemokine ligand (CCL) 2, C-X-C chemokine ligand (CXCL) 10, and CXCL12 migratory response as previously described (20). Briefly, T cells were stimulated with 1 mg/L PHA in the presence of 10 μg/L IL-2 for 72 h, then 3 × 105 cells were resuspended in RPM1 1640 supplemented with 0.5% BSA and placed in Transwell inserts with 5-μm pores (Corning). The inserts were then placed in a 24-well plate at 37°C containing varying concentrations of murine chemokines (as indicated in Fig. 5) (CXCL10 and CXCL12, Peprotech; CCL2, Biolegend). After 3 h, microsphere beads (Bangs Laboratories) were added for normalization and T cells from the bottom chamber were harvested and counted using a flow cytometer. Percent migration was calculated as the ratio of T cells found in the lower chamber to the total cell number added to the upper chamber. The chemotactic index was determined by taking the ratio of percentage migration between wells with chemokines added to wells and wells that did not contain chemokines.
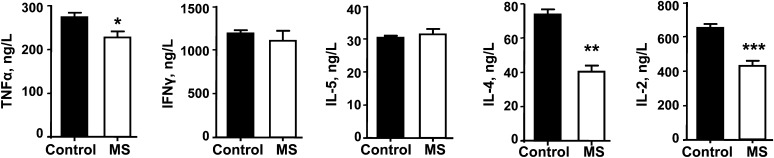
FIGURE 5.
Cytometric bead array detection of TNFα, IFNγ, IL-5, IL-4, and IL-2 cytokines secreted from 3-mo-old F1 control and MS CD4+ T cells after 48-h stimulation. Values are mean ± SEM, n = 3. Different from control where indicated: *P < 0.05, **P < 0.01, ***P < 0.001. MS, micronutrient-supplemented diet.
In vivo cutaneous chemotaxis.
Four-week-old F1 mice were s.c. injected in the loose skin around the neck and shoulder area with a range of concentrations of CCL2. The injection site was marked with ink and the skin was harvested 24 h later then fixed in 10% paraformaldehyde. Paraffin sections were stained with hematoxylin and eosin. Cellular infiltrates were counted at 40× magnification in 5 random fields by a pathologist (R.W.).
Measurement of Th1/Th2 cytokine production.
Freshly isolated CD4+ cells from 3-mo-old F1 mice were cultured in the presence of plate-bound 1 mg/L anti-CD3e and soluble 1 mg/L anti-CD28 in a humidified incubator at 37°C for 48 h. Cytokine concentrations were determined using the Mouse Th1/Th2 Cytokine kit (BD). Capture beads were detected with a FACSVantage flow cytometer (BD) and data analysis was performed using BD CBA Software by the University of Michigan Flow Cytometry Core.
Splenocyte populations.
To characterize changes in the immune cell populations of the F1 spleen, freshly isolated splenocytes were labeled with PE antibodies to CD3, CD19, CD11b, CD11c, and NK1.1, or their recommended isotypes: Rat IgG2b,K, Rat IgG2a,K, Hamster IgG,K, or Mouse IgG2a,K (BD). Flow cytometry was performed using a FACScaliber machine and results were analyzed with FCS Express software.
Statistical analysis.
Significance was determined using a 1-tailed Student’s t test for single comparison. P < 0.05 was considered significant except for the microarray results. Statistical methods used for the microarray experiment are detailed in the relevant “Materials and Methods” section. Values are mean ± SEM.
Results
Dams consume a similar amount of the control and MS diet.
Unmated female mice consumed ad libitum an equal amount of the control or MS diet over a 10-wk observation period (Supplemental Fig. 2A). In wk 1, both groups lost a similar amount of weight (control, 7.9%; MS 5.8%; nonsignificant). During the ensuing weeks, both groups gained comparable amounts of weight (Supplemental Fig. 2B). After mating, weight gain did not differ between the diet groups among pregnant mice (Supplemental Fig. 2C). Pup litter size also did not differ between the 2 diet groups (Supplemental Fig. 2D).
F1 offspring from dams fed the MS diet are smaller in size.
Despite similar macronutrient intake, dams exposed to the MS diet during pregnancy gave birth to pups that, at 4 wk, weighed 22% less than those exposed to the control diet in utero (P < 0.005) (Table 1). An older cohort of 13-wk-old F1 MS mice also weighed 7.0% less than the control mice (P < 0.005), whereas a 52-wk-old cohort did not reach significance. We determined the body composition of the F1 mice by NMR (Supplemental Table 3). As a percentage of overall weight, the F1 offspring from the MS group had similar lean muscle mass, fat, and fluid to the control offspring.
F1 MS mice have increased global CpG methylation.
We initially interrogated the methylation status of mouse repetitive elements B1 (Supplemental Fig. 3A) and Iap (Supplemental Fig. 3B) in T cells by pyrosequencing. The results revealed highly methylated CpG sites at all sites interrogated. Four-week-old F1 control and MS mice did not differ in repetitive element methylation. 5-Azacytidine treatment, used as a test of assay sensitivity and positive control, lowered the mean methylation in B1 and Iap relative to PHA-stimulated T cells from wild-type mice.
Because of the high methylation status of the repetitive elements in T cells, we next estimated global methylation using the LUMA method. Interrogated CpG sites from 4-wk-old F1 control and MS mice were similarly methylated (Supplemental Fig. 3C). To validate the LUMA protocol, we also incorporated several controls in our experiment. Treatment with 5-azacytidine, a DNA-demethylating agent, lowered the mean methylation relative to PHA-stimulated T cells from wild-type mice. λ phage DNA was completely demethylated as expected, whereas Sss1 methylase-treated λ phage DNA was highly methylated. Also, DNA from 3-mo- and 18- to 20-mo-old, wild-type, C57Bl/6 murine T cells did not differ in methylation level.
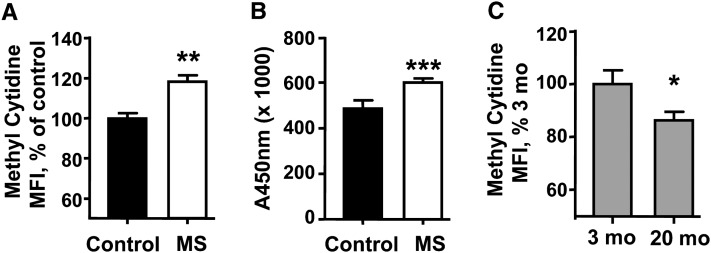
We next interrogated global methylation by flow cytometry using an antibody specific to 5-methylcytosine. Whereas the LUMA and repetitive element analyses are restricted to specific sites within the genome, using an antibody allowed the interrogation of CpG sites independent of DNA sequence. Four-week-old F1 MS mice had 20% greater median fluorescence intensity (MFI) than the F1 control mice (P < 0.005), suggesting that a prenatal diet rich in methionine metabolites may contribute to a higher mean methylation of cellular DNA (Fig. 1A). Treatment with 5-azacytidine lowered MFI by 37% relative to the PHA-stimulated T cells from wild-type mice (P < 0.05) (data not shown). Aging is known to be associated with T cell DNA hypomethylation (21, 22). Flow cytometry detection of methyl-cytidine showed that T cells from 18- to 20-mo-old wild-type mice had 14% lower MFI than 3-mo-old mice (P < 0.05) (Fig. 1C).
FIGURE 1.
Global methylation detected by antimethylcytidine antibody staining (A) or ELISA (B) in murine T cells from 4-wk-old F1 control and MS mice; antimethylcytidine antibody staining of 3-mo- and 20-mo-old, wild-type C57Bl/6 murine T cells (C). Results are mean ± SEM, n = 5 (A), 7 (B), or 6 (C). Means differ where indicated: *P < 0.05, **P < 0.02, ***P < 0.01. MFI, median fluorescence intensity; MS, micronutrient-supplemented diet.
Given the discrepancies between the pyrosequencing/LUMA and the flow cytometry results, we examined the total methylation using a commercially available, nonbiased, antimethylcytosine ELISA. Consistent with the flow cytometry approach, T cells from 4-wk-old F1 MS mice had an 11% greater amount of methylated cytosines than those from the control group (P < 0.05) (Fig. 1B).
MS diet is associated with reduced mRNA expression of selected T cell chemokine receptors.
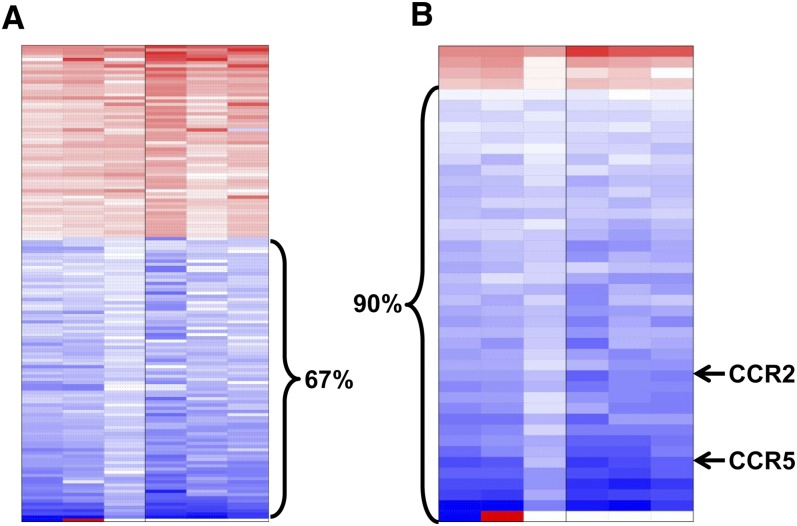
We performed microarray analyses of RNA isolated from the T cells from the F1 progeny of dams exposed to the control and MS diets. Because we are interested in immune function genes, we subsetted the data by preselecting only those probe sets that mapped to the following GO terms: immune, T cell, lymphocyte, chemokine, apoptosis, methylation, and signaling. We used the resulting 6248 probe sets to fit a linear model that accounted for sex, allowing a gender neutral analysis of diet effects on gene expression. A total of 256 probe sets were differentially expressed between the control and MS groups. Prenatal MS diet exposure decreased expression of the majority of the 256 probe sets, which was consistent with prior observations that methylation generally results in the suppression of gene expression (Fig. 2A) (4). When we increased the stringency of the selection criteria by specifying probe sets that had fold-changes >50%, the percentage of probe sets that has decreased expression in the MS group increased to 90% (Fig. 2B).
FIGURE 2.
Heat map of RNA microarray data from splenic T cells of F1 control and MS mice at 4 wk (A) and when the variance is set at <0.02 (B). Red, overexpressed in the MS group compared with controls; blue, reduced expression in the MS group, n = 6 (B). CCR, C-C chemokine receptor; MS, micronutrient-supplemented diet.
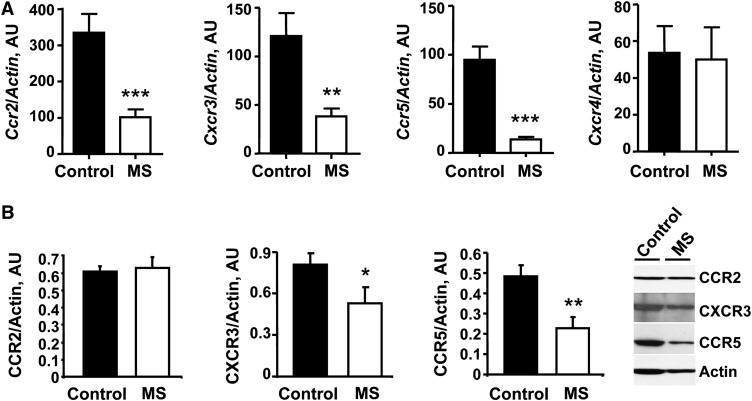
With regard to chemokine receptor genes, 4-wk-old F1 MS mice expressed lower Ccr5 [67% reduction (P < 0.001)], Ccr2 [37% reduction (P < 0.002)], Ccr6 [39% reduction (P < 0.004)], and Cxcr3 [49% reduction (P < 0.001)] relative to controls. To confirm the microarray chemokine receptor results, we quantified the relative gene expression in control and MS F1 mice using qRT-PCR. MS mice had a 70% reduction in Ccr2 expression (P = 0.0005), an 85% reduction in Ccr5 expression (P = 0.0002), and a 68% reduction in Cxcr3 (P = 0.006) compared with control mice (Fig. 3A). Cxcr4, which did not significantly differ in the microarray, showed no expression difference using qRT-PCR. At the protein level, CCR5 and CXCR3 from 4-wk-old F1 MS mice were downregulated, whereas CCR2 did not differ in the offspring of the 2 diet groups (Fig. 3B).
FIGURE 3.
Real-time qRT-PCR (A) and Western-blot analysis (B) of T cell chemokine receptors from 4-wk-old F1 control and MS mice. Values are mean ± SEM, n = 6 (A) or 3 (B). Different from control where indicated: *P < 0.05, **P < 0.02, ***P < 0.01. AU, arbitrary unit; CCR, C-C chemokine receptor; CXCR, C-X-C chemokine receptor; MS, micronutrient-supplemented diet.
T cells from MS F1 mice show impaired chemotaxis in vitro.
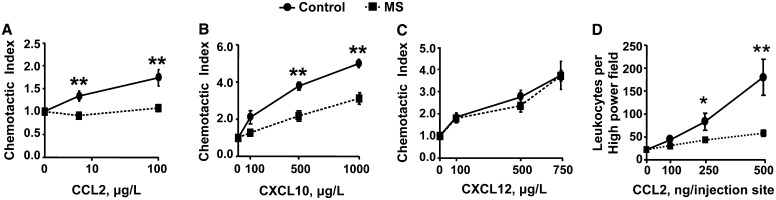
We next determined if the reduction in T cell chemokine receptor mRNA expression was associated with change in in vitro chemotactic activity of T cells from 4-wk-old F1 control and MS mice to CCL2, CXCL10, and CXCL12, ligands for CCR2, CXCR3, and CXCR4, respectively. Four-week-old F1 MS T cells had lower chemotaxis index to 4 μg/L (32% reduction) and 100 μg/L (38% reduction) CCL2 (Fig. 4A) and 500 μg/L (42% reduction) and 1000 μg/L (38% reduction) CXCL10 (Fig. 4B) relative to controls (P < 0.01). The responses to CXCL12 did not differ between the F1 diet groups (Fig. 4C).
FIGURE 4.
In vitro dual chamber T cell chemotactic assay to CCL2 (A), CXCL10 (B), and CXCL12 (C) and in vivo cutaneous leukocyte chemotaxis assay (D) from 4-wk-old F1 control and MS mice. Values are mean ± SEM, n = 3 (A), 4 (B), 4 (C), or 7 control, 6 MS mice (D). Different from control where indicated: *P < 0.05, **P < 0.01. CCL, C-C chemokine ligand; CXCL, C-X-C chemokine ligand; MS, micronutrient-supplemented diet.
In vivo cutaneous chemotaxis shows impaired leukocyte migration to CCL2 in MS F1 mice.
Twenty-four hours after s.c. injection of CCL2, 48 and 68% fewer leukocytes infiltrated the injection sites of the MS group progeny at a dose of 250 ng (P < 0.05) and 500 ng CCL2 (P < 0.01), respectively (Fig. 4D), confirming an in vivo functional role of the observed gene expression data.
Low cell migration is accompanied by a decrease in cytokine production by CD4+ T cells from MS F1 mice.
Given the decreased migratory potential in response to chemotactic signals observed in activated MS T cells, we examined the secretion of cytokines by CD4+ T cells in culture from 3-mo-old control and MS F1 mice. TNFα, IL-4, and IL-2 secreted from MS CD4+ T cells had respective reductions of 17, 45, and 34% compared with T cells from controls (P < 0.05) (Fig. 5). IFNγ and IL-5 did not differ between the 2 diet regimens.
MS F1 mice have similar leukocyte subsets in the spleen.
To exclude the possibility that decreases in T cell mediate immune response are a result of an alteration of relative immune cells populations, we interrogated the leukocyte populations of the spleen. T cells as a percentage of total splenocytes did not differ between F1 MS mice and controls (Supplemental Table 4). Other leukocytes such as B cells, NK cells, and dendritic cells were also similar as a percentage of total splenocytes, except CD11b+ cells, which were lower in F1 MS mice (P < 0.05).
Discussion
The idea that the prenatal nutritional environment has an important impact on late-life disease development was first raised by Barker and Osmond (23). Since then, most investigations have focused on the fetal effect of macronutrient exposure (e.g., protein intake) during pregnancy (24, 25). Although the role of maternal micronutrients in fetal health is incompletely understood, maternal diet supplemented with folic acid has been shown to reduce the incidence of neural tube defects in the offspring. Folic acid, which feeds into the methionine and DNA methylation cycles, has also been found to increase PPARγ methylation in rats fed a protein-restricted diet during pregnancy (26). Mice fed very low levels of methionine throughout life live longer and have a relative preservation of their immune function (27). A recent report also suggested that the offspring of dams fed a similar supplemented diet have enhanced reactive airway response in part related to methylation of Runt-related transcription factor 3 (Runx3) (28), supporting our hypothesis that in utero micronutrient exposure can have important effects on the progeny’s immune system. An important limitation of these previous studies is that the micronutrient intake was poorly controlled, because the animals were allowed to consume grain-based diets that had highly variable micronutrient content. These works also did not report total diet consumption, raising the possibility that the observed effects may in part relate to differences in energy or macronutrient intake.
Unlike previously published studies that added micronutrients to a standard commercial diet, we customized synthetic diets to more precisely control both the macro- and micronutrients in the control and supplemented diets. The diet was constructed based on published data showing that supplementing the key maternal micronutrients in the folate, methionine, and DNA methylation cycles has measurable effects on the health of the progeny in the agouti mouse model (8, 9). Our results showed that the control and MS diets were equally palatable, because the mice consumed similar amounts of the synthetic diet and gained similar weight throughout the experiment. Surprisingly, although the control and MS-fed dams consumed similar amounts of protein, fat, and energy during pregnancy, the F1 progeny of the MS group were smaller in size and the differences persisted until they are at least 1 y of age. NMR studies demonstrated that the mice had similar body fat and muscle composition. Diet-restricted mice and long-lived mutant mice, including Ames dwarf, Snell dwarf, and growth hormone receptor/binding protein knockout mice, all have a small body size. Studies are underway to determine if the small-bodied progeny of the MS-fed dams live longer than their control counterparts. One potential confounding variable due to the small size of MS offspring is that they must be weaned later than the typical 21 d. As such, MS weanlings may be directly exposed to the supplement diet briefly. However, we do not think it is likely that such a very brief exposure would lead to long-term impacts on the immune system (e.g., inflammatory cytokine downregulation).
In the agouti mouse model, variable methylation of the Iap retrotransposon inserted upstream of the agouti gene results in yellow coat color, diabetes, obesity, and increased susceptibility to tumors (8, 9). The pleiotropic effects are presumably due to paracrine signals on other tissues. Classes of elements in the genome may be especially sensitive to nutritional manipulation, including transposons that lie adjacent to genes with metastable epialleles (e.g., the Avy mutation) and imprinted genes (e.g., Igf2 and H19). However, whether similar dietary manipulation will affect normal genes is less clear. We showed for the first time, to our knowledge, that prenatal micronutrient manipulation alters the expression of the progeny’s T cell Ccr2 and Ccr5, 2 critical chemokine receptors central to the pathogenesis of common proinflammatory conditions, including coronary artery disease and autoimmune arthritis. mRNA transcripts were equally downregulated in both male and female MS F1 mice. At the protein level, CCR5 and CXCR3 were downregulated in resting MS T cells, and stimulated MS T cells had decreased chemotaxis in response to the ligands of CCR2 and CXCR3.
We further examined the mechanisms linking the prenatal MS diet and F1 T cell chemokine receptor gene expression. A previous report estimated T cell methylation in F1 mice exposed to a similar diet by examining repetitive elements and LUMA assay did not find any global methylation changes (28). We also did not observe any difference in the 2 study groups when we interrogated the methylation status of the T cell repetitive B1 and Iap elements or used LUMA. However, these methods do not measure methylation of the entire T cell genome. Therefore, we measured global methylation MFI by direct antibody staining and ELISA. These nonbiased assays demonstrated that MS F1 T cells were hypermethylated relative to the control group. Importantly, T cells from aged animals, known to be hypomethylated relative to young mice (29), also showed the expected decrease in methylation using antimethylcytidine staining but showed no difference using LUMA. The disparity between sequence-dependent and sequence-independent assays suggests that the prenatal diet may exert its effect in diverse regions rather than at retrotransposon or CCGG-rich loci. DNA methylation can regulate gene expression at proximal promoters and upstream and downstream enhancers, and there are many examples of methylation changes altering chromatin structures, affecting expression over long distances (30). Therefore, detecting subtle variations in the epigenome may require a sequence-independent approach. Future studies are underway to elucidate the specific regions of the methylome that alter cytokine and chemokine receptor expression and give rise to the antiinflammatory phenotype observed in MS F1 mice.
Although the mechanisms for the transgenerational effect are unclear, our current study used a novel synthetic diet to more precisely examine for the effect of prenatal MS diet on T cell immune functions. We show for the first time, to our knowledge, that F1 offspring have reduced expression of T cell Ccr2 and Ccr5, key players in the pathogenesis of diverse inflammatory diseases. Chemokine receptor changes are accompanied by corresponding changes in T cell migration in vitro and in vivo as well as global T cell hypermethylation. Total splenic population analysis suggests that the observed decrease in T cell-mediated immune response is likely not attributable to changes in splenocyte populations in vivo. Although we have observed a difference in CD11b+ cells, we do not attribute a substantial role in the lower immune response to this disparity, because all in vitro T cell stimulations were performed in the absence of CD11b+ cells. In addition to the suppression of chemotaxis, the prenatal MS diet lowered the proinflammatory cytokine secretion of TNFα as well as IL-2 and IL-4. Because IL-2 is associated with a Th1 response and IL-4 is a signature cytokine in the Th2 phenotype, our diet may not preferentially select for a specific T cell subset. Future studies will elucidate if these changes in T cell function due to prenatal micronutrient exposure alter the course of development of inflammatory diseases in which the chemokine/cytokine axes are known to play a pathogenic role.
Supplementary Material
Acknowledgments
C.D., M.H., S.G., K.J., and R.Y. designed research; C.D. and R.W. performed experiments; and R.Y. designed research and wrote the paper. All authors read and approved the final manuscript.
Footnotes
Supported in part by NIH (grants AG020628, AG028268, and P30 ES017885); University of Michigan (Claude D. Pepper Older American Independence Center, Nathan Shock Center for the Basic Biology of Aging, Rheumatic Disease Clinical Center, Caner Center Microarray Core, Michigan Diabetes and Research Training Center Animal Phenotyping Core); Geriatrics Research, Education and Clinical Care Center; and the VA Ann Arbor Healthcare System. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supplemental Figures 1–3 and Supplemental Tables 1–4 are available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of content at http://jn.nutrition.org.
Abbreviations used: CCL, C-C chemokine ligand; CCR, C-C chemokine receptor; CD, cluster of differentiation; CXCL, C-X-C chemokine ligand; CXCR, C-x-C chemokine receptor; LUMA, luminometric methylation assay; MFI, median fluorescence intensity; MS, micronutrient-supplemented diet; PHA phytohemagglutinin.
Literature Cited
- 1.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J, ALSPAC Study Team Sex-specific, sperm-mediated transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–66 [DOI] [PubMed] [Google Scholar]
- 2.Bygren LO, Kaati G, Edvinsson S. Longevity determined by ancestors’ overnutrition during their slow growth period. Acta Biotheor. 2001;49:53–9 [DOI] [PubMed] [Google Scholar]
- 3.Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet. 2002;10:682–8 [DOI] [PubMed] [Google Scholar]
- 4.Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene expression. Cell Mol Life Sci. 2002;59:241–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136:S1706–10 [DOI] [PubMed] [Google Scholar]
- 6.Finkelstein JD, Martin JJ. Methionine metabolism in mammals. Adaptation to methionine excess. J Biol Chem. 1986;261:1582–7 [PubMed] [Google Scholar]
- 7.Regina M, Korhonen VP, Smith TK, Alakuijala L, Eloranta TO. Methionine toxicity in the rat in relation to hepatic accumulation of S-adenosylmethionine: prevention by dietary stimulation of the hepatic transsulfuration pathway. Arch Biochem Biophys. 1993;300:598–607 [DOI] [PubMed] [Google Scholar]
- 8.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–8 [DOI] [PubMed] [Google Scholar]
- 9.Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc Natl Acad Sci USA. 2006;103:17308–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–16 [DOI] [PubMed] [Google Scholar]
- 11.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–21 [DOI] [PubMed] [Google Scholar]
- 13.Zhao Q. Dual targeting of CCR2 and CCR5: therapeutic potential for immunologic and cardiovascular diseases. J Leukoc Biol. 2010;88:41–55 [DOI] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64 [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004; 3:Article 3 [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300 [Google Scholar]
- 17.Jeong KS, Lee S. Estimating the total mouse DNA methylation according to the B1 repetitive elements. Biochem Biophys Res Commun. 2005;335:1211–6 [DOI] [PubMed] [Google Scholar]
- 18.Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–8 [DOI] [PubMed] [Google Scholar]
- 19.Garg S, Vitvitsky V, Gendelman HE, Banerjee R. Monocyte differentiation, activation, and mycobacterial killing are linked to transsulfuration-dependent redox metabolism. J Biol Chem. 2006;281:38712–20 [DOI] [PubMed] [Google Scholar]
- 20.Mo R, Chen J, Han Y, Bueno-Cannizares C, Misek DE, Lescure PA, Hanash S, Yung RL. T cell chemokine receptor expression in aging. J Immunol. 2003;170:895–904 [DOI] [PubMed] [Google Scholar]
- 21.Yung RL. Julius A. Epigenetics, aging, and autoimmunity. Autoimmunity. 2008;41:329–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grolleau-Julius A, Ray D, Yung RL. The role of epigenetics in aging and autoimmunity. Clin Rev Allergy Immunol. 2010;39:42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker DJP, Osmond C. Infant mortality, childhood nutrition and ischemic heart disease in England and Wales. Lancet. 1986;1:1077–81 [DOI] [PubMed] [Google Scholar]
- 24.Ikenasio-Thorpe BA, Breier BH, Vickers MH, Fraser M. Prenatal influences on susceptibility to diet-induced obesity are mediated by altered neuroendocrine gene expression. J Endocrinol. 2007;193:31–7 [DOI] [PubMed] [Google Scholar]
- 25.Waterland RA, Garza C. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am J Clin Nutr. 1999;69:179–97 [DOI] [PubMed] [Google Scholar]
- 26.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–6 [DOI] [PubMed] [Google Scholar]
- 27.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, Bailey N, Potts EN, Whitehead G, Brass DM, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20:1869–72 [DOI] [PubMed] [Google Scholar]
- 30.Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.