Background: HAMP domains are four-helix bundles that transmit signals from receptor to output domains in signaling proteins/complexes.
Results: Opposite stimulus-induced helix motions with opposite signal output are resolved for two tandem HAMP domains on a phototaxis transducer.
Conclusion: A molecular mechanism for HAMP domain switching and signal transmission is proposed.
Significance: The HAMP switching and relay mechanism is important to many signal transduction proteins.
Keywords: Chemotaxis, Photoreceptors, Prokaryotic Signal Transduction, Protein Structure, Signal Transduction, Phototaxis, Protein Conformational Change
Abstract
The phototaxis receptor complex composed of sensory rhodopsin II (SRII) and the transducer subunit HtrII mediates photorepellent responses in haloarchaea. Light-activated SRII transmits a signal through two HAMP switch domains (HAMP1 and HAMP2) in HtrII that bridge the photoreceptive membrane domain of the complex and the cytoplasmic output kinase-modulating domain. HAMP domains, widespread signal relay modules in prokaryotic sensors, consist of four-helix bundles composed of two helices, AS1 and AS2, from each of two dimerized transducer subunits. To examine their molecular motion during signal transmission, we incorporated SRII-HtrII dimeric complexes in nanodiscs to allow unrestricted probe access to the cytoplasmic side HAMP domains. Spin-spin dipolar coupling measurements confirmed that in the nanodiscs, SRII photoactivation induces helix movement in the HtrII membrane domain diagnostic of transducer activation. Labeling kinetics of a fluorescein probe in monocysteine-substituted HAMP1 mutants revealed a light-induced shift of AS2 against AS1 by one-half α-helix turn with minimal other changes. An opposite shift of AS2 against AS1 in HAMP2 at the corresponding positions supports the proposal from x-ray crystal structures by Airola et al. (Airola, M. V., Watts, K. J., Bilwes, A. M., and Crane, B. R. (2010) Structure 18, 436–448) that poly-HAMP chains undergo alternating opposite interconversions to relay the signal. Moreover, we found that haloarchaeal cells expressing a HAMP2-deleted SRII-HtrII exhibit attractant phototaxis, opposite from the repellent phototaxis mediated by the wild-type di-HAMP SRII-HtrII complex. The opposite conformational changes and corresponding opposite output signals of HAMP1 and HAMP2 imply a signal transmission mechanism entailing small shifts in helical register between AS1 and AS2 alternately in opposite directions in adjacent HAMPs.
Introduction
Phototaxis by the archaeon Natronomonas pharaonis is mediated by the light receptor sensory rhodopsin II (SRII)2 in a 2:2 complex with a transducer protein HtrII (1–3). HtrII contains two transmembrane helices (TM1 and TM2), with which the SRII subunits associate followed by a cytoplasmic signal relay domain (two tandem HAMP modules) and a hairpin-shaped cytoplasmic domain that binds and modulates CheA, a histidine kinase (Fig. 1). HAMP domains are widely distributed among prokaryotic signal-transducing proteins and relay incoming conformational changes in the membrane domain to transducer domains modulating cytoplasmic enzymatic activities (4, 5). The numbers of HAMP modules vary among transducers, but the structure of an individual module characteristically consists of two amphipathic helices, AS1 and AS2, linked through a flexible connector peptide. In all cases studied, dimers of signal transducers produce a four-helix bundle consisting of two AS1 and two AS2 helices. In the HtrII dimer, there are two HAMP domains in tandem forming a biconcatenated HAMP chain connecting the photoreceptive membrane domain to the cytoplasmic kinase-binding domain of the transducer.
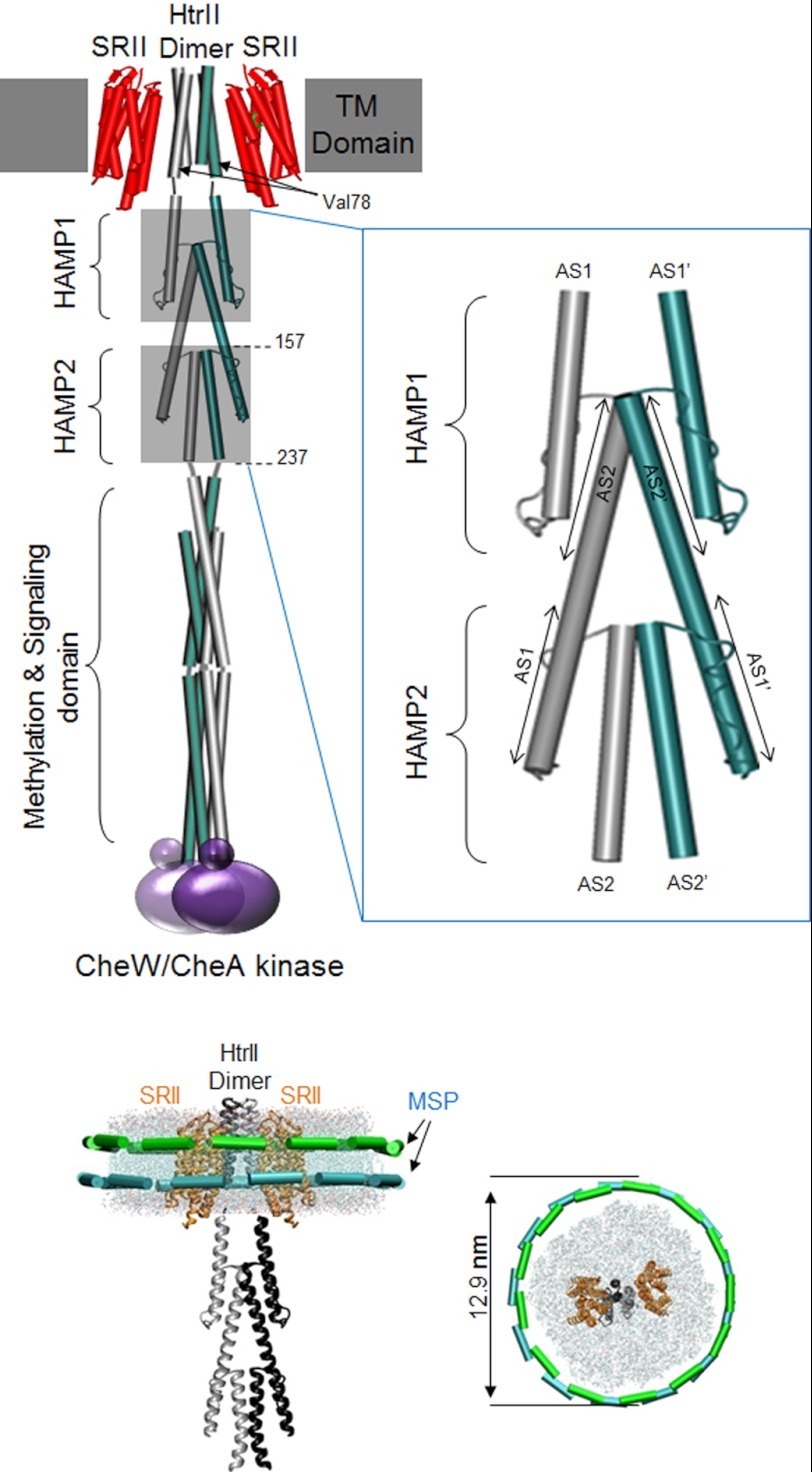
FIGURE 1.
SRII-HtrII dimeric complex in nanodiscs. Top, schematic of the complex with membrane-embedded portion (TM Domain) composed of the transducer subunit HtrII dimer (one protomer colored green and the other gray) flanked by SRII subunits (red) and CheW/CheA kinase (purple) binding at the cytoplasmic distal end of the transducer. The tandem HAMP domains were drawn from the crystal structure of Aer2 (10). HAMP1 and HAMP2 are each composed of two helices named AS1 and AS2 (AS1′ and AS2′ for the other HtrII protomer) linked with a loop. A magnified drawing of the two HAMP domains is shown in the blow-up box on the right to emphasize that (i) the AS2 pair in each HAMP is interior to the AS1 pair, and (ii) the AS2 helices of HAMP1 extend downward and form the AS1 helices of HAMP2; i.e. for each protomer, AS2 of HAMP1 and AS1 of HAMP2 are the membrane-proximal end and membrane-distal end, respectively, of the same α-helix. Truncated versions of HtrII were made at positions 157 and 237 to produce the single HAMP and double HAMP constructs, respectively, used for accessibility labeling. Bottom, drawings of the nanodisc within a lipid bilayer encircled by an MSP dimer viewed from the side and top. The nanodisc particle and SRII-HtrII dimeric complex are drawn to scale; molecular dimensions of the nanodisc are from electron micrographs (Fig. 2) and of the SRII-HtrII complex from Gordeliy et al. (23).
Several mechanisms have been proposed for signal relay by HAMP modules, each assuming a two-state switching function of the HAMP. Based on the NMR structure from a sensor/transducer of unknown function, Af1503 from Archaeoglobus fulgidus, Hulko et al. (6) proposed a gearbox model entailing counter-rotation of the four helices by 26°, triggered by the axial rotation of the input transmembrane helix of Af1503. From a disulfide mapping study of the aspartate chemotaxis receptor, Swain and Falke (7) proposed a possible scissor-like motion of paired helices. From mutation analysis of chemotaxis receptors and structural considerations, Parkinson (5) suggested a dynamic helix destabilization mechanism (8) with stimuli inducing shifts between a tightly packed versus loosely packed four-helix bundle, a concept related to the “frozen-dynamic” model of Sung-Huo Kim (9). Most recently, Crane and co-workers (10) crystallized a poly-HAMP segment of the Pseudomonas aeruginosa aerotaxis receptor, Aer2, which contains three concatenated HAMP modules. This work made a major contribution by revealing two different HAMP conformations. The first and third HAMP modules are similar in structure to each other and to the NMR structure of the Af1503 HAMP, whereas the second differed in helical register, crossing angle, and rotational position of the helices. The authors proposed that the two structures observed are opposing HAMP signaling states and suggested a signal relay mechanism in poly-HAMP chains in which repeating HAMPs would assume alternating conformations and interconvert in a binary fashion (10).
Here we report photostimulus-induced HAMP surface accessibility changes in SRII-HtrII complexes combined with independent assessment of the signal output of HAMP1 and HAMP2 by in vivo photomotility response measurements of mutants. For the accessibility measurements, the HAMPs of functional SRII-HtrII complexes needed to be exposed to solvent to enable open accessibility of a soluble fluorescein probe to the HAMP domain. To this end, we embedded the SRII-HtrII complex into nanodiscs in which the membrane protein complex in a lipid bilayer is encircled by a membrane scaffold protein (MSP1E3D1) (11). After establishing the dimeric state and functional communication between the photoreceptor (SRII) and transducer (HtrII) subunits in the nanodiscs, we performed fluorescein probe accessibility scanning along the AS1 and AS2 helices in the HAMP domain with and without light activation of SRII. The results reveal a stimulus-induced displacement of AS2 with respect to AS1 in the membrane-proximal HAMP1 and the opposite motion in the membrane-distal HAMP2. The physiological assessment of signaling by HAMP1 versus HAMP2 entailed excision of HAMP2 from the wild-type complex, whereas retaining the HAMP1 in functional register with the output kinase-modulating domains. The results show that the photosignal output from HAMP1, like the AS2 to AS1 displacement, is opposite to that from HAMP2, indicating that signal transmission through the HAMP domain region is through alternating conformational changes with alternating sign of the signal.
EXPERIMENTAL PROCEDURES
Reagents
Alexa Fluor 660 was from Invitrogen, 1,2-dimyristoyl-sn-glycero-3-phosphocholine lipid (DMPC) was from Avanti Polar Lipids, Inc., Bio-Beads SM-2 adsorbent was from Bio-Rad Laboratories, and n-dodecyl-β-d-maltopyranoside (DDM) was from Affymetrix Inc.
Plasmid Constructions and Site-directed Mutagenesis
The gene for MSP1E3D1 with codon usage optimized for Escherichia coli expression was synthesized and incorporated in the pUC57 vector between NcoI and HindII sites (Integrated DNA Technologies, Inc., Coralville, IA). The gene was then ligated into an E. coli expression vector pET28a between the NcoI and HindIII sites and was transformed into BL21(DE3) cells.
Single cysteine-encoding mutations were introduced using a site-directed mutagenesis kit, QuikChange XL (Stratagene, Wilmington, DE), on each of residue of the AS1 helix and AS2 helices on HAMP1 and selected residues (162–165, 200–204) on HAMP2 of HtrII in the pET21 E. coli expression vector containing the gene for the fusion protein of N. pharaonis SRII-HtrII in which the C terminus of SRII is joined through a flexible peptide linker (ASASNGASA) to the N terminus of HtrII truncated at residue 157 (single HAMP construct, SRII-HtrII157) or 237 (double HAMP construct, SRII-HtrII237). The HAMP1 is defined from 85 to 135, and HAMP2 is defined from 156 to 210. We truncated at 157 to include the helical linker (135–157) immediately following the HAMP1 region. Similarly, the double HAMP construct was truncated at 237 to include the helical linker (210–237) immediately beyond HAMP2.
HAMP2-deleted versions were designed to excise 136–211, 137–211, and 138–211 from the full-length HtrII. The HAMP2-deleted sequences of the SRII-HtrII fusion protein were transferred into the Halobacterium salinarum expression plasmid pPR5 plasmid (12) for cell motility analysis. All mutations were confirmed by sequencing from either the University of Texas at Houston Medical School Sequencing Facility or Genewiz (Genewiz, South Plainfield, NJ).
Protein Expression and Purification
For expression of MSP1E3D1, BL21(DE3) cells transformed with pET28a_MSP1E3D1 were cultured in LB medium containing 10 μg/ml kanamycin at 37 °C to optical density ∼0.3 at 600 nm, and expression was induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside. 1 h after induction, the temperature was lowered to 25 °C, and the cells were harvested after 4 h. The cells were suspended in Buffer A (50 mm Tris-HCl, pH 7.4, 1 mm PMSF, 1% v/v Triton X-100) and disrupted by sonication. After centrifugation at 20,000 × g for 30 min, the lysate was mixed with Ni-NTA resin for 1 h to allow binding of His-tagged MSP1E3D1, and then the resin was packed in a column and washed with 4 bed volumes of 1% v/v Triton X-100 in Buffer B (50 mm Tris-HCl, pH 7.4 + 150 mm NaCl), 50 mm deoxycholate in Buffer B, and 50 mm imidazole in Buffer B, and finally eluted with 300 mm imidazole in Buffer B. The imidazole was removed by dialysis against Standard Buffer (10 mm Tris-HCl, 0.1 m NaCl, 0.5 mm EDTA, pH 7.4). Purified MSP1E3D1 protein was stored with 0.02% w/v azide at −20 °C.
The cysteine mutants of SRII-HtrII were expressed in E. coli BL21(DE3). Cultures grown at 37 °C to optical density ∼0.3 at 600 nm were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and 5 μm all-trans retinal. After induction, the culture was incubated at 18 °C for 6–14 h. Cells were harvested by centrifugation at 4,000 × g with an SLC-4000 rotor and disrupted with a Microfluidizer (Microfluidics Corp., Newton, MA). Cell debris was pelleted at 6,000 × g for 30 min. Membranes were harvested by centrifuging the supernatant at 200,000 × g for 30 min with an OptimaTM L-100 XP ultracentrifuge from Beckman Coulter followed by resuspending with a buffer containing 50 mm Tris-HCl at pH 7.5 and 300 mm NaCl. The pellet was then solubilized in 2% w/v DDM in the same buffer. SRII-HtrII157 or SRII-HtrII237 mutants were purified through Ni-NTA affinity columns as described earlier (13) and dialyzed against buffer containing 50 mm Tris-HCL at pH 7.5 and 300 mm NaCl.
Nanodisc Reconstitution and Purification
Reconstitution of nanodiscs encircling SRII-HtrII in DMPC lipid bilayers with MSP1E3D1 was performed in accordance with the previous optimization of the ratio of the three components (14–16). A best yield was obtained when the solutions of 50 μm SRII-HtrII157 in 0.02% w/v DDM in 50 mm Tris-HCl at pH 7.5 with 150 mm NaCl, 150 μm MSP1E3D1, and 50 mm DMPC in 10% w/v DDM were combined in a molar ratio of SRII-HtrII157:MSP:lipids = 1:1:100 followed by a 20-min incubation at 23 °C and elimination of DDM using SM-2 Bio-Beads (Bio-Rad) under gentle shaking at 23 °C for 4 h. The lysate after removing precipitated aggregates by low speed centrifugation was concentrated and applied to a size-exclusion column using Superdex 200. The nanodisc samples with the target complex protein omitted eluted at retention volume of 12.5 ml in good agreement with the previous investigation with MSP1E3D1 nanodisc reported earlier (see Fig. 2a) (17). The sample containing SRII-HtrII complex, on the other hand, contains two populations of different sizes eluting at retention volumes of ∼8 and ∼11 ml (see Fig. 2a). The 8-ml fraction is a void volume attributable to aggregates, whereas the 11-ml fraction is the identical retention volume as obtained previously for MSP1E3D1 nanodiscs enclosing the trimer bacteriorhodopsin (18), which is of approximately the same molecular weight as an SRII-HtrII157 dimer, arguing that this fraction contains nanodiscs with dimeric complexes.
FIGURE 2.

Nanodisc isolation. a, chromatography of SRII-HtrII157 nanodiscs. Nanodiscs were prepared from SRII-HtrII fusion protein, MSP, and DMPC lipid as detailed under “Experimental Procedures,” and in the final step, injected into a Superdex 200 sizing column for further purification. Fractions corresponding to elution volume 10.5–11.5 ml were used for electron microscopy. b, electron micrograph images of SRII-HtrII nanodiscs from top view (upper panels) and side view (middle panels) and examples of stacked nanodiscs (bottom panels). The nanodisc diameter is 12.9 ± 0.7 nm, consistent with the size of 12.6 nm from the use of MSP1E3D1 (17). c, SDS-PAGE of SRII-HtrII157 nanodiscs. Molecular masses of SRII-HtrII and MSP based on the molecular mass standards are 42 and 33 kDa, respectively.
Furthermore, the molar ratio between SRII-HtrII157 and MSP1E3D1 as assessed from the intensities of the bands in SDS-PAGE gels shows that the 8-ml fraction is dominated by SRII-HtrII157, whereas the 11-ml fraction contains approximately the same staining intensity between the two proteins (see Fig. 2c). Further confirming nanodiscs with dimeric complexes, the expected A280 nm/A490 nm absorbance ratio of nanodiscs with a 1:1 molar content of SRII-HtrII157 complex and MSP1E3D1 protein is 2.1 (calculated from the measured ratio of 1.7 for the SRII-HtrII157 complex and adding the calculated molar extinction contribution at 280 nm from MSP1E3D1 protein). We consistently measured A280 nm/A490 nm values of 2.1–2.2 in our nanodisc preparations, establishing that the SRII-HtrII157 complexes are dimeric because there are two MSP1E3D1 protein molecules per nanodisc.
Spin Labeling and EPR Measurements
Spin labeling was performed during the purification process on Ni-NTA-agarose His-binding resin. Briefly, harvested E. coli membrane containing the single cysteine substitution V78C mutant of SRII-HtrII157, named SRII-HtrII157_V78C, was solubilized and adsorbed on Qiagen Ni-NTA-agarose His-binding resin. The resin was washed with 6× bed volume of 1% w/v DDM in 50 mm Tris-HCl buffer at pH 7.5 with 150 mm NaCl and subsequently suspended in the spin-labeling buffer containing 0.01% w/v DDM, 200 mm (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (designated R1, Toronto Research Chemicals, North York, Ontario, Canada) dissolved in the same buffer by gently shaking it at 4 °C overnight. Excess labeling reagent was removed by washing with 10 bed volumes of the same buffer containing 0.01% w/v DDM. The purified labeled hexahistidine-tagged proteins were eluted with a buffer containing 0.01% w/v DDM, 150 mm NaCl, 250 mm imidazole, and 25 mm Tris-HCl (pH 7.5) and finally dialyzed to remove imidazole against the same buffer without imidazole.
EPR spectra were recorded on a Bruker EMX spectrometer using a modulation amplitude of 2.0 G, a modulation frequency of 100 kHz, and microwave power of 1 milliwatt. Data acquisition was conducted using WinEPR. 25-μl samples (∼100 μm) were loaded into EPR glass capillaries (0.9-mm inner diameter). Light for photoactivation of the sample was from a fiber optic illuminator transmitting a 250-watt tungsten halogen lamp (product ID FOI-250 from Titan Tool Supply Inc, Buffalo, NY) beam passed through a 500 ± 5-nm interference filter plus a heat-absorbing filter through the side window of the sample chamber (resonator). The spectral data represented in Fig. 3 are averages of four individual measurements for the DDM-solubilized sample and 12 individual measurements for nanodisc samples and proteoliposome samples, respectively.
FIGURE 3.
EPR spectra of spin labeled SRII-HtrII157 at position 78 of HtrII. a, in nanodiscs (green line), DDM-solubilized sample (dotted black line), and HPL proteoliposome sample (solid black line). b, EPR spectra of nanodisc embedded SRII-HtrII157 measured in the dark (green) and during the illumination with 500-nm light at 23 °C (red). Gray shaded spectrum, light spectrum minus dark spectrum. The EPR frequency was 9.30 gHz and centered at 3320 G with a scan length of 250 G.
Electron Microscopy (EM)
SRII-HtrII nanodisc samples were purified through a Superdex 200 sizing column before their examination by EM. The nanodiscs were diluted to 0.05 mg/ml and applied to EM grids and treated with uranyl acetate. The grids were imaged using a Polara G2 electron microscope (FEI Co.) equipped with a field emission gun and a 4000 × 4000 charge-coupled device camera (TVIPS GmbH) as described (19).
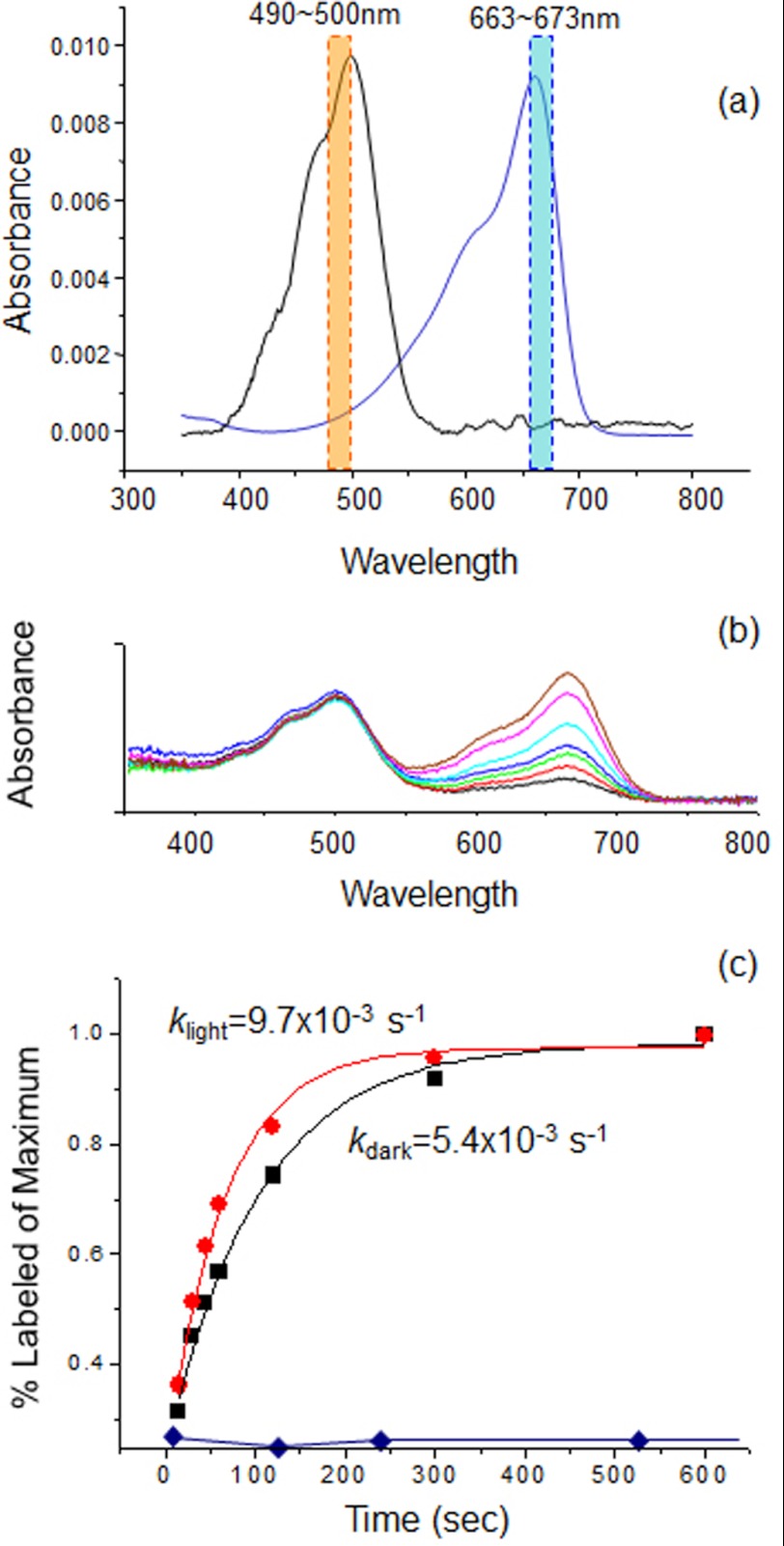
Fluorescein Labeling Efficiency Analysis
The SRII-HtrII complexes with monocysteine mutations in nanodiscs were reacted with Alexa Fluor 660 by rapidly mixing 140 μl of nanodisc sample containing 10 μm SRII-HtrII with 2.8 μl of 1 mm Alexa Fluor 660 (1:2 molar ratio) in the dark or light condition (illumination through an interference filter at 492 ± 6 nm and a heat-absorbing filter) followed by transfer of 20-μl aliquots into tubes for quenching with 20 μl of 1 m 2-mercaptoethanol at 15 s, 30 s, 45 s, 60 s, 2 min, 5 min, and 10 min. The reaction mixtures were dialyzed against buffer or diluted/concentrated with Amicon Ultra 0.5-ml centrifugal filter tubes (Millipore, Billerica, MA) more than six times to eliminate free fluorescein dye. The labeled nanodisc samples were finally concentrated to ∼25 μl and diluted to 0.5 ml for spectrophotometric analysis with a Cary 4000 UV-visible spectrometer (Varian, Cary, NC). The labeling efficiency was defined as the absorbance peak ratio of Alexa Fluor 660 at 667 nm to the SRII peak at 490 nm. A cysteine-less SRII-HtrII sample showed negligible nonspecific labeling background when compared with a typical monocysteine mutant (see Fig. 4). We measured in each case the maximum amplitude of the reaction by adding 50-fold concentrated (500 μm) fluorescence probe for 2 h during which the labeling efficiency reaches plateau. Four time points measured within the initial 60 s of the reaction in each of the samples and the maximum amplitude were subjected to a single exponential curve fit to obtain the rate constant (k). The k values were used as a measure of the accessibility of the cysteine at each position. The analysis of disulfide labeling on HAMP2 takes into account the native Cys-173 HtrII on the SRII-HtrII237 construct. This residue shows identical reaction rates with or without illumination, and its contribution to labeling was subtracted from the data before curve fitting.
FIGURE 4.
Accessibility assay analysis. a, spectra of SRII-HtrII and Alexa Fluor 600. Averages of absorbance in the orange and blue bands were used for determining sample contents of SRII-HtrII and Alexa Fluor 660, respectively. b, example of absorption spectra evolution during the labeling reaction for a representative monocysteine mutant S91C. Spectra from bottom (black) to top (brown) were measured at 15, 30, 45, 60, 120, 300, and 600 s, respectively. A constant amount of SRII-HtrII (at 490 nm) and an increase of the Alexa Fluor labeling (at 670 nm) with time are evident. c, time course of the labeling reactions in dark (black) and in the 500-nm light (red) for the mutant, fit as first-order reactions. Labeling of non-cysteine-containing SRII-HtrII (blue diamonds) is negligible.
Cell Motility Measurements
Transformed H. salinarum Pho81Wr− (20) was grown in complex medium with 1 mg/ml mevinolin as described (12). Exponential growth phase cultures were diluted 10-fold with fresh complex medium and incubated at 37 °C for 1 h before use. Cell trajectories were captured in real time AVI format with a FlashBus Spectrim Lite video capture PCI card on a computer running the VirtualDub 1.6.19.0 AVI encoder software. Swimming reversal frequencies were calculated from 50 cells. Illumination was delivered from a Nikon 100-watt He/Xe short arc lamp beam passed through a 500 ± 20-nm interference filter.
RESULTS
Nanodisc-inserted SRII-HtrII Complex Is a Proper Model for Studying Signal Transmission
Biochemical and structural studies require membrane proteins prepared in liposomes or detergent micelles to maintain their structures and activities. Many membrane proteins alter their structures and functions in the detergent-solubilized state. Conventional reconstituted proteoliposomes have several limitations for many of the techniques to manipulate or analyze membrane proteins because of their enclosed interior, which hinders accessibility to aqueous reagents. Nanodiscs made from purified components are extremely stable, soluble, well defined, and homogeneous. Their nonvesicular structure permits accessibility from both sides, advantageous for study of SRII-HtrII structural changes of transmembrane domains and cytoplasmic domains by probe labeling. Furthermore, the photochemical reaction cycle properties of SRII-HtrII, as well as of SRI-HtrI, complexes in nanodiscs are similar to those in native membranes (data not shown).
Uranyl acetate negative-stained nanodiscs from fraction 11 (Fig. 2a) show homogeneous particles with a disk shape and a disc diameter of 12.9 ± 0.7 nm (Fig. 2b), matching within experimental error the previously measured size of 12.6 nm for nanodiscs made from MSP1E3D1 (21). We observe 40 ± 2% of our nanodiscs stacked after uranyl acetate staining with 2–8 nanodiscs per stack (mean of 3.7). Stacking appears to be caused by the negative staining process because a similar result was reported for bacteriorhodopsin in nanodiscs, in which the nanodiscs were almost entirely stacked after negative staining with phosphotungstate and formed long rods(18), whereas entirely homogeneous single nanodisc particles were observed by scanning probe microscopy (17, 18).
SRII-HtrII157 Is Parallel Dimerized in the Nanodiscs
Dimerization of HtrII in reconstituted proteoliposomes was demonstrated previously by the dipolar interaction of spin probes labeled at membrane-embedded residue 78 near the cytoplasmic end on the TM2 helix of HtrII as observed by EPR spectroscopy (22), confirming adjacent positions of the two residues at position 78 in the dimer pair as expected from the crystal structure (23). Dipolar interaction of the two spin probes causes spectral broadening when compared with the spectrum of noninteracting probes when the distance between the two probes is within 8–25 Å (24), not expected to occur in the monomer or non-native aggregates of the complex. Accordingly, we introduced a cysteine substitution of Val-78 on NpHtrII157 (designated SRII-HtrII157_V78C) and prepared R1 spin-labeled SRII-HtrII157_V78C to test whether SRII-HtrII157 embedded in the nanodisc is in a parallel dimer.
We compared the EPR spectra of the spin-labeled SRII-HtrII157_V78C in 0.02% w/v DDM, in Halobacterium polar lipid proteoliposomes, and in nanodiscs with the amplitude of the spectra normalized by equalizing the total spin concentrations by double integration (Fig. 3a). The absence of spectral broadening in the DDM-solubilized sample shows that SRII-HtrII157 is monomerized in the detergent as reported previously (25). On the other hand, the proteoliposome and nanodisc samples exhibit marked broadening in their spectral shapes when compared with the detergent-solubilized sample, demonstrating the presence of parallel dimerization for these two samples. The lesser broadening in the proteoliposome sample when compared with the nanodisc sample is likely attributable to partial contribution of monomer or incorrectly inserted SRII-HtrII157 in the proteoliposome sample, which is consistent with the relatively weak binding (Kd ∼16 μm) of truncated HtrII in proteoliposomes (26). The nanodisc sample exhibited the highest degree of dipolar coupling, indicating that the nanodisc environment most effectively stabilizes dimerization of SRII-HtrII157.
Illumination Induces Separation at Position 78 on Dimeric TM2 Helices of HtrII
Photoactivation of SRII results in conformational changes in HtrII in the TM domain (27), including light-induced narrowing of the EPR spectrum attributed to separation of the helices at position 78 (22). We observe a similar light-induced narrowing of the spectrum (Fig. 3b) in nanodiscs consistent with helix separation in the HtrII dimer interface.
Fluorescein Probe Labeling Efficiency Analysis on HAMP Domains
We assessed conformational changes of the HAMP domain upon photostimulation of SRII by measuring the rate of labeling of engineered cysteine residues with the water-soluble alkylating reagent Alexa Fluor 660. The Alexa Fluor 660 maleimide group specifically reacts with thiol groups to form stable thioether bonds. Its absorption spectrum shows little overlap with that of SRII (Fig. 4a), enabling an unambiguous evaluation of the amounts of attached fluorescein probe and of SRII in the reaction.
Reaction conditions were optimized so that nonspecific labeling of non-cysteine-engineered SRII-HtrII157 in the nanodisc was negligible when compared with thiol-specific labeling at the cysteine group engineered on the HAMP in NpHtrII157 (Fig. 4b). The labeling reactions were conducted both in the dark and under illumination at 492 nm. Samples from different time points produced a nearly invariant amount of SRII-HtrII complex according to the SRII absorption bands (Fig. 4b), providing confidence in the nondenaturing reaction conditions and the consistent processing of the samples. First-order rate constants (k values) were obtained by fitting the time course of labeling of the fluorescein obtained from the absorbance at 667 nm and used as a measure of the accessibility of the residue position. At position 91, differences are observed in the dark and light k values (Fig. 4c), showing changes in the accessibility of this site resulting from the photoactivation of SRII.
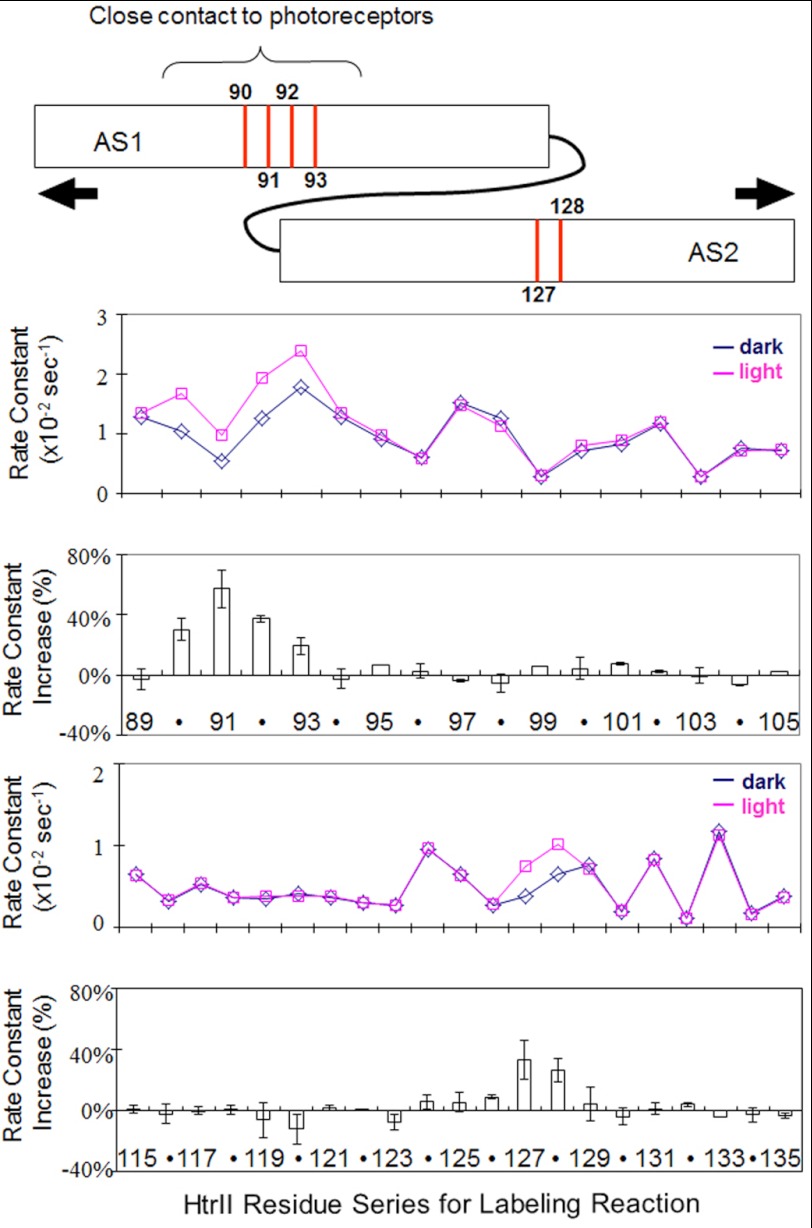
The k values at each engineered position on AS1 and AS2 of HAMP1 (Fig. 5) exhibit periodicity of accessibility along AS1 with minimum k values appearing every 3–4 residues as expected from the α-helical structure of AS1 and its association with AS2 from the other subunit in the known structures (6, 10). In AS2, the accessibility is consistently low in the upper portion between positions 114 and 123 with the rest of the portion of AS2 exhibiting periodic increase and decrease in accessibility. Because the HAMP structures show the upper portion of AS2 in the other HAMP domains exposed to the outside, this flat region in AS2 may be attributable to the contact of SRII with the AS2 upper region of HtrII HAMP1.
FIGURE 5.
Cys accessibility scanning of HAMP1. Alexa Fluor labeling reaction rate constants in dark (blue) and light (red) plotted for positions 89–105 (AS1 helix) and 115–135 (AS2 helix) of HAMP1 are shown. Light-minus-dark changes, which are localized light-induced increases, are shown in the bottom panels as the mean ± S.D. calculated from 2–5 independent measurements. The graphic at the top shows the positions of the most significant changes with arrows indicating a light-induced separation of AS1 and AS2 suggested to cause the increases (see under “Results”).
Light activation of SRII causes accessibility change only in localized portions of the HAMP domain (Fig. 5), as is also evident in the light-dark difference plot. Increases in accessibility are observed with AS1 between residues 90 and 93, a short stretch that has been reported to be in contact with SRII both by Förster resonance energy transfer (27) and by EPR measurements (28). AS2 exhibits reproducible and large changes only at residues 127 and 128.
A salt-dependent equilibrium of two conformations of HAMP1 in the SRII-HtrII complex was suggested to be responsible for phototaxis signal transmission through HtrII; however, the effects of photostimuli were not examined in the study (29). Our data do not support that photostimuli induce shifts between the compact and highly dynamic conformations reported at high versus low salt concentration in that study. Light-induced changes in accessibility of several critical residues on HAMP1 in low salt (300 mm NaCl) and high salt (4 m NaCl) are closely similar (see Fig. 8), indicating that a similar conformational change occurs at both salt concentrations we tested.
FIGURE 8.
Molecular model of di-HAMP region of the HtrII dimer. a, sequence alignment of HAMP1, HAMP2 from NpHtrII, and HAMP1, HAMP2, and HAMP3 from Aer2. Conserved hydrophobic residues are labeled in black, and glycine residues are in gray. b, model built from concatenated HAMP1 and HAMP2 from the Aer2 crystal structure (10). Light-induced increases of accessibility occur at red regions of HAMP1, and light-induced decreases of accessibility occur at green regions of HAMP2. c, light-induced increases of accessibility at selected positions on HAMP1 in low salt (300 mm NaCl) and high salt (4 m NaCl), in the construct that contains HAMP2 (i.e. HtrII truncated at position 237).
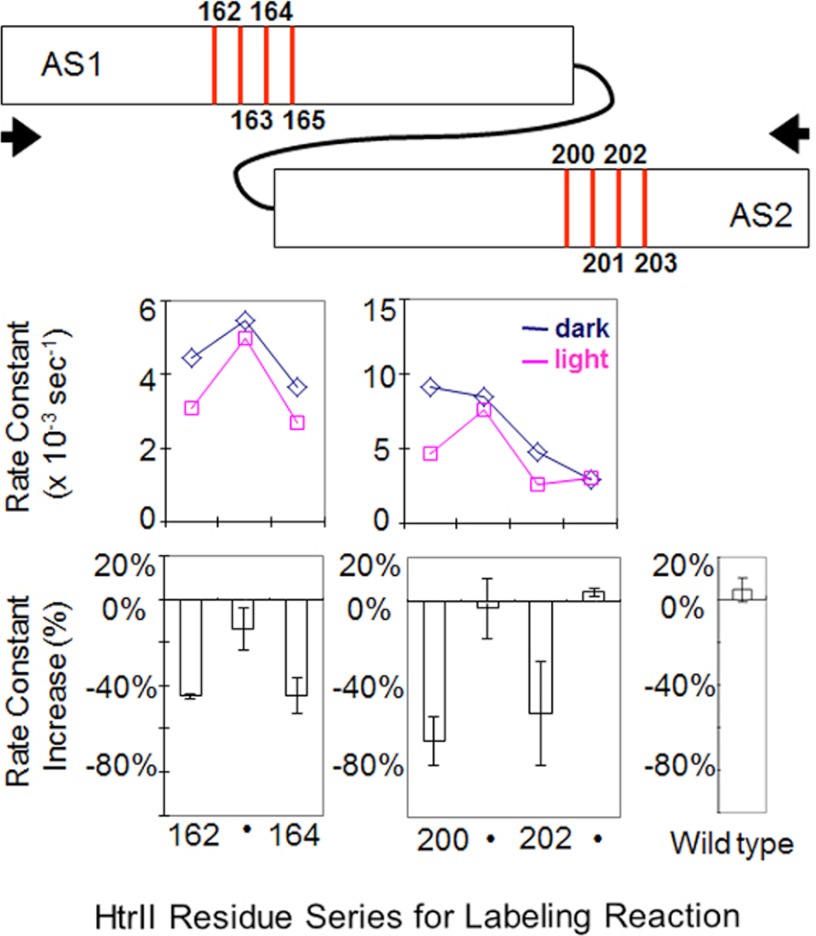
The second HAMP domain, HAMP2, is also composed of AS1 and AS2 helices linked through a flexible peptide linker. To test whether similar conformational changes like those in HAMP1 take place in HAMP2, we constructed a fusion protein of SRII and HtrII with the latter truncated after HAMP2 at position 237 (SRII-HtrII237) and incorporated the protein into nanodiscs with the same procedure as for SRII-HtrII157. Based on the primary sequence alignment (see Fig. 8a), we introduced cysteines into HAMP2 at positions 162–165 in AS1 and 200–203 in AS2, corresponding to the critical positions 90–93 and 126–129 in HAMP1, respectively. A native cysteine exists on wild-type HAMP2 (Cys-173). Because no difference in reaction rate was observed in wild-type SRII-HtrII237 in the dark and under illumination (Fig. 6), we subtracted its constant extent of labeling for the calculation of k values. In contrast to HAMP1, the accessibilities in these same positions decreased upon photostimulation (Fig. 6), implying that conformational changes elicited in HAMP2 are opposite to those in HAMP1. In contrast to HAMP1, the accessibilities in these same positions decreased upon photostimulation (Fig. 6), implying that conformational changes elicited in HAMP2 are opposite to those in HAMP1 (Fig. 7). Accessibility measurements on 90–93 and 126–128 on HAMP1 in the same construct containing HAMP2 (see Fig. 8c) confirm that the accessibility changes are opposite in HAMP1.
FIGURE 6.
Cys accessibility at selected positions of HAMP2. Positions on HAMP2 in the region of the increases on HAMP1 were applied to the labeling study. The R165C mutant failed to express. A distant native cysteine at position 173 on wild-type SRII-HtrII237 shows negligible response to light (bottom right) and therefore provides a constant labeling background in the mutants. The graphic at the top shows the positions of the light-induced changes, which are decreases, with arrows indicating a compaction motion of AS1 and AS2, opposite to that of HAMP1 (Fig. 5), suggested to cause the decreases (see under “Results”). Error bars, mean ± S.D. calculated from 2–5 independent measurements.
FIGURE 7.
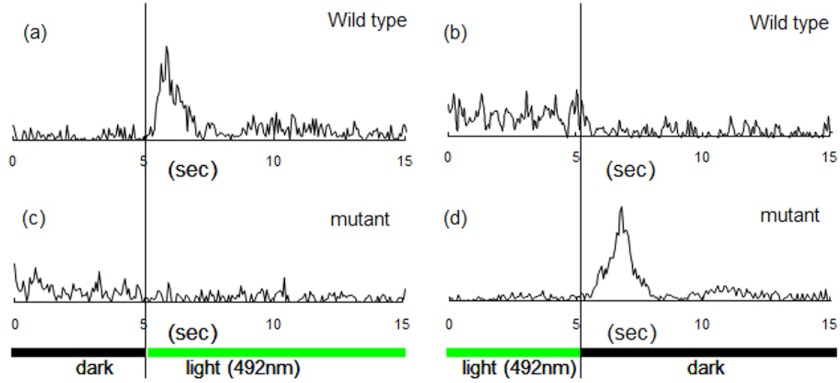
Phototaxis responses by cells expressing wild-type and HAMP2-deleted SRII-HtrII. a–d, cell population swimming reversal frequencies measured by computer-assisted motion analysis during a step-up stimulus in 492-nm light (a and c) and step-down stimulus (b and d) for cells containing wild-type SRII-HtrII (a and b) or a HAMP2 excision mutant of SRII-HtrII (c and d). The illumination regime was cycles of 60 s of dark and 60 s of light. Frequencies are based on 50 cells for each condition.
We observe photostimulus-induced exposure of two residues (positions 127 and 128) on AS2 and four residues on AS1 (positions 90–93). Such a localized exposure could result from a short displacement of AS1 with respect to AS2 (i.e. a sliding or “piston” motion). Such a translational shift (Fig. 5) would explain exposure of ∼2 residues on each of helices AS1 and AS2 without causing changes in exposure at other positions. The results do not rule out that other types of motion also occur but have minimal effects on labeling efficiency. The 90–93 region of AS1 has been shown to be in close contact with SRII (27), and changes in the SRII-HtrII interface may also contribute to exposure of residues on AS1.
If sliding causes the localized accessibility changes, a strong prediction is that in the four-helix bundle, the N-terminal end of the AS2 helix must be positioned in the 90–93 region of AS1 and the C-terminal end of the AS1 helix must be at residues 127 and 128 in AS2 (Fig. 5). To test this possibility, we aligned the HtrII HAMP structure (HAMP1 and HAMP2) with the first two HAMP modules of the Aer2 poly-HAMP domain (10) (Fig. 8a). As shown on the structure of the HtrII HAMP modeled on that of Aer2 (Fig. 8b), residues 90–93 on AS1 and 127–128 on AS2 (shown with red) are positioned at the helix termini of the companion helices precisely as predicted by the sliding model. A second structural prediction from the sliding model is that the continuity of AS2 from HAMP1 with AS1 from HAMP2 would ensure that downward (i.e. away from the membrane) sliding of AS2 relative to AS1 in HAMP1 would result in upward sliding of AS2 relative to AS1 in HAMP2. Therefore, the model predicts that the HAMP2 residues corresponding to those that exhibit light-induced increases of accessibility in HAMP1 will exhibit light-induced decreases in accessibility in HAMP2, i.e. the opposite conformational change. Indeed, monocysteine mutants in the corresponding positions in HAMP2 exhibit light-induced decreases in accessibility (Fig. 8), which is also consistent with the proposal of Crane and co-workers (10) that the two tandem HAMP domains undergo opposite conformational changes. Other residue positions in HAMP2 show negligible light-induced accessibility changes, as do their corresponding positions in HAMP1 (data not shown).
Cell Motility Analysis Indicates the Output Signal from HAMP1 Is Opposite to That of the Output Signal from HAMP2
The hypothesis that HAMP1 and HAMP2 alternatively interconvert between two opposite states during signaling (see “Discussion”) predicts that the output signal from HAMP1 would be opposite from the output signal from HAMP2. To excise HAMP2 from HtrII to allow HAMP1 to directly signal the HtrII output domains, we prepared three HAMP2-deleted constructs, removing residues 136–211, 137–211, or 138–211 in the expectation that two of these would be nonfunctional because only one would be in proper register with the downstream helices. Indeed, we observed that H. salinarum cells expressing SRII-HtrII with the HAMP2 domain excised at 137–211 or 138–211 exhibit no response to light-on or light-off stimuli (data not shown).
However, cells expressing the 136–211 HAMP2-deleted construct exhibit transient swimming reversal frequency responses, and these are indeed opposite to the responses exhibited by cells containing wild-type SRII-HtrII (Fig. 7). Wild-type SRII-HtrII is a repellent receptor, so when light is turned on, the population exhibits a transient increase in swimming reversal behavior (caused by activation of the CheA kinase) (30). Conversely, when light is turned off, the cells exhibit a decrease in reversal frequency. These elemental responses to light-on and light-off stimuli cause cells to reverse their swimming direction more frequently when swimming into higher intensity light, and therefore, they exhibit net migration toward lower light intensity regions. Cells expressing the HAMP2-deleted SRII-HtrII swim more smoothly (exhibit lower reversal frequency) after illumination at 492 nm and, most dramatically, exhibit a strong reversal frequency transient when the light is turned off. This physiological behavior is exactly opposite to the repellent responses of cells expressing wild-type SRII-HtrII, i.e. the HAMP2-deleted complex mediates attractant responses. We conclude that the output of HAMP1 is an attractant signal (i.e. CheA kinase-activating when light is turned off) in an otherwise wild-type receptor complex lacking HAMP2.
DISCUSSION
The pattern of light-induced accessibility changes on the HAMP1 domain helices is notable in being localized to a 4-residue region on AS1 and 2 adjacent residues on AS2. The discrete localized changes we observed are not consistent with the gearbox model, which predicts changes along the entire length of the helices, nor with large-scale destabilization and unbundling of the helices.
Our results support a model in which short displacements of AS2 relative to AS1 alternately in opposite directions in adjacent HAMPs transmit the signal. In addition to the structural evidence for the model from accessibility scanning, a strong functional prediction can be made in terms of phototaxis behavior. If HAMP1 and HAMP2 undergo opposite conformational shifts during signaling, then the output signal from HAMP1 must be opposite to that of HAMP2, i.e. cells expressing a HAMP2-deleted HtrII should have opposite phototaxis behavior. The wild-type SRII-HtrII complex is a strong photorepellent receptor, and no mutations nor alterations of conditions have been previously reported in which the complex acts instead as a photoattractant receptor. Therefore, we interpret our finding that HAMP2 excision uncovers an attractant signal output of HAMP1 that confers robust attractant responses by the complex (Fig. 7) as compelling support for the model.
Prior disulfide mapping of the Salmonella typhimurium aspartate chemotaxis receptor HAMP domain by Butler and Falke (31) also showed only subtle shifts in residue accessibility following receptor activation, and previous study of SRII-HtrII complexes with truncation at different positions in the HAMP1 domain showed light-induced diffusion coefficient changes only when both AS1 and AS2 helices were present (32). These prior findings are consistent with signal transmission from a relative AS1-AS2 interface change.
In summary, our labeling measurements fit well with signal transmission by small displacements of AS2 versus AS1 shortening and lengthening the AS1-AS2 interface. The half-helix turn distance of sliding may locally disrupt the “knob-into-hole” interface between AS1 and AS2 in HAMP1 and form a more dynamic state upon illumination, i.e. upon converting to the dark HAMP2 conformation, a possibility raised for the Aer2 tandem HAMP structures (10).
Lateral interactions, in particular the assembly of bacterial chemotaxis receptors into trimers of dimers, have been found to be responsible for amplification of kinase modulation by chemoeffectors (33–35). A cryoelectron microscopy study of chemotaxis receptor arrays in E. coli cells that have been engineered to overproduce the serine chemoreceptor Tsr shows an increase in electron density indicating significant expansion of the region of the Tsr HAMP domain (36) not expected from the short sliding movement reported here. Possibly, large changes result from conformational changes associated with the amplification of the signal that occurs only in the assembly of taxis receptors (or transducers) into trimer-of-dimer arrays, which the nanodisc is unable to accommodate in our study of the large SRII-HtrII complex. However, the same HAMP domain of Tsr in the single dimer is functional in a chimera joining the adenylyl cyclase output domain at the C terminus of the Tsr HAMP domain (37), excluding the necessity of cooperative amplification for the elemental functioning of HAMP domains.
Our results indicate that small displacements of half a helical turn between AS1 and AS2 in both HAMP1 and HAMP2 domains correlate with positive phototaxis signal output and displacements in the opposite direction with negative signal output. The most notable difference between the two HAMP conformations in the Aer2 crystal structure (10) is a difference in the helical register between the AS1 and AS2 helices by half a helical turn (∼2–3 Å), which staggers the side chains that form the buried core. The displacements would have the effect of altering the AS1/AS2 interface in HAMP1 to make it more similar to that of HAMP2 and vice versa, in agreement with the suggestion by Crane and co-workers (10) of alternating conformational changes in the signal transmission process. However, the displacements alone are not sufficient to interconvert the tandem HAMP structures because they exhibit other differences in side chains, helical tilt, and orientation, in addition to their AS1/AS2 helical register. These other differences may explain our finding that the light-induced increases on the two patches of HAMP1 are not mirror image symmetric in amplitude to the light-induced decreases on the two corresponding patches of HAMP2. The accessibility changes we observe may be caused by AS1/AS2 sliding displacements accompanied by other motions that influence the effect of the displacements on accessibility.
Our data indicate that the N-terminal portion of the HAMP1 AS2 helix is buried, possibly by the SRII subunit, because it is expected to be highly accessible based on the published structures of HAMP domains. FRET measurements (27) indicated that the region spanning residues 91–95 on AS1 of HAMP1 contacts the NpSRII E-F loop, and EPR measurements (28) gave similar results. This region of AS1 is also in contact with the N-terminal portion of AS2, and therefore, the SRII E-F loop may be responsible for the low accessibility of this portion of AS2.
Acknowledgment
We thank Dr. Angel Paredes for expert preparation of electron micrographs.
This work was supported, in whole or in part, by National Institutes of Health Grants R37GM027750 from the NIGMS (to J. L. S.) and RO1HL095820 (A. L. T.). This work was also supported by Endowed Chair AU-0009 from the Robert A. Welch Foundation (to J. L. S.).
- SRII
- sensory rhodopsin II
- HtrII
- transducer for SRII
- HAMP
- a domain named for its presence in histidine kinases, adenylyl cyclases, methyl-accepting proteins, and phosphatases
- DDM
- n-dodecyl-β-d-maltopyranoside
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- MSP
- membrane scaffold protein
- TM
- transmembrane
- Ni-NTA
- nickel-nitrilotriacetic acid
- Np
- N. pharaonis.
REFERENCES
- 1. Spudich J. L., Yang C. S., Jung K. H., Spudich E. N. (2000) Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392 [DOI] [PubMed] [Google Scholar]
- 2. Klare J. P., Gordeliy V. I., Labahn J., Büldt G., Steinhoff H. J., Engelhard M. (2004) The archaeal sensory rhodopsin II/transducer complex: a model for transmembrane signal transfer. FEBS Lett. 564, 219–224 [DOI] [PubMed] [Google Scholar]
- 3. Sasaki J., Spudich J. L. (2008) Signal transfer in haloarchaeal sensory rhodopsin-transducer complexes. Photochem. Photobiol. 84, 863–868 [DOI] [PubMed] [Google Scholar]
- 4. Hazelbauer G. L., Falke J. J., Parkinson J. S. (2008) Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parkinson J. S. (2010) Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64, 101–122 [DOI] [PubMed] [Google Scholar]
- 6. Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. (2006) The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
- 7. Swain K. E., Falke J. J. (2007) Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46, 13684–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Q., Ames P., Parkinson J. S. (2009) Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signaling in chemoreceptors. Mol. Microbiol. 73, 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim S. H. (1994) “Frozen” dynamic dimer model for transmembrane signaling in bacterial chemotaxis receptors. Protein Sci. 3, 159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Airola M. V., Watts K. J., Bilwes A. M., Crane B. R. (2010) Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 18, 436–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Denisov I. G., Baas B. J., Grinkova Y. V., Sligar S. G. (2007) Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 282, 7066–7076 [DOI] [PubMed] [Google Scholar]
- 12. Spudich E. N., Zhang W., Alam M., Spudich J. L. (1997) Constitutive signaling by the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base-Asp-73 interhelical salt bridge. Proc. Natl. Acad. Sci. U.S.A. 94, 4960–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krebs M. P., Spudich E. N., Spudich J. L. (1995) Rapid high-yield purification and liposome reconstitution of polyhistidine-tagged sensory rhodopsin I. Protein Expr. Purif. 6, 780–788 [DOI] [PubMed] [Google Scholar]
- 14. Boldog T., Grimme S., Li M., Sligar S. G., Hazelbauer G. L. (2006) Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U.S.A. 103, 11509–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boldog T., Li M., Hazelbauer G. L. (2007) Using nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 423, 317–335 [DOI] [PubMed] [Google Scholar]
- 16. Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G. (2004) Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 17. Bayburt T. H., Grinkova Y. V., Sligar S. G. (2002) Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins Nanoletters 2, 853–856 [Google Scholar]
- 18. Bayburt T. H., Sligar S. G. (2003) Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 12, 2476–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu J., Howell J. K., Bradley S. D., Zheng Y., Zhou Z. H., Norris S. J. (2010) Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryoelectron tomography. J. Mol. Biol. 403, 546–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yao V. J., Spudich E. N., Spudich J. L. (1994) Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J. Bacteriol. 176, 6931–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nath A., Atkins W. M., Sligar S. G. (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 22. Wegener A. A., Klare J. P., Engelhard M., Steinhoff H. J. (2001) Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. EMBO J. 20, 5312–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gordeliy V. I., Labahn J., Moukhametzianov R., Efremov R., Granzin J., Schlesinger R., Büldt G., Savopol T., Scheidig A. J., Klare J. P., Engelhard M. (2002) Molecular basis of transmembrane signaling by sensory rhodopsin II-transducer complex. Nature 419, 484–487 [DOI] [PubMed] [Google Scholar]
- 24. Rabenstein M. D., Shin Y. K. (1995) Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. U.S.A. 92, 8239–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klare J. P., Bordignon E., Doebber M., Fitter J., Kriegsmann J., Chizhov I., Steinhoff H. J., Engelhard M. (2006) Effects of solubilization on the structure and function of the sensory rhodopsin II/transducer complex. J. Mol. Biol. 356, 1207–1221 [DOI] [PubMed] [Google Scholar]
- 26. Kriegsmann J., Brehs M., Klare J. P., Engelhard M., Fitter J. (2009) Sensory rhodopsin II/transducer complex formation in detergent and in lipid bilayers studied with FRET. Biochim. Biophys. Acta 1788, 522–531 [DOI] [PubMed] [Google Scholar]
- 27. Yang C. S., Spudich J. L. (2001) Light-induced structural changes occur in the transmembrane helices of the Natronobacterium pharaonis HtrII transducer. Biochemistry 40, 14207–14214 [DOI] [PubMed] [Google Scholar]
- 28. Bordignon E., Klare J. P., Doebber M., Wegener A. A., Martell S., Engelhard M., Steinhoff H. J. (2005) Structural analysis of a HAMP domain: the linker region of the phototransducer in complex with sensory rhodopsin II. J. Biol. Chem. 280, 38767–38775 [DOI] [PubMed] [Google Scholar]
- 29. Doebber M., Bordignon E., Klare J. P., Holterhues J., Martell S., Mennes N., Li L., Engelhard M., Steinhoff H. J. (2008) Salt-driven equilibrium between two conformations in the HAMP domain from Natronomonas pharaonis: the language of signal transfer? J. Biol. Chem. 283, 28691–28701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoff W. D., Jung K. H., Spudich J. L. (1997) Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 26, 223–258 [DOI] [PubMed] [Google Scholar]
- 31. Butler S. L., Falke J. J. (1998) Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37, 10746–10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inoue K., Sasaki J., Spudich J. L., Terazima M. (2008) Signal transmission through the HtrII transducer alters the interaction of two α-helices in the HAMP domain. J. Mol. Biol. 376, 963–970 [DOI] [PubMed] [Google Scholar]
- 33. Kim K. K., Yokota H., Kim S. H. (1999) Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400, 787–792 [DOI] [PubMed] [Google Scholar]
- 34. Studdert C. A., Parkinson J. S. (2004) Cross-linking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U.S.A. 101, 2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li M., Khursigara C. M., Subramaniam S., Hazelbauer G. L. (2011) Chemotaxis kinase CheA is activated by three neighboring chemoreceptor dimers as effectively as by receptor clusters. Mol. Microbiol. 79, 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khursigara C. M., Wu X., Zhang P., Lefman J., Subramaniam S. (2008) Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. U.S.A. 105, 16555–16560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanchan K., Linder J., Winkler K., Hantke K., Schultz A., Schultz J. E. (2010) Transmembrane signaling in chimeras of the Escherichia coli aspartate and serine chemotaxis receptors and bacterial class III adenylyl cyclases. J. Biol. Chem. 285, 2090–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]