An adaptor protein complex, AP-2, is involved in the endocytosis of β-secretase (BACE1) via the clathrin-dependent machinery. Endosomal targeting of either the amyloid precursor protein (APP) and/or BACE1 is expendable for the amyloidogenic processing of APP.
Abstract
The β-site amyloid precursor protein (APP)–cleaving enzyme 1 (BACE1) is a transmembrane aspartyl protease that catalyzes the proteolytic processing of APP and other plasma membrane protein precursors. BACE1 cycles between the trans-Golgi network (TGN), the plasma membrane, and endosomes by virtue of signals contained within its cytosolic C-terminal domain. One of these signals is the DXXLL-motif sequence DISLL, which controls transport between the TGN and endosomes via interaction with GGA proteins. Here we show that the DISLL sequence is embedded within a longer [DE]XXXL[LI]-motif sequence, DDISLL, which mediates internalization from the plasma membrane by interaction with the clathrin-associated, heterotetrameric adaptor protein 2 (AP-2) complex. Mutation of this signal or knockdown of either AP-2 or clathrin decreases endosomal localization and increases plasma membrane localization of BACE1. Remarkably, internalization-defective BACE1 is able to cleave an APP mutant that itself cannot be delivered to endosomes. The drug brefeldin A reversibly prevents BACE1-catalyzed APP cleavage, ruling out that this reaction occurs in the endoplasmic reticulum (ER) or ER–Golgi intermediate compartment. Taken together, these observations support the notion that BACE1 is capable of cleaving APP in late compartments of the secretory pathway.
INTRODUCTION
Alzheimer's disease (AD) is a neurodegenerative disorder characterized by accumulation of intraneuronal fibrillary tangles and extracellular amyloid plaques in the brain (Hardy and Selkoe, 2002). The main constituent of amyloid plaques is an overlapping set of peptides known as amyloid-β (Aβ), which are generated by sequential cleavage of a type I transmembrane protein, the amyloid precursor protein (APP), by transmembrane β-secretase and γ-secretase enzymes. Increased Aβ levels have been linked to both familial and sporadic forms of AD. The rate-limiting enzyme for the production of Aβ in neurons is the β-secretase, β-site APP cleaving enzyme 1 (BACE1; Rossner et al., 2001). The levels of BACE1 increase with age (Fukumoto et al., 2004) and are particularly elevated in the brain cortex of patients with AD (Fukumoto et al., 2002, 2004; Holsinger et al., 2002; Yang et al., 2003), making BACE1 a prime target for therapeutic intervention (Vassar et al., 2009; Willem et al., 2009; Evin et al., 2010).
BACE1 is an aspartyl protease having two signature D(T/S)G(T/S) sequences (i.e., DTGS and DSGT) within its ectodomain (Figure 1A), which catalyzes the cleavage of several transmembrane substrates, including APP (Hunt and Turner, 2009; Vassar et al., 2009). BACE1 cleaves APP at two alternative sites, β and β′, in the APP ectodomain, generating transmembrane C-terminal fragments (CTFs) named CTFβ (C99) and CTFβ′ (C89), respectively (see later discussion of Figure 7A). Subsequent cleavage of C99 and C89 by γ-secretase produces Aβ 1-40/42 and Aβ 11-40/42 peptides, respectively (see Figure 7A later in the paper; Vassar et al., 1999; Liu et al., 2002). These peptides are secreted into the extracellular space, where they form oligomers, insoluble fibrils, and amyloid plaques if they are insufficiently cleared (Selkoe, 2002; Mawuenyega et al., 2010). Although critical to the pathogenesis of AD, cleavage by BACE1 is a minor pathway for the processing of APP. Under normal conditions, the majority of APP is cleaved by α-secretases, giving rise to CTFα (C83) (see Figure 7A later in the paper). Cleavage of C83 by γ-secretase generates a peptide known as p3, which neither forms amyloid plaques nor is pathogenic (Haass et al., 1993; Nunan and Small, 2000). The likelihood of progression to AD is thus determined by the proportion of APP that is processed by the pathways initiated by BACE1 (i.e., amyloidogenic) and α-secretases (i.e., nonamyloidogenic), which in turn depends on the intracellular itinerary of APP and the secretases (Hunt and Turner, 2009; Marks and Berg, 2010; Zhi et al., 2011).
FIGURE 1:
Analysis of the interaction of the BACE1 cytosolic tail with the AP-2 α-σ2 hemicomplex. (A) Schematic representation of BACE1, indicating its topological domains. CT, cytosolic tail; PP, propeptide; SP, signal peptide; TM, transmembrane domain. Also indicated are amino acid numbers, DTGS (residues 93–96), and DSGT (residues 289–292) that are necessary for β-secretase activity, and sequence of the C-terminal portion of the CT (residues 477–501), including overlapping DISLL (residues 496–500) and DDISLL signals (residues 495–500) that bind to GGA and AP-2, respectively. (B) Y3H interaction of the BACE1 cytosolic tail, HIV-1 Nef, and mouse tyrosinase (Tyr) tail with the AP-2 α-σ2 hemicomplex (Chaudhuri et al., 2007; Mattera et al., 2011). (C) Y3H analysis of the interaction of the BACE1 tail (left) and HIV-1 Nef (right) with the AP-1 γ-σ1A, AP-2 α-σ2, and AP-3 δ-σ3A hemicomplexes. (D) Y3H analysis of the interaction of BACE1 tail mutants with the AP-2 α-σ2 hemicomplex. The SV40 large T antigen (T-Ag) and p53 were used as controls. Growth in the absence of histidine (–His) is indicative of interactions.
FIGURE 7:
BACE1-catalyzed APP cleavage is independent of BACE1 localization to endosomes. (A) Schematic representation of APP-EFGP, indicating the position of the Aβ peptide, N- and C-terminal fragments (NTF and CTF, respectively), transmembrane domain (TM), the α-, β-, β′- and γ-secretase, caspase-cleavage sites and the resulting fragments, and mutations or inhibitors that block cleavage. Underlined is the region in APP recognized by the 6E10 antibody used in this study to identify C99; all other CTFs and APP-FL were detected using anti–GFP-HRP antibody. The γ-secretase inhibitor DAPT was added to the media posttransfection for experiments in B–F. (B) Cleavage of an APP construct with mutations in the α-secretase and caspase-cleavage sites (APP-F/P-D/A) was analyzed in cells cotransfected with constructs encoding wild-type BACE1, an inactive form of BACE1 (D289N), and wild-type BACE1 in the presence of β-secretase inhibitor (β-INH) or without BACE1 (–). Cell extracts were analyzed by SDS–PAGE and immunoblotting for α-, β-, and β′-secretase cleavage products. C89 was the prominent band detected upon expression of wild-type BACE1 but not under the other conditions. (C) H4 cells were treated with siRNA to AP-2 α or mock treated, and 72 h after RNAi cells were transfected with a wild-type BACE1 construct. Analysis was performed as in B. (D) H4 human neuroglioma cells were transiently cotransfected with APP-F/P-D/A and with BACE1-wt or endocytosis-defective BACE1 mutants or in the presence of BACE1 mutant deficient in β-cleavage constructs, as indicated. A control was carried out in parallel in the absence of BACE1 overexpression as shown. Analysis was performed as in B, using antibodies to detect APP-FL and C89 fragment as mentioned, and BACE1 was detected using anti–HA-HRP antibody. (E) Densitometric quantification of bands was performed on three independent experiments such as that shown in D. The mean ± SD of the ratio of C89 fragment to full-length APP was calculated. (F) Rat cortical neurons were cotransfected with APP-F/P-D/A and BACE1-wt and BACE1 mutant constructs at DIV-4. Controls involving coexpression of APP-F/P-D/A with inactive BACE1 D289N mutant or wild-type BACE1 in the presence of β-secretase inhibitor were included in the experiment. Analysis was performed on DIV-7 as described in D. (G) Quantification from three independent experiments was performed as in E and represented as mean ± SD.
There is conflicting evidence in the literature regarding the intracellular compartment(s) where the amyloidogenic processing of APP takes place. Whereas some studies favor the notion that Aβ is generated in the endosomal–lysosomal system (Haass et al., 1992; Koo and Squazzo, 1994; Perez et al., 1999; He et al., 2005; Wahle et al., 2005; Tesco et al., 2007), other studies indicate that Aβ generation occurs in the endoplasmic reticulum/intermediate compartment (ER/IC; Hartmann et al., 1997), the Golgi complex/trans-Golgi network (TGN; Thinakaran et al., 1996; Xu et al., 1997; Xia et al., 2000), or the plasma membrane (Chyung and Selkoe, 2003). At steady state, the majority of BACE1 localizes to endosomes and the TGN, reflecting its cycling between these compartments (Vassar et al., 1999; Huse et al., 2000; He et al., 2005). The lumen of both endosomes and the TGN is moderately acidic (pH 5–6; Yamashiro and Maxfield, 1984; Demaurex et al., 1998), providing an appropriate, albeit not optimal, environment for the enzymatic activity of BACE1, which has a pH optimum of ∼4.5 (Fukumoto et al., 2002). The intracellular localization and trafficking of BACE1 is controlled by the sequence DISLL (residues 496–500), which fits the consensus motif DXXLL (X is any amino acid; Bonifacino and Traub, 2003), in the cytosolic tail of the protein (Figure 1A). This sequence controls BACE1 recycling between the TGN and endosomes (Walter et al., 2001; Huse et al., 2002; Pastorino et al., 2002; Shiba et al., 2002; He et al., 2005; Tesco et al., 2007) through recognition by the Golgi-associated, γ-adaptin–homologous, ADP-ribosylation factor–interacting (GGA) proteins (Bonifacino, 2004). The dileucine (L499 and L500) that is part of the DXXLL motif also mediates BACE1 internalization from the plasma membrane (Huse et al., 2000; Pastorino et al., 2002), but the mechanism by which it participates in internalization and the machinery involved remains to be elucidated.
In this study, we show that the DISLL sequence is part of a longer sequence, DDISLL (residues 495–500), which fits another consensus motif, [DE]XXXL[LI], for endocytic and lysosomal-, melanosomal-, and basolateral-targeting signals (Bonifacino and Traub, 2003). We demonstrate that the DDISLL sequence mediates internalization of BACE1 by virtue of an interaction with the clathrin-associated, heterotetrameric adaptor protein-2 (AP-2) complex. The DDISLL sequence is therefore a dual-function motif encompassing both DXXLL and [DE]XXXL[LI] signals that alternatively interact with the GGAs and AP-2, respectively. Mutation of the [DE]XXXL[LI] signal or knockdown (KD) of AP-2 or clathrin reduces internalization of BACE1. Remarkably, coexpression of BACE1 and APP mutants that are unable to reach endosomes from either the plasma membrane or the TGN does not prevent BACE1-catalyzed APP cleavage, supporting the notion that this cleavage can occur along the late secretory pathway, en route to the plasma membrane.
RESULTS
BACE1 interacts with the α-σ2 hemicomplex of AP-2
Inspection of the BACE1 tail sequence revealed that the GGA-interacting DISLL sequence (Bonifacino, 2004) is part of a longer DDISLL sequence that fits the [DE]XXXL[LI] motif for dileucine-based sorting signals (Figure 1A). In general, [DE]XXXL[LI]-type signals are recognized by the heterotetrameric, clathrin-associated adaptor protein complexes AP-1 (γ-β1-μ1-σ1), AP-2 (α-β2-μ2-σ2), and AP-3 (δ-β3-μ3-σ3) (subunit composition shown in parentheses; Bonifacino and Traub, 2003). The binding site for [DE]XXXL[LI] signals is found on the corresponding γ-σ1, α-σ2, and δ-σ3 hemicomplexes (Bonifacino and Traub, 2003; Janvier et al., 2003; Chaudhuri et al., 2007; Kelly et al., 2008; Mattera et al., 2011). To analyze whether the role of the dileucine sequence (L499 and L500) in BACE1 internalization (Huse et al., 2000; Pastorino et al., 2002) is due to its ability to interact with the plasma membrane–localized AP-2 complex, we tested for binding of the BACE1 tail to the α-σ2 hemicomplex using a yeast three-hybrid (Y3H) system (Mattera et al., 2011). We found that the BACE1 tail indeed interacted with α-σ2, albeit more weakly than other proteins having [DE]XXXL[LI] signals such as HIV-1 Nef and the tyrosinase tail (Figure 1B; Chaudhuri et al., 2007; Mattera et al., 2011). This interaction was specific to AP-2, as we could not detect interaction of the BACE1 tail with the homologous γ-σ1 and δ-σ3 hemicomplexes (Figure 1C). This was in contrast to HIV-1 Nef, which interacted with all three hemicomplexes (Figure 1C), as previously described (Chaudhuri et al., 2007; Lindwasser et al., 2007; Mattera et al., 2011). In addition, we found that the interaction of the BACE1 tail with α-σ2 was dependent on the dileucine and, to a lesser extent, the first aspartate (D495) residues of the DDISLL sequence (Figure 1D). The serine residue (S498) within this sequence can undergo phosphorylation, affecting its interaction with the VHS domain of GGA proteins and the intracellular localization of BACE1 (Walter et al., 2001; Shiba et al., 2004a). However, we observed that mutation of S498 to alanine had no effect on the interaction of the BACE1 tail with the α-σ2 hemicomplex (Figure 1D). These experiments thus demonstrated that the DDISLL sequence from BACE1 behaves as a [DE]XXXL[LI] signal that specifically interacts with AP-2.
Involvement of AP-2 in the transport of BACE1 to endosomal compartments
We conducted functional analyses to examine the role of the DDISLL sequence and AP-2 in the endosomal targeting of BACE1. Immunofluorescence microscopy of human neuroglioma H4 cells transfected with a construct encoding wild-type (wt) BACE1 tagged with the hemagglutinin (HA) epitope (BACE1-wt) showed predominant staining of punctate cytoplasmic structures (Figure 2, A and C), which corresponded to endosomes and lysosomes, as shown by colocalization with transferrin receptor–positive sorting/recycling endosomes, Rab7-positive maturing endosomes, and Lamp-1–positive late endosomes and lysosomes (Supplemental Figure S1). Mutation of the dileucine (L499 and L500) to two alanine residues decreased endosomal localization and caused accumulation of BACE1 at the plasma membrane and in the juxtanuclear area of the cytoplasm (Figure 2, A and C, and Supplemental Figure S2), as previously reported (Pastorino et al., 2002). Mutation of D495 (part of the AP-2–binding motif but not the GGA-binding motif) to arginine had a similar effect (Figure 2, A and C). In contrast, mutation of S498 to alanine had little or no effect on the distribution of BACE1 (Figure 2, A and C), indicating that phosphorylation of this residue is unlikely to determine the steady-state localization of the protein. Similar results were obtained using rat hippocampal neurons (Figure 2, B and D). These experiments thus demonstrated that the DDISLL sequence indeed behaves as a canonical [DE]XXXL[LI]-type signal that mediates endosomal localization of BACE1.
FIGURE 2:
Requirement of the [DE]XXXL[LI] signal for BACE1 localization to endosomes. (A) H4 human neuroglioma cells were transiently transfected with BACE1-wt and BACE1 mutant constructs for 12 h as indicated. Cells were fixed with 4% paraformaldehyde and processed as described (Burgos et al., 2010). Localization was determined by immunostaining using 1:100 dilution of monoclonal antibody (mAb) to HA, followed by incubation with a 1:1000 dilution of Alexa Fluor 594–labeled anti–mouse immunoglobulin G (IgG) antibody. Coverslips were mounted using Fluoromount-G and examined with an Olympus FluoView FV1000 laser scanning confocal unit attached to an Olympus IX81 motorized inverted microscope. (B) Rat hippocampal neurons were transfected with the same constructs on DIV-4. Cells were fixed with 4% paraformaldehyde and processed on DIV-7 as described in A. (C) Quantification of images from A was performed and scored as cytoplasmic puncta, plasma membrane, or intermediate phenotype (both plasma membrane and cytoplasmic puncta) and plotted as percentage of cells showing the respective phenotype. Values are the mean ± SD from six independent experiments. (D) Quantification of images from B was performed as in C. Values represent the mean ± SD from three independent experiments. ***p < 0.001. Bars, 10 μm.
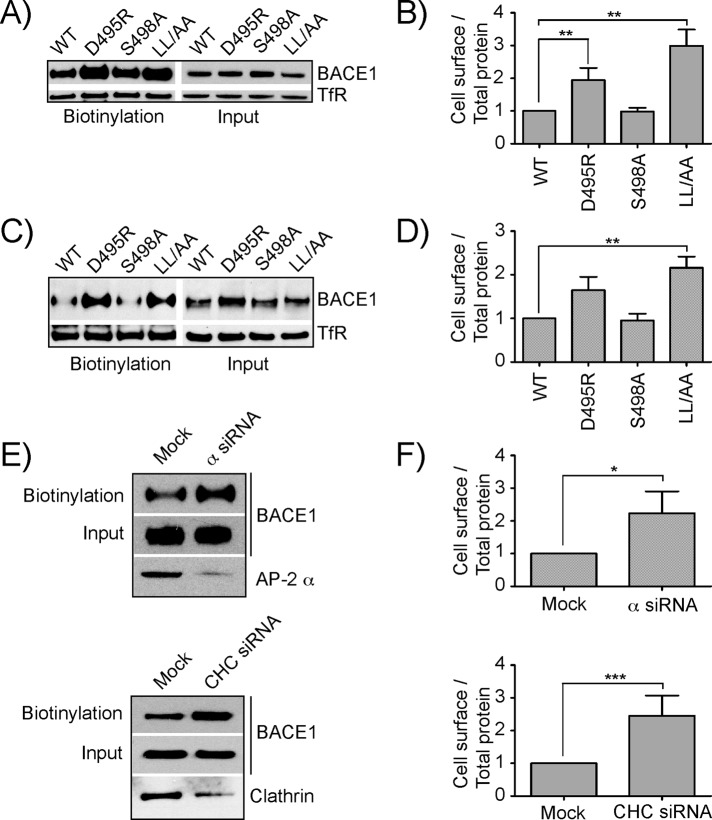
Treatment of H4 cells stably expressing wild-type BACE1 with small interfering RNAs (siRNAs) targeting the α subunit of AP-2 (Figures 3, A and B, and 4) or clathrin heavy chain (CHC; Figure 3, C and D) caused partial redistribution of BACE1 from endosomes to the plasma membrane, as observed by immunofluorescence staining of fixed-permeabilized cells (Supplemental Figure S3). In contrast, siRNA-mediated KD of the γ subunit of AP-1 had no effect on BACE1 distribution (Supplemental Figure S4). Therefore the [DE]XXXL[LI]-mediated targeting of BACE1 to endosomes is dependent on the AP-2 complex and its binding partner, clathrin. Staining for BACE1 on the surface of nonpermeabilized cells revealed localization of this protein to puncta that also contained the transferrin receptor (TfR; Figure 4), characteristic of endocytic clathrin-coated pits (Hansen et al., 1992; Liu et al., 2010). KD of AP-2 α resulted in higher and more diffuse staining for both BACE1 and TfR at the cell surface (Figure 4). These findings indicated that AP-2 is required for colocalization of BACE1 and TfR to endocytic clathrin-coated pits at the plasma membrane.
FIGURE 3:
Redistribution of BACE1 in H4 human neuroglioma cells upon siRNA-mediated knockdown of AP-2 or clathrin. Increased localization of BACE1 to the plasma membrane upon AP-2 α (A) or clathrin heavy chain (CHC) RNAi (C). Left, BACE1 localization, right, AP-2 α or CHC staining. BACE1 was immunostained with pAb to BACE1, whereas localization of clathrin and AP-2 were determined by staining with mAB to CHC (Thermo Scientific) and α-adaptin (Affinity Bioreagents), respectively. Bar, 10 μm. (B, D) Quantification of the phenotype carried out for images in A and C, respectively, by scoring for staining in various compartments, such as cytoplasmic puncta, plasma membrane, and intermediate (both plasma membrane and cytoplasmic puncta). Values are the mean ± SD from three experiments. ***p< 0.001.
FIGURE 4:
Colocalization of BACE1 and TfR upon depletion of AP-2. Mock or AP-2 α siRNA-treated HeLa cells were transiently transfected with a plasmid encoding BACE1-wt. After 12 h of transfection, cells were incubated in culture medium deprived of serum for 1 h at 37°C to deplete the cells of endogenous transferrin. A subset of cells was the incubated either with 40 μg/ml Alexa Fluor 594–labeled human TfR or 1:100 dilution of mouse anti-HA antibody in culture medium without serum for 30 min at 4°C to stain cell surface BACE1-wt and transferrin receptor. Another subset of cells was incubated in culture medium without serum for 30 min at 37°C to allow labeled transferrin to reach a steady state. After treatment, cells were fixed with 4% paraformaldehyde and processed as in Figure 2A. Only cells treated at 37°C were further permeabilized and incubated with a 1:500 dilution of mouse anti-HA antibody for 1 h at 37°C. Both subsets of cells were incubated with a 1:1000 dilution of Alexa Fluor 488–labeled anti-mouse IgG antibody (Invitrogen) for 1 h at 37°C. Notice an almost complete colocalization between BACE1-wt and labeled transferrin at clathrin-coated pits at the cell surface in the presence of AP-2 (mock cells) and the disappearance of the punctate pattern in the AP-2 KD cells. Bar, 10 μm.
Cell surface expression and internalization of BACE1 mutants
The increased surface expression of BACE1 mutants defective in AP-2 binding was corroborated biochemically using a surface biotinylation assay. In both H4 cells (Figure 5, A and B) and cortical neurons (Figure 5, C and D), mutation of the LL or D495 residues of the signal resulted in a twofold-to-threefold increase in the levels of BACE1 at the cell surface. Using the same assay, we demonstrated that siRNA-mediated KD of AP-2 α or CHC (Figure 5, E and F), but not AP-1 γ, AP-3 δ, or AP-4 ε (unpublished data), in H4 cells increased surface BACE1 levels by approximately threefold. These effects were similar to those on the TfR (unpublished data), a prototypic endocytic receptor that is internalized from the cell surface in an AP-2–dependent manner (Motley et al., 2003; Boucrot et al., 2010). These results confirmed that levels of BACE1 at the cell surface are controlled by the [DE]XXXL[LI] signal and the AP-2 endocytic adaptor.
FIGURE 5:
Cell surface expression and internalization of wild-type and mutant BACE1 constructs. (A) H4 human neuroglioma cells transiently transfected either with BACE1-wt or BACE1 mutant constructs were surface labeled with Sulfo-NHS-LC-Biotin at 4°C. After lysis, the extracts were incubated with NeutrAvidin-agarose, and bound proteins were analyzed by SDS–PAGE and immunoblotting using antibodies to an extracellular HA epitope tag and the TfR. (B) Densitometric quantification of bands was performed on three independent experiments as shown in A. Values are the mean ± SD from three independent experiments after normalization to wt in each experiment. (C) Rat cortical neurons were transfected with wild-type and mutant BACE1 constructs on DIV-4 and subjected to surface labeling by biotinylation and isolation of biotinylated proteins on DIV-7. Analysis was performed as described in A. (D) Quantification for three independent experiments as in B. (E) H4 human neuroglioma cells transiently transfected with wild-type BACE1 construct were subjected to KD of either the large subunit of AP-2 (α siRNA) or clathrin heavy chain (CHC siRNA). Bottom, KD efficiency, using specific antibodies to the α-adaptin subunit for AP-2 complex and CHC for clathrin. Cell surface-biotinylation against total protein was calculated and processed as described in A. (F) Quantification for three independent experiments as in B. *p < 0.05, **p < 0.01, ***p < 0.001.
We next examined whether alterations in the intracellular distribution and surface expression of BACE1 were due to changes in endocytosis. This was performed using a protection assay that consisted of labeling proteins at the cell surface with biotin, chasing for different times at 37°C, and cleaving the biotin at the cell surface with glutathione, thus determining the amount of endocytosed biotinylated proteins. As shown in Figure 6A, a substantial fraction of BACE1 was protected from glutathione cleavage after 30 and 60 min of internalization. Mutation of LL or D495 greatly decreased the amount of protected BACE1 at these times, whereas mutation of S498 had no effect (Figure 6A). An antibody uptake assay also showed that mutation of the LL or D495 residues (Figure 6, B and C) or KD of AP-2 α or CHC (Figure 6D) inhibited internalization of BACE1.
FIGURE 6:
Internalization of wild-type and mutant BACE1. (A) H4 human neuroglioma cells transiently transfected with BACE1-wt or mutant constructs were surface labeled with Sulfo-NHS-LC-Biotin at 4°C and chased for 0, 30, and 60 min at 37°C in complete medium. After the chase period, the remaining surface label was specifically cleaved with non–cell-permeable glutathione. Isolation of biotinylated protein and analysis was performed as described in the legend to Figure 4. (B) H4 cells transiently transfected with wild-type and mutant BACE1 constructs were surface labeled at 4°C with polyclonal antibodies to the extracellular HA epitope tag, washed, and then chased for 20 min in complete medium before fixation. Internalization was analyzed by indirect immunofluorescence using Alexa 594–conjugated secondary antibody. (C) Images from B were quantified for localization of internalized BACE1 to cytoplasmic puncta (endosomal), plasma membrane, or intermediate phenotype (both plasma membrane and cytoplasmic puncta) and plotted as percentage of cells. Values are the mean ± SD from three different experiments. ***p < 0.001. (D) Antibody uptake assay carried out upon knockdown on AP-2 α and CHC as described in B. Bars, 10 μm.
Taken together, these experiments indicated that interaction of the [DE]XXXL[LI] signal with AP-2 mediates internalization of BACE1, contributing to the steady-state localization of BACE1 to endosomes.
BACE1-catalyzed cleavage of APP is independent of BACE1 localization to endosomes
The ability to manipulate the endosomal localization of BACE1 by mutations in its cytosolic tail allowed us to investigate the importance of this localization for BACE1-catalyzed APP cleavage. BACE1 cleaves full-length APP at two sites (β and β′) to generate the transmembrane CTFs C99 and C89 (Figure 7A; Sinha et al., 1999; Vassar et al., 1999; Yan et al., 1999; Creemers et al., 2001; Liu et al., 2002). Alternatively, full-length APP, as well as C99, can be cleaved by α-secretase to yield C83 (Figure 7A). Further cleavage of C99 and C83 by γ-secretase yields the secreted Aβ and p3 peptides, respectively, in addition to the cytosolic AICD fragment (Figure 7A; Hunt and Turner, 2009; Burgos et al., 2010). Furthermore, caspases can cleave the cytosolic tail of the CTFs to yield C31 (Figure 7A), a reaction that is enhanced by inhibition of γ-secretase with the compound N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester (DAPT; Burgos et al., 2010). Because of the complexity of APP processing, it is difficult to assess BACE1-catalyzed cleavage in isolation from the other reactions. To facilitate analysis of BACE1 activity, we generated an APP construct (APP-F/P-D/A) with mutations that inhibit cleavage by both α-secretase (F615P; Haass et al., 1994; Jäger et al., 2009) and caspases (D664A; Lu et al., 2000; Supplemental Figure S5). The APP-F/P-D/A mutant was expressed in combination with or without BACE1 constructs, and cleavage products were analyzed by immunoblotting with appropriate antibodies (Figure 7B).
We observed that expression of APP-F/P-D/A alone or in the presence of DAPT resulted in the appearance of two full-length species corresponding to the Golgi-processed (upper band) and ER (lower band) forms of APP (Figure 7B). In addition, we observed the generation of C99 and C83, products of endogenous β- and α-secretase cleavage, respectively (Figure 7B). The presence of C83 indicated that the inhibition of α-secretase by the F615P mutation was incomplete, a phenomenon that was enhanced by mutation of the caspase cleavage site (Supplemental Figure S5). Coexpression with BACE1 resulted in complete loss of the Golgi form, but not the ER form, of the full-length APP mutant, pointing to a post-ER location of the β-secretase cleavage (Figure 7B). Of interest, the most prominent cleavage product under these conditions was C89 rather than C99 (Figure 7B), probably due to the high levels of β-secretase activity resulting from transgenic BACE1 expression. Loss of the Golgi form of full-length APP and appearance of C89 were prevented by a mutation in the BACE1 catalytic site (D289N; Schmechel et al., 2004) or by incubation with BACE1 inhibitor IV (Stachel et al., 2004; Figure 7B), corroborating that changes were due to BACE1 catalytic activity.
We used this assay to examine the generation of C89 upon AP-2 α-adaptin KD (Figure 7C) or mutation of the [DE]XXXL[LI] signal in BACE1 (Figure 7, D and E). We observed that α-adaptin KD did not prevent the generation of C99 by endogenous β-secretase or C89 by transgenic BACE1 (Figure 7C). Moreover, mutation of D495 or L499-L500 in the [DE]XXXL[LI] signal had no effect on C89 production after correction for the different expression levels of the APP constructs in both H4 cells (Figure 7, D and E) and cortical neurons (Figure 7, F and G). From these experiments, we concluded that delivery of BACE1 to endosomes by virtue of interactions between the [DE]XXXL[LI] signal and AP-2 is dispensable for the BACE1-catalyzed APP cleavage.
BACE1-catalyzed cleavage occurs even when both BACE1 and APP are incapable of reaching endosomes
The results shown in Figure 7 indicated that transport of BACE1 to endosomes is not required for BACE1-catalyzed APP cleavage. To test whether transport of APP to endosomes is required for BACE1-catalyzed APP cleavage, we generated an APP-F615P/D664A construct with additional mutations in all three tyrosine residues (Y653, Y682, and Y687) in the cytosolic tail (APP-3Y-F615P/D664A). These mutations are expected to prevent delivery of APP from the plasma membrane (Ono et al., 1997; Zheng et al., 1998; Tarr et al., 2002; Schettini et al., 2010), as well as AP-4–mediated transport of APP from the TGN to endosome (Burgos et al., 2010). Indeed, immunofluorescence microscopy showed that this construct localized to the ER, TGN, and plasma membrane, in contrast to the APP-F615P/D664A construct, which predominantly localized to endosomes (Figure 8, A and B). Surface biotinylation showed increased levels of APP-3Y-F615P/D664A relative to APP-F615P/D664A at the cell surface, consistent with its inability to internalize (Figure 8C). The APP-3Y-F615P/D664A construct was next coexpressed with BACE1-wt and mutants of the [DE]XXXL[LI] signal that cannot be internalized, and BACE1 activity was assessed by generation of C89 in cortical neurons incubated with DAPT to prevent cleavage by γ-secretase. We observed that similar amounts of C89 were produced by coexpression with internalization-competent and internalization-defective BACE1 mutants (Figure 8D). These results indicated that BACE1-catalyzed cleavage of APP occurs even when both BACE1 and APP cannot be delivered to endosomes. Thus, cleavage is likely to occur along the secretory pathway, before delivery of APP and BACE1 to endosomes.
FIGURE 8:
APP is cleaved by BACE1 independent of endosomal localization. (A) H4 human neuroglioma cells were transfected with constructs encoding APP-F/P-D/A or APP-F/P-D/A with additional mutation of all three tyrosine residues in the cytosolic tail (APP 3Y-F/P-D/A), both expressing C-terminal GFP. Cells were stained for TGN46 and examined by fluorescence microscopy. Merging red and green channels generated the third picture on each row; yellow indicates overlapping localization. Insets show 3× magnifications. (B) Immunofluorescence images from A were quantified for Golgi localization. Values are the mean ± SD from three different experiments. ***p < 0.001. (C) Cells were biotinylated on the cell surface as described in Figure 5A and proteins analyzed by immunoblotting with an anti–GFP-HRP antibody. (D) Rat cortical neurons were cotransfected with APP 3Y-F/P-D/A and wild-type or mutant BACE1 constructs on DIV-4 and analyzed on DIV-7 as described in Figure 7E. Bar, 10 μm.
Cleavage of APP by BACE1 occurs in post-ER compartments
To determine whether BACE1-catalyzed APP cleavage occurs in the early or late secretory pathway, we used the drug brefeldin A (BFA). This drug redistributes proteins from the Golgi complex to the ER and interrupts export of newly synthesized proteins from the ER (Lippincott-Schwartz et al., 1989). BFA treatment of cells coexpressing APP-F615P/D664A with wild-type and BACE1 mutants in the presence of DAPT completely abrogated generation of C89 (Figure 9A). This treatment also abolished C99 generation by endogenous β-secretase (unpublished data). Washout of BFA restored C89 generation in cells coexpressing APP-F615P/D664A and BACE1-wt (Figure 9B). These experiments thus demonstrated that BACE1-catalyzed APP cleavage requires export from the ER.
FIGURE 9:

Cleavage of APP by BACE1 occurs in post-ER compartments. (A) H4 cells were transiently cotransfected with constructs encoding APP-F/P-D/A and BACE1-wt or mutants. Cells were incubated with DAPT in the presence or absence of BFA for 10 h posttransfection as indicated. Total extracts were subjected to SDS–PAGE and analyzed by immunoblotting with specific antibodies. Notice the generation of the C89 fragment by BACE1-wt and BACE1 mutants in the absence of BFA but not in the presence of BFA. (B) H4 cells were cotransfected with APP-F/P-D/A and BACE1-wt construct and maintained for 10 h in the presence of DAPT plus or minus BFA as indicated. BFA was washed out, and cells were further incubated for 0, 5, and 10 h in the presence of DAPT. Total extracts were analyzed by SDS–PAGE and immunoblotting as in A.
DISCUSSION
The amounts of pathogenic Aβ peptide that are generated in the brain depend on not only the levels and activity but also the intracellular itinerary of APP and the α-, β- and γ-secretases. All of these proteins have been proposed to cycle between the TGN, the plasma membrane and endosomes (Zhi et al., 2011), but the mechanisms that regulate their cycling and steady-state levels in each compartment are incompletely defined. Of particular importance to the pathogenesis of AD is the understanding of the trafficking of β-secretase, the key enzyme for Aβ production.
Previous studies identified a DISLL sequence in the cytosolic tail of the β-secretase, BACE1, which, like other DXXLL-type motifs (Bonifacino and Traub, 2003), is recognized by the GGA adaptor proteins (Shiba et al., 2004a; von Arnim et al., 2004; He et al., 2005). Phosphorylation of the serine residue (S498) in this sequence enhances binding of GGA1 to BACE1 (Shiba et al., 2004b; von Arnim et al., 2004). This phosphorylation event was proposed to promote binding of BACE1 to GGA1 at the TGN for recycling of BACE1 to the plasma membrane (von Arnim et al., 2004), as well as in early endosomes for transport to the TGN and/or late endosomes (Walter et al., 2001; Wahle et al., 2005). However, the significance of BACE1 phosphorylation for APP processing remains unclear, as mutation of S498 to alanine or aspartate had no effect on APP cleavage (Walter et al., 2001).
The results of our study show that the DISLL sequence is embedded within a DDISLL sequence that fits the [DE]XXXL[LI] consensus AP-binding motif (Bonifacino and Traub, 2003) and interacts with the α-σ2 hemicomplex of the plasma membrane–localized AP-2 complex (Figure 1, B and C). This interaction is dependent on not only the dileucine (L499 and L500), but also the first aspartate (D495) of the sequence (Figure 1D), and is independent of the phosphorylatable serine (S498), thus distinguishing the requirements for binding to AP-2 from those of the GGAs (Bonifacino and Traub, 2003; Doray et al., 2008). The use of D instead of E at position −4 in the [DE]XXXL[LI] consensus motif likely explains why this sequence binds to AP-2 but not AP-1 (Figure 1C), as previously demonstrated for other signals (Doray et al., 2008). L499, L500, and D495, but not S498, are also required for internalization of BACE1 from the plasma membrane and contribute to the steady-state localization of BACE1 to endosomes (Figures 2 and 6; Huse et al., 2000; Pastorino et al., 2002). Accordingly, RNA interference (RNAi)–mediated depletion of AP-2 reduces BACE1 internalization and endosomal localization (Figures 3, 4, and 6), supporting the physiological significance of the BACE1–AP-2 interaction. Therefore the DDISLL sequence is a dual-specificity signal that can alternatively interact with the GGAs at the TGN and/or endosomes and with AP-2 at the plasma membrane. Two bifunctional GGA/AP-2–interacting signals have also been reported to occur in the cytosolic tail of the lipoprotein receptor–related protein LRP9 (Doray et al., 2008). The proteins LRP3 (Takatsu et al., 2001), consortin (del Castillo et al., 2010), and SorLA (Jacobsen et al., 2002) also have cytosolic tail sequences that fit this dual consensus motif, but it is not known whether they function as bifunctional GGA/AP-2 interactions signals. Together with previous findings (Huse et al., 2000; Kinoshita et al., 2003; He et al., 2005), our observations indicate that BACE1 engages in a short cycling loop between the TGN and endosomes that is regulated by the GGAs and a long cycling loop involving the TGN, the plasma membrane, and endosomes in which AP-2 mediates the internalization step. Thus, BACE1 might be capable of reaching endosomes both from the TGN via the GGAs and from the plasma membrane via AP-2. The fact that the GGAs and AP-2 are clathrin adaptors (Bonifacino and Traub, 2003) is consistent with the observation that depletion of clathrin reduces the endosomal localization of BACE1 (Figure 3, C and D). These results indicate that a substantial pool of BACE1 is delivered to endosomes by AP-2/clathrin–dependent internalization from the plasma membrane.
While this article was in preparation, another report (Sannerud et al., 2011) was published confirming that endosomal targeting of BACE1 is dependent on the LL residues of the DDISLL sequence (Huse et al., 2000; Pastorino et al. 2002) but arguing that BACE1 internalization is independent of clathrin and AP-2 and instead mediated by Arf6. We do not know the reason for the discrepancy with our findings but speculate that it could be due to differences in the methods used for interfering with clathrin and AP-2 function. In our study we ablated clathrin function by siRNA-mediated depletion of the clathrin heavy chain, whereas Sannerud et al. (2011) interfered with clathrin function by overexpression of an AP180c dominant-negative mutant. In addition, we performed siRNA-mediated depletion of α-adaptin, a subunit involved in the recognition of dileucine-based sorting signals (Chaudhuri et al., 2007; Doray et al., 2007; Kelly et al., 2008), whereas Sannerud et al. (2011) performed siRNA-mediated depletion of μ2, a subunit that recognizes tyrosine-based sorting signals (Ohno et al., 1995; Owen and Evans, 1998). Moreover, using siRNAs we routinely get >80% depletion of α-adaptin, in contrast to the 60% depletion of μ2 reported by Sannerud et al. (2011). In any event, although demonstrating a role for clathrin and AP-2 in BACE1 endocytosis, our studies do not rule out the additional occurrence of clathrin-independent endocytosis of BACE1.
APP has been proposed to travel through short and long cycling loops similar to those of BACE1, in the course of which APP becomes proteolytically processed by the secretases. In principle, processing could occur in any or all of the compartments of the cycling pathways, although the predominant site of processing remains controversial (Hunt and Turner, 2009; Zhi et al., 2011). To assess the importance of BACE1 endosomal localization for β-secretase cleavage of APP, we examined the effect of mutating the BACE1 DDISLL sequence (Figure 7, D and F) or depleting AP-2 (Figure 7C) under conditions in which processing by α-secretase, γ-secretase, and caspases was reduced by mutation of the corresponding cleavage sites or use of pharmacological inhibitors. It is worth pointing out that mutation of the LL sequence within DDISLL prevents interactions with both GGAs and AP-2, impairing sorting to endosomes from both the TGN and the plasma membrane, whereas AP-2 depletion only inhibits endocytosis form the plasma membrane. All of these perturbations reduced endosomal localization of BACE1 but had no effect on β-secretase cleavage of APP (Figure 7, D and F). This was also the case for an APP construct (APP-3Y-F615P/D664A) that was incapable of undergoing endosomal targeting because of mutations in three tyrosine residues in its cytosolic tail (Figure 8D). These residues include Y687, which is part of an NPTY signal that mediates internalization (Ono et al., 1997; Zheng et al., 1998; Tarr et al., 2002; Schettini et al., 2010), most likely through interactions with PTB domain–containing adaptors, as well as of an overlapping YKFFE signal that mediates transport from the TGN to endosomes by virtue of an interaction with the AP-4 complex (Burgos et al., 2010). On the basis of these findings, we conclude that BACE1-catalyzed APP cleavage does not require localization of these proteins to endosomes. We also found that arresting protein export from the ER/Golgi complex by treatment with BFA blocked BACE1-catalyzed APP cleavage (Figure 9), as previously shown (Martin et al., 1995). Our results are therefore in line with previous studies that argued for β-secretase cleavage and Aβ generation occurring at the TGN, the plasma membrane, and/or in transit between these compartments (Hartmann et al., 1997; Kinoshita et al., 2003). Although we cannot rule out that biosynthetic transport of BACE1 involves passage through an endosomal compartment en route to the plasma membrane, the ability of BACE1 constructs to cleave the endosomally excluded APP-3Y-F615P/D664A mutant supports the conclusion that endosomal localization is not required for BACE1-mediated APP cleavage. A caveat in our experiments is that transgenic BACE1 constructs were overexpressed relative to endogenous BACE1, with the result that most APP was cleaved to the shorter C89 form. However, depletion of AP-2 also failed to prevent APP cleavage to C99 by endogenous BACE1 (Figure 7C), indicating that the dispensability of AP-2/clathrin–dependent BACE1 endocytosis for APP cleavage is independent of BACE1 expression levels.
BACE1 has an acidic optimum pH (Sinha et al., 1999; Vassar et al., 1999; Yan et al., 1999; Lin et al., 2000), ideally suited for cleavage in late endosomes and lysosomes. However, BACE1 is likely to be partially active at the moderately acidic pH of the TGN and early endosomes (Yamashiro and Maxfield, 1984; Demaurex et al., 1998). Moreover, unlike lysosomal hydrolases with a similar acidic optimum pH, BACE1 is active from the Golgi complex on, independent of the furin-dependent cleavage of its propeptide sequence (Creemers et al., 2001). Indeed, most of the known physiological substrates of BACE1 are plasma membrane proteins that mediate intercellular adhesion or signaling (Lichtenthaler et al., 2003; von Arnim et al., 2005; Hemming et al., 2009). Other BACE1 substrates include Golgi sialyltransferases (Kitazume et al., 2001, 2006) and plasma membrane voltage-gated sodium channels (Wong et al., 2005; Kim et al., 2007). Taken together, our findings indicate that the main role of BACE1 is to cleave substrates in transit to the plasma membrane, consistent with our observations on APP processing.
MATERIALS AND METHODS
Antibodies
We used mouse monoclonal antibodies to the following antigens: AP-1 γ and AP-4 ε (BD Biosciences, San Diego, CA), AP-2 α (BD Biosciences, and Affinity Bioreagents, Golden, CO), AP-3 δ (BD Biosciences, and Developmental Studies Hybridoma Bank, Iowa City, IA), clathrin heavy chain (BD Biosciences, and Thermo Scientific, Rockford, IL), transferrin receptor (Invitrogen, Carlsbad, CA), HA epitope (Covance, Dedham, MA), and APP C99 fragment (6E10; Sigma-Aldrich, St. Louis, MO). We also used rabbit polyclonal antibodies to the HA epitope (Covance) and BACE1 (kind gift from Robert W. Doms, University of Pennsylvania, Philadelphia, PA). Sheep anti-human TGN46 from Serotech (Raleigh, NC) was used. Anti–green fluorescent protein (GFP) and anti-HA antibodies conjugated to horseradish peroxidase (HRP) were from Miltenyi Biotech (Auburn, CA). Fluorophore-conjugated secondary antibodies were obtained from Invitrogen, and HRP-conjugated secondary antibodies were from GE Healthcare (Piscataway, NJ).
Recombinant DNA procedures, site-directed mutagenesis, and yeast three-hybrid assays
For expression in cells, human BACE1 cDNA (GeneCopoeia, Germantown, MD) was cloned in pEGFP-N1 (Clontech, Mountain View, CA). A single HA tag was inserted after the propeptide cleavage site and a stop codon was introduced after residue K501 to stop the expression of GFP using site-directed mutagenesis (Stratagene, La Jolla, CA). This construct was considered as the wild type and used as template to introduce subsequent amino acid substitutions. An HA-tagged APP-EGFP fusion construct was generated by PCR amplification of the sequence encoding HA-APP695 (Burgos et al., 2010) and cloned in-frame into the HindIII and SalI sites of pEGFP-N1. The substitutions F615P, D664Am and 3Y/A (Y653A, Y683Am and Y687A) in the cytosolic tail of APP were introduced using site-directed mutagenesis as described. For Y3H assays, the cytosolic tail of BACE1 was amplified by PCR and cloned into multiple cloning site I of pBRIDGE (Clontech) in fusion with the DNA-binding domain of Gal4, whereas individual σ subunits such as AP-1 σ1, AP-2 σ2, and AP-2 σ3A were cloned into multiple cloning site II of the same vector. Activation domain constructs for AP-2 α, AP-1 γ, and AP-3 δ were generated in pGADT7 vector as previously described (Chaudhuri et al., 2007; Mattera et al., 2011). Amino acid substitutions were introduced by site-directed mutagenesis as described. Y3H assays were performed as previously described (Chaudhuri et al., 2007; Mattera et al., 2011).
Cell culture, transfection, and RNAi
H4 human neuroglioma cells were obtained from the American Type Culture Collection (Manassas, VA). Transfections were carried out using Lipofectamine 2000 (Invitrogen). Cells were analyzed 12–16 h after transfection, unless otherwise stated. Where indicated, cells were treated with 1 μM β-secretase inhibitor IV (EMD Chemicals, Gibbstown, NJ), 250 nM DAPT (Sigma-Aldrich), or 5 μg/ml brefeldin A (Sigma-Aldrich). For RNAi experiments, we used siRNAi GAGCAUGUGCACGCUGGCCA to the coding region of AP-2 αC and UCCAAUUCGAAGACCAAUU to the coding region of clathrin heavy chain. The respective siGENOME SMARTpools were used to knock down AP-1 γ, AP-3 δ, and AP-4 ε (Dharmacon, Lafayette, CO). siRNAs were transfected using Oligofectamine (Invitrogen). Cells were transfected twice at 24-h intervals and analyzed 72 h after the second transfection. Hippocampal and cortical neurons were obtained from Sprague Dawley rats at embryonic day 18 (Kaech and Banker, 2006). Neurons were transfected on in vitro day 4 (DIV-4) using Lipofectamine 2000 and analyzed between DIV-6 and DIV-8 as mentioned in the text.
Antibody uptake assay and immunofluorescence microscopy
H4 cells grown on coverslips were labeled with a 1:100 dilution of rabbit anti-HA polyclonal antibodies in DMEM containing 10% fetal bovine serum at 4°C for 1 h, washed with phosphate-buffered saline (PBS), and chased with complete medium at 37°C for the indicated times. Cell fixation, permeabilization, indirect immunofluorescence staining, and image acquisition were performed as previously described (Rojas et al., 2008; Burgos et al., 2010). All images within one panel were captured with the same settings.
Cell surface biotinylation and endocytosis assays
Cell surface biotinylation was performed at 4°C. Cells were rinsed twice in PBS and exposed to 1 mM Sulfo-NHS-LC-Biotin reagent (Pierce, Rockford, IL) in PBS for 1 h. The cells were subsequently washed, incubated with 50 mM Tris-HCl for 10 min to quench unreacted ester, rinsed twice with PBS, and lysed in cell lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% [vol/vol] Triton X-100, and protease inhibitors [Roche Applied Science, Indianapolis, IN]). Biotinylated proteins were isolated by incubation with NeutrAvidin-agarose (Pierce) in a rotary shaker for 2 h at 4°C, washed twice with cell lysis buffer and once with PBS, and analyzed by SDS–PAGE and immunoblotting with HRP-conjugated antibody to HA. For internalization experiments, cells were biotinylated as described but with 1 mM Sulfo-NHS-SS-Biotin reagent (Pierce). After quenching the unreacted ester, we subjected the cells to chase at 37°C for different time intervals. The cells were treated with non–cell-permeable 50 mM glutathione (Sigma-Aldrich) in cleavage buffer (Huse et al., 2000) for 1 h, followed by treatment with 50 mM iodoacetamide for 30 min, washed twice with PBS, and extracted with cell lysis buffer in the presence of 2.5 mM iodoacetamide. SDS–PAGE and immunoblotting were performed as described (Rojas et al., 2008).
Densitometry and statistical analysis
Immunoblots were scanned and densitometric analysis performed using the Quantity One software (Bio-Rad, Hercules, CA). Intensity values were corrected for background. All statistical analyses were performed using an analysis of variance test provided by Prism5 software (GraphPad, La Jolla, CA).
Supplementary Material
Acknowledgments
We thank X. Zhu and H. Tsai for expert technical assistance, R. W. Doms for kind gift of reagents, R. Chaudhuri for his valuable suggestions in the initial phases of this work, V. C. Padmakumar for help with the figures, and G. Mardones for image quantification and critical discussion of the manuscript. This research was supported by the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, and by Grant 1100027 from the Fondo Nacional de Desarro-llo Científico y Tecnológico (P.V.B.).
Abbreviations used:
- AD
Alzheimer's disease
- APP
amyloid precursor protein BACE1, β-site amyloid precursor protein (APP)-cleaving enzyme 1
- Aβ
amyloid beta
- CTF
C-terminal fragment
- GGA
Golgi-associated, γ-adaptin homologous, ADP-ribosylation factor (ARF)-interacting protein
- KD
knockdown
- Y3H
yeast three-hybrid
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0944) on May 2, 2012.
REFERENCES
- Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Saffarian S, Zhang R, Kirchhausen T. Roles of AP-2 in clathrin-mediated endocytosis. PLoS One. 2010;5:e10597. doi: 10.1371/journal.pone.0010597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos PV, Mardones GA, Rojas AL, daSilva LL, Prabhu Y, Hurley JH, Bonifacino JS. Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex. Dev Cell. 2010;18:425–436. doi: 10.1016/j.devcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyung JH, Selkoe DJ. Inhibition of receptor-mediated endocytosis demonstrates generation of amyloid beta-protein at the cell surface. J Biol Chem. 2003;278:51035–51043. doi: 10.1074/jbc.M304989200. [DOI] [PubMed] [Google Scholar]
- Creemers JW, Ines Dominguez D, Plets E, Serneels L, Taylor NA, Multhaup G, Craessaerts K, Annaert W, De Strooper B. Processing of beta-secretase by furin and other members of the proprotein convertase family. J Biol Chem. 2001;276:4211–4217. doi: 10.1074/jbc.M006947200. [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, Cohen-Salmon M, Charollais A, Caille D, Lampe PD, Chavrier P, Meda P, Petit C. Consortin, a trans-Golgi network cargo receptor for the plasma membrane targeting and recycling of connexins. Hum Mol Genet. 2010;19:262–275. doi: 10.1093/hmg/ddp490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N, Furuya W, D'souza S, Bonifacino JS, Grinstein S. Mechanism of acidification of the trans-Golgi network (TGN). In situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J Biol Chem. 1998;273:2044–2051. doi: 10.1074/jbc.273.4.2044. [DOI] [PubMed] [Google Scholar]
- Doray B, Knisely JM, Wartman L, Bu G, Kornfeld S. Identification of acidic dileucine signals in LRP9 that interact with both GGAs and AP-1/AP-2. Traffic. 2008;9:1551–1562. doi: 10.1111/j.1600-0854.2008.00786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Lee I, Knisely J, Bu G, Kornfeld S. The gamma/sigma1 and alpha/sigma2 hemicomplexes harbor the dileucine recognition site. Mol Biol Cell. 2007;18:1887–1896. doi: 10.1091/mbc.E07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evin G, Barakat A, Masters CL. BACE: therapeutic target and potential biomarker for Alzheimer's disease. Int J Biochem Cell Biol. 2010;42:1923–1926. doi: 10.1016/j.biocel.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Beta-secretase activity increases with aging in human, monkey, and mouse brain. Am J Pathol. 2004;164:719–725. doi: 10.1016/s0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Hung AY, Schlossmacher MG, Teplow DB, Selkoe DJ. beta-Amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J Biol Chem. 1993;268:3021–3024. [PubMed] [Google Scholar]
- Haass C, Hung AY, Selkoe DJ, Teplow DB. Mutations associated with a locus for familial Alzheimer's disease result in alternative processing of amyloid beta-protein precursor. J Biol Chem. 1994;269:17741–17748. [PubMed] [Google Scholar]
- Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- Hansen SH, Sandvig K, van Deurs B. Internalization efficiency of the transferrin receptor. Exp Cell Res. 1992;199:19–28. doi: 10.1016/0014-4827(92)90457-j. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hartmann T, et al. Distinct sites of intracellular production for Alzheimer's disease A beta40/42 amyloid peptides. Nat Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Identification of beta-secretase (BACE1) substrates using quantitative proteomics. PLoS One. 2009;4:e8477. doi: 10.1371/journal.pone.0008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- Hunt CE, Turner AJ. Cell biology, regulation and inhibition of beta-secretase (BACE-1) FEBS J. 2009;276:1845–1859. doi: 10.1111/j.1742-4658.2009.06929.x. [DOI] [PubMed] [Google Scholar]
- Huse JT, Liu K, Pijak DS, Carlin D, Lee VM, Doms RW. Beta-secretase processing in the trans-Golgi network preferentially generates truncated amyloid species that accumulate in Alzheimer's disease brain. J Biol Chem. 2002;277:16278–16284. doi: 10.1074/jbc.M111141200. [DOI] [PubMed] [Google Scholar]
- Huse JT, Pijak DS, Leslie GJ, Lee VM, Doms RW. Maturation and endosomal targeting of beta-site amyloid precursor protein-cleaving enzyme. The Alzheimer's disease beta-secretase. J Biol Chem. 2000;275:33729–33737. doi: 10.1074/jbc.M004175200. [DOI] [PubMed] [Google Scholar]
- Jacobsen L, Madsen P, Nielsen MS, Geraerts WP, Gliemann J, Smit AB, Petersen CM. The sorLA cytoplasmic domain interacts with GGA1 and -2 and defines minimum requirements for GGA binding. FEBS Lett. 2002;511:155–158. doi: 10.1016/s0014-5793(01)03299-9. [DOI] [PubMed] [Google Scholar]
- Jäger S, et al. alpha-Secretase mediated conversion of the amyloid precursor protein derived membrane stub C99 to C83 limits Abeta generation. J Neurochem. 2009;111:1369–1382. doi: 10.1111/j.1471-4159.2009.06420.x. [DOI] [PubMed] [Google Scholar]
- Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Venkatesan S, Bonifacino JS. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes. J Cell Biol. 2003;163:1281–1290. doi: 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;5:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Honing S, Owen DJ. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, et al. BACE1 regulates voltage-gated sodium channels and neuronal activity. Nat Cell Biol. 2007;9:755–764. doi: 10.1038/ncb1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- Kitazume S, Tachida Y, Oka R, Nakagawa K, Takashima S, Lee YC, Hashimoto Y. Screening a series of sialyltransferases for possible BACE1 substrates. Glycoconj J. 2006;23:437–441. doi: 10.1007/s10719-006-6671-x. [DOI] [PubMed] [Google Scholar]
- Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y. Alzheimer's beta-secretase, beta-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc Natl Acad Sci USA. 2001;98:13554–13559. doi: 10.1073/pnas.241509198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- Lichtenthaler SF, Dominguez DI, Westmeyer GG, Reiss K, Haass C, Saftig P, De Strooper B, Seed B. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J Biol Chem. 2003;278:48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
- Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser OW, Chaudhuri R, Bonifacino JS. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr Mol Med. 2007;7:171–184. doi: 10.2174/156652407780059177. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AP, Aguet F, Danuser G, Schmid SL. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J Cell Biol. 2010;191:1381–1393. doi: 10.1083/jcb.201008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Doms RW, Lee VM. Glu11 site cleavage and N-terminally truncated A beta production upon BACE overexpression. Biochemistry. 2002;41:3128–3136. doi: 10.1021/bi015800g. [DOI] [PubMed] [Google Scholar]
- Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, Salvesen GS, Koo EH, Bredesen DE. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- Marks N, Berg MJ. BACE and gamma-secretase characterization and their sorting as therapeutic targets to reduce amyloidogenesis. Neurochem Res. 2010;35:181–210. doi: 10.1007/s11064-009-0054-1. [DOI] [PubMed] [Google Scholar]
- Martin BL, Schrader-Fischer G, Busciglio J, Duke M, Paganetti P, Yankner BA. Intracellular accumulation of beta-amyloid in cells expressing the Swedish mutant amyloid precursor protein. J Biol Chem. 1995;270:26727–26730. doi: 10.1074/jbc.270.45.26727. [DOI] [PubMed] [Google Scholar]
- Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J Biol Chem. 2011;286:2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunan J, Small DH. Regulation of APP cleavage by alpha-, beta- and gamma-secretases. FEBS Lett. 2000;483:6–10. doi: 10.1016/s0014-5793(00)02076-7. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Ono Y, Kinouchi T, Sorimachi H, Ishiura S, Suzuki K. Deletion of an endosomal/lysosomal targeting signal promotes the secretion of Alzheimer's disease amyloid precursor protein (APP) J Biochem. 1997;121:585–590. doi: 10.1093/oxfordjournals.jbchem.a021625. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Ikin AF, Nairn AC, Pursnani A, Buxbaum JD. The carboxyl-terminus of BACE contains a sorting signal that regulates BACE trafficking but not the formation of total A(beta) Mol Cell Neurosci. 2002;19:175–185. doi: 10.1006/mcne.2001.1065. [DOI] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Apelt J, Schliebs R, Perez-Polo JR, Bigl V. Neuronal and glial beta-secretase (BACE) protein expression in transgenic Tg2576 mice with amyloid plaque pathology. J Neurosci Res. 2001;64:437–446. doi: 10.1002/jnr.1095. [DOI] [PubMed] [Google Scholar]
- Sannerud R, et al. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc Natl Acad Sci USA. 2011;108:E559–E568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schettini G, Govoni S, Racchi M, Rodriguez G. Phosphorylation of APP-CTF-AICD domains and interaction with adaptor proteins: signal transduction and/or transcriptional role—relevance for Alzheimer pathology. J Neurochem. 2010;115:1299–1308. doi: 10.1111/j.1471-4159.2010.07044.x. [DOI] [PubMed] [Google Scholar]
- Schmechel A, Strauss M, Schlicksupp A, Pipkorn R, Haass C, Bayer TA, Multhaup G. Human BACE forms dimers and colocalizes with APP. J Biol Chem. 2004;279:39710–39717. doi: 10.1074/jbc.M402785200. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Deciphering the genesis and fate of amyloid beta-protein yields novel therapies for Alzheimer disease. J Clin Invest. 2002;110:1375–1381. doi: 10.1172/JCI16783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Kametaka S, Kawasaki M, Shibata M, Waguri S, Uchiyama Y, Wakatsuki S. Insights into the phosphoregulation of beta-secretase sorting signal by the VHS domain of GGA1. Traffic. 2004a;5:437–448. doi: 10.1111/j.1600-0854.2004.00188.x. [DOI] [PubMed] [Google Scholar]
- Shiba T, et al. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature. 2002;415:937–941. doi: 10.1038/415937a. [DOI] [PubMed] [Google Scholar]
- Shiba Y, Katoh Y, Shiba T, Yoshino K, Takatsu H, Kobayashi H, Shin HW, Wakatsuki S, Nakayama K. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J Biol Chem. 2004b;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- Sinha S, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- Stachel SJ, et al. Structure-based design of potent and selective cell-permeable inhibitors of human beta-secretase (BACE-1) J Med Chem. 2004;47:6447–6450. doi: 10.1021/jm049379g. [DOI] [PubMed] [Google Scholar]
- Tarr PE, Roncarati R, Pelicci G, Pelicci PG, D'Adamio L. Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem. 2002;277:16798–16804. doi: 10.1074/jbc.M110286200. [DOI] [PubMed] [Google Scholar]
- Takatsu H, Katoh Y, Shiba Y, Nakayama K. Golgi-localizing, γ-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J Biol Chem. 2001;276:28541–28545. doi: 10.1074/jbc.C100218200. [DOI] [PubMed] [Google Scholar]
- Tesco G, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS. Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the Golgi apparatus. J Biol Chem. 1996;271:9390–9397. doi: 10.1074/jbc.271.16.9390. [DOI] [PubMed] [Google Scholar]
- Vassar R, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim CA, et al. The low density lipoprotein receptor-related protein (LRP) is a novel beta-secretase (BACE1) substrate. J Biol Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- von Arnim CA, Tangredi MM, Peltan ID, Lee BM, Irizarry MC, Kinoshita A, Hyman BT. Demonstration of BACE (beta-secretase) phosphorylation and its interaction with GGA1 in cells by fluorescence-lifetime imaging microscopy. J Cell Sci. 2004;117:5437–5445. doi: 10.1242/jcs.01422. [DOI] [PubMed] [Google Scholar]
- Wahle T, Prager K, Raffler N, Haass C, Famulok M, Walter J. GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol Cell Neurosci. 2005;29:453–461. doi: 10.1016/j.mcn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Walter J, Fluhrer R, Hartung B, Willem M, Kaether C, Capell A, Lammich S, Multhaup G, Haass C. Phosphorylation regulates intracellular trafficking of beta-secretase. J Biol Chem. 2001;276:14634–14641. doi: 10.1074/jbc.M011116200. [DOI] [PubMed] [Google Scholar]
- Willem M, Lammich S, Haass C. Function, regulation and therapeutic properties of beta-secretase (BACE1) Semin Cell Dev Biol. 2009;20:175–182. doi: 10.1016/j.semcdb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Wong HK, Sakurai T, Oyama F, Kaneko K, Wada K, Miyazaki H, Kurosawa M, De Strooper B, Saftig P, Nukina N. Beta subunits of voltage-gated sodium channels are novel substrates of beta-site amyloid precursor protein-cleaving enzyme (BACE1) and gamma-secretase. J Biol Chem. 2005;280:23009–23017. doi: 10.1074/jbc.M414648200. [DOI] [PubMed] [Google Scholar]
- Xia W, Ray WJ, Ostaszewski BL, Rahmati T, Kimberly WT, Wolfe MS, Zhang J, Goate AM, Selkoe DJ. Presenilin complexes with the C-terminal fragments of amyloid precursor protein at the sites of amyloid beta-protein generation. Proc Natl Acad Sci USA. 2000;97:9299–9304. doi: 10.1073/pnas.97.16.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sweeney D, Wang R, Thinakaran G, Lo AC, Sisodia SS, Greengard P, Gandy S. Generation of Alzheimer beta-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc Natl Acad Sci USA. 1997;94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro DJ, Maxfield FR. Acidification of endocytic compartments and the intracellular pathways of ligands and receptors. J Cell Biochem. 1984;26:231–246. doi: 10.1002/jcb.240260404. [DOI] [PubMed] [Google Scholar]
- Yan R, et al. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- Yang LB, et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- Zheng P, Eastman J, Vande PS, Pimplikar SW. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc Natl Acad Sci USA. 1998;95:4745–4750. doi: 10.1073/pnas.95.25.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi P, Chia C, Gleeson PA. Intracellular trafficking of the β-secretase and processing of amyloid precursor protein. IUBMB Life. 2011;63:721–729. doi: 10.1002/iub.512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.