Abstract
Histone modification is an important mechanism of gene regulation in eukaryotes. Why many histone modifications can be stably maintained in the midst of genetic and environmental changes is a fundamental question in evolutionary biology. We obtained genome-wide profiles of three histone marks, H3 lysine 4 tri-methylation (H3K4me3), H3 lysine 4 mono-methylation (H3K4me1), and H3 lysine 27 acetylation (H3K27ac), for several cell types from human and mouse. We identified histone modifications that were stable among different cell types in human and histone modifications that were evolutionarily conserved between mouse and human in the same cell type. We found that histone modifications that were stable among cell types were also likely to be conserved between species. This trend was consistently observed in promoter, intronic, and intergenic regions for all of the histone marks tested. Importantly, the trend was observed regardless of the expression breadth of the nearby gene, indicating that slow evolution of housekeeping genes was not the major reason for the correlation. These regions showed distinct genetic and epigenetic properties, such as clustered transcription factor binding sites (TFBSs), high GC content, and CTCF binding at flanking sides. Based on our observations, we proposed that TFBS clustering in or near a histone modification plays a significant role in stabilizing and conserving the histone modification because TFBS clustering promotes TFBS conservation, which in turn promotes histone modification conservation. In summary, the results of this study support the view that in mammalian genomes a common mechanism maintains histone modifications against both genetic and environmental (cellular) changes.
Keywords: histone modification, transcription factor binding site, evolution of chromatin state
Introduction
An important question in evolutionary biology is how organisms regulate biological processes reproducibly in the midst of genetic and environmental changes (Wagner 2007). For faithful execution of gross cellular processes, such as development, cell cycle, and reproduction, gene expression must be regulated properly at the molecular level. How stable gene regulation is mechanistically achieved is an ongoing area of research in evolution (Masel and Siegal 2009).
Eukaryotic genomes are organized into chromatins, in which DNA sequences wrap around histone octamers to form nucleosomes. Posttranslational modification of histones (histone modification) at different regulatory regions can activate or repress gene expression (Wang et al. 2008; Heintzman et al. 2009; Ernst et al. 2011; Negre et al. 2011). Their importance in eukaryotic gene expression is supported by the fact that misregulation of chromatins can result in lower fitness of the organism, developmental defects, or diseases (Hendrich and Bickmore 2001). Histone modifications can be influenced by many factors, such as DNA mutations, cellular environmental variations, and external environmental variations (Bernstein et al. 2005; Ku et al. 2008; Mikkelsen et al. 2010; Cain et al. 2011; Ernst et al. 2011; Negre et al. 2011; Wang et al. 2011). Because stabilizing mechanisms have a genetic basis and can evolve (Waddington 1942), we wondered if eukaryotic genomes have mechanisms to stably maintain histone modifications.
The robustness or stability of biological systems depends on the source of perturbation, which means that the effectiveness of stabilizing mechanisms depends on the source of perturbation. For example, a phenotype that is robust to genetic mutations may not be robust to environmental changes (Masel and Siegal 2009). However, there are many phenotypes that are robust against both genetic mutations and environmental changes (Szollosi and Derenyi 2009; Price et al. 2011), indicating that a single mechanism can stabilize phenotypes against multiple sources of perturbations. An outstanding question is whether there is a common mechanism for maintaining histone modifications against different types of environmental and genetic changes.
In this study, we investigated the relationship between the conservation of histone modifications between species and the stability of histone modification among different cell types. We compared genome-wide histone modification profiles between mouse and human in the same cell types or among different human cell types to identify genomic sites with conserved or stable histone modifications, respectively. We found a strong association between the stability and the conservation of histone modifications, suggesting a common mechanism that maintains histone modifications against both genetic and environmental (cellular) changes. These regions showed distinct genetic and epigenetic properties, such as clustered transcription factor binding sites (TFBSs), high GC content, and CTCF binding at flanking sides. TFBS clustering provides an intuitive explanation for maintaining histone modifications against genetic and environmental changes.
Materials and Methods
Histone Modification Data Sets
We obtained genome-wide histone modification maps for three well-studied histone marks, H3 lysine 4 mono-methylation (H3K4me1), H3 lysine 4 tri-methylation (H3K4me3), and H3 lysine 27 acetylation (H3K27ac), which were generated by Chromatin Immunoprecipitation coupled with next-generation sequencing (ChIP-seq) (Johnson et al. 2007) (supplementary table S1, Supplementary Material online). The data sets were obtained as processed by the original study unless noted otherwise. First, to compare histone modifications between different species in matching cell types, we collected histone modification data sets from mouse and human livers, embryonic stem (ES) cells, undifferentiated preadipocytes, and differentiated adipocytes (supplementary table S1, Supplementary Material online). For the histone modification data in mouse ES and liver samples, the data in wig format was converted to enriched regions (“peaks”) using the Cistrome analysis platform (http://cistrome.org). Second, to compare histone modifications between different cell types in the same species, we obtained genome-wide modification maps for the histone marks from human cell lines of nine different cell types: B lymphoblastoid cells (GM12878), ES cells (H1), epidermal keratinocytes (NHEK), erythrocytic leukemia cells (K562), hepatocellular carcinoma cells (HepG2), lung fibroblasts (NHLF), mammary epithelial cells (HMEC), skeletal muscle myoblasts (HSMM), and umbilical vein endothelial cells (HUVEC) (Ernst et al. 2011). We used Model-based Analysis for ChIP-Seq (MACS) (Zhang et al. 2008) to detect significantly enriched regions (default parameters used except for “—tsize 36—mfold 10”) from ChIP sequences aligned to the human genome (GSE26386). Enriched regions were merged between biological replicates.
Histone Modification Conservation and Stability
Histone modification conservation was determined as follows. For all regulatory sites that incurred histone modifications in the human genome (the reference genome), we identified the fraction of the sites whose orthologous region in the mouse genome incurred the same histone modification (the test genome). Histone modification stability was determined as the number of cell types in which the regulatory site incurs histone modifications. We determined the histone modification status around a regulatory site as having either 1) a peak that encompasses ±75-bp regions around the site or 2) a peak in each of two flanking regions that are ±75–2,000 bp from the site because a regulatory site could be 1) encompassed by a single histone modification region (a monomodal distribution) or 2) flanked by two modification regions (a bimodal distribution), respectively.
Transcription Factor Binding Sites
Genome-wide maps of in vivo TFBSs were obtained from the ENCODE project (track name, “Txn Factor ChIP”) (The ENCODE Project Consortium 2011). Genome-wide maps of TF binding sequence motifs were obtained from the University of California Santa Cruz (UCSC) genome browser (“tfbsCons”). Note that this sequence motif data represent motifs that are evolutionarily conserved among human, mouse, and rat, and were used only for testing whether the regions containing clustered in vivo TFBSs were caused by nonspecific bindings. We used only in vivo TFBS maps for testing conservation or stability of TFBSs or histone modifications.
Gene Expression Data
We obtained gene expression data for the nine cell types generated using Affymetrix GeneChip U133A (GSE26312). Because the H1 ES cell type was assayed using a different GeneChip, U133 plus 2, the expression data for this cell type was not included. For some of the analyses, we used histone modification data for the remaining eight cell types. Expression breadth was defined as the number of cell types in which the gene shows an expression level above a threshold (log2-transformed rma expression value from Affymetrix GeneChip > 5) (Irizarry et al. 2003).
Regulatory Sites
We first obtained 105,204 regulatory sites across the genome as follows. First, we obtained sites that were predicted to be bound by TFs using multiple lines of experimental and computational evidence in the human genome (Pique-Regi et al. 2011). We combined sites across all cell types tested. Second, we filtered out sites which do not show any overlap with in vivo TFBSs from the ENCODE project (“Txn Factor ChIP”) (The ENCODE Project Consortium 2011). Third, to sample uniformly across the genome, we walked through the genome, selecting one site at each 2,000-bp window. Fourth, for the mouse–human comparison, we used the Galaxy platform to map orthologous regions in the mouse genome based on the pairwise multiple alignment between the mouse and the human genome.
We classified each site by their location relative to gene transcription: intronic, promoter (0–1,000 bp upstream of TSS), proximal promoter (1,000–5,000 bp upstream of TSS), and intergenic regions (>5,000 bp upstream of TSS), where TSS refers to the transcription start site. We used gene definitions from three different annotations: GENCODE (The ENCODE Project Consortium 2011), Refseq, and UCSC gene annotations. The regions that were classified consistently across all three annotations were used. For intronic regulatory sites, we removed regulatory sites located within 200 bp of exon–intron junctions, so as to avoid inclusion of splicing-related chromatin marks. We used the canonical gene start site, which refers to the 5′ upstream boundary of the transcript start, as an approximate location of TSS, a reasonably accurate and practical approach used by previous authors (McLean et al. 2010).
Data Analysis
The genome assembly build versions used in the study were hg18, for human, and mm9, for mouse. To examine conservation of TF-binding events between species, we used the UCSC liftOver tool for aligning the genomes of different species. For reliable mapping between orthologous regions of distant (not closely related) species, we used 0.2 instead of 0.1 (the default value) for minimum match and used the mapped region only if the intervals mapped to the same orthologous region after adding 100 or 500 bp to the flanking regions. We repeated the analysis for some TFs using MAF alignments in the Galaxy platform and reached the same conclusion (data not shown).
All high-level analyses were conducted using R (http://www.rprojects.org). The R/Intervals package and BEDtools were used for genomic interval calculations (Quinlan and Hall 2010). The R function glm was used to perform logistic or Poisson regression analyses with the binomial or Poisson family distribution. We calculated nucleotide composition information using R/Biostrings and R/BSgenome.Hsapiens.UCSC.hg18, obtained from the bioconductor repository (http://www.bioconductor.org). We restricted the analysis to the major autosomes.
Results
Stability and Conservation of Histone Modifications
We compared genome-wide histone modification profiles between mouse and human in matching cell types (conservation) or among different human cell types (stability) as follows. First, we obtained genome-wide histone modification maps for histone marks, H3K4me1, H3K4me3, and H3K27ac (supplementary table S1, Supplementary Material online). All three histone marks are well characterized and similar in overall function, but they are sufficiently different from each other, so we can draw a broad conclusion about the dynamics of active histone modifications. Second, we selected ∼100,000 putative regulatory sites across the human genome, which were classified into four types of regions (hereafter termed “genomic region type”): intronic, promoter (<1 kb from TSS), proximal promoter (1–5 kb from TSS), or intergenic (>5 kb from TSS) regions. We determined the histone modification status around each of the sites and around its orthologous site in the mouse genome. Third, we identified sites with the histone modifications maintained between species (conservation) or among cell types (stability).
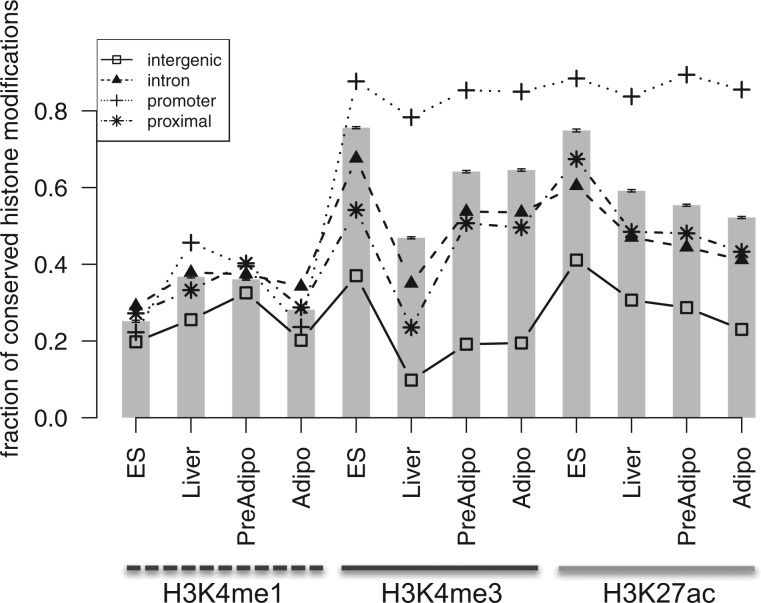
We tested if conservation of histone modifications would depend on the type of genomic regions and/or the histone mark. We found that the fraction of conserved histone modifications was the highest in promoter regions and the lowest in intergenic regions (fig. 1), indicating that the type of genomic region is an important determinant of histone modification conservation. The trend was found for all three histone marks examined. However, the strength of the trend was different between histone marks. For H3K4me3, the conserved fraction was much higher in the promoter region, consistent with previous studies (Ku et al. 2008; Cain et al. 2011). For H3K27ac, the conserved fraction was also high in the promoter region. For H3K4me1, there was no clear pattern (fig. 1). Next, we tested if stability of histone modifications would depend on the type of genomic regions and/or the histone mark. Stable histone modifications were more frequent in the promoter than in nonpromoter regions for H3K4me3 and HK27ac, but not for H3K4me1 (fig. 2; supplementary fig. S1, Supplementary Material online). It seems that the genomic region type influences both the conservation and stability of histone modifications in a histone mark-specific way.
Fig. 1.
Conservation of histone modifications between mouse and human, in different genomic regions. For all sites with histone modifications in the human genome (the reference genome), we identified the fraction (y axis) of the sites whose orthologous region in the mouse genome incurred the same histone modification (the test genome). x axis: results are shown for three histone marks (H3K4me1, H3K4me3, and H3K27ac) in four cell types (ES cells, liver cells, undifferentiated adipocytes, and differentiated adipocytes). Individual lines indicate the type of genomic region: intronic, promoter (<1 kb from TSS), proximal promoter (1–5 kb from TSS), or intergenic (>5 kb from TSS) regions, respectively. The bars indicate the overall conserved fraction for all types of genomic regions.
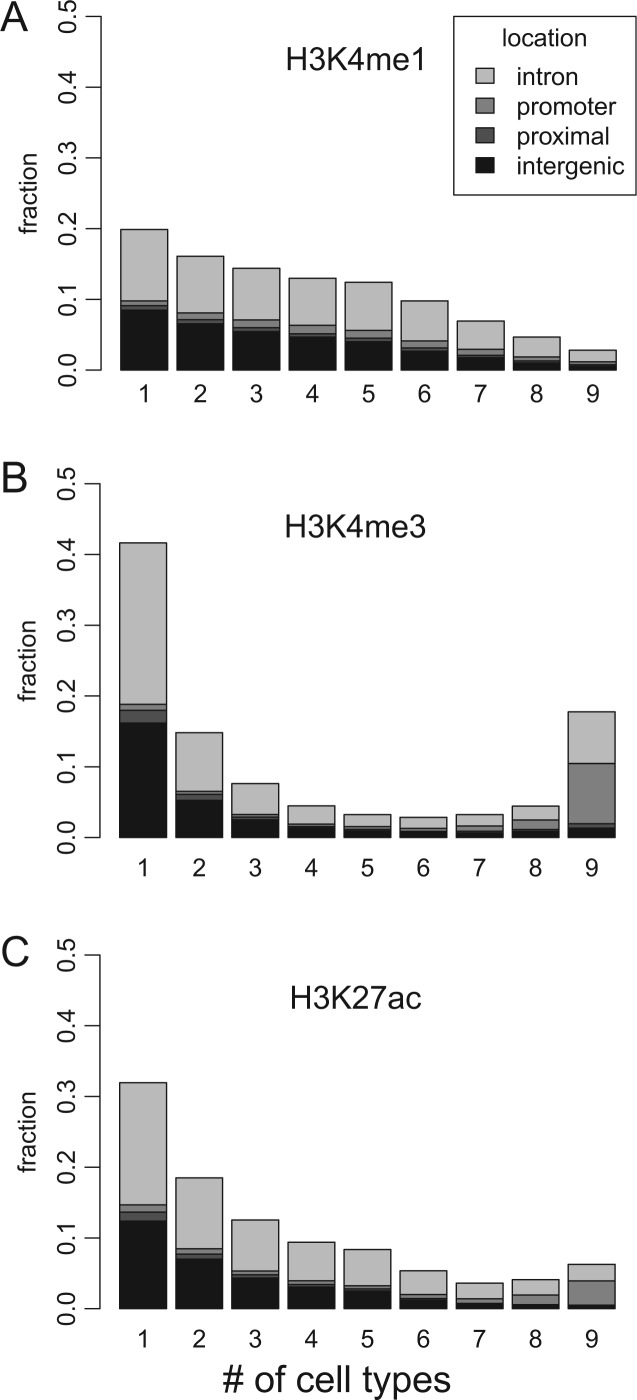
Fig. 2.
Stability of histone modifications among nine cell types in different genomic regions. (A) Histogram showing the fraction of the sites (y axis) in each stability class (x axis), that is, the number of cell types with histone modifications. A higher value on the x axis reflects a higher histone modification stability against changes in cellular conditions. The fractions sum up to one. We included only regulatory sites with histone modification in at least one cell type. The shade of the bar denotes the type of genomic region: intergenic, intronic, promoter, and proximal promoters. Note that most sites with H3K4me3 modifications (B) are found at x = 9, that is, ubiquitous or present among all cell types. The histone marks tested were H3K4me1 (A), H3K4me3 (B), and H3K27ac (C).
Correlation between Stability and Conservation of Histone Modifications
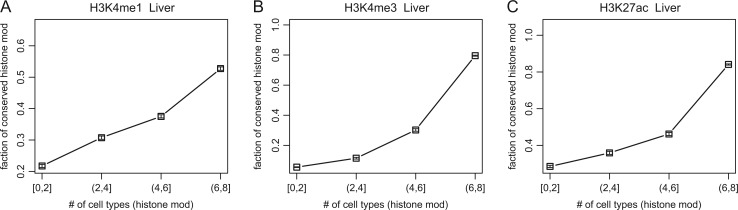
We examined the relationship between stability of histone modification among cell types and conservation between species. We found that the fraction of conserved histone modifications increased as their stability among cell types increased (fig. 3; supplementary fig. S2, Supplementary Material online). Of regulatory sites with H3K4me1 modifications in human liver, ∼20% were conserved between mouse and human when it occurred in less than or equal to two human cell types (that is, liver + another cell type); in contrast, more than 50% were conserved when it occurred in more than six human cell types (liver + more than five other cell types). The correlation between stability and conservation of histone modifications suggests a shared general stabilizing mechanism against different types of changes.
Fig. 3.
Correlation between conservation of histone modifications across species and stability of histone modifications among cell types. x axis: the number of cell types with histone modifications. y axis: the fraction of evolutionarily conserved histone modifications. Higher values indicate higher stability (x axis) or conservation (y axis). Error bars indicate the standard error based on a binomial distribution. Histone marks tested were H3K4me1 (A), H3K4me3 (B), and H3K27ac (C).
We considered the possibility that these trends were due to higher stability and conservation in promoter regions than in nonpromoter regions (figs. 1, 2B, and 2C). We repeated the analysis separately for the sites in intronic, promoter, proximal promoter, and intergenic regions and still detected a strong association between stability and conservation of histone modifications (supplementary fig S3, Supplementary Material online). If the differences between promoter and nonpromoter regions drove the correlation, such correlations would have disappeared after the stratification. It seems that the shared stabilizing mechanism is general, not limited to promoter regions.
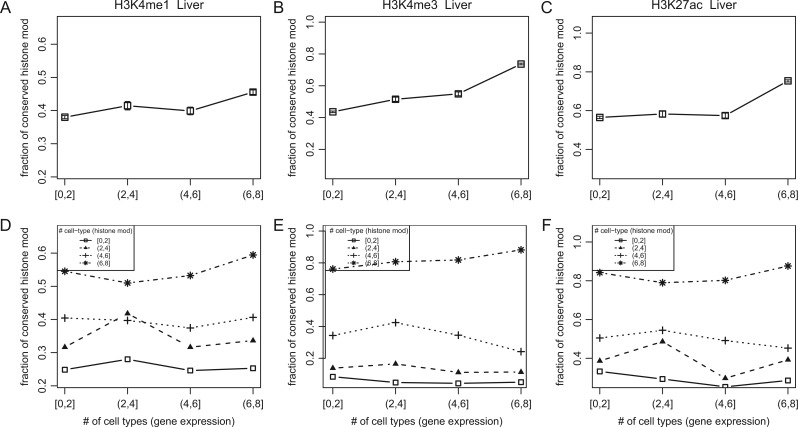
We studied if the association between stability and conservation of histone modifications can be explained by functional constraints of the nearby genes. Because broadly expressed genes tend to evolve slowly (Zhang and Li 2004; Yang et al. 2005), it was possible that the higher conservation of the histone modification was due to functional constraints of broadly expressed (housekeeping) genes. Indeed, conservation of histone modifications was high when the nearby gene is broadly expressed (fig. 4A–C; supplementary fig. S4, Supplementary Material online). The increase was, however, modest, suggesting that functional constraints by broadly expressed genes are not the major reason for the conservation of histone modifications. Importantly, the correlation between stability and conservation of histone modification was strong regardless of whether the site was near narrowly or broadly expressed genes (fig. 4D and F; supplementary fig. S5, Supplementary Material online). It seems that histone modification stability influences histone modification conservation independently of the functional constraints imposed by broadly expressed genes.
Fig. 4.
Evolutionary conservation of histone modifications, stability of histone modifications among cell types, and gene expression breadth of the nearby genes. y axis: the fraction of evolutionarily conserved histone modifications. x axis: gene expression breadth of the nearby genes. We removed sites that did not have a known gene within 10 kb. Expression breadth was defined as the number of cell types in which the gene showed an expression level above a threshold (log2-transformed rma expression value from Affymetrix GeneChip > 5). Error bars indicate the standard error based on a binomial distribution. In (A–C), all regulatory sites were used together for the analysis; in (D–F), the regulatory sites were stratified based on their histone modification stability (lines). The histone marks tested were H3K4me1 (A,D), H3K4me3 (B,E), and H3K27ac (C,F).
Genomic Sequence Composition and Conservation of Histone Modifications
We studied the mechanistic basis for high conservation and stability of the histone modifications. We examined the relationship between histone modification stability/conservation and nucleotide composition of the region. GC-rich and GC-poor isochores exhibit different nucleosome binding and regulatory properties in the human genome. We found that the fraction of G or C nucleotides in the region (“GC content”) around the ∼100,000 regulatory sites increased as histone modification conservation and stability increased (fig. 5A and B; supplementary fig. S6, Supplementary Material online). A likely explanation is that stable or conserved histone modifications of a nucleosome would depend on stable binding of histones to the DNA sequence. Supporting this, intrinsic sequence preferences for nucleosomes, which were determined from an in vitro nucleosome DNA reconstitution experiment (Valouev et al. 2011), was positively associated with increased GC content (linear regression, t-statistic, P < 2 × 10−16). We also examined the regional density of CpG dinucleotides because regions with high CpG density, that is, CpG islands, exhibit unique regulatory and chromatin properties (Guenther et al. 2007). The strength of association between CpG density and the histone modification stability and conservation was generally strong, but especially so for H3K4me3 (supplementary fig. S7, Supplementary Material online), consistent with previous findings that CpG islands readily incur H3K4me3 modifications (Guenther et al. 2007). It seems that the nucleotide composition of a genomic region influences the stability and conservation of the region's histone modifications in a histone mark-specific way.
Fig. 5.
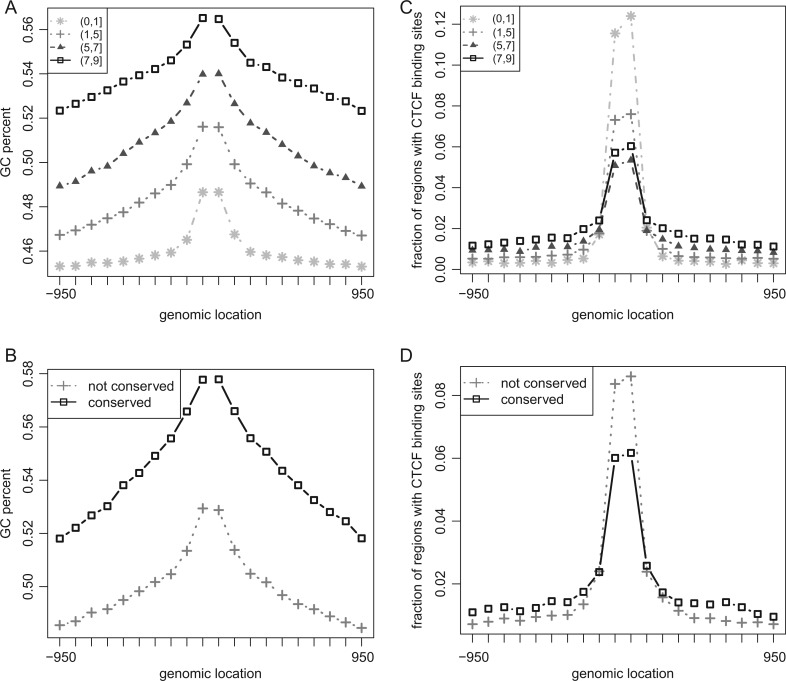
Evolutionary conservation of histone modifications, stability of histone modifications among cell types, nucleotide composition, and CTCF binding distribution. x axis: genomic distance (base pairs) from regulatory sites. Each point represents a 100-bp interval; for example, the leftmost position (x = −950 bp) would indicate genomic regions that are 900–1,000 bp upstream of the regulatory site on the positive genomic strand. y axis: the fraction of G and C nucleotides (A,B) or the fraction of sites bound with CTCF in HepG2 (C,D). In (A) and (C), the lines indicate sites with varying histone modification stability, that is, the number of cell types with histone modifications. In (B) and (D), dark and light lines indicate sites whose H3K4me1 modifications are conserved or not in HepG2 (liver cell type), respectively. Results for all three histone marks (H3K4me1, H3K4me3, and H3K27ac) in four cell types (ES, liver, preadipocyte, and adipocyte) are shown in supplementary figs. S6 and S8 (Supplementary Material online).
CTCF Binding Sites Demarcate Stable and Conserved Histone Modifications
We examined the distribution of CTCF binding sites, given its unique versatile roles as a transcription repressor and a chromatin regulator (Ohlsson et al. 2001). We found a complex relationship between distribution of CTCF binding sites and histone modification stability and conservation. Regulatory sites bound by CTCF, that is, <100 bp from the site, did not show higher histone modification stability and conservation. However, when bound CTCFs were hundreds of bases away, the regions exhibited higher histone modification stability and conservation (Fig. 5C and D; supplementary fig. S8, Supplementary Material online). The increase in histone modification stability and conservation was observed even when bound CTCFs were kilobases away; however, the degree of increase decreased as the genomic distance increased (supplementary fig. S9, Supplementary Material online). A likely explanation is that CTCF would stabilize the chromatin state around the regulatory site by demarcating active and repressive chromatin states (Cuddapah et al. 2009) or by positioning nucleosomes at the flanking regions (Fu et al. 2008). This explanation is supported by the observation that the stability and the conservation were significantly higher when CTCF was bound at both flanking sides than at one side alone (supplementary fig. S10, Supplementary Material online).
TFBS Clustering, TFBS Conservation, and Histone Modification Conservation
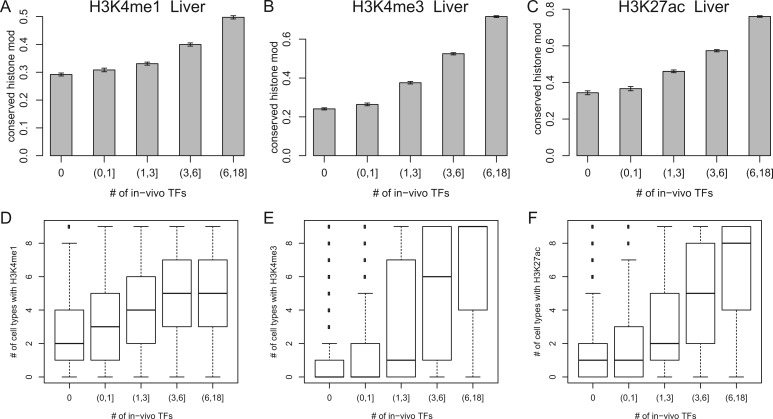
We hypothesized that TFBS clustering plays a significant role in stabilizing and conserving the histone modification for the following reasons. First, TF binding events are important for establishing histone modifications in the region (Graf and Enver 2009; Bonasio et al. 2010). Second, TFBSs are often clustered with each other with specific intersite spaces (Cai et al. 2010; He et al. 2011). From genome-wide in vivo TFBS maps for HepG2, a human hepatocarcinoma cell line (The ENCODE Project Consortium 2011), we calculated the number of TFBSs in the 1-kb region centered on each regulatory site. With an increase in the number of TFBSs around the site, there was an increase in the fraction of conserved (fig. 6A–C; supplementary fig. S11, Supplementary Material online) and stable (fig. 6D–F) histone modifications. We reached the same conclusion using larger or smaller genomic window sizes, for example, 200 bp or 2 kb (data not shown). Regions that contain clusters of in vivo TFBSs also contained clusters of sequence motif groups; note that multiple sequence motifs recognized by the same TF were not counted (supplementary fig. S12, Supplementary Material online). So, the observed trend was not likely to be caused by nonspecific TF bindings in open chromatin regions (Gerstein et al. 2010; modENCODE Consortium et al. 2010). We explore possible mechanisms in Discussion.
Fig. 6.
Evolutionary conservation of histone modifications, stability of histone modifications among cell types, and TFBS clustering. x axis: the number of in vivo TFBSs within a 1-kb window. We obtained in vivo TFBSs from the HepG2 cell line generated as a part of the ENCODE project (The ENCODE Project Consortium 2011). We counted the number of unique TFs that are bound instead of the total number of TFBSs, to remove the effect of homotypic clustering, that is, multiple bindings of the same TF. y axis: (A–C): the fraction of evolutionarily conserved histone modifications between human and mouse; (D–F): the number of cell types with the histone modifications. The histone marks tested were H3K4me1 (A,D), H3K4me3 (B,E), and H3K27ac (C,F).
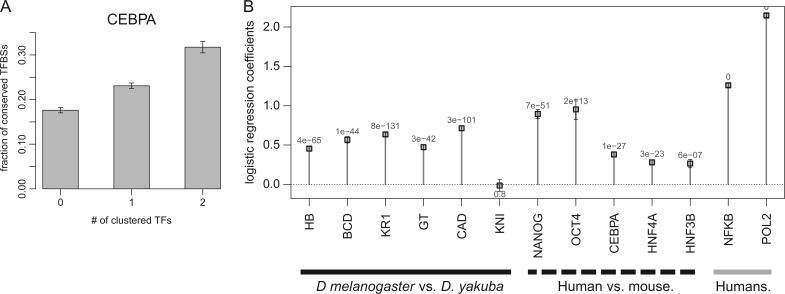
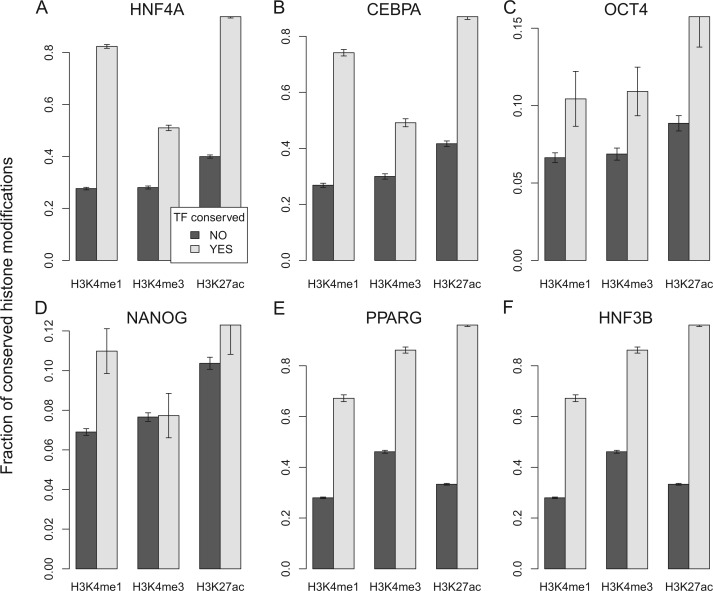
We hypothesized that clustering of TFBSs promotes histone modification conservation by increasing conservation of TF binding. We therefore examined 1) if TFBS clustering promotes conservation of TF binding and 2) if their conservation is accompanied by histone modifications. First, we tested for the relationship between conservation and clustering of TFBSs. We obtained cross-species TFBS maps for CCAAT/enhancer-binding protein alpha (CEBPA), hepatocyte nuclear factor 4 alpha (HNF4A), and hepatocyte nuclear factor 3 beta (HNF3B), in mouse and human liver samples; octamer-binding transcription factor 4 (OCT4) and homeobox (NANOG) in mouse and human ES cells; and six developmental TFs in Drosophila melanogaster and D. yakuba embryos. We found that the fraction of conserved TFBSs increased as the number of clustered TFBSs increased (fig. 7A; supplementary fig. S13, Supplementary Material online). Logistic regression analyses revealed that the trend was statistically significant (P < 0.05) in all cases except for one (fig. 7B). Together with previous studies (Kasowski et al. 2010; Schmidt et al. 2011), these observations support the view that a TFBS is better conserved between species when clustered with other TFBSs. Second, we tested for the relationship between conservation of TF bindings and conservation of histone modifications. We obtained matching mouse and human histone modification profiles for CEBPA, HNF4A, and HNF3B in liver and OCT4 and NANOG in ES cells. We also included histone modification profiles and TFBSs for peroxisome proliferator-activated receptor gamma (PPARG), a major regulator of fatty acid storage. For all of the six TFs tested, histone modification conservation drastically increased with TF binding conservation (fig. 8). For example, conservation of H3K4me1 between mouse and human liver was less than 30% when HNF4A binding was not conserved, but it increased to more than 80% when HNF4A binding was conserved. Together, these observations support our hypothesis that TFBS clustering promotes TFBS conservation, which leads to conservation of histone modification.
Fig. 7.
TF binding clustering and TF binding conservation. (A) y axis: the fraction of CEBPA binding sites in human that are conserved in mouse. x axis: the number of TFs, that is, HNF3B and/or HNF4A, that are bound nearby (<500 bp between midpoints of the binding peak). The results for the other TFs are shown in supplementary fig. S13 (Supplementary Material online). (B) y axis: the coefficient from the logistic regression, which measures the extent of changes in conservation when clustered with other TFBSs. Positive and negative values indicate increases or decreases in the conserved fraction when clustered with other TFSBs, respectively. An error bar indicates the standard error of the coefficient. The P value for the statistical significance of the coefficient is given at each x value. x axis: TFs. Supplementary table S2 (Supplementary Material online) provides details of each TF.
Fig. 8.
TF binding conservation and histone modification conservation. A barplot showing the fraction of conserved histone modifications (y axis) as a function of TF binding conservation (x axis). y axis: the fraction of TF binding sites whose orthologous regions in the mouse showed conservation. We used mouse and human binding maps for six TFs: HNF4A (A), CEBPA (B), OCT4 (C), NANOG (D), PPARG (E), and HNF3B (F), and histone modification data in matching cell types: ES cells (C,D), adipocyte (E), and liver (A,B,F).
We examined the relationship between TFBS clustering, TF binding stability, and histone modification stability between two human cell types, HepG2 and K562. Similar to the trend for conservation between species, we found that TF binding events were maintained between the two cell types more often when clustered with other TFBSs (supplementary fig. S14, Supplementary Material online), and the TF binding retention was coupled with histone modification stability (supplementary figs. S15 and S16, Supplementary Material online). The trend was observed for the majority, though not all, of the TFs. Deviations from the trend perhaps represent a complex intricate control of cell-type specific gene regulation by different combinatorial TFBSs. It seems that TFBS clustering promotes stability of TF binding, leading to the stability of histone modifications, reminiscent of conservation of TF binding between species.
Discussion
In this study, we showed that histone modifications that are maintained among many cell types are also likely to be conserved between species. We also showed several genomic features, such as GC content, CTCF binding at flanking sides, and clustering of TFBSs, provide a common genomic architecture for the conservation and the stability of histone modifications.
Our observations suggest that a common mechanism maintains histone modifications against both genetic mutations and environmental variations. How did this mechanism evolve? In biological systems, stability against one type of perturbation can evolve as a by-product of evolved stability against another type of perturbation (Haldane 1930; van Nimwegen et al. 1999; Szollosi and Derenyi 2009; Price et al. 2011). The mechanism to conserve histone modifications between species probably evolved as a by-product of the evolution of a mechanism to stabilize histone modifications against nongenetic variations.
TFBS clustering provides an intuitive explanation for conserving and stabilizing histone modifications against genetic and environmental changes. Binding events of individual TFs may change during mammalian evolution due to genetic mutations of the binding motifs (Schmidt et al. 2010). Loss of a TFBS would be less likely to affect the histone modification landscape of the region in the presence of other nearby TFBSs, reminiscent of duplicate genes providing robustness against null mutations of a single gene (Gu et al. 2003). TFBSs could also be influenced by many nongenetic factors. For example, the level of TF proteins could fluctuate stochastically or due to changes in internal or external cellular environment. Intuitively, such fluctuations would be less likely to lead to loss of TF binding events because the chromatin of the region is readily accessible due to other TFs bound to the region. We propose that clustering of TFBSs is not only a combinatorial logic for gene regulation but also a stabilizing mechanism, complementing and expanding the functional role of cis-regulatory modules (Istrail and Davidson 2005).
Which biological process underlies stable and conserved histone modifications? The process of transcription is a major mechanism for regulating the chromatin state (Ernst et al. 2011; Kharchenko et al. 2011). Noncoding RNA transcription occurs extensively in the mammalian genome, around promoters of protein-coding genes and enhancers in intergenic regions (Core et al. 2008; Wang et al. 2011). We have previously shown that head-to-head clustering of genes, whose transcription occurs divergently or bidirectionally, can promote stability of the gene expression (Woo and Li 2011). We speculate that bidirectional transcription can promote stability of the histone modification in the region, in the promoter and nonpromoter regions. Supporting this, our preliminary analysis revealed the pattern of bidirectional transcription in regions with stable and conserved histone modifications (Woo YH, Li W-H, unpublished data). This interesting hypothesis warrants further investigations.
We examined the effect of TFBS clustering on stability and conservation of histone modifications for a varying range of clustering distances (base pairs). We found that the increase in conservation was the highest for short clustering distances and decreased with increasing distances (supplementary fig. S17, Supplementary Material online). It is likely that there are different mechanisms for short-range and long-range clustering. For short-range clustering that span over one, two, or three nucleosomes (e.g., <500 bp), neighboring TF binding events could be coupled, so that their evolution would be slower due to the constraints. An example of such constraints would be functional constraints of cis-regulatory modules, in which specific TFBS combinations give rise to a functionally important regulatory code. Also, certain clustering might not provide specific regulatory functions, but could still increase collaborative effects; short-range clustering (<200 bp) of TFBSs would be evolutionarily stable because such simultaneous binding could compete with and evict the nucleosome by “collaborative competition” (Miller and Widom 2003). For long-range clustering beyond neighboring nucleosomes (e.g., >500 bp), an intuitive explanation is that having many TFBSs spread over multiple nucleosomes would result in mutual sharing of histone modifications in the region. Theoretical simulations would predict that multiple loci for histone modification initiation would achieve a more stable histone modification state in the region than a linear progression of modification from a single locus (Dodd et al. 2007). Such multiple TFBSs would provide multiple redundant venues for histone modification processes, reminiscent of multiple gene copies providing robustness against null mutations (Gu et al. 2003). Supporting this, we found a strong association between clustering of TFBSs and overlaps of multiple histone modifications, which was associated with increased chromatin accessibility and increased conservation of the histone modifications (Woo YH, Li W-H, in preparation). It is an interesting hypothesis whether the clustering of TFBSs can also occur across genomic loci that are far in the linear genome but close in the 3D nucleus (Woo et al. 2010).
How TF binding coevolves with histone modification is difficult to study because of a complex causal relationship between TF binding and histone modification. In many cases, TF binding conservation would be the ultimate cause of histone modification conservation. Two lines of evidence support this view. First, in prokaryotes, TF regulatory networks can be inherited to subsequent generations without chromatins, suggesting that TF binding events can form gene regulatory networks without histone modifications (Bonasio et al. 2010). Second, in cellular reprogramming experiments, changing the TF gene expression level can cause histone modification changes (Graf and Enver 2009). However, histone modifications could also influence TF binding to a certain extent, depending on the TF and the histone mark. A positive-feedback loop can have stabilizing effects in biology (Masel and Siegal 2009; Bonasio et al. 2010). We envisage that the feedbacks between TF binding and histone modification have evolved in eukaryotes in response to increased complexity in cellular processes and, at the same time, provide stability to the regulatory system in the midst of genetic and environmental changes.
Supplementary Material
Supplementary figures S1–S17 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Institutes of Health grant GM30998. The data reported in the paper are obtained from publicly available data repositories and are listed in the Supporting Information. We thank the ENCODE and the modENCODE consortium for making the data available.
References
- Bernstein BE, Kamal M, Lindblad-Toh K, et al. (12 co-authors) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hou L, Su N, Hu H, Deng M, Li X. Systematic identification of conserved motif modules in the human genome. BMC Genomics. 2010;11:567. doi: 10.1186/1471-2164-11-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain CE, Blekhman R, Marioni JC, Gilad Y. Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics. 2011;187:1225–1234. doi: 10.1534/genetics.110.126177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- doi: 10.1371/journal.pbio.1001046. The ENCODE Project Consortium. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9:e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, et al. (14 co-authors) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, et al. (131 co-authors) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. A note on Fisher's theory of the origin of dominance, and on a correlation between dominance and linkage. Am Nat. 1930;64:87–90. [Google Scholar]
- He BZ, Holloway AK, Maerkl SJ, Kreitman M. Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules. PLoS Genet. 2011;7:e1002053. doi: 10.1371/journal.pgen.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, et al. (21 co-authors) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bickmore W. Human diseases with underlying defects in chromatin structure and modification. Hum Mol Genet. 2001;10:2233–2242. doi: 10.1093/hmg/10.20.2233. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci U S A. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, et al. (17 co-authors) Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, et al. (30 co-authors) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, et al. (17 co-authors) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium, Roy S, Ernst J, et al. (97 co-authors) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, et al. (40 co-authors) A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N, Cartwright RA, Sabath N, Graur D, Azevedo RB. Neutral evolution of robustness in Drosophila microRNA precursors. Mol Biol Evol. 2011;28:2115–2123. doi: 10.1093/molbev/msr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, et al. (13 co-authors) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SF, Jorgensen M, Chen Y, Nielsen R, Sandelin A, Mandrup S. Cross species comparison of C/EBPalpha and PPARgamma profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC Genomics. 2011;12:152. doi: 10.1186/1471-2164-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi GJ, Derenyi I. Congruent evolution of genetic and environmental robustness in micro-RNA. Mol Biol Evol. 2009;26:867–874. doi: 10.1093/molbev/msp008. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E, Crutchfield JP, Huynen M. Neutral evolution of mutational robustness. Proc Natl Acad Sci U S A. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The canalization of development and the inheritance of acquired characters. Nature. 1942;150:563. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Wagner A. Robustness and evolvability in living systems. Princeton (NJ): Princeton University Press; 2007. [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, et al. (13 co-authors) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, et al. (11 co-authors) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YH, Li WH. Gene clustering pattern, promoter architecture, and gene expression stability in eukaryotic genomes. Proc Natl Acad Sci U S A. 2011;108:3306–3311. doi: 10.1073/pnas.1100210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YH, Walker M, Churchill GA. Coordinated expression domains in mammalian genomes. PLoS One. 2010;5:e12158. doi: 10.1371/journal.pone.0012158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Su AI, Li WH. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol Biol Evol. 2005;22:2113–2118. doi: 10.1093/molbev/msi206. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, et al. (11 co-authors) Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.