Background: The master regulator of cysteine metabolism-CymR influences S. aureus stress resistance and virulence.
Results: Mutation of the sole cysteine residue Cys-25 to Ser eradicates the redox sensing ability of the protein.
Conclusion: CymR is a new thiol-based oxidation-sensing regulator.
Significance: Elucidating the oxidation-sensing mechanism of CymR is important for understanding oxidation sensing by S. aureus.
Keywords: Bacteria, Cell Signaling, Crystal Structure, Metabolism, Staphylococcus aureus, CymR, Cysteine Metabolism, Oxidation Sensing, Sulfenic Acid
Abstract
As a human pathogen, Staphylococcus aureus must cope with oxidative stress generated by the human immune system. Here, we report that CymR utilizes its sole Cys-25 to sense oxidative stress. Oxidation followed by thiolation of this cysteine residue leads to dissociation of CymR from its cognate promoter DNA. In contrast, the DNA binding of the CymRC25S mutant was insensitive to oxidation and thiolation, suggesting that CymR senses oxidative stress through oxidation of its sole cysteine to form a mixed disulfide with low molecular weight thiols. The determined crystal structures of the reduced and oxidized forms of CymR revealed that Cys-25 is oxidized to Cys-25-SOH in the presence of H2O2. Deletion of cymR reduced the resistance of S. aureus to oxidative stresses, and the resistance was restored by expressing a C25S mutant copy of cymR. In a C25S substitution mutant, the expression of two genes, tcyP and mccB, was constitutively repressed and did not respond to hydrogen peroxide stress, whereas the expression of the genes were highly induced under oxidative stress in a wild-type strain, indicating the critical role of Cys-25 in redox signaling in vivo. Thus, CymR is another master regulator that senses oxidative stress and connects stress responses to virulence regulation in S. aureus.
Introduction
Staphylococcus aureus is an important Gram-positive human pathogen that causes a variety of ailments ranging from soft tissue infections to life-threatening diseases such as toxic shock syndrome to endocarditis to necrotizing pneumonia (1, 2). The human host has developed defense systems such as macrophages to fight against pathogen infections like S. aureus (3). Macrophages use toxic reactive oxygen species (ROS)3 to destroy phagocytosed bacteria during active infection (4, 5). To cope with ROS, S. aureus is equipped with multiple defensive systems to sense and defend against ROS (5). In particular, thiol-based oxidation-sensing regulatory proteins such as MgrA and SarZ play major regulatory roles in S. aureus. Recent work also suggests that another master regulator, SarA, utilizes the same thiol-based redox sensing to control gene expression in S. aureus (6, 7). These regulators belong to the OhrR family of proteins (8, 9). Similar redox-sensitive regulators also exist in Gram-negative pathogens such as OspR and MexR in Pseudomonas aeruginosa and OxyR in Escherichia coli (8, 10–16).
Previously, we have shown that MgrA and SarZ sense oxidative stress through the oxidation of the sole cysteine residue conserved in these proteins to form a sulfenic acid intermediate. A subsequent thiolation of the generated sulfenic acid with cellular LMW thiols yields mixed disulfide, which has been thought to lead to dissociation of the modified proteins from DNA (10, 11, 17). For example, MgrA uses this mechanism to regulate antibiotic resistance and virulence. It controls more than 300 genes that cover a broad range of functions (17, 18). SarZ is used to control expression of genes involved in detoxification of ROS and metabolic switching (11). CymR is the master regulator of cysteine metabolism in S. aureus, and it has been shown to affect expression of over 300 genes (19). It also plays important roles in stress resistance and bacterial virulence (20). Perhaps not surprisingly, deletion of cymR makes S. aureus more sensitive to hydrogen peroxide, tellurite, and other stresses (20), which can be attributed to its function as a regulator of ROS detoxification genes such as ahpFC, as well as cysteine biosynthesis genes such as cysM and mccAB. In S. aureus, cysteine is considered to be one of the major components of the cellular reducing buffer because of the absence of GSH biosynthesis (21–23). Despite past studies of CymR, the exact sensing and regulatory mechanism of this master regulator have yet to be elucidated. An inspection of the CymR sequence revealed to us that CymR has only one cysteine, a feature characteristic of the MgrA family proteins (supplemental Fig. S1). This observation prompted us to hypothesize that CymR could employ a thiol-based oxidation-sensing mechanism similar to that of MgrA and OhrR to regulate gene expression. In this study, we show that oxidation of CymR leads to its dissociation from the cognate promoter DNA. The oxidation intermediate, Cys-25-SOH, was captured and characterized by crystallography. We also demonstrate that the sole cysteine residue is critical for its oxidation-sensing regulation inside bacterium.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in supplemental Table S2. E. coli strains were grown in LB broth. Staphylococci were grown in tryptic soy broth (TSB) except transduction procedures, for which heart infusion broth supplemented with 5 mm CaCl2 was used. When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml. Vector pMCSG7 was used for expressing His-tagged proteins. Vector pKOR1 was used for deletion of cymR gene in S. aureus. E. coli-S. aureus shuttle vector pCL55-FLAG was used for complementation.
Construction of cymR Deletion Mutant and Complementation
To delete the cymR gene, the allelic exchange plasmid pKOR1 was used. Four primers were used into order to generate flanking DNA fragments for deletion (supplemental Table S1): upcymRFor and upcymRRev for the upstream fragment and downcymRFor and downcymRRev for the downstream fragment. The fragments were cloned into pKOR1 by using Gateway BP Clonase Technology (Invitrogen) to generate the plasmid pKOR1::1528UD. Integration and excision of the plasmid were performed as previously described (24). cymRFor and cymRRev-Flag complementation primers amplified the cymR gene for complementation experiments. The PCR product was introduced into pCL55-FLAG to generate cymR-FLAG plasmid. CymR was expressed from its own promoter in S. aureus. To make cymRC25S-FLAG plasmid, two primers cymRFor Cys-25 to Ser and cymRRev Cys-25 to Ser were used, and the mutagenesis was carried out by following QuikChange site-directed mutagenesis (Stratagene). These plasmids were introduced into RN4220 first by electroporation and then introduced into ΔcymR strain by phage transduction.
Protein Expression and Purification
The 423-bp ORF of cymR was PCR-amplified from Newman chromosomal DNA by using the primers cymRFor-LIC and cymRRev-LIC. The PCR product was introduced into pMCSG7 by ligation-independent cloning (LIC) to generate the plasmid pMCSG7-His-cymR (25). To make pMCSG7-His-cymRC25S plasmid, the QuikChange site-directed mutagenesis (Stratagene) strategy was used as described above. The resulting plasmids were transformed first into DH5α and then into BL21. The BL21 strain carrying the plasmids was grown in LB to A600 = 0.6 at 37 °C, cell cultures were cooled down to 16 °C, and then 1 mm of isopropyl β-d-1-thiogalactopyranoside was added for overnight induction of the proteins. The expressed protein was purified with a nickel-nitrilotriacetic acid column (Qiagen) and further purified by gel filtration column (GE Healthcare) with high salt buffer (10 mm Tris·HCl, pH 7.4, 2 m NaCl, 1 mm DTT). The fractions containing the proteins were buffered with 100 mm NaCl, 10 mm Tris·HCl, pH 7.4, and 1 mm DTT by desalting column (GE Healthcare).
Electrophoretic Mobility Shift Assays
The 291-bp mccAB promoter region was PCR-amplified with the primers mccABFor and mccABRev. The amplified DNA probe was phosphorylated by polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. CymR and CymRC25S were serially diluted to 0, 0.0125, 0.025, 0.05, 0.1, and 0.8 μm in the buffer (10 mm HEPES, pH 8.0, 1 mm EDTA, 50 mm KCl, 0.05% Triton X-100, 5% glycerol, 10 μg/ml salmon sperm DNA), followed by the addition of 0.5 nm 32P-labeled probe and 1 mm H2O2. After the reaction mixtures (20 μl each) were incubated at room temperature for 30 min, 0.2 mm coenzyme A was added and further incubated at room temperature for 30 min. The resulting reaction mixture was analyzed by 6% polyacrylamide gel electrophoresis (100 V, 30 min for pre-run, 100 V, and 80 min for sample separation). The gels were dried and subjected to autoradiography. The dissociation constant estimation was based on this gel shift assay, and the protein concentration that made 50% DNA shift was used as the Kd. The assay was repeated at least twice with similar results.
Crystallization and Structure Determination
Reduced CymR in the buffer (100 mm NaCl, 10 mm Tris·HCl, pH 7.4, 1 mm DTT) was crystallized with 0.1 m lithium sulfate monohydrate, 0.1 m sodium citrate tribasic dehydrate, pH 5.6, and 12% (w/v) polyethylene glycol 6000 buffer by the hanging drop method. The crystals were cryoprotected by the reservoir solution containing 20% glycerol and frozen in liquid N2. The data were collected to 1.7 Å at the macromolecular crystallography for life science Beamline 21-ID-D at the Advanced Photon Source, Argonne National Laboratory, and processed with HKL2000. The phases were determined by using Molrep (model molecule Protein Data Bank code 3LWF, identities 64%) from CCP4i software suite, and the model was built and improved by using Coot. The final structures have been validated though software Procheck in the CCP4i suit. The Ramachandran distribution shows 99.2 and 0.8% in favored and allowed region for wild-type CymR structure, and 96.6 and 3.4% distribution for CymRC25-SOH structure, respectively. No residue in either structure is located in the outlier region. The final structure was visualized by PyMol software.
To achieve the crystallization of the oxidized CymR, after the reduced CymR was desalted with desalting buffer (100 mm NaCl, 10 mm Tris·HCl, pH 7.4), 1 mm H2O2 was added to the protein solution and incubated at room temperature for 1 h. The protein solution was further purified by gel filtration with a desalting buffer (100 mm NaCl, 10 mm Tris·HCl, pH 7.4). Oxidized CymR in desalting buffer crystallized in the same conditions as described above for the reduced CymR. The crystals also diffracted to 1.7 Å resolution at the macromolecular crystallography microbeam for life science Beamline 23-ID-B at the Advanced Photon Source, Argonne National Laboratory. The structure of the protein was determined by the same method described above for the reduced CymR.
Western Blot
The strains were grown overnight in TSB. The bacteria were collected and suspended in buffer A (500 mm NaCl, 10 mm Tris·HCl, pH 7.4, 1 mm DTT, 5% glycerol), followed by bead beater (FastPrep) treatment to lysis the bacteria. The 20-μl supernatants were loaded into 12% SDS page for separation. After standard Western blot procedures, the proteins were detected by anti-FLAG antibody.
Disk Diffusion Assay
Four different strains were grown overnight in TSB and diluted to A600 = 0.2 in fresh TSB. The diluted culture (400 μl) was mixed with 20 ml of tryptic soy agar (TSA) and used as an overlay on a TSA plate. Sterile 6-mm filter paper was placed on the top of the plate, and 10 μl of 200 mm CuSO4 was added to the filter paper. The plates were incubated at 37 °C overnight and photographed with a Sony DSC-W210 camera. The assay was repeated at least twice with similar results.
Blood Plate Assay
Four different strains were grown overnight in TSB. 1-μl cell cultures were added onto the sheep blood plate and incubated at 37 °C overnight before being photographed with a Sony DSC-W210 camera.
Quantitative RT-PCR
Four different strains were grown overnight in TSB with 2 mm cysteine. The cell cultures were 1:100 diluted into TSB with 2 mm cysteine and grown to A600 = 0.6. The resulting culture was divided into two groups, one of which was treated with 20 mm H2O2 for at 37 °C for 10 min. Total RNA were isolated with RNeasy mini kit (Qiagen) by following the manufacturer's recommendations. The purified RNA (5 ng) was used for qRT-PCR, which was performed with SuperScript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen). The mRNA level of the test genes was normalized by 16 S rRNA. The following primers were used for qRT-PCR: 16 S rRNAFor and 16 S rRNARev, mccBFor and mccBRev gene, tcyPFor, and tcypRev. The assay was repeated at least twice with similar results.
4-Chloro-7-nitrobenzo-2-oxa-1,3-diazole Assay
The wild-type CymR and CymRC25S mutant protein were purified with 1 mm DTT. To generate the sulfenic acid form, both of the proteins (50 μm) were exchanged into buffer containing 100 mm KH2PO4/K2HPO4, 200 mm NaCl, and 1 mm EDTA at pH 7.0. Afterward, the proteins were treated with 1 mm H2O2 and incubated at room temperature for 30 min followed by washing with the above buffer three times. To generate the thiolated complex, the oxidized proteins were incubated with 1 mm free cysteine or 1 mm CoA for another 30 min and washed with the buffer three times to remove any unreacted LMW thiols. At last, all the samples were incubated with 1 mm 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole in the dark for 1 h. After extensive washing, the spectra were taken using an Agilent 8453 UV-visible spectrophotometer. The assay was repeated at least twice with similar results.
RESULTS
Cys-25 Is Redox-active Cysteine That Affects DNA Binding Ability of CymR
Cys-25 is the sole cysteine in CymR. Helix-turn-helix domain prediction (26) showed that this cysteine residue is very close to the winged helix-turn-helix DNA-binding domain (supplemental Fig. S1), implying that oxidation and potential further modification of Cys-25 may influence the DNA binding ability of the protein. CymR has been reported to be the master regulator of cysteine homeostasis in S. aureus; deletion of cymR renders S. aureus more sensitive to H2O2, diamide, and CuSO4 stress (19, 20). All of this information suggests that CymR might be an oxidation-sensing regulator that functions through Cys-25, its sole cysteine residue.
To test the hypothesis, we examined whether Cys-25 oxidation affects the DNA binding of CymR by employing an electrophoretic mobility shift assay using recombinantly expressed CymR (supplemental Fig. S2). The mccAB promoter that was reported to be recognized by CymR was used (19). As expected, CymR binds the mccAB promoter sequence (CymR in Fig. 1A). Oxidation of CymR with 1 mm H2O2 weakened its DNA binding affinity (CymR + H2O2 in Fig. 1A) from Kd of ∼25–50 nm (Table 1). When the oxidized CymR was further modified by coenzyme A, which could form a mixed disulfide bond with oxidized CymR, the DNA binding affinity of the protein was dramatically decreased to Kd of ∼800 nm (Table 1). To further examine the role of Cys-25 in DNA binding by CymR, we constructed a CymRC25S mutant with Cys-25 mutated to Ser and employed it in the same DNA binding assay. CymRC25S exhibited a similar binding affinity as the wild-type CymR (Fig. 1B). However, binding was not affected by either H2O2 or H2O2/coenzyme A treatment. These results strongly suggest that Cys-25 plays an important sensing and regulatory role in CymR.
FIGURE 1.
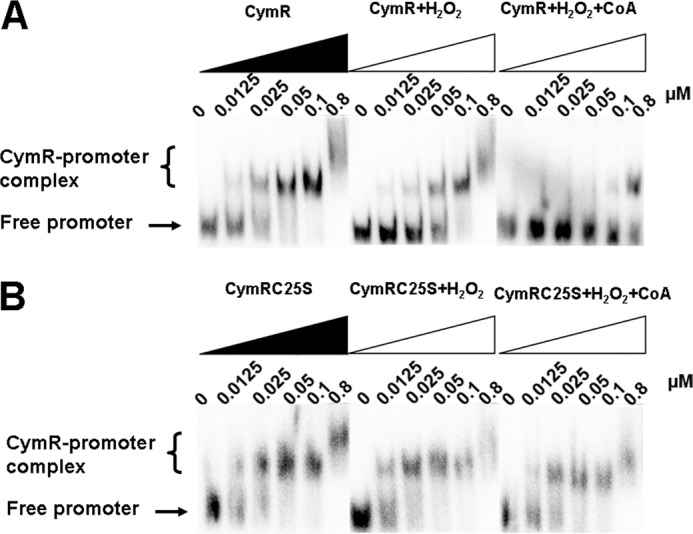
Oxidation effects of CymR on DNA binding affinity. Purified CymRWT (A) and CymRC25S (B) were incubated with the mccAB promoter. H2O2 was added to the reaction and incubated for 30 min. Then CoA was added and further incubated for 30 min. The concentrations of reaction components are as follows: mccAB promoter, 0.5 nm; CymR, 0, 0.0125, 0.025, 0.05, 0.1, and 0.8 μm; H2O2, 1 mm; CoA, 0.2 mm.
TABLE 1.
Dissociation constant (Kd) of CymRWT and CymRC25S from mccAB promoter in different treatment conditions
| Treatment protein |
Kd |
||
|---|---|---|---|
| Native | H2O2 | H2O2 + CoA | |
| nm | |||
| CymRWT | 25 | 50 | 800 |
| CymRC25S | 12.5–25 | 12.5–25 | 25 |
Cys-25-SOH in Oxidized CymR Was Characterized by X-ray Crystallography
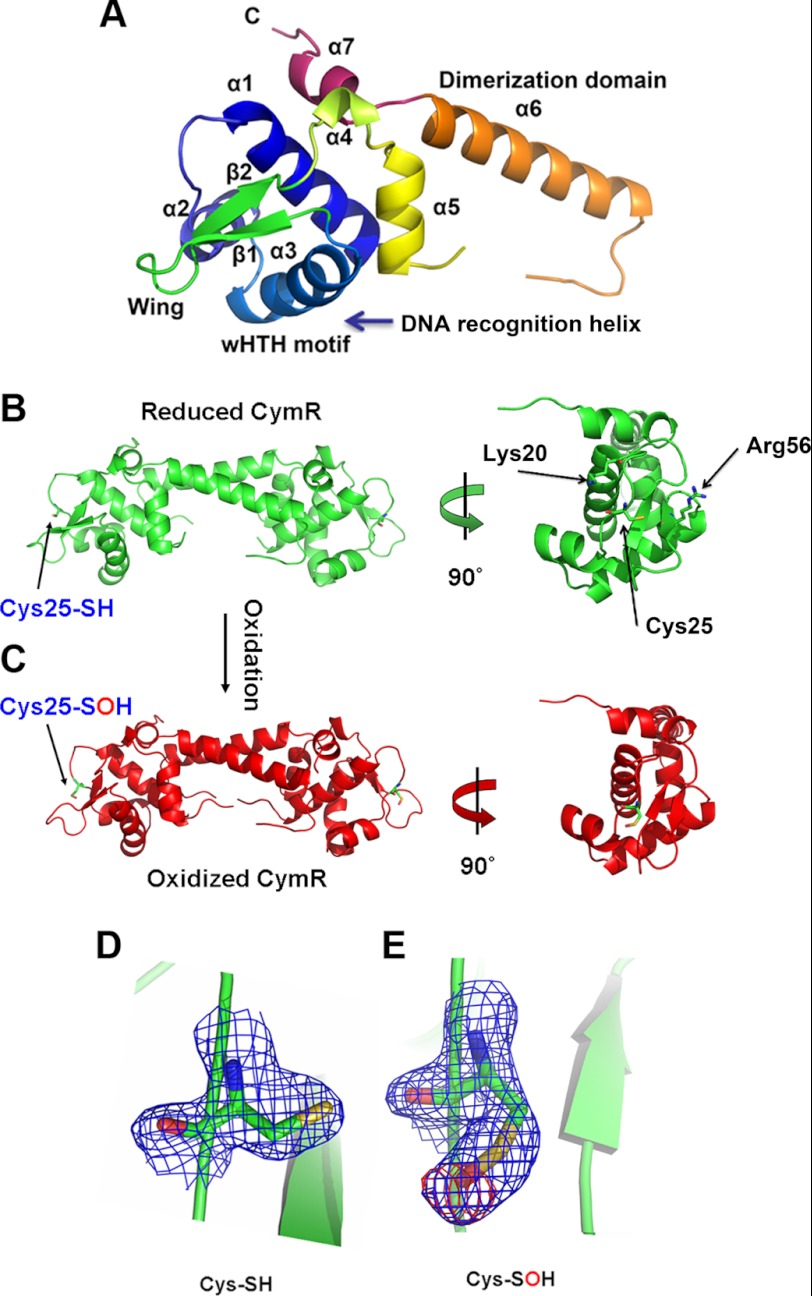
To understand the role of Cys-25 oxidation in the oxidized CymR compared with the reduced form, we crystallized and solved protein structures of both the reduced and the oxidized CymR (Table 2). Crystals of both protein forms diffract to 1.7 Å resolution. The reduced CymR structure revealed that CymR forms a biologically active homodimer, which consists of two domains: one winged helix-turn-helix domain and one long dimerization domain (Fig. 2, A and B), similar to a recently reported structure of B. subtilis CymR (27) except that Cys-25 is not conserved in B. subtilis CymR.
TABLE 2.
Data collection and refinement statistics for CymRWT and CymRCys25-SOH
The highest resolution shell is shown in paretheses.
| Data collection and crystal parameters | CymRWT | CymRCys25-SOH |
|---|---|---|
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 77.10, 52.50, 33.44 | 77.33, 52.58, 33.45 |
| α, β, γ (°) | 90.00, 98.52, 90.00 | 90.00, 98.12, 90.00 |
| Resolution (Å) | 20-1.70 (1.76-1.70) | 20-1.75 (1.81-1.75) |
| Rmerge (%) | 4.4 (28.9) | 8.7 (62.1) |
| I/σI | 25.2 (2.6) | 17.1 (1.5) |
| Completeness (%) | 96.6 (79.3) | 99.9 (99.2) |
| Redundancy | 3.4 (2.6) | 5.5 (5.0) |
| Refinement | ||
| Resolution (Å) | 20-1.70 (1.76-1.70) | 20-1.75 (1.81-1.75) |
| Rwork/Rfree (%) | 21.9/25.9 (26.6/29.2) | 21.7/25.1 (28.9/34.1) |
| Root mean square deviation bond lengths (Å) | 0.006 | 0.007 |
| Root mean square deviation bond angle (°) | 1.068 | 1.052 |
| Mean B factor (Å2) | 33.216 | 36.175 |
FIGURE 2.
A, ribbon diagram of the CymR monomer, showing secondary structure numbering. B–E, crystal structures of the reduced (B) and oxidized (C) CymR (both at 1.7 Å resolution) and electron density of the reduced (D) and sulfenic acid (E) forms of Cys-25 in CymR. CymR was purified with 1 mm DTT for the reduced CymR crystal. To crystallize oxidized CymR, the reduced CymR was desalted with a buffer devoid of DTT and then treated with 1 mm H2O2 at room temperature for 1 h. The 2Fo − Fc map (1.0 σ) of Cys-25 and the Fo − Fc map (3.0 σ) of Cys-25 in the absence of oxygen atom are shown as blue and red mesh, and the atoms are colored green (carbon), dark blue (nitrogen), red (oxygen), and yellow (sulfur).
A close inspection of the active cysteine in CymR revealed three major differences compared with the active cysteine residue in OhrR, which is a prototype of the OhrR/MgrA family thiol-dependent redox regulatory proteins. First, the sole cysteine in CymR is located in the winged helix-turn-helix DNA-binding domain and near the HTH motif (Fig. 2B) (28). Oxidization and further S-thiolation of this cysteine could directly affect DNA binding by the oxidized CymR. However, in the OhrR type proteins, the sole cysteine is located toward the end of the first α helix at the dimerization domain. First, oxidization and thiolation affect the conformation of the dimerization domain, which is transferred to the DNA-binding domains (29, 30). Second, Cys-25 in CymR is quite exposed (Fig. 2B), which makes it very accessible to oxidants and subsequent reactions with LMW thiols. However, in the OhrR type proteins, the active cysteine is partially embedded in the dimerization domain (29). Third, there are numerous basic residues near the active cysteine of CymR (Fig. 2B). Lys-20 and Arg-56 are less than 10 Å away from Cys-25, which may contribute to lower sulfhydryl pKa and increase its reactivity (30). In contrast, there are fewer basic residues surrounding the sole cysteine in OhrR, and the high reactivity of the active cysteine in OhrR was thought to be a result of the positive macrodipole of the first α helix and hydrogen bonding to nearby Tyr-29 and Tyr-40, which helps to stabilize the negatively charged thiolate (29).
Upon oxidation, Cys-25-SH is oxidized to Cys-25-SOH (Fig. 2C). A 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)) assay supported formation of sulfenic acid in the oxidized CymR in the absence of small-molecule thiols (supplemental Fig. S3). A close inspection revealed that the carbon-sulfur bond of Cys-25 rotates about 180 degrees after oxidation (Fig. 2, D and E). This rotation directs the sulfur-oxygen bond toward the DNA-binding domain (supplemental Fig. S4). As a result, intracellular LMW thiols such as Cys or CoA could easily attack the sulfur on Cys-25-SOH from the opposite side to form a mixed disulfide through a nucleophilic reaction (supplemental Fig. S4). Superimposition of the reduced and sulfenic acid forms of CymR showed almost identical conformations, which is consistent with the gel shift data that yielded similar DNA binding affinities for these two forms of CymR (Kd of 25 and 50 nm, respectively). However, after S-thiolation, their DNA binding ability is dramatically decreased (Kd increases to 800 nm), which indicates that S-thiolation significantly alters the DNA-binding domain structure of CymR. This behavior is similar to the OhrR type proteins and has already been confirmed by a thiol-modified structure of SarZ, in which a dramatic conformational change was induced by S-thiolation (30).
C25S Substitution Mutant Showed Similar Phenotypes as Wild-type Strain
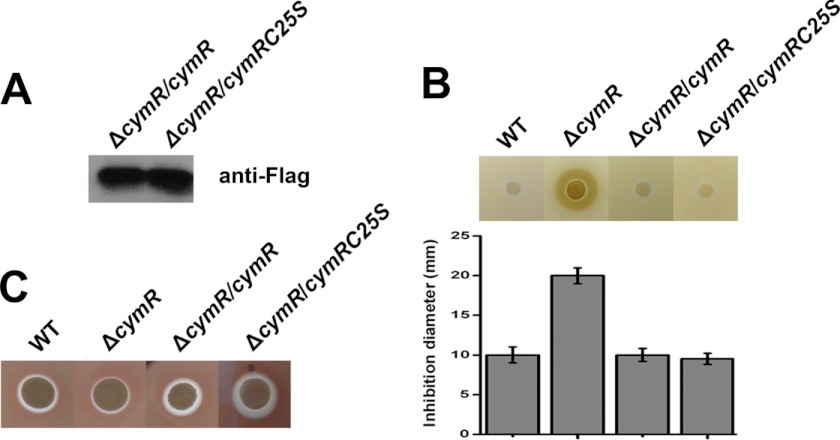
To investigate the contribution of Cys-25 to the function of CymR, this residue was mutated to serine (CymRC25S). The FLAG tag expression vector pCL55-FLAG was used for complementation. As a result, four strains were constructed: WT (wild-type Newman with the empty vector), ΔcymR (mutant with the empty vector), ΔcymR/cymR (mutant complemented with wild-type cymR-FLAG), and ΔcymR/p-cymRC25S (mutant complemented with cymRC25S-FLAG mutant). We used CuSO4 to examine the resistance of the strains to oxidative stress and blood plate assay to test α-hemolysin expression on the above four strains. As shown in Fig. 3A, the ΔcymR/cymR and ΔcymR/cymRC25S stains can effectively express the FLAG-tagged proteins. In the stress resistance assay, the ΔcymR mutant showed decreased stress resistance to CuSO4, confirming the previous report (20). At the same time, both of the introductions of the wild-type cymR and cymRC25S to theΔcymR mutant can restore the resistance (Fig. 3B), demonstrating that CymRC25S protein is a biologically active protein in vivo. In the blood plate assay, the ΔcymR mutant showed decreased α-hemolysin expression, confirming the previous report (20). Introduction of cymRC25S to the ΔcymR mutant showed even higher expression of α-hemolysin than that in both the WT and ΔcymR mutant strains (Fig. 3C), thereby indicating the critical role of Cys-25 in the overall function of CymR.
FIGURE 3.
A, Western blot showing that CymR-FLAG and CymRC25S-FLAG can be effectively expressed in vivo. B, disk diffusion assay showing the effect of mutation of Cys-25 on S. aureus stress resistance. Four strains were grown in TSB overnight. They were diluted to A600 = 0.2 and mixed with TSA to be used as an overlay on the plates. Disks were placed on the top of the overlay, and 200 mm CuSO4 was added. The plates were incubated overnight. C, blood plate assay showing the effect of mutation of Cys-25 on S. aureus α-hemolysin expression.
Monitoring Role of Cys-25 in Regulating Gene Expression
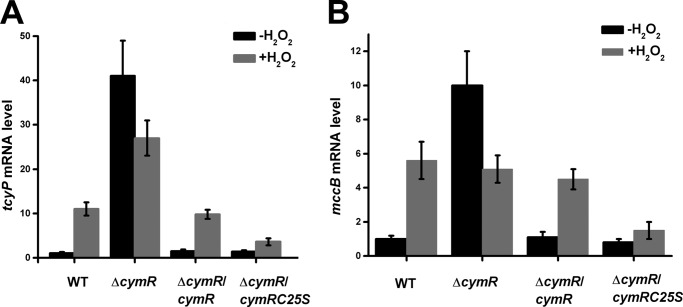
To provide further in vivo support to the mechanism, quantitative real time PCR was performed on the four aforementioned strains. Bacteria were grown to mid-log growth phase and treated with H2O2 for 10 min. Then the transcript levels of two genes, tcyP and mccB, were analyzed by quantitative RT-PCR. The tcyP gene functions as a cysteine transporter and contributes to the redox potential equilibrium. The mccB gene functions in reverse transsulfuration and both of the tcyP and mccB were directly regulated by CymR (20). In the absence of H2O2, the transcript levels of both genes were low in the wild-type strain. Upon the addition of H2O2, a close to 10-fold (tcyP) and 5-fold (mccB) induction were observed, respectively. However, in the cymR mutant, the transcript levels of both genes were high even in the absence of H2O2, and the addition of H2O2 decreased the transcript levels of both genes, showing that CymR plays a key role in redox signaling. In addition, complementation of the cymR mutant strain with the wild-type copy of cymR led to the wild-type behavior to H2O2 treatment, but the complementation of the mutant strain with the C25S mutant copy of cymR failed to yield the wild-type behavior (Fig. 4), resulting in a redox-silent protein. These experiments clearly indicate that Cys-25 is a key residue that impacts redox regulation of CymR.
FIGURE 4.
The role of Cys-25 in the expression of genes in CymR regulon. Test strains were grown in TSB supplemented with 2 mm cysteine to mid-log growth phase and divided into two copies. One of the cultures was treated with 20 mm H2O2 for 10 min, and total RNA was purified. The mRNA levels of two genes, tcyP (A) and mccB (B), were analyzed by qRT-PCR with normalization with 16 S rRNA.
DISCUSSION
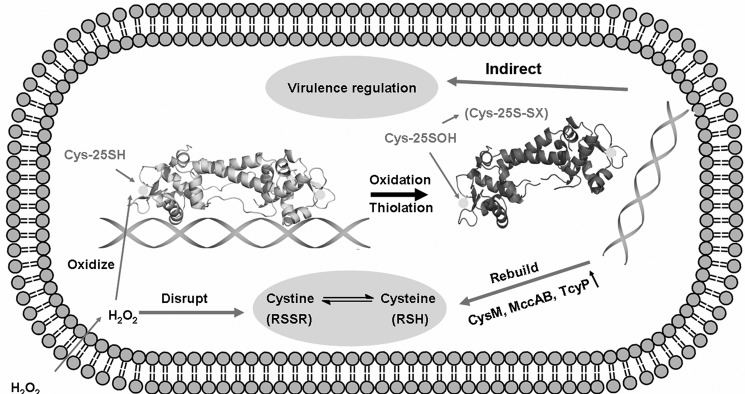
In this study, we show that CymR is a thiol-based, oxidative stress-sensing regulator in S. aureus. CymR directly senses ROS by oxidation of the sole cysteine to afford a sulfenic acid intermediate. The sulfenic acid intermediate may quickly react with intracellular LMW thiols to form a mixed disulfide. This thiolation will lead to dissociation of CymR from its cognate DNA promoters and impact the expression of the cymR regulon, including cysteine biosynthesis genes, ROS detoxifying genes, and virulence genes, thus resulting in a rebalance of the disturbed redox equilibrium and a modulation of virulence (Fig. 5).
FIGURE 5.
A proposed model of CymR-based oxidation stress sensing, response, and virulence regulation in S. aureus. When S. aureus is exposed to ROS, the redox potential equilibrium will be disrupted. Meanwhile, CymR senses the oxidative stress through oxidation of the active cysteine and then dissociates from DNA, which may activate two distinct pathways: cysteine biosynthesis/ROS detoxification pathway and the virulence regulation pathway. In the cysteine biosynthesis and ROS detoxification pathway, mccAB, tcyP, cysM, and other genes with similar functions are induced and restore the redox equilibrium in cytoplasm. In the virulence regulation pathway, upon sensing oxidative stress by CymR, S. aureus may turn down its virulence gene expression and convert itself into a form more resistant to host killing.
As a human pathogen, S. aureus has to cope with the oxidative stress generated by the human immune system. Evolutionary adaption enriches the oxidation-sensing system of S. aureus. For example, at least three Bacillus subtilis OhrR type oxidation-sensing regulators, MgrA, SarZ, and SarA (6, 10, 11) were identified in S. aureus. Sequence alignment of CymR in S. aureus with its homologues in B. subtilis and Listeria innocua revealed that the key cysteine residue in S. aureus is unique, which may also be attributed to this evolutionary difference, promoting its survivability in the host body.
In S. aureus, coenzyme A and cysteine are considered to be the major LMW thiols for the cellular reducing buffer (21–23). In addition to these two thiols, bacilli thiol was discovered to form a mixed disulfide with the well known thiol-based sensor OhrR in B. subtilis (31, 32), which raises the possibility that the BSH-like molecule may also serve as a substitute for glutathione in Gram-positive bacteria. The OhrR-type proteins show two distinct types of redox-sensing mechanisms. In the first type, the proteins sense oxidative stress by forming a mixed disulfide with LMW thiols through the sulfenic acid intermediate, such as OhrR, MgrA, and SarZ (8, 10, 11, 29). In the second type the proteins sense oxidative stresses by forming intermonomer disulfide, such as Xanthomonas campestris OhrR (33, 34), OspR, and MexR (13, 14). The oxidation-sensing mechanism of CymR resembles the first type of OhrR proteins. However, CymR has its own unique features. Unlike the cysteine in the OhrR proteins, Cys-25 in CymR is located in the DNA-binding domain, suggesting that the modification of Cys-25 would directly affect the DNA binding affinity of the protein.
An interesting result we found is that the cymR mutant is more sensitive to oxidative stress (Fig. 3), despite the fact that the genes for ROS detoxification are highly expressed in the cymR mutant (20). In fact, a previous study also reported that a cymR deletion mutant shows 1.4–5.8-fold higher transcription of multiple genes involved in stress response (e.g., ahpC, ahpF, dps, sodA, sodM, and perR). These results imply that high expression of stress response genes is not sufficient for resistance to oxidative stress. Because the knock-out of cymR increases the intracellular cysteine pool by ∼68-fold and the cysteine to cystine ratio by ∼36-fold, it was suggested that the imbalance of thiol redox status and the elevated generation of ROS by Fenton reaction caused the higher susceptibility of the cymR mutant (20). In the wild-type cells, when oxidative stress causes oxidation of cysteine to cysteine and significantly alters the thiol redox balance, Cys-25 of CymR will also be oxidized, which induces the expression of genes involved in stress responses, as well as cysteine uptake and cysteine biosynthesis, in a controlled manner (Fig. 4), the result of which will lead to rebalance of the thiol equilibrium without permanent disruption of cysteine metabolism. Therefore, CymR can be regarded as a key regulator of intracellular redox homeostasis.
Because the human immune system generates ROS to kill invaded pathogens, broad attention has been focused on the study of redox sensing, regulation, and pathogenesis in human pathogens (6, 8, 10–14, 16, 29, 35–42). Previously, we identified the molecular mechanism of two other thiol-based redox-sensing regulators of S. aureus: MgrA and SarZ. MgrA has the ability to sense oxidative stress and regulate virulence (10). SarZ responds to oxidative stresses and induces the expression of genes involved in stress response as well as metabolic switching to complement the MgrA regulon (11). The discovery of the CymR-mediated regulation and ROS-sensing mechanism further connects cellular redox buffering with redox sensing and virulence. The sole cysteine of CymR is quite exposed to the environment, which makes it a potential exposed target for new therapeutic agents to treat S. aureus infection.
Acknowledgments
We thank all beamline staff for data collection support, Catherine B. Poor for CymR reduced form data collection, Dr. Olga Soutourina (Institut Pasteur) for helpful discussion. We thank S. Frank Reichard for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI077564 (to T. B.) and AI074658 (to C. H.). This work was also supported by Scientist Development Grant 0835158N from the American Heart Association (to T. B.), and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (to C. H.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S4.
The atomic coordinates and structure factors (codes 3T8R and 3T8T) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ROS
- reactive oxygen species
- LMW
- low molecular weight
- TSB
- tryptic soy broth
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Archer G. L. (1998) Staphylococcus aureus. A well-armed pathogen. Clin. Infect. Dis. 26, 1179–1181 [DOI] [PubMed] [Google Scholar]
- 2. Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 3. Laarman A., Milder F., van Strijp J., Rooijakkers S. (2010) Complement inhibition by gram-positive pathogens. Molecular mechanisms and therapeutic implications. J. Mol. Med. 88, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maruyama A., Kumagai Y., Morikawa K., Taguchi K., Hayashi H., Ohta T. (2003) Oxidative-stress-inducible qorA encodes an NADPH-dependent quinone oxidoreductase catalysing a one-electron reduction in Staphylococcus aureus. Microbiology 149, 389–398 [DOI] [PubMed] [Google Scholar]
- 5. Chang W., Small D. A., Toghrol F., Bentley W. E. (2006) Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188, 1648–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujimoto D. F., Higginbotham R. H., Sterba K. M., Maleki S. J., Segall A. M., Smeltzer M. S., Hurlburt B. K. (2009) Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage λ integrase-mediated excision/recombination. Mol. Microbiol. 74, 1445–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ballal A., Manna A. C. (2010) Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J. Bacteriol. 192, 336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. W., Soonsanga S., Helmann J. D. (2007) A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ballal A., Ray B., Manna A. C. (2009) sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J. Bacteriol. 191, 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen P. R., Bae T., Williams W. A., Duguid E. M., Rice P. A., Schneewind O., He C. (2006) An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2, 591–595 [DOI] [PubMed] [Google Scholar]
- 11. Chen P. R., Nishida S., Poor C. B., Cheng A., Bae T., Kuechenmeister L., Dunman P. M., Missiakas D., He C. (2009) A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 71, 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuangthong M., Helmann J. D. (2002) The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U.S.A. 99, 6690–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan L., Murray T. S., Kazmierczak B. I., He C. (2010) Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol. Microbiol. 75, 76–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen H., Hu J., Chen P. R., Lan L., Li Z., Hicks L. M., Dinner A. R., He C. (2008) The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl. Acad. Sci. U.S.A. 105, 13586–13591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aslund F., Zheng M., Beckwith J., Storz G. (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 96, 6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S. O., Merchant K., Nudelman R., Beyer W. F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J. S. (2002) OxyR. A molecular code for redox-related signaling. Cell 109, 383–396 [DOI] [PubMed] [Google Scholar]
- 17. Luong T. T., Newell S. W., Lee C. Y. (2003) Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185, 3703–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luong T. T., Dunman P. M., Murphy E., Projan S. J., Lee C. Y. (2006) Transcription Profiling of the mgrA Regulon in Staphylococcus aureus. J. Bacteriol. 188, 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soutourina O., Poupel O., Coppée J. Y., Danchin A., Msadek T., Martin-Verstraete I. (2009) CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73, 194–211 [DOI] [PubMed] [Google Scholar]
- 20. Soutourina O., Dubrac S., Poupel O., Msadek T., Martin-Verstraete I. (2010) The pleiotropic CymR regulator of Staphylococcus aureus plays an important role in virulence and stress response. PLoS Pathog. 6, e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lithgow J. K., Hayhurst E. J., Cohen G., Aharonowitz Y., Foster S. J. (2004) Role of a cysteine synthase in Staphylococcus aureus. J. Bacteriol. 186, 1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pöther D. C., Liebeke M., Hochgräfe F., Antelmann H., Becher D., Lalk M., Lindequist U., Borovok I., Cohen G., Aharonowitz Y., Hecker M. (2009) Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 191, 7520–7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. delCardayre S. B., Stock K. P., Newton G. L., Fahey R. C., Davies J. E. (1998) Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J. Biol. Chem. 273, 5744–5751 [DOI] [PubMed] [Google Scholar]
- 24. Bae T., Schneewind O. (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 [DOI] [PubMed] [Google Scholar]
- 25. Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Combet C., Blanchet C., Geourjon C., Deléage G. (2000) NPS@. Network protein sequence analysis. Trends Biochem. Sci. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 27. Shepard W., Soutourina O., Courtois E., England P., Haouz A., Martin-Verstraete I. (2011) Insights into the Rrf2 repressor family. The structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 278, 2689–2701 [DOI] [PubMed] [Google Scholar]
- 28. Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M. (2005) The many faces of the helix-turn-helix domain. Transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262 [DOI] [PubMed] [Google Scholar]
- 29. Hong M., Fuangthong M., Helmann J. D., Brennan R. G. (2005) Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20, 131–141 [DOI] [PubMed] [Google Scholar]
- 30. Poor C. B., Chen P. R., Duguid E., Rice P. A., He C. (2009) Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 284, 23517–23524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Newton G. L., Rawat M., La Clair J. J., Jothivasan V. K., Budiarto T., Hamilton C. J., Claiborne A., Helmann J. D., Fahey R. C. (2009) Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 5, 625–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Helmann J. D. (2011) Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox. Signal 15, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Soonsanga S., Lee J. W., Helmann J. D. (2008) Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J. Bacteriol. 190, 5738–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newberry K. J., Fuangthong M., Panmanee W., Mongkolsuk S., Brennan R. G. (2007) Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28, 652–664 [DOI] [PubMed] [Google Scholar]
- 35. Hausladen A., Privalle C. T., Keng T., DeAngelo J., Stamler J. S. (1996) Nitrosative stress. Activation of the transcription factor OxyR. Cell 86, 719–729 [DOI] [PubMed] [Google Scholar]
- 36. Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., Ryu S. E. (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105, 103–113 [DOI] [PubMed] [Google Scholar]
- 37. Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W. S., Yu M. H., Storz G., Ryu S. E. (2004) Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 38. Lee J. W., Helmann J. D. (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367 [DOI] [PubMed] [Google Scholar]
- 39. Poole L. B., Karplus P. A., Claiborne A. (2004) Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 40. Chander M., Demple B. (2004) Functional analysis of SoxR residues affecting transduction of oxidative stress signals into gene expression. J. Biol. Chem. 279, 41603–41610 [DOI] [PubMed] [Google Scholar]
- 41. Klomsiri C., Karplus P. A., Poole L. B. (2011) Cysteine-based redox switches in enzymes. Antioxid. Redox. Signal. 14, 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jönsson T. J., Ellis H. R., Poole L. B. (2007) Cysteine reactivity and thiol-disulfide interchange pathways in AhpF and AhpC of the bacterial alkyl hydroperoxide reductase system. Biochemistry 46, 5709–5721 [DOI] [PMC free article] [PubMed] [Google Scholar]