Abstract
Susceptibility to thrombosis varies in human populations as well as many in inbred mouse strains. The objective of this study was to characterize the genetic control of thrombotic risk on three chromosomes. Previously, utilizing a tail-bleeding/rebleeding assay as a surrogate of hemostasis and thrombosis function, three mouse chromosome substitution strains (CSS) (B6-Chr5A/J, Chr11A/J, Chr17A/J) were identified (Hmtb1, Hmtb2, Hmtb3). The tailbleeding/rebleeding assay is widely used and distinguishes mice with genetic defects in blood clot formation or dissolution. In the present study, quantitative trait locus (QTL) analysis revealed a significant locus for rebleeding (clot stability) time (time between cessation of initial bleeding and start of the second bleeding) on chromosome 5, suggestive loci for bleeding time (time between start of bleeding and cessation of bleeding) also on chromosomes 5, and two suggestive loci for clot stability on chromosome 17 and one on chromosome 11. The three CSS and the parent A/J had elevated clot stability time. There was no interaction of genes on chromosome 11 with genes on chromosome 5 or chromosome 17. On chromosome 17, twenty-three candidate genes were identified in synteny with previously identified loci for thrombotic risk on human chromosome 18. Thus, we have identified new QTLs and candidate genes not previously known to influence thrombotic risk.
Introduction
Thrombosis plays a critical role in the development of cardiovascular diseases (Sturm 2004) and may be caused by the upregulation of the procoagulant pathway, or the downregulation of anticoagulant and fibrinolytic pathways (Mackman 2005). Thrombotic risk has been associated with genetic variation in these three pathways (Grant 2003; Williams and Bray 2001). Mutations may directly alter components of these pathways (Hong and Kwaan 1999; Zoller et al. 1997) or may indirectly alter their regulation or the proteins required for their processing (Zhang and Ginsburg 2004). Family history has long been associated with susceptibility to thrombosis, but these known mutations account for only a small portion of the variation. The genetic complexity of human populations and the huge influence of environmental factors on phenotypes make finding genes that may have small effects on thrombotic risk a difficult task.
Mouse models have been used to determine the function of individual genes and uncover new genes underlying complex diseases such as atherosclerosis and obesity. To date, only a few studies (Lemmerhirt et al. 2007; Mohlke et al. 1996) have reported the use of quantitative trait locus (QTL) mapping in mouse models to characterize the genetic control of thrombotic risk. The two inbred mouse strains, C57BL/6J (B6) and A/J, differ markedly in susceptibility to thrombosis and fibrinolysis and are ideally suited to identify genes associated with thrombotic risk. Previously, we reported that the arterial occlusion time in the ferric chloride-induced vascular injury model in the A/J mice was twofold less than in B6 mice, and clot stability time in a tailbleeding/rebleeding assay was threefold longer in the A/J mice (Hoover-Plow et al. 2006). A panel of chromosome substitution strains (CSS; B6-Chr1-19, X, YA/J), with an individual A/J chromosome in a B6 background, was utilized for this study. This approach has advantages over genome-wide scans, including detection of more QTLs, the requirement of fewer mice, and simplification of subsequent fine-mapping. The panel was screened with the bleeding/rebleeding assay as a surrogate marker of thrombosis and hemostasis. The bleeding/rebleeding assay has been used to identify functional changes in hemostasis and thrombosis in several genetically engineered mice (Broze et al. 2001; Hamilton et al. 2004; Kato et al. 2004; Sweeney et al. 1990). We demonstrated that the assay reports on platelet defects and fibrinolytic component deficiencies and corresponds well with a carotid injury assay (Hoover-Plow et al. 2006). Although bleeding time was not different between A/J and B6 mice (Hoover-Plow et al. 2006), a reduced bleeding time was identified in five of the CSSs, including strains with A/J chromosomes 5, 6, 8, 14, 15, and Y. In addition, three CSS with A/J chromosomes 5, 11, and 17 were identified with increased clot stability time that was similar to the elevated values of the A/J parent strain compared to the B6 strain. The purpose of this study was to characterize the genetic control of bleeding and clot stability time on chromosomes 5, 11, and 17.
Materials and methods
Mice
The inbred mouse strains C57BL/6J (B6, #000664) and A/J (#000646) were purchased from The Jackson Laboratory (Bar Harbor, ME). The CSSs were previously described (Nadeau et al. 2000; Singer et al. 2004). CSS-5 (female), CSS-11 (female), or CSS-17 (male) were crossed with B6 to produce F1 progeny that were intercrossed to generate F2 progeny. Mice were housed in sterilized isolator cages with a 14-h/10-h light/dark cycle and were provided sterilized food and water ad libitum. The bleeding/rebleeding assay was performed on mice (both males and females) at 6–8 weeks of age. This study was approved by the Institutional Animal Care and Use Committee and procedures were followed in accordance with institutional guidelines.
Genotyping
Genomic DNA was prepared from ear punches of the mice and genotyping was performed using polymerase chain reaction (PCR) for microsatellite markers (Mouse Mappairs, Invitrogen, Carlsbad, CA) and primers for restriction fragment length polymorphism (RFLP) markers (Operon, Huntsville, AL). PCR was performed using HotstarTaq Master Mix Kit (Qiagen, Valencia, CA). The PCR products were detected by electrophoresis on 10% polyacrylamide gel (National Diagnostics, Atlanta, GA) or on 1.5% agarose gel after digestion with restriction endonucleases (New England Biolabs, Beverly, MA) and visualized by ethidium bromide staining. Markers were selected 10–15 cM apart on each chromosome, and markers that clearly distinguished A/J and B6 genotypes were selected. The location of these markers is identified in Table 1 and Table 2.
Table 1.
Microsatellite markers and their chromosomal positiona
| Chromosome 5 |
Chromosome 11 |
Chromosome 17 |
||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Position (cM) | Position (Mb) | Marker | Position (cM) | Position (Mb) | Marker | Position (cM) | Position (Mb) |
| rs16809655 | 5.0 | 20.3 | D11Mit74 | 0 | 0.5 | D17Mit113 | 6.5 | 12.0 |
| D5Mit13 | 20 | 35.9 | D11Mit80 | 10 | 20.0 | D17Mit133 | 10.4 | 24.5 |
| D5Mit394 | 34 | 53.2 | D11Mit179 | 52 | 90.0 | D17Mit198 | 16 | 27.5 |
| D5Mit197 | 36 | 63.4 | D11Mit123 | 58 | 100.0 | D17Mit28 | 18.4 | 33.5 |
| rs6297441 | 54 | 98.0 | D11Mit258 | 65 | 107.5 | D17Mit178 | 24.5 | 48.0 |
| D5Mit338 | 59 | 107.7 | D11Mit61 | 70 | 110.5 | D17Mit20 | 34.3 | 57.0 |
| D5Mit320 | 70 | 125.5 | D11Mit336 | 75 | 112.0 | rs13483077 | 39.0 | 64.8 |
| rs13478553 | 77 | 135.7 | rs6380626 | 44.0 | 72.0 | |||
| D5Mit409 | 84 | 144.8 | D17Mit39 | 45.3 | 74.0 | |||
| D17Mit76 | 54.6 | 85.5 | ||||||
| D17Mit155 | 55.7 | 84.5 | ||||||
Position of markers obtained from Mouse Genome Database (Mouse Genome Database 2007)
Table 2.
RFLP markers, their chromosomal positions, primers, and restriction endonucleases
| SNP ID | Position (bp) | Forward primer | Reverse primer | Enzyme |
|---|---|---|---|---|
| rs16809655 | Chr 5: 20274741 | GCAACCCAGATCAAGCATAAGA | ATGATGAGAAGGTCCCCACA | SalI |
| rs6297441 | Chr 5: 98820203 | TAAGGCTGGGGAATGGTTTG | GGATTGGGTCTGACAACATAGG | ApalI |
| re13478449 | Chr 5: 107393510 | CCGTAGGTTCGTACCCACC | GTCCCATCATATTCCACAAAGTGC | EcoRV |
| rs13478553 | Chr 5: 135724469 | CATAGCCCAGCCCTCTGC | GGAGACACCACAAGCAGAATTG | XhoI |
| rs13483077 | Chr 17: 64806259 | GAAGGTACTGTCCCCGAGTC | TGGCGACGACTAAGCTACTT | XhoI |
| rs6380626 | Chr 17: 71942738 | TCCTGCTACCTCTCCTAGGAC | CTGTGAGTCTGTGTGTGGGT | HinfI |
Phenotyping
Phenotyping was performed using the bleeding/rebleeding assay as previously described (Hoover-Plow et al. 2006). Briefly, mice were anesthetized, and prewarmed tails were clipped and placed in saline. Bleeding time was measured as the time between the start of the bleeding and cessation of the bleeding. Clot stability time was measured as the time between the cessation of the bleeding and the start of the second bleeding.
Statistics
The linkage analysis was performed with MapManager QTX program (Manly et al. 2001). Ten thousand permutations of the trait values were used to define significant and suggestive thresholds and corresponded to the 95th and 37th percentiles, respectively. Kruskal-Wallis nonparametric ANOVA and the Mann-Whitney test were used to determine statistical differences in bleeding and clot stability times between genotype alleles.
Results
Chromosome 5
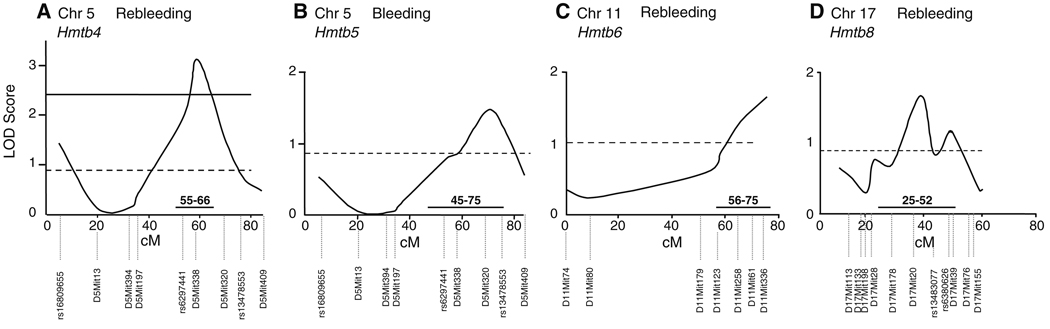
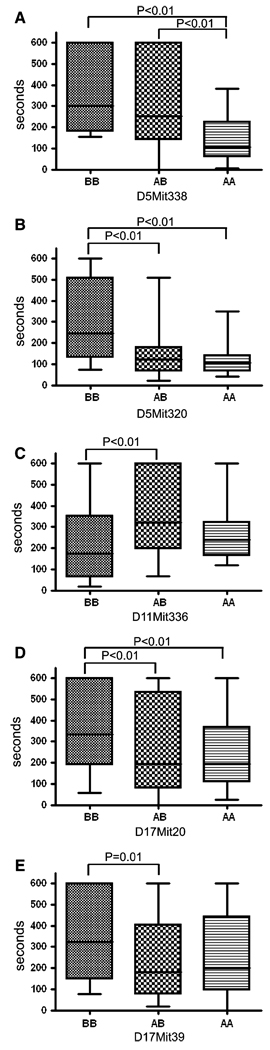
QTL analysis was carried out in F2 progeny (n = 79) from the CSS-5 × B6 intercross. CSS mapping is typically more efficient than traditional genome-wide scanning and requires fewer animals. This was illustrated by Singer et al. (2004) (see Supplementary Material) and noted in other studies (Belknap 2003; Matin et al. 1999). We estimated that QTL mapping for clot stability time using recombinant inbred F2 intercrosses requires nearly three times more mice than using CSSs to detect the same effect. In F2 progeny, bleeding/clot stability times were measured and a chromosome 5 genome scan was performed. For clot stability time, a significant locus named Hmtb4 (hemostasis thrombosis 4) was obtained at marker D5Mit338 (59 cM) with a LOD score of 3.1 (significant threshold = 2.4) and p = 0.0009; p = 0.008 with Bonferroni correction (Bland and Altman 1995) (Fig. 1a). This locus explained 16% of the variance in clot stability time in the F2 mice (Table 3). When clot stability time was plotted according to the genotypes at D5Mit338, a significant 2.9-fold increase (p = 0.04) was found for F2 mice homozygous for the B6 allele compared to the F2 mice homozygous for the A/J allele (Fig. 2a). This was unexpected because the B6 parental strain had a shorter clot stability time than A/J or CSS-5 mice (Hoover-Plow et al. 2006) (see Supplementary Material). Linkage analysis was also performed for bleeding time (Fig. 1b) for chromosome 5 and a suggestive peak was identified at marker D5Mit320 (70 cM) with a LOD score of 1.5 (suggestive threshold = 0.9, significant threshold = 2.4), which accounted for 8% of the variance (Table 3). This QTL was designated as Hmtb5. Mice with the homozygous B6 allele had a 2.5-fold longer bleeding time compared with mice with the A/J homozygous allele (Fig. 2b). There was no gender difference (p > 0.05) in the parent strains, CSS mice, and F2 progeny for clot stability and bleeding (data not shown). The F1 mice from the crosses of CSS-5 with B6 or CSS-17 with B6 had short clot stability time similar to the B6 mice (Hoover-Plow et al. 2006). However, the F1 mice from the cross of CSS-5 with CSS-17 conferred long clot stability time similar to A/J mice, indicating interactions between chromosome 5 and chromosome 17 (see Supplementary Material).
Fig. 1.
QTL analysis. (a) Hmtb4, peak marker D5Mit338 (59 cM). (b) Hmtb5, peak marker D5Mit320 (70 cM). (c) Hmtb6, peak marker D11Mit336 (75 cM). (d) Hmtb8, peak marker D17Mit 20 (34.3 cM); Hmtb9, peak marker D17Mit39 (45.3 cM). The linkage analysis was performed with MapManager QTX program. (Manly et al. 2001). Solid line indicates significant threshold (0.05). Dashed line indicates suggestive threshold (0.63). The 95% confidence intervals for each QTL are indicated
Table 3.
QTL analysis
| Locus name | Marker | Phenotype | Position (cM)a | Position (Mb) | LOD score | 95% CI | Variance % | p valueb | Significance |
|---|---|---|---|---|---|---|---|---|---|
| Hmtb4 | D5Mit338 | Clot stability | 59 | 107.7 | 3.1 | 55–66 | 16 | 0.0009 | Significant |
| Hmtb5 | D5Mit320 | Bleeding | 70 | 125.5 | 1.5 | 45–75 | 8 | 0.02 | Suggestive |
| Hmtb6 | D11Mit336 | Clot stability | 75.0 | 110.5 | 1.7 | 56–75 | 10 | 0.02 | Suggestive |
| Hmtb8 | D17Mit20 | Clot stability | 34.3 | 57.0 | 1.7 | 25–52 | 6 | 0.02 | Suggestive |
| Hmtb9 | D17Mit39 | Clot stability | 45.3 | 74.0 | 1.2 | 0–56 | 4 | 0.06 | Suggestive |
From Mouse Genome Database (2007). Number of F2 mice analyzed: chromosome 5, n = 79; chromosome 11, n = 76; chromosome 17, n = 130
Chromosome-wide
Fig. 2.
The allele distributions at peak markers in the F2 mice. (a) Hmtb4, clot stability time, chromosome 5, D5Mit338 BB, n = 14; AB, 45; AA, 20. (b) Hmtb5, bleeding time, chromosome 5, D5Mit320 BB, n = 10; AB, 47; AA, 22. (c) Hmtb6, clot stability time, chromosome 11, D11Mit336 BB, n = 27; AB, 34; AA, 15. (d) Hmtb8, clot stability time, chromosome 17, D17Mit20 BB, n = 39; AB, 60; AA, 31. (e) Hmtb9, clot stability time, chromosome 17, D17Mit39 BB, n = 35; AB, 48; AA, 47. A = A/J; B = B6. The lines at the middle indicate the medians. The boxes show the 25th and 75th percentiles. The whiskers show the ranges. Statistical differences are indicated between bars
Chromosome 11
A CSS-11 strain that had only the A/J-derived segments from D11Mit70 (0 cM) to D11Mit20 (20 cM) and from D11Mit4 (37 cM) to D11Mit336 (75 cM) was used for the chromosome 11 QTL analysis (n = 76). Using F2 mice, a suggestive locus, named Hmtb6, for clot stability time (Fig. 1c) was identified at marker D11Mit336 (75 cM) with LOD score of 1.7 (suggestive threshold = 1.0, significant threshold = 4.3), explaining 10% of the variance in clot stability time (Table 3). Clot stability time of the mice with the homozygous BB genotype at the marker D11Mit336 (Fig. 2c) was similar to the parental B6 strain (see Supplementary Material). However, the clot stability time of the mice with the heterozygous BA genotype (Fig. 2c) was significantly (p < 0.05) longer than the mice with the homozygous B6 genotype. The longer clot stability time in the heterozygous mice suggests overdominance (Smith et al. 2006). No gender difference was found in clot stability time in the F2 mice. Unlike the F1 mice from the cross CSS-5 × CSS-17, the clot stability times of the F1 mice from the crosses of CSS-11 with CSS-5 (264 ± 48 sec, n = 16) or CSS-17 (320 ± 63 sec, n = 15) were not different than the value for B6 mice, suggesting no interactions of chromosome 11 with chromosome 5 or chromosome 17.
Chromosome 17
QTL analysis was performed in F2 mice (n = 130) from the CSS-17 × B6 intercross (Fig. 1d). For clot stability time, two suggestive loci were identified. One, named Hmtb8, was at marker D17Mit20 (34.3 cM) with a LOD score of 1.7 (suggestive threshold = 0.8, significant threshold = 2.3), which explained 6% of the variance (Table 3). Another locus (Hmtb9) for clot stability was identified at marker D17Mit39 (45.3 cM) with a LOD score of 1.2 (suggestive threshold = 0.8, significant threshold = 2.3) that explained 4% of the variation (Table 3). As with QTLs on chromosome 5, at both Hmtb8 and Hmtb9 the homozygous B6 genotype conferred a longer clot stability time (Fig. 2d, e) than for the homozygous A/J genotype. This was unexpected since the A/J strain has prolonged clot stability time. No gender difference in bleeding and clot stability time was found in the F2 mice. The QTL interval on chromosome 17 is in synteny with a human QTL for protein C resistance (Hasstedt et al. 1998; Soria et al. 2003) on the short arm of chromosome 18. This conserved region is 6.4 Mb and 23 homologous genes were identified.
Discussion
In this study we used CSSs derived from A/J and B6 inbred strains to study the genetic control of thrombosis. Specific CSSs have been shown to have many phenotypic differences in response to vascular injury (Hoover-Plow et al. 2006) and marked differences in a tail-bleeding/rebleeding assay. We identified four QTLs for clot stability on chromosomes 5, 11, and 17, and one for bleeding on chromosomes 5. These five QTLs account for 44% of the total variance in the bleeding and clot stability phenotype, suggesting that other QTLs determining thrombotic risk remain to be identified. In this study, in addition to the five QTLs, 23 candidate genes, not previously suggested as thrombotic risk factors, were identified in the syntenic region in the Hmtb8 QTL for clot stability on chromosome 17.
In a previous study (Hoover-Plow et al. 2006), the observation was made of the possible interaction of chromosome 5 and chromosome 17. The F1 mice heterosomic for either chromosome 5 or chromosome 17 had similar rebleeding times compared to the B6 parental strain. Nevertheless, the F1 mice heterosomic for both chromosome 5 and chromosome 17 conferred prolonged rebleeding time similar to the A/J parental strain, suggesting that gene interactions between the two chromosomes had an additive effect. In contrast, no interactions of chromosome 11 with chromosome 5 or chromosome 17 were found. F1 mice from the crosses (CSS-11 × CSS-5) or (CSS-11 × CSS-17) had short clot stability time similar to the F1 mice hetersomic for chromosome 5 or chromosome 17.
The Hmtb4, Hmtb8, and Hmtb9 B6 alleles had prolonged clot stability times. A similar phenomenon has also been found in other studies that investigated atherosclerosis susceptibility in mice (Dansky et al. 2002; Ishimori et al. 2004). The prolonged times in our study did not coincide with phenotypes in the parental strains and suggest two possibilities: The parental strains bring a composite of allelic variants with contrasting and independent effects, or are from gene interactions. This paradox was present only in the loci on the two interacting chromosomes, chromosome 5 and chromosome 17, but not chromosome 11. A gene-gene interaction is supported by F1 data from different crosses. Chromosome 17 carries two loci for the clot stability trait, Hmtb8 and Hmtb9, and while clot stability time was recessive in F1 for chromosome 17, both of the two loci had dominant effects in F2. This could be explained by an inhibitory interaction between the two loci on chromosome 17 and suggests that A/J Hmtb8 or A/J Hmtb9 contributes to the dominant effect and that A/J Hmtb8 and A/J Hmtb9 have an inhibitory effect on each other. As a consequence, the combination of the two loci is recessive. Our study suggests the complexity of thrombosis and hemostasis.
In the Mouse Genome Database (2007) there are over 300 genes listed in the chromosome 5 QTL, so the next step is to generate a congenic strain with the QTL region and perform fine mapping (Armstrong et al. 2006; Christians and Keightley 2004; Wang et al. 2007). Interestingly, one QTL (Mvwf) associated with plasma von Willebrand factor (vWF) level was previously identified at the distal region of mouse chromosome 11 (Mohlke et al. 1996). The Mvwf candidate interval is between Ngfr and Hoxb9 at 56 cM within the clot stability locus Hmtb6. Whether the Mvwf locus and QTL for clot stability identified in this study are related remains to be determined. In addition, four genes known to modify thrombosis are located on chromosomes 5, 11, and 17: Serpin1 (plasminogen activator inhibitor-1) on chromosome 5; Serpinf2 (alpha-2 antiplasmin) on chromosome 17; Plg (plasminogen) on chromosome 17; and Mcfd2 (multiple coagulation deficiency 2) on chromosome 17. All four of these genes are outside the 95% CI of the respective locus suggesting that these genes are not the causative genes for the bleeding/clot stability trait.
A QTL on human chromosome 18 was reported to influence protein C resistance and thrombotic risk (Hasstedt et al. 1998; Soria et al. 2003). This human QTL region (18p11.32-11.23) coincides with the mouse QTL region on chromosome 17. Whether these two QTLs in different species result from variation of the same causative gene(s) for thrombosis remains to be verified. Twenty-three candidate genes are located in the conserved chromosome segment.
Susceptibility to thrombosis is a major risk factor for cardiovascular disease (CVD) and family history has been long associated with this risk, but the genetic determinants of thrombotic risk identified thus far do not account for the observed variation in human populations. To our knowledge, this is the first study to use CSSs for mapping QTLs for thrombosis susceptibility. Identifying the causative genes in these mouse QTLs could lead to the identification of new thrombotic risk factors in humans and ultimately to new therapeutic approaches to prevent and treat thrombosis.
Supplementary Material
Acknowledgments
The authors thank Drs. Jonathan Smith, Lindsey Burrage, and David Sinasac for their helpful discussions and Robin Lewis and Nadine Klimczak for assistance with preparation of the manuscript. This study was supported by grants from NIH, HL17964, HL65204, HL078701 (JHP), and RR12305 (JHN).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-008-9122-0) contains supplementary material, which is available to authorized users.
Contributor Information
Qila Sa, Department of Molecular Cardiology, Joseph J. Jacobs Center for Thrombosis and Vascular Biology, Cleveland, OH 44195, USA; Department of Cardiovascular Medicine, Cleveland Clinic Lerner Research Institute, Cleveland, OH 44195, USA.
Erika Hart, Department of Molecular Cardiology, Joseph J. Jacobs Center for Thrombosis and Vascular Biology, Cleveland, OH 44195, USA; Department of Cardiovascular Medicine, Cleveland Clinic Lerner Research Institute, Cleveland, OH 44195, USA.
Annie E. Hill, Department of Genetics, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA
Joseph H. Nadeau, Department of Genetics, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA
Jane L. Hoover-Plow, Department of Molecular Cardiology, Joseph J. Jacobs Center for Thrombosis and Vascular Biology, Cleveland, OH 44195, USA Department of Cardiovascular Medicine, Cleveland Clinic Lerner Research Institute, Cleveland, OH 44195, USA; Department of Molecular Cardiology, NB50, Cleveland Clinic Lerner Research Institute, Cleveland, OH 44195, USA hooverj@ccf.org.
References
- Armstrong NJ, Brodnicki TC, Speed TP. Mind the gap: analysis of marker-assisted breeding strategies for inbred mouse strains. Mamm Genome. 2006;17:273–287. doi: 10.1007/s00335-005-0123-y. [DOI] [PubMed] [Google Scholar]
- Belknap JK. Chromosome substitution strains: some quantitative considerations for genome scans and fine mapping. Mamm Genome. 2003;14:723–732. doi: 10.1007/s00335-003-2264-1. [DOI] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broze GJ, Yin ZF, Lasky N. A tail vein bleeding time model and delayed bleeding in hemophiliac mice. Thromb Haemost. 2001;85:747–748. [PubMed] [Google Scholar]
- Christians JK, Keightley PD. Fine mapping of a murine growth locus to a 1.4-cM region and resolution of linked QTL. Mamm Genome. 2004;15:482–491. doi: 10.1007/s00335-004-3046-0. [DOI] [PubMed] [Google Scholar]
- Dansky HM, Shu P, Donavan M, Montagno J, Nagle DL, et al. A phenotype-sensitizing Apoe-deficient genetic background reveals novel atherosclerosis predisposition loci in the mouse. Genetics. 2002;160:1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PJ. The genetics of atherothrombotic disorders: a clinician’s view. J Thromb Haemost. 2003;1:1381–1390. doi: 10.1046/j.1538-7836.2003.00276.x. [DOI] [PubMed] [Google Scholar]
- Hamilton JR, Cornelissen I, Coughlin SR. Impaired hemostasis and protection against thrombosis in protease-activated receptor 4-deficient mice is due to lack of thrombin signaling in platelets. J Thromb Haemost. 2004;2:1429–1435. doi: 10.1111/j.1538-7836.2004.00783.x. [DOI] [PubMed] [Google Scholar]
- Hasstedt SJ, Bovill EG, Callas PW, Long GL. An unknown genetic defect increases venous thrombosis risk, through interaction with protein C deficiency. Am J Hum Genet. 1998;63:569–576. doi: 10.1086/301947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JJ, Kwaan HC. Hereditary defects in fibrinolysis associated with thrombosis. Semin Thromb Hemost. 1999;25:321–331. doi: 10.1055/s-2007-994934. [DOI] [PubMed] [Google Scholar]
- Hoover-Plow J, Shchurin A, Hart E, Sha J, Hill AE, et al. Genetic background determines response to hemostasis and thrombosis. BMC Blood Disord. 2006;6:6. doi: 10.1186/1471-2326-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimori N, Li R, Kelmenson PM, Korstanje R, Walsh KA, et al. Quantitative trait loci analysis for plasma HDL-cholesterol concentrations and atherosclerosis susceptibility between inbred mouse strains C57BL/6 J and 129S1/SvImJ. Arterioscler Thromb Vasc Biol. 2004;24:161–166. doi: 10.1161/01.ATV.0000104027.52895.D7. [DOI] [PubMed] [Google Scholar]
- Kato K, Martinez C, Russell S, Nurden P, Nurden A, et al. Genetic deletion of mouse platelet glycoprotein Ibb produced a bernard-soulier phenotype with increased a-granule size. Blood. 2004;104:2339–2344. doi: 10.1182/blood-2004-03-1127. [DOI] [PubMed] [Google Scholar]
- Lemmerhirt HL, Broman KW, Shavit JA, Ginsburg D. Genetic regulation of plasma von Willebrand factor levels: quantitative trait loci analysis in a mouse model. J Thromb Haemost. 2007;5:329–335. doi: 10.1111/j.1538-7836.2007.02325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackman N. Tissue-specific hemostasis in mice. Arterioscler Thromb Vasc Biol. 2005;25:2273–2281. doi: 10.1161/01.ATV.0000183884.06371.52. [DOI] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH, Jr, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- Matin A, Collin GB, Asada Y, Varnum D, Nadeau JH. Susceptibility to testicular germ-cell tumours in a 129.MOLF-Chr 19 chromosome substitution strain. Nat Genet. 1999;23:237–240. doi: 10.1038/13874. [DOI] [PubMed] [Google Scholar]
- Mohlke KL, Nichols WC, Westrick RJ, Novak EK, Cooney KA, et al. A novel modifier gene for plasma von Willebrand factor level maps to distal mouse chromosome 11. Proc Natl Acad Sci U S A. 1996;93:15352–15357. doi: 10.1073/pnas.93.26.15352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Database (MGD) Mouse Genome Informatics website. Bar Harbor, ME: The Jackson Laboratory; 2007. Sep 15, Available at http://www.informatics.jax.org. [Google Scholar]
- Nadeau JH, Singer JB, Matin A, Lander ES. Analysing complex genetic traits with chromosome substitution strains. Nat Genet. 2000;24:221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- Singer JB, Hill AE, Burrage LC, Olszens K, Song J, et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Smith JD, Bhasin JM, Baglione J, Settle M, Xu Y, et al. Atherosclerosis susceptibility loci identified from a strain intercross of apolipoprotein E-deficient mice via a high-density genome scan. Arterioscler Thromb Vasc Biol. 2006;26:597–603. doi: 10.1161/01.ATV.0000201044.33220.5c. [DOI] [PubMed] [Google Scholar]
- Soria JM, Almasy L, Souto JC, Buil A, Martinez-Sanchez E, et al. A new locus on chromosome 18 that influences normal variation in activated protein C resistance phenotype and factor VIII activity and its relation to thrombosis susceptibility. Blood. 2003;101:163–167. doi: 10.1182/blood-2002-06-1792. [DOI] [PubMed] [Google Scholar]
- Sturm AC. Cardiovascular genetics: are we there yet? J Med Genet. 2004;41:321–323. doi: 10.1136/jmg.2003.017475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney JD, Novak EK, Reddington M, Takeuchi KH, Swank RT. The RIIIS/J inbred mouse strain as a model for von Willebrand disease. Blood. 1990;76:2258–2265. [PubMed] [Google Scholar]
- Wang M, Zhang Z, Zhang Z, Vikis H, Yan Y, et al. Fine mapping and candidate gene analyses of pulmonary adenoma resistance 1, a major genetic determinant of mouse lung adenoma resistance. Cancer Res. 2007;67:2508–2516. doi: 10.1158/0008-5472.CAN-06-3157. [DOI] [PubMed] [Google Scholar]
- Williams MS, Bray PF. Genetics of arterial prothrombotic risk states. Exp Biol Med (Maywood) 2001;226:409–419. doi: 10.1177/153537020122600504. [DOI] [PubMed] [Google Scholar]
- Zhang B, Ginsburg D. Familial multiple coagulation factor deficiencies: new biologic insight from rare genetic bleeding disorders. J Thromb Haemost. 2004;2:1564–1572. doi: 10.1111/j.1538-7836.2004.00857.x. [DOI] [PubMed] [Google Scholar]
- Zoller B, Hillarp A, Berntorp E, Dahlback B. Activated protein C resistance due to a common factor V gene mutation is a major risk factor for venous thrombosis. Annu Rev Med. 1997;48:45–58. doi: 10.1146/annurev.med.48.1.45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.