Whereas biopolymers, such as proteins, are ubiquitous for well-defined secondary, tertiary and quaternary structures,[1] it remains a fundamental challenge to design synthetic polymers that can fold into predictable high structures. One major research thrust in our laboratory is aimed at programming weak forces into polymers to guide their assembly into well-defined molecular and nano-structures.[2] Previously we reported biomimetic multi-domain polymers following modular design of titin.[3] In this study, our attention was drawn to β-sheet polymers. Despite significant progress has been made in the area of designing discrete short peptidomimetic oligomers with β-sheet structures,[4] it remains largely illusive to materials chemists to design synthetic high polymers that can fold into well-defined β-sheets and hierarchical nanostructures. β-Sheet is not only a basic secondary structure in proteins, but also an important structural motif in many fibril biomaterials such as amyloids[5] and silks.[6] Their hierarchical nanostructures and excellent mechanical properties have inspired biomemetic material designs.[7] Despite a number of peptide-related systems were reported to form β-sheet based fibrils, most of them are short peptides[8] or peptide-polymer conjugates.[9] The self-assembly in these systems usually proceeds intermolecularly, forming relatively weak structures. Genetically engineered polypeptides via recombinant DNA technology were reported to form β-sheets and various nanostructures,[10] however, the efficiency and versatility are limited by the biosynthetic pathway. Both for fundamental interest and for advanced materials designs, it is highly desirable to develop efficient synthesis to access well-defined covalently bonded β-sheet polymers. Herein we describe a new strategy of constructing covalent synthetic polymers that fold into well-defined β-sheets and further assemble into hierarchical nanofibrils (Fig. 1).
Figure 1.
Concept of cycloaddition-induced folding and self-assembly: (a) [2+3] cycloaddition polymerization of a protected peptide monomer; (b) upon deprotection polypeptides fold into well-defined antiparallel β-strands; (c) self-assembly of multiple β-sheets forms hierarchical nanofibrils.
As illustrated in Fig. 1, we employed Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC or commonly referred to as “click” chemistry) for polymerization of a peptide monomer. CuAAC is a versatile methodology because of its efficiency, functional group tolerance and applicability to a broad range of substrates.[11] Since the initial reports of this methodology,[11a,b] this reaction has been employed in a wide range of applications including selective ligations,[12] bioconjugation,[13] molecular recognition,[14] and material and polymer synthesis.[15,16] Based on structural similarities, 1,4- and 1,5-disubstituted 1,2,3-triazole rings formed by azide–alkyne cycloaddition have been used as biomimetic surrogates of peptides [17] in α-helical coil,[18] β-strands,[19] β-turn mimics,[20,21] and prosthetic proteins.[22] Despite these developments, it should be noted that the work described here represents the first example of applying this chemistry to induce high order structure formation in synthetic high polymers.
Our design is based on a convergent β-turn mimic our group has developed recently based on CuAAC reaction. We have shown that cycloaddition between two short peptide strands terminated with azide and alkyne forms 1,4-disubstituted 1,2,3-triazole ring that induces β-turn formation.[21] 1H NMR, FT-IR and molecular mechanics calculations reveal that three carbon linkers for 1,4- disubstituted triazole are optimal for the formation of β-turn structure in non-protic media. We reasoned that if an AB peptide monomer is prepared (A = azide, B = acetylene), [2+3] dipolar cycloaddition will not only efficiently polymerize the monomer, but should form β-turn to induce folding into antiparallel β-strands. Essentially, the cycloaddition polymerization induces folding of an encoded polymer into extensive β-sheets which further self-assemble into nanofibrils (Fig. 1).
For our initial concept demonstration, we installed alkyne and azide moieties onto a simple alanylglycine (AG)3 hexapeptide as the monomer unit (6) because AG repeats are common motifs for antiparalell β-sheet formation in silk[6] and biosynthetic poly- peptides.[10a,c] Based on our previous model studies, [21] three-carbon linkers were chosen to attach alkyne and azide to the C- and N- termini of the monomer 6 to maximize β-turn formation. One challenge in the syntheses and studies of well-defined β-sheet systems is their general poor solubility. Protecting strategy and switch-peptide concept have been utilized to circumvent this problem.[9a] In our design, we introduced an acid-cleavable 2,4-dimethoxy-benzyl (DMB) protecting group on one amide to prevent premature aggregation during the polymerization and facilitate polymer processing and characterizations. 2,4-dimethoxy-benzyl (DMB) has widely been used in peptide synthesis to inhibit excessive H-bonding.[23] In the end, deprotection of DMB triggers intramolecular folding and intermolecularly self-assembly (Fig. 1).
The synthesis of the monomer and polymer are illustrated in Scheme 1. The peptide monomer was prepared by combining standard solution and solid phase peptide coupling reactions. The preparation of the acetylene half of the monomer (4) began with Gabriel synthesis of 5-amino-1-pentyne followed by solution coupling to Boc-glycine to give Boc-Gly-pentyne 2. Boc deprotection of 2 followed by coupling with Boc-alanine afforded 3. Following Boc removal, DMB group was installed by reductive amination to give the DMB protected Ala-Gly-pentyne 4. The azide half of the monomer (azido-Ala-Gly-Ala-Gly-OH, 5) was prepared by solid phase peptide synthesis using 2-chlorotrityl chloride resin and standard Fmoc protocols.[24] The final step of segment coupling between the secondary amine in 4 and the terminal carboxylic acid in 5 was effected by using HATU as the coupling reagent in DMF (with 5% DMSO). The final monomer 6 was purified by column chromatography and characterized by 1H, 13C NMR, FTIR, HRMS and analytical HPLC. The full synthesis and characterization details for the monomer and polymers can be found in the Supporting Information (Scheme S1).
Scheme 1.
Synthesis of the β-sheet mimic polypeptide 8. Reaction conditions: (a) phthalimide, K2CO3, DMF, 70 °C, 24 h, (96%); (b) hydrazine hydrate, EtOH, 70 °C, 2 h, (74%); (c) Boc-glycine, EDC, HOBt, iPt2EtN, DCM, rt, 12 h, (98%); (d) TFA, DCM, rt, 3 h; (e) Boc-alanine, EDC, HOBt, iPt2EtN, DCM, rt, 12 h, (90%); (f) TFA, DCM, rt, 3 h; (g) 2,4-dimethoxybenzaldehyde, NaCNBH3, MeOH, rt, 12 h, (83%); (h) 7, HATU, iPt2EtN, DMF (with 5% DMSO), 48 h, (46%); (i) 2 mol% CuOAc, DMF, 80 °C, 2 h, (85%); (j) TFA, DCM, 2 h.
With the monomer 6 in hand, [2+3] cycloaddition polymerization was then carried out using a modified procedure[16] to afford polymer 7 with Mn of 11500 g/mol, Mw of 21800 g/mol and PDI of 1.89, determined by gel permeation chromatography (GPC) using poly(ethylene glycol) (PEG) as the molecular weight standards. The structure of polymer 7 was confirmed by NMR and FTIR. FTIR spectrum of 7 showed a sharp signal at ~2100 cm−1 corresponding to the azide functionality, indicating that polymer 7 still carries active azide end groups amenable for further reactions. DMB protecting groups successfully prevent premature aggregation to render polymer 7 fully soluble in polar organic solvent like DMF and 2,2,2-trifluroethanol (TFE). This enables the preparation of a soluble and processable prepolymer with encoded information for subsequent folding and self-assembly. Upon complete removal of the DMB groups in a 1/5 (v/v) TFA/methanol mixture (confirmed by 1H NMR, see Fig. S1 in Supporting Information), the resulting polymer 8 started folding and self-assembling into nanofibrils.
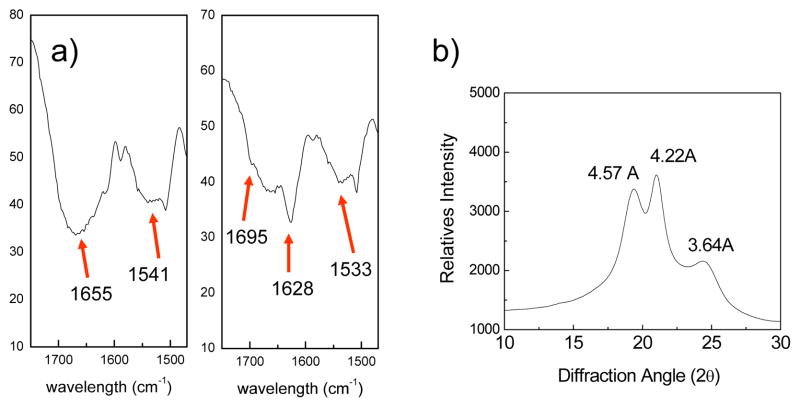
The β-sheet structure of polymer 8 was subsequently confirmed by FTIR, far-UV circular dichroism (CD) spectroscopy, and wide-angle powder X-ray diffraction (WXRD). FTIR spectrum (Fig. 2a) of the protected polymer 7 shows strong amide I band at ~1655 cm−1 and amide II band ~1541 cm−1, characteristic of random coil conformation.[25] Upon DMB deprotection and nanofibril formation, polypeptide 8 exhibits a significant enhancement at amide I band ~1628 cm−1 and amide II band ~1533 cm−1, supporting a β-sheet conformation.[25] Additionally, the weak component at ~1695 cm−1 is indicative of antiparallel β-sheets.[25] CD spectrum of 8 in hexafluoroisopropanol (HFIP) solution exhibits a minimum at 206 nm and maximum at 195 nm (see Fig. S2 in Supporting Information), confirming the β-sheet conformation because similar CD spectra have also been observed in β-sheet forming poly(AG)3YG or poly(AG)3HG made via genetic process.[10] In addition, wide-angle powder X-ray diffraction (WXRD) pattern of polymer 8 reveals major reflections at d spacings of 4.57, 4.22, and 3.64 Å, respectively (see Fig. 2b), which are similar to those observed for antiparallel β-sheet d spacings reported for the unoriented silk fibroin film [26] and the biosynthetic poly(AG)n.[10a]
Figure 2.
(a) FTIR spectra of 7 (left) and 8 (right) in amide I, II band region; (b) Wide angle X-ray diffraction of polymer 8.
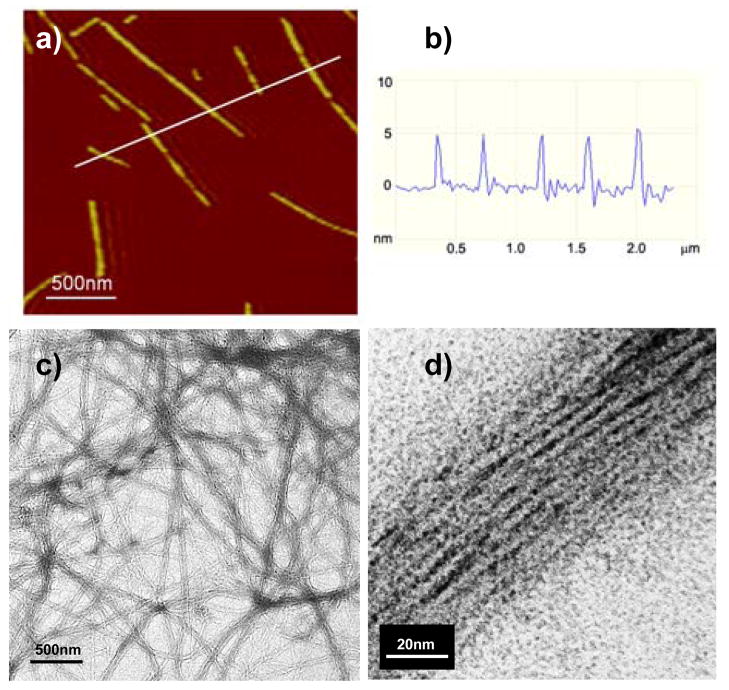
Finally, transmission electron microscopy (TEM) and atomic force microscopy (AFM) were employed to directly visualize the self-assembled nanostructures. For TEM studies, polymer 8 nanofibril suspensions were deposited on carbon-coated grid and a 2% uranyl acetate stain was used to increase edge contrast of the nanofibrils. Indeed, TEM images show assemblies of polymer 8 into long linear nanofibrils (Fig. 3c). Examination of the micrograph at higher magnification indicates the nanofibrils are composed of a stack of molecular fibrils formed from the β-sheet polymer. The average width of the molecular fibril was determined to be 3.8 ± 0.4 nm (see Fig. S4 in Supporting Information), which is in good agreement with the estimated width of the single β-sheet (see Fig. 4). The average width of the nanofibrils was found to be around 32 nm, indicating in average ~ 8 β-sheets stack laterally in one nanofibril. In order to investigate further the fibrillar morphology and texture of β-sheet fibrils, we analyzed the fibril dimensions by AFM for samples spin-coated on mica surfaces. AFM images (Fig. 3a and S6) revealed a similar morphology for the nanofiberils as shown by TEM. In addition, AFM analysis provides the height values of nanofibrils in the range of 1.7 to 7 nm with the most probable height at 5.0 ± 0.5 nm (Fig. 3b and Fig. S8 in Supporting Information). Both TEM and AFM images clearly demonstrated that the intramolecularly folded β-sheets of polymer 8 further self-assemble intermolecularly into amyloid-like nanofibrils.
Figure 3.
(a) A representative AFM micrograph for nanofibrils; (b) Height profile for 5 nanofibrils across the white line; (c) A representative TEM image of nanofibrils; (d) A zoom up view of one nanofibril.
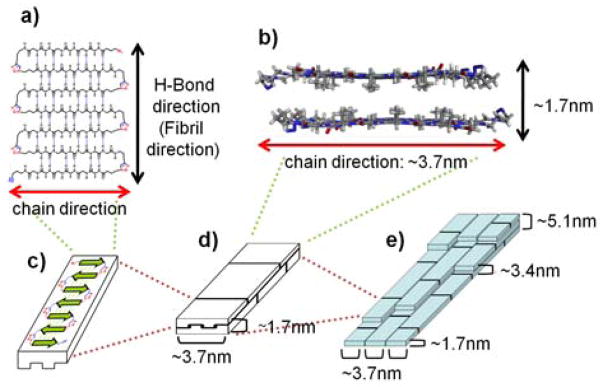
Figure 4.
Proposed model for hierarchical self-assembly of 8 to form nanofibrils: (a) Top-view of an antiparallel single β-sheet; (b) Side-view of a face-to-face stacked double layer of two β-sheets; (c) A single β-sheet; (d) Face-to-face stacked double layer of two β-sheets; (e) Stacking of many double layers forms the hierarchical nanofibrils.
On the basis of the TEM and AFM results and previous models,[10a,c] we propose here a model for the hierarchical self-assembly of polymer 8 into extensive b-sheets and nanofibrils (Fig. 4). In previous studies of poly(AG)n systems,[10a,c] it was shown that all alanine methyl groups orient toward the same face and the β-sheets arrange in the way that two like surfaces are in contact. We assume this occurs the same way in our system. The polymer 8 first folds into individual antiparallel β-sheets (Fig. 4a,c), then stack face-to-face into bilayers (Fig. 4b,d), and finally assemble both horizontally and vertically into nanofibrils (Fig. 4e). The β-strands run perpendicular to the fibril axis, resembling the cross-β structure of amyloid fibrils.[5] This model agrees with our experimental data. The observed width of individual molecular fibril (3.8 ± 0.4 nm) is consistent with the width of the β-strand estimated from the model (~3.7 nm). The nanofibril height measured by AFM falls in the range of 1.7 – 7 nm, with most probable height at 5.0 ± 0.5 nm. This agrees with the stacking of 1 – 4 layers of β-sheet bilayers with the most common ones of 3 layers (1.7nm × 3). Due to polydispersity of polymer 8, the longitudinal length of β-sheets varies so different β-sheet bilayers are interdigitated (Fig. 4d,e).
In summary, we describe here the first example of a synthetic polymer that can fold into well-defined β-sheets and further self-assemble into hierarchical nanostructures. The polymer with DMB protecting group was efficiently synthesized via Cu(I)-catalyzed azide–alkyne cycloaddition. Upon deprotection of DMB, the polymer was triggered to folds into well-defined β-sheets structure. The β-sheet structure of 8 was confirmed by FTIR, CD and WXRD data. TEM and AFM images show that the β-sheets further assemble into hierarchical amyloid-like nanofibrils. A key design element here is that the [2+3] dipolar cycloaddition not merely serves as the polymerization method but also induces the folding and self-assembly of the formed polymer. This demonstrates a unique example in which a polymerization leads to intramolecular folding to a secondary structure (β-sheet) and further intermolecular organization into hierarchical nanostructures. The efficiency and versatility of the click chemistry should allow for further design of more complex polymer materials. In our continuing studies, β-sheet motif will be combined with other folding motifs for the design of novel hierarchical biomaterials with advanced physical properties and specific functions on the nanometer scale.
Experimental Section
Monomer 6
In a 250 mL round bottom flask, compound 4 (0.365 g, 1.01 mmol) and peptide 5 (0.39 g, 1.01 mmol) were dissolved in 15 mL of DMF:DMSO (95:5) mixed solvent containing 0.265 mL DIPEA. Following addition of HATU (0.461g, 1.2 mmol), the mixture was stirred at room temperature for 48 hours. After addition of 100 mL of H2O to the completed reaction solution, the mixture was extracted with EtOAc (50 mL × 2). The combined organic layer was washed with H2O (100 mL × 1), 1M HCl (100 mL × 1), sat. NaHCO3 (100 mL × 1), and brine, then dried over anhydrous MgSO4, filtered, and finally concentrated on a rotary evaporator. The residue was purified by flash chromatography with MeOH/DCM (1:9) to give 6 as white solid (349 mg, 46%).1H NMR (500 MHz, MeOD-d4) δ 7.16 (d, J = 8.3, 1H), 6.53 (s, 1H), 6.48 (d, J = 8.3, 1H), 4.62 (d, J = 15.7, 1H), 4.40–4.37 (m, 2H), 4.22 (s, 2H), 4.17 (d, J = 7.2, 1H), 4.05 (d, J = 7.2, 1H), 3.89-3.58 (m, 10H), 3.43 (q, J = 7.0, 1H), 3.32-3.27 (m, 2H), 3.19-3.14 (m, 1H), 2.28 (t, J = 7.2, 2H), 2.16-2.10 (m, 3H), 1.86-1.81 (m, 2H), 1.72-1.70 (m, 2H), 1.36–1.10 (m, 9H); 13C NMR (125MHz, DMSO-d6) δ 173.3, 172.7,172.4, 172.0, 171.5, 171.1, 169.9, 169.2, 168.9, 168.6, 161.8, 160.6, 159.2, 158.2, 132.8, 129.3, 117.4, 105,2, 104.9, 98.8, 98.7, 84.6, 84.5, 71.9, 71.8, 56.1, 55.9, 55.8, 55.7, 55.3, 50.7, 39.0, 48.5, 48.5, 45.6, 44.0, 43.0, 42.4, 41.5, 38.1, 32.4, 31.2, 28.5, 24.8, 18.7, 18.2, 17.1, 15.83, 15.77, 15.0; HRMS (ESI), m/z calcd for [C33H48N10O9 + Na]+ = 751.3503; found 751.3494.
Polymer 7
7 was prepared according to a modified literature procedure.[25] Peptide monomer 6 (182 mg, 0.25 mmol), copper acetate (4 mg, 0.03 mmol), and 0.25 mL of N2-degassed DMF were introduced into a small vial. Under stirring the mixture was heated at 80°C in an oil bath for 2 hours. The initial clear solution turned into a dark green gel. After cooling down the polymerization mixture with an ice bath, the gel was dissolved with additional DMF and then polymer was precipitated into 0.1 N HCl (20 mL). The precipitate was purified by three consecutive times of centrifugation and re-dispersion with 0.1 N HCl. A white solid precipitate was finally isolated and dried under vacuum to give 155 mg of product 7 (yield: 85%). The molecular weight of the polymer was measured with GPC using poly(ethylene glycol) as standards: Mn = 11500, and Mw = 21750. 1H NMR (500 MHz, d6-DMSO and d-TFA) δ 8.37-8.26 (m, 1H), 7.02 (s, 1H), 6.39-6.33(m, 2H), 4.42-4.07 (m, 7H), 3.83–3.20 (m, 11H), 2.73 (s, 2H), 2.17-2.08 (m, 4H), 1.80 (s, 4H), 1.19-1.10 (m, 9H).
Full experimental details, including the syntheses and characterization, NMR, HRMS, GPC, FTIR, CD, WXRD, TEM and AFM experiments, can be found in the Supporting Information.
Supplementary Material
Footnotes
We acknowledge the financial support from the National Institutes of Health (R01EB004936) and the Department of Energy (DE-FG02-04ER46162). We thank Prof. Alexander McPherson in the Department of Molecular Biology & Biochemistry for assistance with AFM, Prof. James Nowick in the Department of Chemistry for assistance with HPLC, Dr. Jian-Guo Zheng at the Materials Characterization Center for assistance with TEM, and Drs. Wytze Van der Veer, Phil Dennison and John Greaves in the Department of Chemistry for assistance with CD, NMR and MS. We also thank Dr. Youli Li at the Materials Research Lab of UCSB for WXRD instrument assistance.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Stryer L. Biochemistry. 4. Spektrum; Heidelberg, Germany: 1996. [Google Scholar]
- 2.Guan Z. Polym Int. 2007;56:467–473. [Google Scholar]
- 3.a) Guan Z, Roland JT, Bai J, Ma S, McIntire T, Nguyen M. J Am Chem Soc. 2004;126:2058–2065. doi: 10.1021/ja039127p. [DOI] [PubMed] [Google Scholar]; b) Roland JT, Guan Z. J Am Chem Soc. 2004;126:14328–14329. doi: 10.1021/ja0448871. [DOI] [PubMed] [Google Scholar]; c) Kushner AM, Gabuchian V, Johnson EG, Guan Z. J Am Chem Soc. 2007;129:14110–14111. doi: 10.1021/ja0742176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Nowick JS. Acc Chem Res. 2008 doi: 10.1021/ar800064f. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hughes RM, Waters ML. Curr Opin Struct Biol. 2006;16:514–524. doi: 10.1016/j.sbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Guerette PA, Ginzinger DG, Weber BHF, Gosline JM. Science. 1996;272:112–115. doi: 10.1126/science.272.5258.112. [DOI] [PubMed] [Google Scholar]
- 7.a) Cherny I, Gazit E. Angew Chem. 2008;120:4128–4136. doi: 10.1002/anie.200703133. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:2–10. [Google Scholar]; b) König HM, Kilbinger AFM. Angew Chem. 2007;119:8484–8490. [Google Scholar]; Angew Chem Int Ed. 2007;46:8334–8340. doi: 10.1002/anie.200701167. [DOI] [PubMed] [Google Scholar]
- 8.a) Lashuel HA, LaBrenz SR, Woo L, Serpell LC, Kelly JW. J Am Chem Soc. 2000;122:5262–5277. doi: 10.1021/ja9937831. [DOI] [PubMed] [Google Scholar]; b) Lopez de la Paz M, Goldie K, Zurdo J, Lacroix E, Dobson CM, Hoenger A, Serrano L. Proc Natl Acad Sci USA. 2002;99:16052–16057. doi: 10.1073/pnas.252340199. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J Am Chem Soc. 2002;124:15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]; d) Reches M, Porat Y, Gazit E. J Biol Chem. 2002;277:35475–35480. doi: 10.1074/jbc.M206039200. [DOI] [PubMed] [Google Scholar]; e) Jung JP, Jones JL, Cronier SA, Collier JH. Biomaterials. 2008;29:2143–2151. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Hentschel J, Krause E, Börner HG. J Am Chem Soc. 2006;128:7722–7723. doi: 10.1021/ja060759w. [DOI] [PubMed] [Google Scholar]; b) Boerner HG, Smarsly BM, Hentschel J, Rank A, Schubert R, Geng Y, Discher DE, Hellweg T, Brandt A. Macromolecules. 2008;41:1430–1437. [Google Scholar]; c) Jahnke E, Lieberwirth I, Severin N, Rabe JP, Frauenrath H. Angew Chem. 2006;118:5510–5513. doi: 10.1002/anie.200600610. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2006;45:5383–5386. doi: 10.1002/anie.200600610. [DOI] [PubMed] [Google Scholar]; d) Klok HAJ. J Polym Sci Part A. 2005;43:1–17. [Google Scholar]; e) Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]; f) Burkoth TS, Benzinger TLS, Urban V, Lynn DG, Meredith SC, Thiyagarajan P. J Am Chem Soc. 1999;121:7429–7430. [Google Scholar]; g) Qu Y, Payne SC, Apkarian RP, Conticello VP. J Am Chem Soc. 2000;122:5014–5015. [Google Scholar]; h) Rathore O, Sogah DY. J Am Chem Soc. 2001;123:5231–5239. doi: 10.1021/ja004030d. [DOI] [PubMed] [Google Scholar]; i) Smeenk JM, Otten MBJ, Thies J, Tirrell DA, Stunnenberg HG, van Hest JCM. Angew Chem. 2005;117:2004–2007. doi: 10.1002/anie.200462415. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2005;44:1968–1971. [Google Scholar]; j) Zhang S, Marini DM, Hwang W, Santoso S. Curr Opin Chem Biol. 2002;6:865–871. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 10.a) Krejchi MT, Atkins EDT, Waddon AJ, Fournier MJ, Mason TL, Tirrell DA. Science. 1994;265:1427–1432. doi: 10.1126/science.8073284. [DOI] [PubMed] [Google Scholar]; b) West MW, Wang W, Patterson J, Mancias JD, Beasley JR, Hecht MH. Proc Natl Acad Sci USA. 1999;96:11211–11216. doi: 10.1073/pnas.96.20.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Topilina NI, Higashiya S, Rana N, Ermolenkov VV, Kossow C, Carlsen A, Ngo SC, Wells CC, Eisenbraun ET, Dunn KA, Lednev IK, Geer RE, Kaloyeros AE, Welch JT. Biomacromolecules. 2006;7:1104–1111. doi: 10.1021/bm0509016. [DOI] [PubMed] [Google Scholar]
- 11.a) Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]; b) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem. 2002;114:2708–2711. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; c) Kolb HC, Finn MG, Sharpless KB. Angew Chem. 2001;113:2056–2075. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; d) Meldal M, Tornoe CW. Chem Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 12.Bock VD, Hiemstra H, van Maarseveen JH. Eur, J Org Chem. 2006:51–68. [Google Scholar]
- 13.Moses JE, Moorhouse AD. Chem Soc Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 14.Meudtner RM, Hecht S. Angew Chem. 2008;120:5004–5008. doi: 10.1002/anie.200800796. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2008;47:4926–4930. doi: 10.1002/anie.200800796. [DOI] [PubMed] [Google Scholar]
- 15.Meudtner RM, Hecht S. Macromol Rapid Comm. 2008;29:347–351.Lutz J-F. Angew Chem. 2007;119:1036–1043.Angew Chem, Int Ed. 2007;46:1018–1025. doi: 10.1002/anie.200604050.Lutz J-F, Boerner HG, Weichenhan K. Australian J Chem. 2007;60:410–413.Lutz J-F, Boerner HG. Progress Polym Sci. 2008;33:1–39.Wu P, Feldman AK, Nugent AK, Hawker CJ, Scheel A, Voit B, Pyun J, Frechet JMJ, Sharpless KB, Fokin VV. Angew Chem. 2004;116:4018–4022. doi: 10.1002/anie.200454078.Angew Chem, Int Ed. 2004;43:3928–3932. doi: 10.1002/anie.200454078.also see Macromol Rapid Comm. 2008;29(12–13)for a series of reviews on applications of CuAAC to polymer synthesis.
- 16.a) Liu Y, Diaz DD, Accurso AA, Sharpless KB, Fokin VV, Finn MG. J Polym Sci Part A: Polym Chem. 2007;45:5182–5189. [Google Scholar]; b) Geng J, Lindqvist J, Mantovani G, Haddleton DM. Angew Chem. 2008;120:4248–4251. doi: 10.1002/anie.200800179. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:4180–4183. doi: 10.1002/anie.200800179. [DOI] [PubMed] [Google Scholar]; c) Hilf S, Hanik N, Kilbinger AFM. J Polym Sci Part A: Polym Chem. 2008;46:2913–2921. [Google Scholar]; d) Fournier D, Hoogenboom R, Schubert US. Chem Soc Rev. 2007;36:1369–1380. doi: 10.1039/b700809k. [DOI] [PubMed] [Google Scholar]; e) van Dijk M, Mustafa K, Dechesne AC, van Nostrum CF, Hennink WE, Rijkers DTS, Liskamp RMJ. Biomacromolecules. 2007;8:327–330. doi: 10.1021/bm061010g. [DOI] [PubMed] [Google Scholar]; d) van Dijk M, Nollet ML, Weijers P, Dechesne AC, van Nostrum CF, Hennink WE, Rijkers DTS, Liskamp RMJ. Biomacromolecules. 2008 doi: 10.1021/bm8005984. ASAP. [DOI] [PubMed] [Google Scholar]; f) Riva R, Schmeits S, Jerome C, Jerome R, Lecomte P. Macromolecules. 2007;40:796–803. [Google Scholar]; g) Lutz J-F, Boerner HG, Weichenhan K. Macromolecules. 2006;39:6376–6383. [Google Scholar]
- 17.Angell YL, Burgess K. Chem Soc Rev. 2007;36:1674–1689. doi: 10.1039/b701444a. [DOI] [PubMed] [Google Scholar]
- 18.Horne WS, Yadav MK, Stout CD, Ghadiri MRM. J Am Chem Soc. 2004;126:15366–15367. doi: 10.1021/ja0450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angelo NG, Arora PS. J Am Chem Soc. 2005;127:17134–17135. doi: 10.1021/ja056406z. [DOI] [PubMed] [Google Scholar]
- 20.a) Angell YL, Burgess K. J Org Chem. 2005;70:9595–9598. doi: 10.1021/jo0516180. [DOI] [PubMed] [Google Scholar]; b) Angell Y, Chen D, Brahimi F, Uri Saragovi H, Burgess K. J Am Chem Soc. 2008;130:556–565. doi: 10.1021/ja074717z. [DOI] [PubMed] [Google Scholar]
- 21.Oh K, Guan Z. Chem Comm. 2006:3069–3071. doi: 10.1039/b606185k. [DOI] [PubMed] [Google Scholar]
- 22.Tam A, Arnold U, Soellner MB, Raines RT. J Am Chem Soc. 2007;129:12670–12671. doi: 10.1021/ja075865s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zahariev S, Guarnaccia C, Zanuttin F, Pintar A, Esposito G, Maravic G, Krust B, Hovanessian AG, Pongor S. J Pept Sci. 2005;11:17–28. doi: 10.1002/psc.577. [DOI] [PubMed] [Google Scholar]
- 24.NovaBiochem Catalog. 2003 [Google Scholar]
- 25.a) Miyazawa T, Blout ER. J Am Chem Soc. 1961;83:712–719. [Google Scholar]; b) Haris PI, Chapman D. Biopolymers. 1995;37:251–263. doi: 10.1002/bip.360370404. [DOI] [PubMed] [Google Scholar]; c) Safar J, Roller PP, Ruben GC, Gajdusek DC, Jr, Gibbs CJ. Biopolymers. 1993;33:1461–1476. doi: 10.1002/bip.360330915. [DOI] [PubMed] [Google Scholar]
- 26.a) Fraser RDB, MacRae TP. Conformation in Fibrous Proteins. Academic Press; New York: 1973. [Google Scholar]; b) Asakura T, Sugino R, Okumura T, Nakazawa Y. Protein Sci. 2002;11:1873–1877. doi: 10.1110/ps.0208502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.