Abstract
The vast majority of proteins are believed to have one specific function. Throughout the course of evolution, however, some proteins have acquired additional functions to meet the demands of a complex cellular milieu. In some cases changes in RNA or protein processing allow the cell to make the most of what is already encoded in the genome to produce slightly different forms. The eukaryotic Elongation Factor 1 (eEF1) complex subunits, however, have acquired such moonlighting functions without alternative forms. Here, we discuss the canonical functions of the components of the eEF1 complex in translation elongation as well as the secondary interactions they have with other cellular factors outside of the translational apparatus. The eEF1 complex itself changes in composition as the complexity of eukaryotic organisms increases. Members of the complex are also subject to phosphorylation, a potential modulator of both canonical and non-canonical functions. Although alternative functions of the eEF1A subunit have been widely reported, recent studies are shedding light on additional functions of the eEF1B subunits. A thorough understanding of these alternate functions of eEF1 is essential for appreciating their biological relevance.
Keywords: translation, elongation, actin, virus, phosphorylation
Introduction
The process of eukaryotic protein synthesis is traditionally defined as occurring in three phases: initiation, elongation and termination. Each phase requires the action of not only the ribosome, mRNA and aminoacylated-tRNAs (aa-tRNA) but also a series of soluble protein factors that facilitate each step. Once translation is initiated at the first codon by the assembled 80S ribosome, it continues onto the elongation phase wherein the peptide chain increases its length cyclically one amino acid at a time. The aa-tRNA delivery step is catalyzed by the eukaryotic Elongation Factor 1 (eEF1) complex. In bacteria two factors, EF-Tu and EF-Ts, are involved in making the appropriate aa-tRNA available to the elongating ribosome. The eukaryotic Elongation Factor 2 (eEF2, the homolog of bacterial EF-G) acts as the translocase, allowing the ribosome to move down the mRNA one codon at a time. Peptide elongation continues until the ribosome reaches the stop codon at which point termination is triggered by the release factors. Aside from their function in translation elongation, components of the eEF1 complex have been implicated in a wide variety of cellular and viral processes. In this review, we present data from a number of laboratories in support of the secondary functions of the eEF1 subunits and consider their biological significance.
THE eEF1 COMPLEX
The search for eukaryotic counterparts involved in the first step of translation elongation has led to the isolation of various multi-protein macromolecular complexes in different species. For the sake of clarity and functional consistency across different kingdoms, we are adopting the modified nomenclature as proposed by Le Sourd et al (Table 1) to describe the various players involved in the first step of translational elongation.1
Table 1.
Components of the eEF1 complex:
| Current Nomenclature | Prior Nomenclature | Canonical Function | |
|---|---|---|---|
| eEF1A | eEF1α | aa-tRNA delivery to the ribosome |
|
| eEF1B | eEF1Bα | eEF1β (yeast and metazoan) eEF1β’(plant) |
GEF for eEF1A |
| eEF1Bβ (plant) | eEF1β (plant) | GEF for eEF1A | |
| eEF1Bγ | eEF1γ | Structural component | |
| eEF1Bδ (metazoan) |
eEF1δ (metazoan) | GEF for eEF1A | |
| Val-RS (metazoan) | Val-RS | Valyl tRNA synthetase | |
The eukaryotic elongation factor 1 (eEF1) complex consists of eEF1A (EF-Tu in bacteria), the Gprotein involved in delivering aa-tRNA to the elongating ribosome and eEF1B (EF-Ts in bacteria), a multiprotein complex that acts as the guanine nucleotide exchange factor (GEF). In animals, Val-RS is found bound to the eEF1 complex and this complex is often called the eEF1H complex. In mammals, multiple aminoacyl-tRNA synthetases have been suggested to interact with eEF1 complex.87
Canonical Function of eEF1 in Translation Elongation
eEF1A in its GTP-bound form binds and delivers aa-tRNAs to the A site of the ribosome.2 Upon formation of a correct codon-anticodon pair, a conformational change in the ribosome leads to GTP hydrolysis and release of eEF1A•GDP.3 Since the spontaneous rate of GDP dissociation from eEF1A is very slow with an equilibrium dissociation constant of 10 × 10−7 M, GDP must be actively exchanged for GTP by the guanine nucleotide exchange factor (GEF) eEF1B in order for eEF1A to participate in another round of elongation4 In the yeast Saccharomyces cerevisiae, one of the simplest eukaryotes, the eEF1B complex is made up of two subunits: eEF1Bα and eEF1Bγ. Early work by Moller and colleagues indicated that the C-terminal domain of eEF1Bα was involved in eEF1A binding and the nucleotide exchange activity.5 This observation was later confirmed by the co-crystal structure of the yeast complex.6 The N-terminal domain of eEF1Bα forms a strong binding interface with the eukaryote specific structural protein eEF1Bγ.7 Unlike eEF1A and eEF1Bα, eEF1Bγ is not essential for viability in yeast.7 It has been shown to bind to the endoplasmic membrane and is thought to have an anchoring role, localizing the eEF1B complex to the endoplasmic reticulum (ER), a major site of protein synthesis.8
In higher eukaryotes, the eEF1B complex takes on a third factor that has its own GEF activity, called eEF1Bβ in plants and eEF1Bδ in metazoans. The C-terminal domain of eEF1Bβ and eEF1Bδ is highly similar to the corresponding region of eEF1Bα and accounts for its nucleotide exchange activity.1 Metazoan eEF1B has often been isolated as a complex bound to valyl tRNA synthetase (ValRS) via the metazoan-specific 200 amino acid N-terminal extension on ValRS.9 As tRNA aminoacylation precedes tRNA binding to eEF1A, the presence of ValRS in complex with eEF1B supports the channeling hypothesis according to which translation in eukaryotes is streamlined by coupling the charging of tRNA to its delivery to the ribosome.10 eEF1Bδ seems to be present primarily as two isoforms, eEF1Bδ1 and eEF1Bδ2, both of which can bind to eEF1Bαγ complex.
Phosphorylation of the eEF1 Complex
Studies on the regulation of translation by phosphorylation have traditionally focused on the initiation step. There is, however, growing evidence that the elongation step is also regulated to achieve a more robust control of the cellular translational machinery in response to environmental cues. Seminal work, particularly by the Traugh laboratory, demonstrated that the subunits of the eEF1 complex are phosphorylated by a number of serine/threonine protein kinases (Table 2). Protein kinase C (PKC), a key stimulator of cell proliferation and differentiation, has been shown to upregulate both transcription and translation rates via its kinase activity. PKC phosphorylates eEF1A, eEF1Bα, eEF1Bδ as well as ValRS causing a two fold increase in the GDP/GTP exchange activity.11 This correlates well with the reported two- to three- fold stimulation of elongation activity in response to PKC activation.12 Insulin also causes a rapid upregulation of protein synthesis.13 Studies using serum deprived 3T3-L1 cells showed that insulin stimulates phosphorylation of all the components of the eEF1 complex, possibly through S6 kinase. Thus there is evidence from multiple systems implicating phosphorylation of the eEF1 complex in increased translation rates.
Table 2.
Kinases, their targets in the eEF1 complex and effects of protein synthesis
| Kinases | Effect on Translation | Subunits Phosphorylated |
References |
|---|---|---|---|
| Protein Kinase C | Upregulation | eEF1A, eEF1Bα, eEF1Bδ | Peters et al.11; Venema et al.12 |
| Multipotential S6 Kinase |
Upregulation | eEF1A, eEF1Bα, eEF1Bδ | Chang and Traugh13 |
| Cyclin Dependent Kinase 1 |
Possible Downregulation |
eEF1Bβ, eEF1Bγ, eEF1Bδ |
Janssen et al.18 ; Mulner-Lorillon et al.19; Sivan et al.21 |
| Casein Kinase II | No Effect | eEF1Bα, eEF1Bδ | Sheu and Traugh15 |
| TGF-β Receptor I | Downregulation | eEF1A | Lin et al.14 |
| C-Raf | Unknown | eEF1A | Lamberti et al.68 |
| PAS Kinase | Upregulation | eEF1A | Eckhardt et al.88 |
| DOA Kinase | Unknown | eEF1Bγ | Fan et al.16 |
While most of the studies on eEF1 suggest a stimulatory role for phosphorylation, there are reports that phosphorylation by certain kinases may result in translational repression. eEF1A was identified in a screen as a phosphorylation substrate for type I transforming growth factor β receptor (TβR-I).14 TβR-I was shown to phosphorylate eEF1A1 at Ser300 in vitro and in vivo. Mutation of this residue significantly decreased the amount of phosphorylated eEF1A1 in vitro, while a phosphomimetic substitution resulted in inhibition of cell proliferation. As Ser300 is located near the region of eEF1A proposed to interact with the 3′-end of the aa-tRNA, its phosphorylation could disrupt the interacting surface for the aa-tRNA eventually leading to a reduction in translation and proliferation.14 Although, phosphopeptide mapping of eEF1A has revealed phosphorylation on at least five different sites,13 the exact residues that enhance eEF1A activity when phosphorylated have not been determined. Future experiments should address this issue so that a direct comparison of the similarities or differences among the phosphorylation sites that affect eEF1A function can be made. It has also been reported that casein kinase II phosphorylates both eEF1Bα and eEF1Bδ without an associated effect on eEF1A activity as measured in vitro.15 This phosphorylation event might be required to mediate unknown protein-protein interactions or changes in the distribution of eEF1 complex within the cell. It has also been reported that the C-terminus of eEF1Bγ contains a conserved LAMMER kinase phosphorylation site that is important for its normal function.16
Kinases also play an important role in the tight regulation of cell cycle progression. The plant specific eEF1Bβ has a conserved cell cycle dependent kinase (CDK) phosphorylation site.17 A mutant form of plant eEF1Bβ lacking the CDK phosphorylation site, but not the wild type, was able to complement the loss of eEF1Bα in yeast implying that phosphorylation is most likely used to reduce the GEF activity of eEF1Bβ.17 In metazoan oocytes, eEF1Bγ and eEF1Bδ have been identified as a target for the CDK1.18, 19 Additionally, CDK1 activation during M-phase has been correlated with a decrease in the translation elongation rate.20, 21 eEF1Bδ has been shown to undergo mitosis dependent spatial redistribution which could potentially be driven by CDK1 dependent phosphorylation. A fraction of the eEF1Bδ pool was shown to form a ring around the nucleus and following the breakdown of the nuclear membrane, accumulated around the mitotic spindle pole.20 It is tempting to speculate that this cell cycle specific localization pattern might be the result of isoform specific mobilization and degradation of eEF1Bδ transcripts.22 Changes in the subcellular localization of the translation factors caused by CDK1 phosphorylation may lead to localized decreases in protein synthesis. Wild type eEF1Bδ has been shown to pull down a significantly lower amount of eEF1A when compared to its non-phosphorylatable mutant form, eEF1Bδ-Ser133Ala.21 Such a reduced interaction may result in reduced GEF activity which in turn can decrease eEF1A activity leading to downregulation of translation. Interestingly the same site was shown to be phosphorylated in HSV infected cells by a viral kinase.23 A global reduction of elongation might be advantageous for the translation of viral mRNA in infected cells.
As shown herein, a number of different metabolic signals converge at the level of translational elongation validating it as a target of regulation. Whether the changes in translation observed in response to extracellular signals reflects changes specific to a subset of mRNAs or against the total transcriptome of the cell remains unknown. The use of these modifications to release eEF1 complex members from the translational apparatus to perform other functions is another intriguing possibility.
Architecture of the eEF1 complex
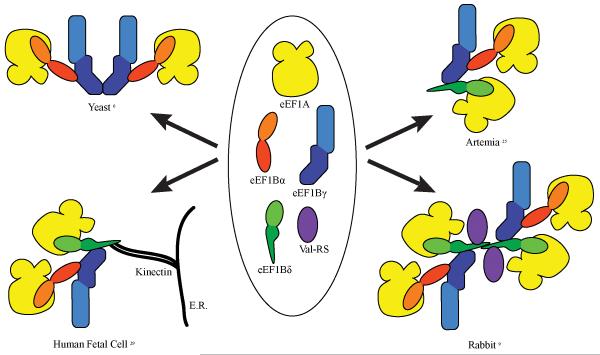
Several models have been proposed to explain the structural organization of the eEF1 complex (Figure 1). Initial attempts to purify eEF1A from calf brain produced a novel form 5 to 20 times heavier than free eEF1A. This complex aggregate of eEF1A contains cholesterol and phospholipids, and was proposed to have a storage or regulatory role .24, 25 Our understanding of the organization of eEF1 complex has changed over the years with the discovery of the multiple subunits of the eEF1B complex. The complete eEF1 complex structure was initially proposed in Artemia salina to consist of the four subunits eEF1A, eEF1Bα, eEF1Bγ and eEF1Bδ in a molar ratio of 2:1:1:1. eEF1Bδ was shown to bind to the N terminal domain of eEF1Bγ which in turn is bound to eEF1Bα. The presence of two molecules of eEF1A in the complex is not surprising since there are the two GEFs in the complex.26 Since the sequences of the eEF1 subunits are largely conserved across various species including humans, the interaction pattern uncovered in A. salina may hold true for mammalian systems as well. In the same study, a second distinct eEF1 complex lacking the eEF1Bδ subunit was also isolated, containing eEF1A, eEF1Bα and eEF1Bγ in a 1:1:1 molar ratio. In S. cerevisiae, an organism lacking eEF1Bδ, the presence of such a 1:1:1 complex was supported by crystallization and gel filtration studies of the N terminal half of eEF1Bγ.6 In addition, eEF1Bγ was determined to facilitate the dimerization of this complex. Such dimerization by eEF1Bγ was also suggested in a rabbit eEF1 complex model proposed by Chang and Traugh.13
Figure 1. Model of eEF1 complex.
The variation in the organization of the eEF1 complex in different organisms illustrates the differences in the composition as well as the interactions of the various subunits. Domain I, the largest domain of eEF1A (gold) is the GTP binding domain. For the eEF1B subunits α (red) γ (blue) δ (green), the N and C terminal domains are indicated in darker and lighter shades respectively. Val RS (purple) interacts with the eEF1 complex in metazoans.
Early characterization of the eEF1 complex in mammalian cells involved in vitro reconstitution experiments with purified rabbit proteins. Waller and colleagues analyzed different combinations of eEF1Bαγ, eEF1Bδ and native or truncated ValRS with each other by chromatography and demonstrated that the N-terminal extension of ValRS was required for binding to eEF1Bδ. eEF1Bαγ was proposed to prevent eEF1Bδ from forming high molecular weight aggregates, thereby giving the complex a defined quaternary structure. The entire complex was also proposed to dimerize via a leucine heptad repeat within the N-terminal region eEF1Bδ.9 The ability of eEF1Bδ to dimerize along with its distinct binding interface with eEF1Bγ indicates that eEF1Bα and eEF1Bδ might have different regulatory roles in metazoans. This model is also supported by the interactions uncovered using the yeast two-hybrid system, which demonstrated eEF1Bγ can interact with eEF1Bδ.27 Finally, 3D reconstruction studies using electron microscopic images of the complex shows the presence of two symmetrical domains allowing two eEF1Bδ subunits to be placed near the center of the complex.28
Work from Yu and colleagues suggest that there is a unique organization of eEF1B complex in human fetal brain cells.29, 30 According to this model, kinectin anchors the eEF1B complex to the ER by binding to eEF1Bδ. Kinectin is an integral membrane protein whose primary role is to anchor the microtubule associated cargo protein kinesin on intracellular organelle membranes. However, the brain specific isoform of kinectin lacks the kinesin binding domain pointing to its possible role in a non-canonical process.29 A conserved 60 amino acid fragment of kinectin was shown to co-immunoprecipitate eEF1Bδ from cell lysates. Moreover, both kinectin and eEF1B subunits were found to co-localize in the ER as would be expected of a spatially organized translation system. It is of note that overexpression of the eEF1Bδ binding domain of kinectin caused displacement of the eEF1B subunits from the ER which argues against eEF1Bγ being the primary membrane binding component of the eEF1B complex. In the absence of eEF1Bδ-kinectin interaction, cytoplasmic protein synthesis was favored of that of membrane proteins.30 The distribution of the elongation factors might, therefore, potentially be used to regulate synthesis of different types of proteins.
Interactions with the Cytoskeleton
Work from a number of laboratories has firmly established a link between cytoskeletal organization and translation. Most of the work bridging these processes has placed actin as the major cytoskeletal component involved. Actin plays a significant role in regulating the efficiency of translation, based on the observation that slight perturbations of the actin cytoskeleton in permeabilized cells produces a profound negative effect on global translation even though the levels of key translation components remained similar to those of intact cells.31
Since the discovery of eEF1A as an actin binding protein,32 an extensive number of in vitro studies have characterized this interaction. Competitive binding experiments with eEF1A, F-actin and aa-tRNA showed that as pH increases, the affinity of eEF1A for F-actin decreased while that for aa-tRNA increased.33 eEF1A has two pH-sensitive actin binding domains that could be important for modulating the cellular response to external stimuli by allowing the reorganization of the actin cytoskeleton and the associated translational machinery.33 pH also influences the ability of eEF1A to inhibit actin polymerization in vitro, which may be important in regulating processes like the transport, localization and translation of mRNAs.34 Such is the case in sea urchins, where an increase in intracellular pH acts as a signal in activating protein synthesis at fertilization.35
Genetic analysis in S. cerevisiae revealed that overexpression of eEF1A disrupts the organization of the actin cytoskeleton without causing appreciable effects on protein synthesis.36 In a genetic screen designed to separate the actin binding/bundling and translation elongation functions of eEF1A, two classes of mutants were identified (Figure 2). The first class of eEF1A mutants exhibited defects in actin cytoskeleton organization due to reduced actin bundling activity while maintaining normal translation activity.37 The second class of mutants displayed severe defects in actin organization and surprisingly translation initiation.38 Several actin associated protein mutants also exhibit reduced initiation providing further evidence that the cytoskeleton and the translational machinery are intimately linked. As eEF1A interacts with Bni1p/She5p, a downstream target of Rho1p that can regulate actin reorganization,39 it is possible that eEF1A acts as bridge between the cytoskeleton and actin modulators. The findings also suggest that regulation of the actin cytoskeleton by eEF1A is crucial for efficient protein synthesis.
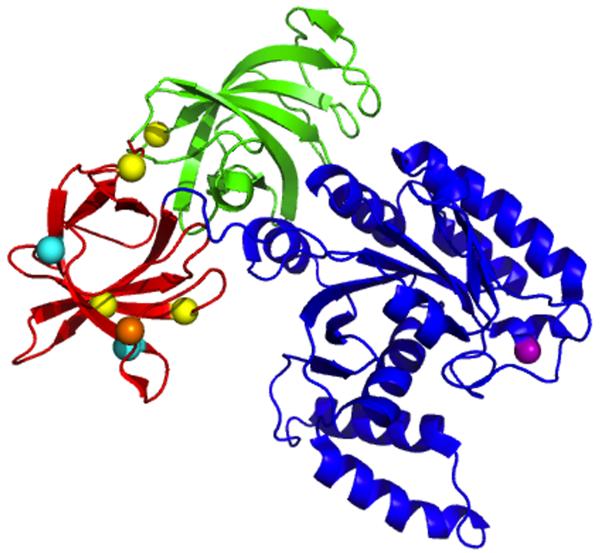
Figure 2. Location of mutations that affect non-canonical functions and known phosphorylation sites on eEF1A.
eEF1A from S. cerevisiae is composed of three well-defined domains as determined from the co-crystal structure with the C terminus of eEF1Bα. Domain I (blue) binds GTP, domain II (red) is proposed to interact with the aminoacyl end of the aa-tRNA, and domains II and III (green) are linked to actin binding and bundling. eEF1Bα binds eEF1A in domains I and II. Colored spheres indicate mutations that affect the non-canonical functions of eEF1A. In domain I, Asp156Asn (purple) affects protein turnover. In domain II, Glu286Lys and Glu291Lys (cyan) affect nuclear transport. Mutations that affect actin organization, Asn305Ser, Asn329Ser, Phe308Lys and Ser405Pro, (yellow), are shown in domains II and III. Phosphorylation of Glu298 (the yeast equivalent of human Ser300) (orange) may lead to downregulation of translation. The figure was prepared with PyMol using Protein Data Bank (PDB) 1F60.86
A number of reports have shown that eEF1A can also interact with microtubules. eEF1A was identified as a component of the mitotic apparatus,40 and was shown to bind, bundle and stabilize microtubules in vitro in a Ca2+/calmodulin dependent manner.41, 42 In contrast, a different study found that eEF1A acts to sever microtubules, as microinjection of eEF1A into cultured fibroblasts led to a rapid decline in tubulin polymers.43 The specific role eEF1A plays in the regulation of microtubule organization remains unclear. The same domains of eEF1A that interact with actin (domains I and III) also interact with microtubules,44 raising the question of how the interactions between eEF1A, actin, microtubules and other eEF1A binding partners are regulated or distributed in the cell.
Genetic and biochemical data suggest that eEF1Bα modulates the equilibrium of eEF1A between translation and actin organization.4, 45 In addition, eEF1Bα itself has been implicated in direct binding to actin. A 17 kDa fragment of eEF1Bα was isolated as a protein that bound tightly with actin on gel filtration and ion-exchange chromatography columns. However, the effect on eEF1Bα on actin filaments is unclear. While intact eEF1Bαγ complex isolated was shown to stimulate initial rate of actin assembly in vitro, indicating a role in the nucleation phase of the reaction, recombinant D. discoideum eEF1Bα exhibited a concentration dependent negative effect on actin assembly.46 This later finding is more consistent with the demonstration that eEF1A bundling of actin bundling is mutually exclusive to eEF1Bα binding.47 It remains to be seen whether eEF1Bγ may alter the confirmation of eEF1Bα and affect actin assembly.
On the basis of a yeast two-hybrid screen, eEF1Bγ has been proposed to bind to keratin, an intermediate filament of the cytoskeleton.48 It has since then been shown to bind and bundle keratin filaments in human epithelial cells, the disruption of which resulted in decreased translation.49 The presence of large amounts of eEF1Bγ in keratin bundle rich hair fibers supports its biological role in the intermediate filament organization. Keratin filaments not only provide mechanical strength to cells, but also regulate cell signaling, apoptosis and the response to stress. The integration of translational factors with the keratin cytoskeleton therefore opens up the potential for pathways that centrally regulate cellular processes by modulating the translational apparatus or vice versa.50
The observation that translation factors associate with the cytoskeleton has led to the hypothesis that protein synthesis and cytoskeletal organization undergo reciprocal regulation. The actin cytoskeleton in particular is proposed to serve as a scaffold for components of the translation machinery, since a number of initiation factors as well as a large proportion of translating ribosomes have been found in association with the actin cytoskeleton.51, 52 However, it remains unclear how changes in actin organization would lead to changes in the rate of translation. One possibility may be that the cytoskeleton provides a spatial ordering of the components such that it allows coordinated interactions between the mRNA, ribosome, and the various translation factors to take place and thus give rise to efficient protein synthesis. The role microtubules would play in such a model remains to be determined. Thus, future experiments designed to separate the effects of the eEF1 complex on the different components of the cytoskeleton are needed.
MOONLIGHTING FUNCTIONS OF eEF1A
Nuclear Export
The role of eEF1A in the nuclear export of tRNAs and the shuttling of eEF1A between the nucleus and cytoplasm have been debated for some time. The initial work on the function of eEF1A in nuclear export of tRNAs was based on a multi-copy suppressor screen of a synthetic lethal los1 arc1 mutant in S. cerevisiae.53 Overexpression of eEF1A was able to restore the growth of this mutant in the tRNA export pathway. Likewise, a reduction in the levels of eEF1A led to a decrease in nuclear export of several aa-tRNAs.53 Mutations in the aa-tRNA binding domain of eEF1A (Glu286Lys or Glu291Lys) show defects in nuclear export of aa-tRNA, leading to an accumulation of tRNAs in the nucleus.54 The abundance of eEF1A in the cell would suggest that all aa-tRNA could be bound by eEF1A as a ternary complex with GTP, thus not only protecting the labile aa-tRNA linkage but also effectively eliminating the possibility of aa-tRNA diffusion into the nucleus. Previous work by the Hopper lab has demonstrated that retrograde import of tRNAs into the nuclei of yeast cells occurs when they are deprived of nutrients.55 However, it remains unclear whether eEF1A is involved in the initial export of tRNAs from the nucleus or their re-export during recovery from suboptimal growth conditions.
Most of the studies examining the subcellular localization of eEF1A have found the protein to be predominantly localized in the cytoplasm.53, 56, 57 Exportin-5, a member of the β-importin family of nuclear transport receptors, has been implicated in keeping eEF1A outside of the nucleus in a manner dependent on the presence of aa-tRNAs.56, 57 However, a recent study demonstrated that eEF1A could be found in the nucleus of the msn5Δ nuclear export mutant.58 Although eEF1A appeared to localize around the nuclear membrane of wild-type cells upon nutrient deprivation, a greater proportion of eEF1A was found to localize within the nucleus of msn5Δ cells under the same conditions. Since eEF1A is found in high levels in cell, localization studies can be challenging. Therefore, more studies using advanced imaging techniques will help address the question of nuclear roles of eEF1A.
Proteolysis
Although there are examples of different isoforms of the same protein performing opposing functions, not many examples are known of a single protein having opposing functions. For eEF1A, this dichotomy exists for the proposed functions in both protein synthesis and protein degradation. Because of its central role in translation elongation, eEF1A has been proposed to be a good candidate for recognizing damaged proteins and shuttling them to the proteosome for degradation. eEF1A was initially identified as a factor required for the degradation of N-α-acetylated proteins.59 Later experiments showed that eEF1A could directly interact with nascent polypeptides while they are being synthesized.60 eEF1A was shown to not only interact with an unfolded protein after its release from the ribosome, but also to help mediate refolding. The idea that eEF1A could possess a chaperone-like activity is not too surprising as the bacterial homolog EF-Tu has been shown to have chaperone properties.61 In vivo evidence supporting a role for eEF1A in protein degradation came from its identification as a high copy suppressor of the slow growth phenotype caused by deletion of the RAD23 and RPN10 genes involved in the proteolytic pathway.62 eEF1A was subsequently shown to interact directly with Rpt1p, a subunit of the 19S regulatory particle. Deletion of RPT1 significantly reduced the interaction between eEF1A and the proteosome. An association between eEF1A and ubiquitinated proteins following ATP depletion was also demonstrated. Interestingly, resistance to the arginine analog canavinine, which leads to misfolding of proteins and subsequent ubiquitin-dependent degradation, was conferred by a single amino acid mutation (Asp156Asn) in the GTP-binding domain of eEF1A.62 Taken together, these data support a model in which eEF1A acts by directing damaged or misfolded proteins from the ribosome to the proteosome.
Apoptosis
The first report of the involvement of eEF1A in apoptosis showed a direct correlation between the levels of eEF1A and the rate of apoptosis.63 In another study, eEF1A was isolated in a cDNA screen to identify factors that give resistance to apoptosis following growth factor withdrawal.64 How is it possible for eEF1A to have both pro- and anti-apoptotic effects? The answer may lie in the fact that higher eukaryotes have two different isoforms of eEF1A. Although roughly 93% identical at the protein level, these two eEF1A isoforms vary not only in their expression pattern but also in their involvement in apoptosis. eEF1A1 is ubiquitously expressed throughout all tissues, whereas eEF1A2 seems to only be expressed in brain, heart and muscle.65, 66 Analysis of the expression levels of the eEF1A isoforms during muscle fiber differentiation revealed that eEF1A1 acts as a pro-apoptotic factor whereas eEF1A2 is anti-apoptotic.67 Interestingly, when the levels of eEF1A1 declined, eEF1A2 levels increased during the differentiation process providing further support for the opposing roles of the two isoforms. This has led to speculation that the switch between eEF1A1 and eEF1A2 in brain, heart and muscle is a critical component of the developmental cycle of these tissues.66, 67 Other studies have shown that increased eEF1A2 expression or decreased eEF1A1 expression results in resistance to apoptosis induced by ER stress.25, 64 Findings from a more recent study suggest that interferon α (IFNα) may induce an anti-apoptotic response by modulating the expression of eEF1A through the serine/threonine kinase C-Raf, pointing to a link between the phosphorylation status of eEF1A and apoptosis.68 Although the details of how phosphorylation affects the role of eEF1A in apoptosis are yet to be determined, the finding further supports the idea that non-canonical functions of eEF1A could be affected by post-translational modifications.
Viral Propagation
Viruses count not only on proteins encoded within their genome but also on host proteins in order to replicate. Given that eEF1A is one of the most abundant cellular proteins, it makes sense that viruses would hijack eEF1A for their own benefit. To date, eEF1A has been shown to affect positive-strand RNA viruses such as Dengue virus,69 turnip yellow mosaic virus (TYMV),70 tobacco mosaic virus (TMV),71 turnip crinkle virus (TCV)72 and West Nile Virus (WNV).73A common theme among these viruses is that eEF1A can interact with the 3′ C-terminal end of the viral RNA. This result most likely stems from the fact that the proposed secondary structure of the 3′-ends of the viral RNA resembles that of tRNA, a canonical eEF1A binding partner. eEF1A can also interact with their respective viral encoded RNA-dependent RNA polymerase (RdRp). eEF1A was able to stimulate the RdRp activity, or minus strand synthesis, of TCV in vitro.72 Interestingly, the efficiency of synthesis appears to increase when the RdRp and eEF1A were combined prior to the addition of the viral RNA suggesting the possibility of a sequential order in the interactions. In the case of TYMV, eEF1A appears to antagonize minus strand synthesis presumably by preventing the binding of RdRp to the viral RNA dependent on the latter’s aminoacylation status70, the tRNA-like structure at the 3′-end of the TYMV RNA resembles that of tRNAVal and has been shown to be valylated in vitro.74 Although eEF1A associates primarily with positive-strand viruses, it is worth noting that it has also been found to interact with the RdRp of vesicular stomatitis virus (VSV), a negative single strand RNA virus.75
Though the role of eEF1A in translation of the viral RNA is straightforward to understand, what could be the significance of the role it plays in the replication of these RNA viruses? Given that the viral replication cycle is composed of a sequential series of events, some have speculated that eEF1A could be involved in helping maintain that order. In the case of TYMV, eEF1A could prevent the viral RNA from being copied by allowing time for the viral proteins like the RdRp to be synthesized first. Once sufficient viral encoded proteins are produced, RdRp competes eEF1A off the 3′-end of the viral RNA positive stand to allow for minus strand synthesis70 (Figure 3). For viruses where eEF1A interacts with both the viral RNA and RdRp, eEF1A may help sequester these components of the replicase complex into vesicles.76 eEF1A could help in anchoring to the ER membrane via a phospholipid modification at Asp306 and possibly mediate vesicle formation.73 Findings from a more recent study suggest that See1p, a methyltransferase involved in vesicular transport, methylates eEF1A providing further support to this idea.75
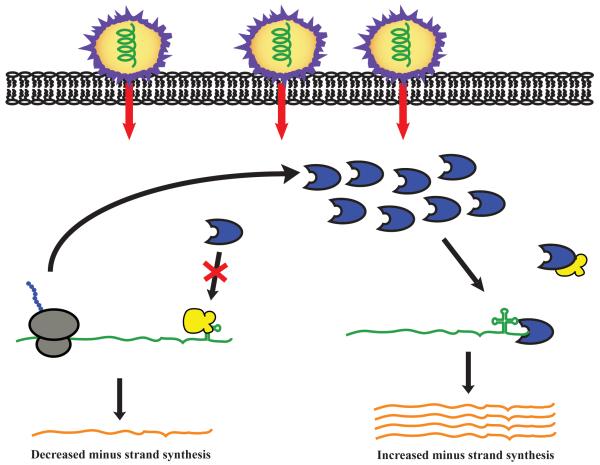
Figure 3. The role of eEF1A in minus strand synthesis of positive strand viral RNAs.
Upon release of the viral RNA (green) into the cytoplasm, eEF1A (yellow) binds at the tRNA-like structure at the 3′-end of the RNA preventing the RNA-dependent RNA polymerase (RdRp, blue) from initiating minus strand synthesis. By inhibiting minus strand synthesis, eEF1A allows viral proteins to be synthesized first. Once sufficient RdRp is made, RdRp competes eEF1A off of the viral end allowing for minus strand synthesis to occur.
THE eEF1B COMPLEX OUTSIDE THE TRANSLATION SYSTEM
Although about 10 fold less abundant than eEF1A, the eEF1B complex is an abundant protein by cellular standards. Though fewer non-canonical functions have been ascribed to the eEF1B complex proteins, more examples are recently emerging. These alternate functions are most apparent for the eEF1Bγ and eEF1Bδ subunits, perhaps separating the highly conserved catalytic GEF function of eEF1Bα from these additional roles.
The primary role attributed to eEF1Bγ in translation elongation is as a structural scaffold for eEF1Bα. Given that the deletion of eEF1Bγ in S. cerevisiae has no significant effect either on total protein synthesis or translational fidelity in vivo,77 it is possible that eukaryotic evolution has selected eEF1Bγ as a part of the translation complex for its additional functions. Expression patterns during embryogenesis also suggest that it might have a role in the formation of anterior/posterior axis.16 The N terminal domain of eEF1Bγ is structurally similar to the theta class of Glutathione S-transferases (GST).78 GSTs catalyze the conjugation of glutathione to electrophilic substrates such as peroxides and hence play an important role in cellular detoxification of reactive oxygen species. While the structural details of the GST catalytic sites seem to be conserved in eEF1Bγ, attempts to detect its GST activity have been largely unsuccessful.6 However rice recombinant eEF1Bγ, as well as the entire eEF1B complex, was shown to possess a low GST activity.79 On a similar note the eEF1B complex isolated from Leishmania major was shown to conjugate a variety of electrophilic substrates to trypanothione, a substrate similar to glutathione. The Trypanothione-S-Transferase pathway is used by L. major in its defense against chemical and oxidative stress.80 An alternative possibility is that eEF1Bγ, with its lower transferase activity, could be involved in coupling the redox balance of the cell to the translation complex.79
eEF1Bγ plays a significant role in the oxidative stress response pathway. Deletion of the two genes encoding eEF1Bγ in S. cerevisiae gave an additive resistance to oxidative stress.77 Total protein analysis by 2D gel electrophoresis indicated a constitutively active stress response pathway in the absence of eEF1Bγ.81 It was also shown to exhibit defective vacuolar morphology resulting from defective ER-Golgi transport and consequentially had a defective protein turnover mechanism.81 Mislocalization of eEF1Bαγ has previously been shown to decrease the synthesis of membrane proteins.30 It is therefore interesting to speculate that in the absence of eEF1Bγ, synthesis of certain membrane proteins essential for vesicle formation is severely affected, which in turn, can lead to defective vacuole and ER-Golgi transport.
eEF1Bγ has also been shown to have a potential role in the transcription, localization and translation of vimentin mRNA.82 Passananti and colleagues were able to show by co-immunoprecipitation (co-IP) and chromatin immunoprecipitation (ChIP) experiments that eEF1Bγ physically interacts with Rpb3p, a subunit of the RNA polymerase II, as well as the promoter region of the vimentin gene. Knockdown of eEF1Bγ, however, resulted in mitochondrial fragmentation and a concomitant increase in cellular superoxide levels as would be expected with its previously established role in cellular stress response. However eEF1Bγ knockdown by siRNA did not seem to have a major effect on vimentin mRNA levels leaving unanswered the role it plays at the vimentin promoter. Although there are reports suggesting that eEF1Bγ binds to the 3′ UTR of vimentin mRNA, studies have indicated that the binding is non-specific.83 Hence, further evidence is required to prove that eEF1Bγ can affect the localization of specific mRNAs in the cell.
In a recent study, eEF1Bδ was revealed to undergo alternate splicing to form a novel heat-shock response (HSR) transcription factor.84 While the common isoforms of eEF1Bδ are around 30-40 kDa a larger splicing variant termed eEF1BδL was detected in mouse brain and testis. Unlike the short isoform, the long isoform includes a putative nuclear localization signal and localization studies indicated that eEF1BδL was found both in the nucleus and the cytoplasm. eEF1BδL expression induced expression from a heat shock element (HSE) containing reporter. The association of eEF1BδL with the HSE was confirmed by ChIP analysis and was shown to regulate HSE responsive gene induction in response to heat shock, thereby implicating eEF1BδL as a HSR transcription factor. In response to heat shock, there was a simultaneous up-regulation of eEF1BδL levels and a downregulation of eEF1Bδ isoform levels. This points to a novel two-pronged effect on cellular metabolism in response to heat shock, in the form of increased molecular chaperone levels and decreased total protein synthesis.
Yeast two hybrid screen and co-immunoprecipitation analysis recently demonstrated that eEF1Bδ also interacts with SIAH-1, an E3 ubiquitin ligase.85 Overexpression of eEF1Bδ caused an inhibition of the E3 ligase activity as evidenced by the decrease in ubiquitination of its substrate HPH2. While it is not immediately obvious the physiological requirement for such an interaction in the cell, It is probable that the endogenous interaction between the two proteins is regulated by specific cellular cues.
Conclusion
Translation elongation factors are among the most abundant proteins in the cell and play an essential role in assuring the proper decoding of mRNA to produce cellular proteins. A common theme emerging from the proposed models of the eEF1 complex is that this system has acquired compositional and organizational complexity with evolution from bacteria to yeast to metazoans. The extensive interactions between the elongation factors and the cytoskeleton also point to the existence of a higher order structural organization of the protein synthesis machinery. This supramolecular organization not only leads to a highly efficient translational system, but also opens up multiple avenues for regulatory control. Although the eEF1 complex can be phosphorylated at multiple sites, many aspects of the in vivo consequences of this modification remain largely unknown. The central role played by translation in producing the appropriate cellular response to external stimuli may have driven the elongation factors into acquiring multiple functions. For eEF1A, sustained work has demonstrated the importance of these alternate functions in viral systems and the actin cytoskeleton. However, far less is known about the non-canonical functions of the eEF1B complex. It will be interesting to see if more roles for these proteins emerge in the future in the context of the various post translational modifications in higher eukaryotes. One of the major challenges that remains is to determine the mechanism underlying these secondary functions. Gathering mechanistic data on the non-canonical functions of the eEF1 subunits, particularly eEF1A, is challenging as they are some of the most abundant proteins in the cell. This makes the post-translational modifications of the eEF1 complex an attractive venue to explore how canonical and non-canonical functions converge to regulate the cellular processes. Future work will provide a better picture of the role of the eEF1 complex within the global cellular context.
Acknowledgments
The authors would like to thank Maria Mateyak and Dongming He for comments on the manuscript and members of the Kinzy lab for helpful feedback. This work was supported by NIH GM57483 to TGK, WBP was partially supported by T32 AI007403.
Contributor Information
Arjun N. Sasikumar, Department of Molecular Genetics, Microbiology and Immunology, Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey, Piscataway, New Jersey 08854-5635.
Winder B. Perez, Department of Molecular Genetics, Microbiology and Immunology, Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey, Piscataway, New Jersey 08854-5635.
Terri Goss Kinzy, Department of Molecular Genetics, Microbiology and Immunology, Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey, Piscataway, New Jersey 08854-5635.
References
- 1.Le Sourd F, Boulben S, Le Bouffant R, Cormier P, Morales J, Belle R, Mulner-Lorillon O. eEF1B: At the dawn of the 21st century. Biochim Biophys Acta. 2006;1759:13–31. doi: 10.1016/j.bbaexp.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho MD, Carvalho JF, Merrick WC. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch Biochem Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 3.Taylor DR, Frank J, Kinzy TG. Structure and Function of the Eukaryotic Ribosome and Elongation Factors. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2007. pp. 59–85. [Google Scholar]
- 4.Pittman YR, Valente L, Jeppesen MG, Andersen GR, Patel S, Kinzy TG. Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha. J Biol Chem. 2006;281:19457–19468. doi: 10.1074/jbc.M601076200. [DOI] [PubMed] [Google Scholar]
- 5.Janssen GM, Moller W. Kinetic studies on the role of elongation factors 1 beta and 1 gamma in protein synthesis. J Biol Chem. 1988;263:1773–1778. [PubMed] [Google Scholar]
- 6.Jeppesen MG, Ortiz P, Shepard W, Kinzy TG, Nyborg J, Andersen GR. The crystal structure of the glutathione S-transferase-like domain of elongation factor 1Bgamma from Saccharomyces cerevisiae. J Biol Chem. 2003;278:47190–47198. doi: 10.1074/jbc.M306630200. [DOI] [PubMed] [Google Scholar]
- 7.Kinzy TG, Ripmaster TL, Woolford JL., Jr. Multiple genes encode the translation elongation factor EF-1 gamma in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:2703–2707. doi: 10.1093/nar/22.13.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders J, Brandsma M, Janssen GM, Dijk J, Moller W. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 beta gamma delta in the endoplasmic reticulum. J Cell Sci. 1996;109(Pt 5):1113–1117. doi: 10.1242/jcs.109.5.1113. [DOI] [PubMed] [Google Scholar]
- 9.Bec G, Kerjan P, Waller JP. Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1 beta gamma delta complex. Essential roles of the NH2-terminal extension of valyl-tRNA synthetase and of the EF-1 delta subunit in complex formation. J Biol Chem. 1994;269:2086–2092. [PubMed] [Google Scholar]
- 10.Negrutskii BS, Stapulionis R, Deutscher MP. Supramolecular organization of the mammalian translation system. Proc Natl Acad Sci U S A. 1994;91:964–968. doi: 10.1073/pnas.91.3.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters HI, Chang YW, Traugh JA. Phosphorylation of elongation factor 1 (EF-1) by protein kinase C stimulates GDP/GTP-exchange activity. Eur J Biochem. 1995;234:550–556. doi: 10.1111/j.1432-1033.1995.550_b.x. [DOI] [PubMed] [Google Scholar]
- 12.Venema RC, Peters HI, Traugh JA. Phosphorylation of valyl-tRNA synthetase and elongation factor 1 in response to phorbol esters is associated with stimulation of both activities. J Biol Chem. 1991;266:11993–11998. [PubMed] [Google Scholar]
- 13.Chang YW, Traugh JA. Insulin stimulation of phosphorylation of elongation factor 1 (eEF-1) enhances elongation activity. Eur J Biochem. 1998;251:201–207. doi: 10.1046/j.1432-1327.1998.2510201.x. [DOI] [PubMed] [Google Scholar]
- 14.Lin KW, Yakymovych I, Jia M, Yakymovych M, Souchelnytskyi S. Phosphorylation of eEF1A1 at Ser300 by TbetaR-I results in inhibition of mRNA translation. Curr Biol. 2010;20:1615–1625. doi: 10.1016/j.cub.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Sheu GT, Traugh JA. A structural model for elongation factor 1 (EF-1) and phosphorylation by protein kinase CKII. Mol Cell Biochem. 1999;191:181–186. [PubMed] [Google Scholar]
- 16.Fan Y, Schlierf M, Gaspar AC, Dreux C, Kpebe A, Chaney L, Mathieu A, Hitte C, Gremy O, Sarot E, et al. Drosophila translational elongation factor-1gamma is modified in response to DOA kinase activity and is essential for cellular viability. Genetics. 2010;184:141–154. doi: 10.1534/genetics.109.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomerening JR, Valente L, Kinzy TG, Jacobs TW. Mutation of a conserved CDK site converts a metazoan Elongation Factor 1Bbeta subunit into a replacement for yeast eEF1Balpha. Mol Genet Genomics. 2003;269:776–788. doi: 10.1007/s00438-003-0888-1. [DOI] [PubMed] [Google Scholar]
- 18.Janssen GM, Morales J, Schipper A, Labbe JC, Mulner-Lorillon O, Belle R, Moller W. A major substrate of maturation promoting factor identified as elongation factor 1 beta gamma delta in Xenopus laevis. J Biol Chem. 1991;266:14885–14888. [PubMed] [Google Scholar]
- 19.Mulner-Lorillon O, Minella O, Cormier P, Capony JP, Cavadore JC, Morales J, Poulhe R, Belle R. Elongation factor EF-1 delta, a new target for maturation-promoting factor in Xenopus oocytes. J Biol Chem. 1994;269:20201–20207. [PubMed] [Google Scholar]
- 20.Boulben S, Monnier A, Le Breton M, Morales J, Cormier P, Belle R, Mulner-Lorillon O. Sea urchin elongation factor 1delta (EF1delta) and evidence for cell cycle-directed localization changes of a sub-fraction of the protein at M phase. Cell Mol Life Sci. 2003;60:2178–2188. doi: 10.1007/s00018-003-3201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivan G, Aviner R, Elroy-Stein O. Mitotic Modulation of Translation Elongation Factor 1 Leads to Hindered tRNA Delivery to Ribosomes. J Biol Chem. 2011;286:27927–27935. doi: 10.1074/jbc.M111.255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka TS, Nishiumi F, Komiya T, Ikenishi K. Characterization of the 38 kDa protein lacking in gastrula-arrested mutant Xenopus embryos. Int J Dev Biol. 2010;54:1347–1353. doi: 10.1387/ijdb.092862tt. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi Y, Kato K, Tanaka M, Kanamori M, Nishiyama Y, Yamanashi Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J Virol. 2003;77:2359–2368. doi: 10.1128/JVI.77.4.2359-2368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon HM, Redfield B, Millard S, Vane F, Weissbach H. Multiple forms of elongation factor 1 from calf brain. Proc Natl Acad Sci U S A. 1973;70:3282–3286. doi: 10.1073/pnas.70.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legocki AB, Redfield B, Liu CK, Weissbach H. Role of phospholipids in the multiple forms of mammalian elongation factor 1. Proc Natl Acad Sci U S A. 1974;71:2179–2182. doi: 10.1073/pnas.71.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janssen GM, van Damme HT, Kriek J, Amons R, Moller W. The subunit structure of elongation factor 1 from Artemia. Why two alpha-chains in this complex? J Biol Chem. 1994;269:31410–31417. [PubMed] [Google Scholar]
- 27.Mansilla F, Friis I, Jadidi M, Nielsen KM, Clark BF, Knudsen CR. Mapping the human translation elongation factor eEF1H complex using the yeast two-hybrid system. Biochem J. 2002;365:669–676. doi: 10.1042/BJ20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang S, Wolfe CL, Warrington JA, Norcum MT. Three-dimensional reconstruction of the valyl-tRNA synthetase/elongation factor-1H complex and localization of the delta subunit. FEBS Lett. 2005;579:6049–6054. doi: 10.1016/j.febslet.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 29.Ong LL, Er CP, Ho A, Aung MT, Yu H. Kinectin anchors the translation elongation factor-1 delta to the endoplasmic reticulum. J Biol Chem. 2003;278:32115–32123. doi: 10.1074/jbc.M210917200. [DOI] [PubMed] [Google Scholar]
- 30.Ong LL, Lin PC, Zhang X, Chia SM, Yu H. Kinectin-dependent assembly of translation elongation factor-1 complex on endoplasmic reticulum regulates protein synthesis. J Biol Chem. 2006;281:33621–33634. doi: 10.1074/jbc.M607555200. [DOI] [PubMed] [Google Scholar]
- 31.Stapulionis R, Kolli S, Deutscher MP. Efficient mammalian protein synthesis requires an intact F-actin system. J Biol Chem. 1997;272:24980–24986. doi: 10.1074/jbc.272.40.24980. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, Demma M, Warren V, Dharmawardhane S, Condeelis J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature. 1990;347:494–496. doi: 10.1038/347494a0. [DOI] [PubMed] [Google Scholar]
- 33.Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray JW, Edmonds BT, Liu G, Condeelis J. Bundling of actin filaments by elongation factor 1 alpha inhibits polymerization at filament ends. J Cell Biol. 1996;135:1309–1321. doi: 10.1083/jcb.135.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler MM, Steinhardt RA, Grainger JL, Minning L. Dual ionic controls for the activation of protein synthesis at fertilization. Nature. 1980;287:558–560. doi: 10.1038/287558a0. [DOI] [PubMed] [Google Scholar]
- 36.Munshi R, Kandl KA, Carr-Schmid A, Whitacre JL, Adams AE, Kinzy TG. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics. 2001;157:1425–1436. doi: 10.1093/genetics/157.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 38.Gross SR, Kinzy TG. Improper organization of the actin cytoskeleton affects protein synthesis at initiation. Mol Cell Biol. 2007;27:1974–1989. doi: 10.1128/MCB.00832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umikawa M, Tanaka K, Kamei T, Shimizu K, Imamura H, Sasaki T, Takai Y. Interaction of Rho1p target Bni1p with F-actin-binding elongation factor 1alpha: implication in Rho1p-regulated reorganization of the actin cytoskeleton in Saccharomyces cerevisiae. Oncogene. 1998;16:2011–2016. doi: 10.1038/sj.onc.1201724. [DOI] [PubMed] [Google Scholar]
- 40.Ohta K, Toriyama M, Miyazaki M, Murofushi H, Hosoda S, Endo S, Sakai H. The mitotic apparatus-associated 51-kDa protein from sea urchin eggs is a GTP-binding protein and is immunologically related to yeast polypeptide elongation factor 1 alpha. J Biol Chem. 1990;265:3240–3247. [PubMed] [Google Scholar]
- 41.Durso NA, Cyr RJ. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1 alpha. Plant Cell. 1994;6:893–905. doi: 10.1105/tpc.6.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore RC, Durso NA, Cyr RJ. Elongation factor-1alpha stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motil Cytoskeleton. 1998;41:168–180. doi: 10.1002/(SICI)1097-0169(1998)41:2<168::AID-CM7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1 alpha. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- 44.Moore RC, Cyr RJ. Association between elongation factor-1alpha and microtubules in vivo is domain dependent and conditional. Cell Motil Cytoskeleton. 2000;45:279–292. doi: 10.1002/(SICI)1097-0169(200004)45:4<279::AID-CM4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen L, Andersen GR, Knudsen CR, Kinzy TG, Nyborg J. Crystallization of the yeast elongation factor complex eEF1A-eEF1B alpha. Acta Crystallogr D Biol Crystallogr. 2001;57:159–161. doi: 10.1107/s0907444900015559. [DOI] [PubMed] [Google Scholar]
- 46.Furukawa R, Jinks TM, Tishgarten T, Mazzawi M, Morris DR, Fechheimer M. Elongation factor 1beta is an actin-binding protein. Biochim Biophys Acta. 2001;1527:130–140. doi: 10.1016/s0304-4165(01)00157-x. [DOI] [PubMed] [Google Scholar]
- 47.Pittman YR, Kandl K, Lewis M, Valente L, Kinzy TG. Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha. J Biol Chem. 2009;284:4739–4747. doi: 10.1074/jbc.M807945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441:362–365. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Kellner J, Lee CH, Coulombe PA. Interaction between the keratin cytoskeleton and eEF1Bgamma affects protein synthesis in epithelial cells. Nat Struct Mol Biol. 2007;14:982–983. doi: 10.1038/nsmb1301. [DOI] [PubMed] [Google Scholar]
- 50.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 51.Howe JG, Hershey JW. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell. 1984;37:85–93. doi: 10.1016/0092-8674(84)90303-9. [DOI] [PubMed] [Google Scholar]
- 52.Vedeler A, Pryme IF, Hesketh JE. The characterization of free, cytoskeletal and membrane-bound polysomes in Krebs II ascites and 3T3 cells. Mol Cell Biochem. 1991;100:183–193. doi: 10.1007/BF00234167. [DOI] [PubMed] [Google Scholar]
- 53.Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- 54.Grosshans H, Simos G, Hurt E. Review: transport of tRNA out of the nucleus-direct channeling to the ribosome? J Struct Biol. 2000;129:288–294. doi: 10.1006/jsbi.2000.4226. [DOI] [PubMed] [Google Scholar]
- 55.Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calado A, Treichel N, Muller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:639–649. doi: 10.1091/mbc.E09-07-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonen H, Smith CE, Siegel NR, Kahana C, Merrick WC, Chakraburtty K, Schwartz AL, Ciechanover A. Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci U S A. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hotokezaka Y, Tobben U, Hotokezaka H, Van Leyen K, Beatrix B, Smith DH, Nakamura T, Wiedmann M. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J Biol Chem. 2002;277:18545–18551. doi: 10.1074/jbc.M201022200. [DOI] [PubMed] [Google Scholar]
- 61.Caldas TD, El Yaagoubi A, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- 62.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duttaroy A, Bourbeau D, Wang XL, Wang E. Apoptosis rate can be accelerated or decelerated by overexpression or reduction of the level of elongation factor-1 alpha. Exp Cell Res. 1998;238:168–176. doi: 10.1006/excr.1997.3819. [DOI] [PubMed] [Google Scholar]
- 64.Talapatra S, Wagner JD, Thompson CB. Elongation factor-1 alpha is a selective regulator of growth factor withdrawal and ER stress-induced apoptosis. Cell Death Differ. 2002;9:856–861. doi: 10.1038/sj.cdd.4401078. [DOI] [PubMed] [Google Scholar]
- 65.Kahns S, Lund A, Kristensen P, Knudsen CR, Clark BF, Cavallius J, Merrick WC. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res. 1998;26:1884–1890. doi: 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee S, Francoeur AM, Liu S, Wang E. Tissue-specific expression in mammalian brain, heart, and muscle of S1, a member of the elongation factor-1 alpha gene family. J Biol Chem. 1992;267:24064–24068. [PubMed] [Google Scholar]
- 67.Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem. 2002;277:5418–5425. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamberti A, Longo O, Marra M, Tagliaferri P, Bismuto E, Fiengo A, Viscomi C, Budillon A, Rapp UR, Wang E, et al. C-Raf antagonizes apoptosis induced by IFN-alpha in human lung cancer cells by phosphorylation and increase of the intracellular content of elongation factor 1A. Cell Death Differ. 2007;14:952–962. doi: 10.1038/sj.cdd.4402102. [DOI] [PubMed] [Google Scholar]
- 69.De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 70.Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 71.Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, Suzuki M, Namba S, Hibi T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 2006;347:100–108. doi: 10.1016/j.virol.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 72.Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 2010;6:e1001175. doi: 10.1371/journal.ppat.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dreher TW, Goodwin JB. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998;26:4356–4364. doi: 10.1093/nar/26.19.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das T, Mathur M, Gupta AK, Janssen GM, Banerjee AK. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc Natl Acad Sci U S A. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thivierge K, Cotton S, Dufresne PJ, Mathieu I, Beauchemin C, Ide C, Fortin MG, Laliberte JF. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology. 2008;377:216–225. doi: 10.1016/j.virol.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 77.Olarewaju O, Ortiz PA, Chowdhury WQ, Chatterjee I, Kinzy TG. The translation elongation factor eEF1B plays a role in the oxidative stress response pathway. RNA Biol. 2004;1:89–94. doi: 10.4161/rna.1.2.1033. [DOI] [PubMed] [Google Scholar]
- 78.Koonin EV, Mushegian AR, Tatusov RL, Altschul SF, Bryant SH, Bork P, Valencia A. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain--study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kobayashi S, Kidou S, Ejiri S. Detection and characterization of glutathione S-transferase activity in rice EF-1betabeta’gamma and EF-1gamma expressed in Escherichia coli. Biochemical and biophysical research communications. 2001;288:509–514. doi: 10.1006/bbrc.2001.5799. [DOI] [PubMed] [Google Scholar]
- 80.Vickers TJ, Wyllie S, Fairlamb AH. Leishmania major elongation factor 1B complex has trypanothione S-transferase and peroxidase activity. J Biol Chem. 2004;279:49003–49009. doi: 10.1074/jbc.M407958200. [DOI] [PubMed] [Google Scholar]
- 81.Esposito AM, Kinzy TG. The eukaryotic translation elongation Factor 1Bgamma has a non-guanine nucleotide exchange factor role in protein metabolism. J Biol Chem. 2010;285:37995–38004. doi: 10.1074/jbc.M110.160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corbi N, Batassa EM, Pisani C, Onori A, Di Certo MG, Strimpakos G, Fanciulli M, Mattei E, Passananti C. The eEF1gamma subunit contacts RNA polymerase II and binds vimentin promoter region. PLoS One. 2010;5:e14481. doi: 10.1371/journal.pone.0014481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jonusiene V, Sasnauskiene S, Juodka B. The cloning and characterization of Tetrahymena pyriformis translation elongation factor 1b alpha and gamma subunits. Cell Mol Biol Lett. 2005;10:689–696. [PubMed] [Google Scholar]
- 84.Kaitsuka T, Tomizawa K, Matsushita M. Transformation of eEF1Bdelta into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011;12:673–681. doi: 10.1038/embor.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu H, Shi Y, Lin Y, Qian W, Yu Y, Huo K. Eukaryotic translation elongation factor 1 delta inhibits the ubiquitin ligase activity of SIAH-1. Mol Cell Biochem. 2011 doi: 10.1007/s11010-011-0891-5. [DOI] [PubMed] [Google Scholar]
- 86.Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 87.Sang Lee J, Gyu Park S, Park H, Seol W, Lee S, Kim S. Interaction network of human aminoacyl-tRNA synthetases and subunits of elongation factor 1 complex. Biochem Biophys Res Commns. 2002;291:158–164. doi: 10.1006/bbrc.2002.6398. [DOI] [PubMed] [Google Scholar]
- 88.Eckhardt K, Troger J, Reissmann J, Katschinski DM, Wagner KF, Stengel P, Paasch U, Hunziker P, Borter E, Barth S, et al. Male germ cell expression of the PAS domain kinase PASKIN and its novel target eukaryotic translation elongation factor eEF1A1. Cell Physiol Biochem. 2007;20:227–240. doi: 10.1159/000104169. [DOI] [PubMed] [Google Scholar]