Background: ATP-binding cassette transporters exist in all life forms and usually require only one substrate-binding domain.
Results: The TRIGALACTOSYLDIACYLGLYCEROL (TGD) complex contains 8–12 substrate-binding proteins.
Conclusion: Multiple substrate-binding proteins may be needed by the TGD complex to enhance its putative lipid transport activity.
Significance: Knowing the subunit stoichiometry of the TGD complex furthers understanding of lipid transfer between chloroplast membranes.
Keywords: ABC Transporter, Chloroplast, Lipid Transport, Protein Complexes, Proteomics, TGD Proteins, TIM17/22/23 Family, Potassium Efflux Antiporter (KEA), Substrate-binding Domain
Abstract
Members of the ATP-binding cassette (ABC) transporter family are essential proteins in species as diverse as archaea and humans. Their domain architecture has remained relatively fixed across these species, with rare exceptions. Here, we show one exception to be the TRIGALACTOSYLDIACYLGLYCEROL 1, 2, and 3 (TGD1, -2, and -3) putative lipid transporter located at the chloroplast inner envelope membrane. TGD2 was previously shown to be in a complex of >500 kDa. We demonstrate that this complex also contains TGD1 and -3 and is very stable because it cannot be broken down by gentle denaturants to form a “core” complex similar in size to standard ABC transporters. The complex was purified from Pisum sativum (pea) chloroplast envelopes by native gel electrophoresis and examined by mass spectrometry. Identified proteins besides TGD1, -2, or -3 included a potassium efflux antiporter and a TIM17/22/23 family protein, but these were shown to be in separate high molecular mass complexes. Quantification of the complex components explained the size of the complex because 8–12 copies of the substrate-binding protein (TGD2) were found per functional transporter.
Introduction
ATP-binding cassette (ABC)3 transporters are an ancient protein family. Its members exist in species from all kingdoms of life, where they play important, frequently essential roles (1, 2). They have evolved to transport a variety of substrates, including inorganic and organic cations and anions, amino acids, peptides, proteins, vitamins, drugs, fatty acids, and lipids (3). Despite their diversity, ABC transporters have a conserved domain structure consisting of two membrane-spanning “permease” domains and two ATPase or nucleotide-binding domains (NBDs). In most prokaryotic ABC importers, these exist as separate polypeptide chains, which assemble as tetramers to form a functional transporter complex. In prokaryotic ABC exporters and most eukaryotic ABC transporters, a single polypeptide strand contains domains for both permease and nucleotide binding and assembles into functional homodimers. In either scenario, when a functional transporter is assembled, physical movements associated with ATP binding and hydrolysis are transmitted through contacts between NBDs and permease domains to catalyze substrate transport (4).
A third domain associated with a subset of ABC transporters is called the substrate-binding domain (SBD). In Gram-negative bacterial importers, SBDs are usually part of soluble periplasmic proteins that bind substrates and deliver them to their transporters (5). In Gram-positive bacteria and archaea, which lack an outer membrane, SBD-containing proteins are anchored by lipid moieties or transmembrane α-helices while performing similar functions (6). In almost every case, one SBD is associated with a functional ABC transporter. The known exceptions to this rule are the OpuA family from Lactococcus lactis, which has two SBDs per transport complex, and the PAO subfamily of transporters in L. lactis and various Streptococcus spp., which has four SBDs per transport complex (5, 7).
TGD1, -2, and -3 (systematically named AtABCI4, -5, and -3) form a putative ABC transporter found in the chloroplast inner envelope membrane of plants and algae, in which TGD1 is similar to an ABC transporter permease, TGD3 to an NBD, and TGD2 to an SBD (8, 9). The TGD genes are most similar to genes encoding an ABC importer of Gram-negative bacteria, probably because chloroplasts were once cyanobacteria acquired by an endosymbiotic event and have many ancestral genes. However, unlike most bacteria in which lipids are transported across a single membrane, chloroplasts transport the bulk of their lipids across two membranes. The major lipid sink in plant cells is the photosynthetic thylakoid membrane, and an essential supply of lipid precursors is provided by the endoplasmic reticulum (ER), where fatty acids are assembled into membrane lipids (10). These lipids must then be transported across the chloroplast outer envelope membrane to the inner envelope membrane, where they are further metabolized into chloroplast-specific lipids, such as monogalactosyldiacylglycerol (11), for eventual transport to the thylakoids. Alternatively, chloroplasts can also synthesize their own lipids, although this pathway is not essential to plant viability (12). Contributions from the ER or chloroplast pathways to total chloroplast lipids can be distinguished by the number of carbons of the fatty acid in the sn-2 position; Chloroplast-derived lipids have 16 carbons, whereas ER-derived lipids have 18 carbons (13). Evidence that TGD1, -2, and -3 function together in lipid transport is derived from the tgd point mutants in Arabidopsis thaliana, which each show a reduction in 18 carbon fatty acids and an increase in 16 carbon fatty acids, implying impaired ER to chloroplast lipid transport (14–16). Furthermore, TGD2, the putative substrate-binding protein, has been shown to bind lipids in vitro, specifically phosphatidic acid (17).
Recently, TGD2 was shown to be part of a complex much larger than the typical ABC transporter (18). Because no direct evidence supported TGD1, -2, and -3 acting in association, the finding shed doubt on whether or not TGD2 was part of a complex with TGD1 and -3. Here we immunoprecipitated tagged versions of TGD1 and -3 and confirmed their association with TGD2 in vivo. The stability of the TGD complex was tested by treatment with a variety of gentle and harsh denaturants and was shown not to break down into a smaller complex. The entire complex was then purified from pea chloroplast envelopes, and components were identified by mass spectrometry. All possible partners were investigated, but those besides TGD1, -2, and -3 were found to be in independent, similarly sized complexes. Finally, the size of the TGD complex was accounted for by label-free quantification, which clearly showed that TGD1, -2, and -3 form a variation of an ABC transporter in which TGD2 is present in 8–12 copies. The biological rationale is discussed.
EXPERIMENTAL PROCEDURES
Plant Growth and Chloroplast Isolations
Arabidopsis of the Columbia ecotype were planted and grown as described (19). Chloroplasts were isolated from 3–4-week-old, plate-grown plants as described (18). Trypsin and thermolysin treatments of intact chloroplasts were as described (20). Pisum sativum (pea) var. Little Marvel (Gurney's Seed and Nursery Co.) were grown, and chloroplasts were isolated from 2–3-week-old plants as described (21). Envelopes of pea chloroplasts were isolated by combining existing methods (22, 23) to give the cleanest envelopes with lowest Rubisco contamination as follows. Pelleted chloroplasts were broken by resuspension in 10 mm Hepes-KOH, 10 mm MgCl2, pH 7.5, at a concentration of 1 mg of chlorophyll/ml. After 10 min incubation on ice, bulk thylakoids were removed by three 1500 × g 5-min centrifugations at 4 °C, and after each centrifugation, the supernatant was moved to a new tube. Crude envelopes were pelleted by a 100,000 × g 30-min centrifugation at 4 °C; resuspended in 10 mm Tricine-HCl, 2 mm EDTA, pH 7.5; and separated on a 0.6–1 m sucrose gradient. Bands containing outer and inner envelope membranes were pooled and washed once with 0.1 m NaHCO3 to help remove peripheral proteins. During no step of the procedure was the complex size altered.
Generation of kea1kea2 Lines
T-DNA insertion lines homozygous for kea1 (CS861469) or kea2 (SALK_045324C) were obtained from the Arabidopsis Information Resource (24, 25). Genotyping of various alleles used the following primers: KEA1 forward (5′-GATCCGTGTATCTCATTCCACATC-3′), wild-type reverse (5′-GCAGACGGGAAGTACTGGC-3′) and T-DNA reverse (5′-GCTTCCTATTATATCTTCCCAAATTACCAATACA-3′); KEA2 forward (5′-GTGATGTAATGGCAGATGGCG-3′), wild-type reverse (5′-CTTACAAGAGCATTTAAGCAGGC-3′), and T-DNA reverse (5′-ATTTTGCCGATTTCGGAAC-3′). kea1 and kea2 plants were crossed, and double homozygous lines were selected by genotyping of the F2 population.
Generation of TGD1 and TGD3 HA Lines
HA tags were added to TGD1 or TGD3 sequences by PCR amplification with the following primers: TGD1, 5′-GCGCAGATCTATGATGCAGACTTGTTGTTCC-3′ and 5′-GCGCGTCGACTCATGCATAATCGGGAACATCATAGGGATAAACACAGTTCTTCAAAGAATCTCC-3′; TGD3, 5′-TGGCGTCGACATGCTTTCGTTATCATGCTCTTC-3′ and 5′-GCCAGGTACCTCATGCATAATCGGGAACATCATAGGGATAGTATCTGATTGGTCCATCGAG-3′. Products were cloned using the Zero® blunt PCR cloning kit (Invitrogen), sequenced, and then digested with BglII and SalI (TGD1) or SalI and KpnI (TGD3) and cloned into pCAMBIAmcs1300. Agrobacteria tumefaciens strain GV3101 bearing the plasmids was used to transform tgd1-1 or tgd3-1 Arabidopsis as described (26). Parental genotype mapping was precisely as described previously (15, 16).
Immunoprecipitation
Isolated chloroplasts were dissolved in 50 mm BisTris-HCl, pH 7.0, 500 mm 6-aminocapronic acid, 150 mm NaCl, 1% dodecylmaltoside with one protease inhibitor chip minus EDTA (Roche Applied Science) per 500 μl. After incubating on ice for 30 min, insoluble materials were removed by centrifugation at 100,000 × g for 10 min at 4 °C. Supernatants were immunoprecipitated using the Pierce HA Tag IP/Co-IP kit (Thermo Fisher Scientific) essentially as per instructions. Resin was washed with buffer similar to dissolving buffer with only 0.1% dodecylmaltoside. Proteins were eluted with non-reducing SDS-PAGE loading buffer.
Lipid Analyses
Lipids were extracted from plant leaves as described (27) and used directly for TLC analysis as described (15) or extracted from the TLC plates, broken down into fatty acid methyl esters, and quantified as described (28).
Native Gel Analyses
Protein complexes from isolated chloroplasts, chloroplast envelopes, or whole leaves were solubilized in 50 mm BisTris-HCl, pH 7.0, 500 mm 6-aminocapronic acid, 10% glycerol, supplemented with reagents described here (detergents obtained from Affymetrix or Sigma). Solubilization of leaf proteins was increased by micropestle homogenization in buffer. When chloroplasts had already been digested by thermolysin or trypsin, solubilization buffer also contained 20 mm EDTA or 0.1 mg/ml trypsin inhibitor, respectively. Insoluble materials were removed by centrifugation at 100,000 × g for 10 min at 4 °C. Supernatants were run on blue native or histidine deoxycholate native gels, denatured, and run in a second dimension on denaturing SDS-PAGE as described (29, 30). Gels were immunoblotted or stained with the Pierce Silver Stain kit for mass spectrometry (Thermo Fisher Scientific) according to instructions.
Antisera
Creation of TGD2 antiserum was described previously (14). TOC159 antiserum was a gift from Masato Nakai (29). TIC110 antiserum was a gift from John Froehlich (31). HA antiserum was obtained from Covance or Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). At3g49560 antiserum was raised against its N-terminal 50 amino acids as follows. cDNA was cloned into pDEST15 using Gateway® technology (Invitrogen) with a PreScission protease site at the C terminus. GST-tagged recombinant proteins were expressed in BL21 cells, bound to GST-Sepharose (Scientifix), and eluted via on-column digestion using PreScission Protease (GE Healthcare). The identity of the recombinant protein was confirmed by mass spectrometry prior to inoculation into rabbits as per the standard protocol (32). For testing the antibody, recombinant proteins of Tim17-1 (At1g20350), Tim17-2 (At2g37410), Tim17-3 (At5g11690), Tim23-1 (At1g17530), Tim23-2 (At1g72750), Tim23-3 (At3g04800), Tim22-1 (At1g18320/At3g10110), At3g25120, At3g49560, and At5g24650 were produced by Gateway® cloning into pDEST17 in BL21 cells. Equal abundance of recombinant proteins was estimated by detection with 6× His antiserum (Qiagen).
Mass Spectrometry Analyses
Gel bands were subjected to in-gel tryptic digestion (33) with modifications. Briefly, gel bands were dehydrated using 100% acetonitrile (ACN) and incubated with 10 mm dithiothreitol (DTT) in 100 mm NH4HCO3, pH ∼8, at 56 °C for 45 min, dehydrated again, and incubated in the dark with 50 mm iodoacetamide in 100 mm NH4HCO3 for 20 min. Gel bands were then washed with 100 mm NH4HCO3 and dehydrated again. Sequencing grade modified trypsin (Promega) was prepared to 0.01 μg/μl in 50 mm NH4HCO3, and ∼50 μl of this was added to each gel band so that the gel was completely submerged. Bands were then incubated at 37 °C overnight. Peptides were extracted by water bath sonication in a solution of 60% ACN, 1% trichloroacetic acid and vacuum dried to ∼2 μl. Peptides were then resuspended in 2% ACN, 0.1% trifluoroacetic acid to 20 μl, and from this, 10 μl were injected by a Waters nanoAcquity Sample Manager and loaded for 5 min onto a Waters Symmetry C18 peptide trap (5 μm, 180 μm × 20 mm) at 4 μl/min in 5% ACN, 0.1% formic acid. Bound peptides were eluted onto a Michrom Bioresources 0.1 × 150-mm column packed with 3u 200A Magic C18AQ material over 35 min with a gradient of 5% B to 30% B in 21 min using a Waters nanoAcquity UPLC system (Buffer A contained 99.9% water, 0.1% formic acid; Buffer B contained 99.9% ACN, 0.1% formic acid) with an initial flow rate of 1 μl/min. Eluted peptides were sprayed into a Thermo Fisher Scientific LTQ linear ion trap mass spectrometer outfitted with a Michrom Bioresources ADVANCE nanospray source. The top five ions in each survey scan were then subjected to data-dependent zoom scans followed by low energy collision-induced dissociation, and the resulting MS/MS spectra were converted to peak lists using the BioWorks Browser version 3.3.1 (Thermo Fisher Scientific) using the default LTQ instrument parameters. Peak lists were searched against a P. sativum cDNA library, previously generated in house (34) using the Mascot searching algorithm, version 2.3 (see the Matrix Science Web site). Mascot parameters were as follows: two missed tryptic sites allowed, fixed modification of carbamidomethyl cysteine, variable modification of oxidation of methionine, peptide tolerance of ±200 ppm, MS/MS tolerance of 0.6 Da, peptide charge state limited to +2/+3. The Mascot output was then analyzed using Scaffold, version 3.3.3, to probabilistically validate protein identifications using the ProteinProphet computer algorithm (35). Assignments validated above the scaffold 95% protein and 99% peptide confidence filters were reported.
RESULTS
Preparation of Tagged TGD1 and TGD3 Arabidopsis Lines
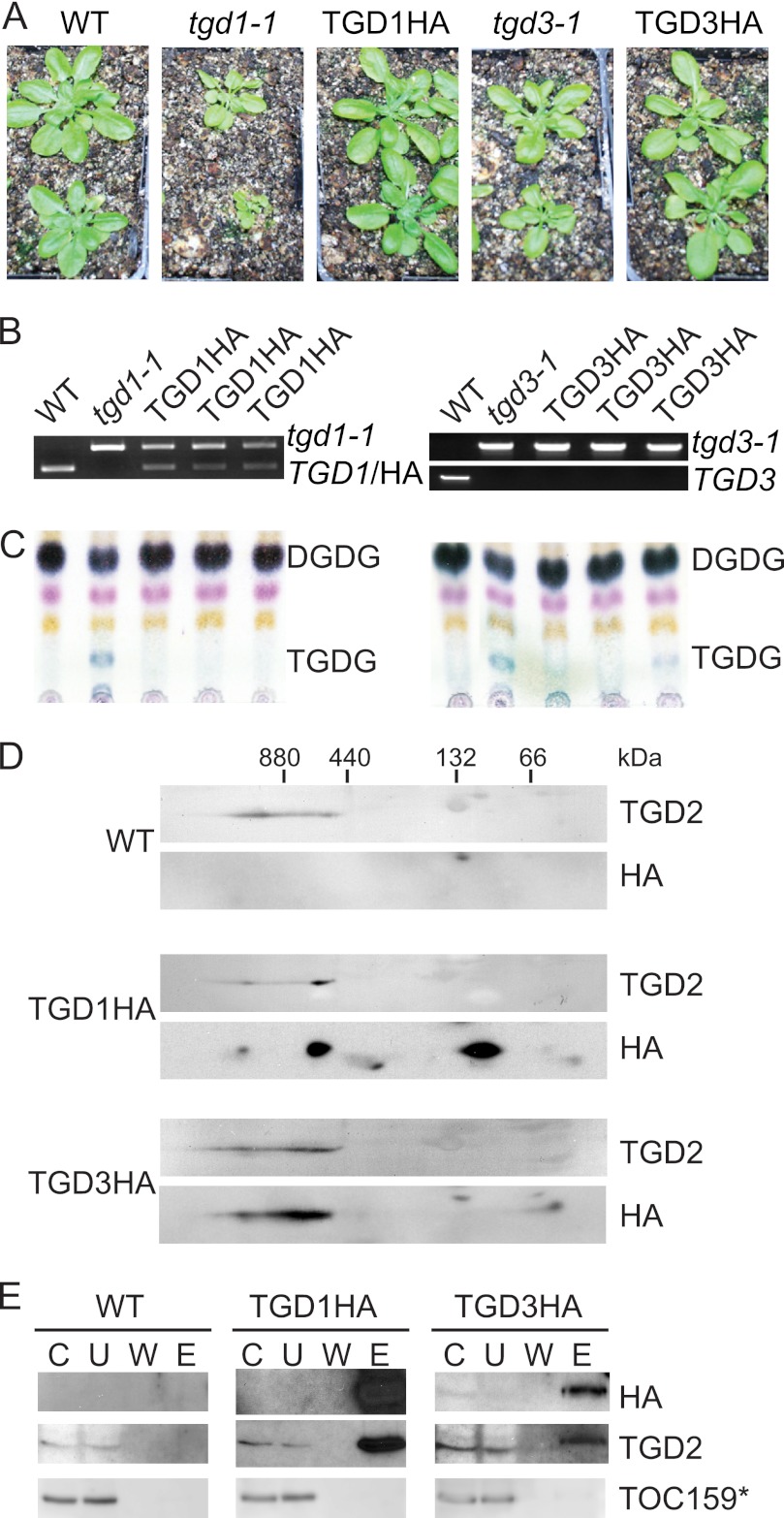
TGD1, -2, and -3 have been proposed to constitute an ABC transporter. However, previous investigations of TGD2 showed its presence in a complex much larger than predicted of a classic ABC, shedding doubt on its association with TGD1 and -3 (18). To investigate the composition of the TGD2-containing complex, tagged lines of TGD1 and -3 were constructed (8) because antisera against these proteins are lacking. Constructs expressing TGD1 or TGD3 with a C-terminal HA tag under control of the constitutive 35S promoter were introduced into tgd1-1 or tgd3-1 Arabidopsis (Fig. 1A). The resulting plants were screened by antibiotic resistance and then confirmed to have the transgene by PCR of genomic DNA (Fig. 1B). TGD1HA lines fully complemented the growth phenotype of tgd1-1 mutant line and reduced levels of diagnostic lipid trigalactosyldiacylglycerol (TGDG) to wild-type levels (Fig. 1C). Similarly, TGD3HA lines complemented growth phenotypes of the tgd3-1 tDNA insertion allele (Fig. 1A), and most lines reversed TGDG accumulation, although TGDG could still be seen in some lines (Fig. 1C). Together, these data indicate that a portion of produced TGD1HA and TGD3HA proteins are acting similarly to their native counterparts and that it is unlikely that the HA tag interferes with their native function.
FIGURE 1.
TGD1, -2, and -3 associate in a large complex. A, 28-day-old Arabidopsis are shown, with genotypes as labeled above (WT indicates wild-type Arabidopsis). TGD1HA or TGD3HA labels indicate plants homozygous for the tgd1-1 or tgd3-1 alleles, which also express TGD1HA or TGD3HA under the control of the 35S promoter, respectively. B, genotyping of plants as labeled showing the presence of the mutant tgd1-1 or tgd3-1 alleles and lack of the endogenous TGD1 or TGD3 alleles. C, α-naphthol-stained, thin layer chromatogram of lipids isolated from Arabidopsis of genotypes given at the top. Digalactosyldiacylglycerol (DGDG) and trigalactosyldiacylglycerol (TGDG) are indicated. D, immunoblots detecting proteins indicated at the right of 20 μg of 1% dodecylmaltoside-solubilized, chlorophyll-equivalent chloroplasts isolated from Arabidopsis of genotypes indicated at the left. Protein complexes were separated in the first dimension on a 4–14% BN-PAGE (marker at top) and then denatured and run on a 12% SDS-PAGE in the second dimension. E, immunoblots detecting proteins listed at the right (TOC159* indicates the 86-kDa fragment of TOC159) of an immunoprecipitation using HA antiserum of solubilized Arabidopsis chloroplasts, genotypes given at the top. C, chloroplast starting material; U, unbound fraction; W, final wash; E, eluate.
TGD1, -2, and -3 Are Parts of the Same Complex
To determine if TGD1 and -3 are in a similarly sized complex as TGD2, chloroplast complexes were separated in the first dimension by blue native PAGE (BN-PAGE), and in the second dimension by SDS-PAGE (Fig. 1D). HA signals matching the size of TGD1 or TGD3 were found in high molecular mass complexes of similar size and pattern to TGD2 in the same plants. Some of the TGD1HA and TGD3HA proteins were also present at lower molecular masses, although this may be due to increased transgene expression compared with that of the native genes. To confirm that TGD1, -2, and -3 interact directly, HA-containing proteins were immunoprecipitated from solubilized chloroplasts isolated from wild-type, TGD1HA-producing, or TGD3HA-producing lines. TGD1HA or TGD3HA proteins efficiently co-precipitated TGD2 (Fig. 1E). Validating the specificity of the co-immunoprecipitation, HA-tagged proteins were not precipitated from wild-type chloroplasts, and neither TGD1HA nor TGD3HA proteins co-precipitated an unassociated chloroplast membrane protein, the translocon at the outer envelope membrane of chloroplasts, 159 kDa (TOC159). That TGD1HA and TGD3HA proteins can each independently pull down TGD2 confirms the hypothesis that TGD1, -2, and -3 are physically associated.
Characterizing TGD Complex
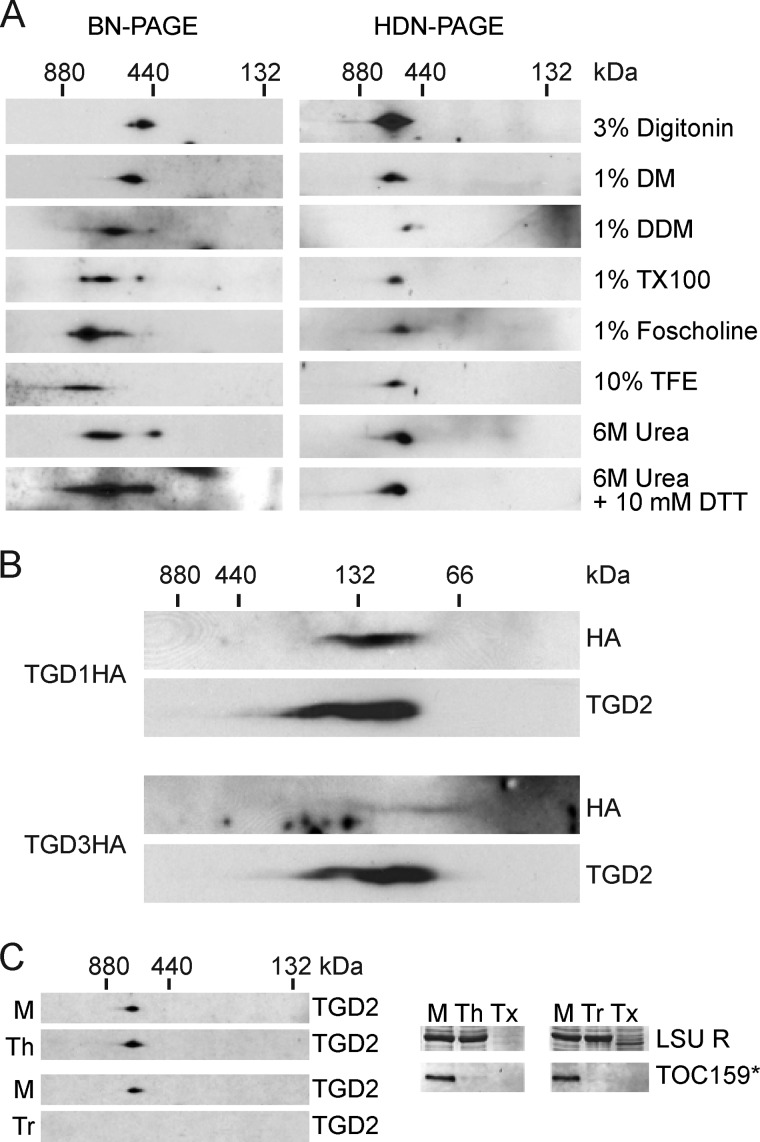
The expected size of a canonical ABC transporter with a TGD1/TGD3/TGD2 subunit ratio of 2:2:1 would be ∼160 kDa; however, the identified TGD complex is larger than 500 kDa (Fig. 1A). We hypothesized that the size difference could be due to additional components surrounding a “core” complex similar to the canonical transporter. To gently break down the large complex and reveal the “core” complex, isolated chloroplasts were treated using mildly disrupting conditions, and the complexes were resolved both by BN-PAGE and by a recently described variation of native gel for chloroplast envelope proteins, histidine deoxychlolate native PAGE (HDN-PAGE) (30). Several gentle detergents known to solubilize membrane protein complexes, including digitonin, decylmaltoside, dodecylmaltoside (DDM), and Triton X-100, were employed to treat isolated chloroplasts. In every case, the resulting TGD2-containing complex was similar in size and stability (Fig. 2A) (36). A harsher detergent that is used in crystallization studies, foscholine-12, was similarly tested and was also found not to disrupt the complex (Fig. 2A) (37). The complex was then challenged with well known denaturants trifluoroethanol or 6 m urea in the presence of 1% DDM, but again, the complex was not reduced in size or stability (Fig. 2A) (38, 39). At higher concentrations of either trifluoroethanol or urea, the complex precipitated in the native gel loading well. To exclude the possibility that disulfide bonds were holding the complex together, reducing agent DTT was included during solubilization with 6 m urea and 1% DDM, but again the complex was not disrupted (Fig. 2A). Throughout each of these trials, the HDN-PAGE showed superior complex resolution and accordingly was used for all further native gel analysis.
FIGURE 2.
The stable TGD1, -2, and -3 complex is present in the chloroplast inner envelope. A, anti-TGD2 immunoblots of two-dimensional gels in which the first dimension is a 4–10% BN-PAGE (left) or HDN-PAGE (right) and the second dimension is a 12% SDS-PAGE. Isolated wild-type Arabidopsis chloroplasts (equivalent to 20 μg of chlorophyll) were solubilized with reagents listed at the right (DM, decylmaltoside; TX100, Triton X-100; TFE, trifluoroethanol). In the case of trifluoroethanol and urea samples, 1% DDM was also present. B, immunoblots detecting proteins indicated at the right, similar to the HDN-PAGEs in A, except the gradient was 4–14%, and chloroplasts isolated from Arabidopsis of genotypes indicated at the left were solubilized in 1% SDS. C, anti-TGD2 immunoblots similar to the HDN-PAGE solubilized with DDM in A, except prior to solubilization the chloroplasts were mock-treated (M), thermolysin-treated (Th), or trypsin-treated (Tr). Immunoblots of single-dimension SDS-PAGEs show digestion of the 86-kDa fragment of TOC159 (TOC159*), and Coomassie Brilliant Blue stain shows protection of the large subunit of Rubisco (LSU R). Tx, trypsin treatment with Triton X-100 present.
Having failed to disrupt the complex with gentle denaturants and reducing agents, and knowing that it was disrupted in SDS-PAGEs, solubilization with 1% SDS in otherwise native conditions was tried. A smaller complex was resolved under these conditions, in which the TGD2 signal overlapped with the HA signal in the TGD1HA-producing plant but not to a large extent with the HA signal from the TGD3HA-producing plant (Fig. 2B). This smaller complex was not well defined, and although it included TGD1HA, most of the TGD3HA was not in the complex, indicating that if a core complex does exist, it is not similar to a canonical ABC transporter.
Localization of TGD Complex
Previous studies localized TGD1-GFP and TGD2-GFP fusion constructs to the chloroplast inner envelope membrane (14, 19) and a TGD3 construct to the chloroplast stroma (16). If additional proteins were present in the complex, they could span the gap between the chloroplast outer and inner envelope membranes, forming a lipid conduit similar to the well studied protein transport conduit formed by translocons at the outer or inner envelope membrane of chloroplasts (TOC or TIC) (40). To determine the location of the complex, thermolysin and trypsin protease treatments were used. Thermolysin cannot penetrate the chloroplast outer envelope membrane, and it digests proteins exposed at the surface of chloroplasts (41), whereas trypsin can pass through the outer envelope membrane and therefore digests all susceptible proteins not protected by the inner envelope membrane (20). Intact chloroplasts were digested with thermolysin or trypsin, the digestion was quenched, and then chloroplasts were solubilized and complexes were analyzed. Both thermolysin and trypsin digested the control outer envelope protein TOC159, but only trypsin was able to affect the TGD complex (Fig. 2C). Neither protease penetrated the chloroplast inner envelope membrane, as evinced by the lack of digestion of stromal protein, large subunit of ribulose bisphosphate carboxylase/oxygenase (Rubisco) (LSU R, Fig. 2C). These data corroborate the hypothesis that the entire TGD complex is located in the chloroplast inner envelope membrane. Notably, no monomers of TGD2 were detectable after thermolysin digestion. TGD2 and TGD3 are known to be resistant to trypsin treatment (14, 16), and TGD2 can homo-oligomerize in vitro (18). Monomers of TGD2 may not exist if trypsin treatment only digests TGD1, presumably resulting in poorly resolved multimeric fragments below the detection limit.
Determining TGD Complex Components
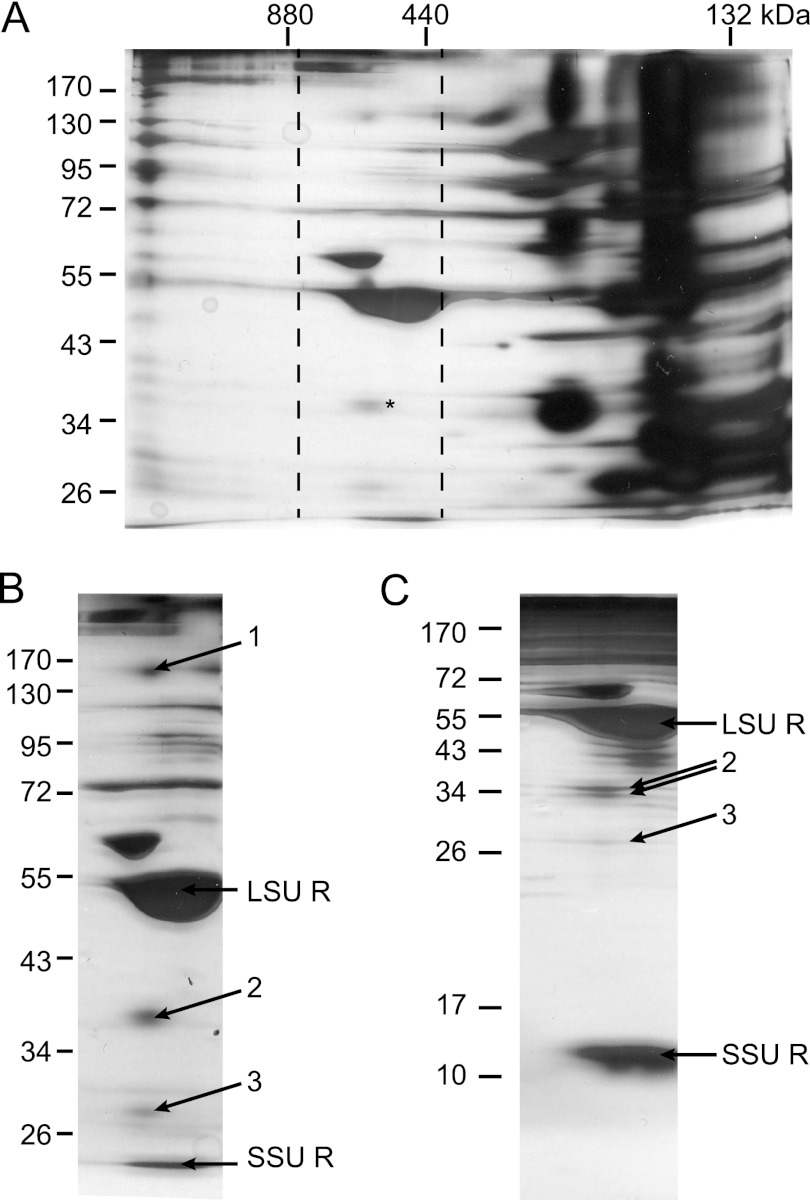
A first attempt at determining members of the TGD complex was a scaled up version of the HA tag immunoprecipitation experiment done with Arabidopsis chloroplasts, as shown in Fig. 1C. However, the only spectra reliably found for this material were assigned to TGD2 (data not shown), and these were few. In order to further scale up the experiment, pea plants, specifically P. sativum var. Little Marvel, were used. This pea model system yields abundant chloroplasts and was appropriate for mass spectrometry analysis because it has an extensive, annotated expressed sequence tag database available (34, 42). Although the Arabidopsis TGD2 antiserum does not recognize its pea homolog, rendering affinity purification of the complex impossible, a standard biochemical chloroplast envelope isolation procedure has the advantage of not creating artificial abundance of any single complex component. To this end, highly purified pea chloroplast envelope membranes were solubilized and separated by HDN-PAGE in the first dimension and SDS-PAGE in the second dimension (Fig. 3A). Few proteinaceous complexes were visible between 880 and 440 kDa, and preliminary mass spectrometry of bands near the size of TGD2 (∼36 kDa based on the Arabidopsis protein) identified the pea homolog (Fig. 3A). To detect other bands in the same vertical line (same complex size) with similar distribution patterns, multiple SDS-PAGEs with varying acrylamide concentration were used to resolve the complex components in the second dimension, 10 and 15% shown in Fig. 3, B and C. Only those bands reproducibly showing distributions similar if not identical to those of the TGD2-containing band were considered, with three bands meeting this criterion: band 1 at 140 kDa, band 2 (TGD2-containing) at 36 kDa, and band 3 at 28 kDa (Fig. 3B). To collect enough protein for sufficiently high spectral counts, four to five replicates of each band were pooled and analyzed by LC MS-MS. Proteins identified by at least one spectral count are displayed in Table 1, with spectral counts, numbers of unique peptides, and percentage of coverage data.
FIGURE 3.
The TGD complex of pea co-migrates with another high molecular mass protein. A, isolated pea chloroplast envelopes solubilized in 3% digitonin and separated by 4–14% HDN-PAGE in the first dimension and 10% SDS-PAGE in the second and then visualized by silver staining. Dashed lines mark the selection of gels displayed in B and C. An asterisk marks location of TGD2. B, as in A, with arrows showing bands submitted for mass spectrometry analysis. The large and small subunits of Rubisco are also labeled (LSU R and SSU R, respectively). C, as in B, except 15% SDS-PAGE in the second dimension. These data were not used in final analysis because band 1 could not be separated under these conditions.
TABLE 1.
Proteins identified by mass spectrometry
Gel bands from which proteins were identified are indicated at the left. The major P. sativum (pea) contig identified is given, as is the annotated Arabidopsis homolog's accession number. Total spectral counts (SpC), unique peptides, molecular masses (kDa), and percentage coverage are given for each of three biological replicates (numbered 1–3), based on 99% protein identification and 95% peptide identifications. Molecular masses and lengths used to calculate percentage coverage are based on predicted mature Arabidopsis homologs to reduce variations due to pea contig lengths and overlap. Pea contigs without Arabidopsis homologs were assigned the molecular mass of the band and the percentage coverage of the contig. APE2, acclimation of photosynthesis to environment 2; CLP, caseinolytic protease; LSU R, large subunit of Rubisco; PORA, protochlorophyllide oxidoreductase; TIC, translocon at the inner envelope membrane of chloroplasts; TIM, translocase at the inner mitochondrial membrane; TOC, translocon at the outer envelope membrane of chloroplasts; WPP, Trp-Pro-Pro motif.
| Band no. | A. thaliana accession no. | Pea contig | Name | SpC |
Mass | Unique peptides |
Percentage coverage |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |||||
| kDa | % | % | % | ||||||||||
| 1 | At4g00630 | Ps066597f | K+ efflux antiporter (KEA2) | 45 | 33 | 21 | 126.2 | 19 | 12 | 8 | 18.7 | 10.7 | 8.4 |
| At5g23890 | Ps000222b | Putative transporter | 21 | 103.9 | 7 | 10.1 | |||||||
| At4g34830 | Ps027447f | Maturation of LSU R 1 | 11 | 119.8 | 5 | 6.6 | |||||||
| At4g02510 | Ps007362a | TOC159 | 1 | 10 | 1 | 160.8 | 1 | 6 | 1 | 0.8 | 8.2 | 1.6 | |
| At1g06950 | Ps023494a | TIC110 | 37 | 112.1 | 18 | 25.7 | |||||||
| At3g46740 | Ps022977c | TOC75 | 1 | 75 | 1 | 2.9 | |||||||
| At2g28000 | Ps010082c | Chaperone, CPN60a | 2 | 62.07 | 2 | 5.6 | |||||||
| At3g13470 | Ps009941b | Chaperone, CPN60b-2 | 2 | 63.34 | 2 | 4.7 | |||||||
| N/A | Ps070470a | No annotation | 6 | 2 | 140 | 4 | 2 | 78.8 | 59.6 | ||||
| N/A | Ps039763e | No annotation | 2 | 3 | 140 | 2 | 2 | 21.0 | 21.0 | ||||
| N/A | Ps054876c | No annotation | 3 | 140 | 2 | 51.4 | |||||||
| N/A | Ps033657a | No annotation | 3 | 140 | 2 | 15.7 | |||||||
| 2 | At3g20320 | Ps000114b | TGD2 | 133 | 92 | 100 | 36 | 43 | 34 | 32 | 77.4 | 68.5 | 73.0 |
| At1g65410 | Ps016716e | TGD3 | 25 | 16 | 27 | 36 | 14 | 7 | 12 | 55.1 | 27.6 | 45.2 | |
| At5g05000 | Ps013223b | TOC34 | 48 | 9 | 34.71 | 18 | 5 | 42.5 | 14.7 | ||||
| At3g47520 | Ps007032f | Malate dehydrogenase | 3 | 10 | 16 | 42.41 | 3 | 5 | 8 | 9.9 | 10.5 | 30.9 | |
| At5g42130 | Ps002665b | Putative transporter | 3 | 44.36 | 2 | 9.2 | |||||||
| At5g54190 | Ps005013a | PORA | 3 | 4 | 43.86 | 2 | 3 | 8.5 | 14.4 | ||||
| At5g01500 | Ps017159a | Thylakoid ATP carrier | 1 | 45.09 | 1 | 5.1 | |||||||
| At4g04640 | Ps017279a | ATP synthase g chain 1 | 3 | 40.91 | 3 | 11.7 | |||||||
| At3g63490 | Ps013200a | Ribosomal L1 family protein | 2 | 7 | 37.63 | 2 | 3 | 9.4 | |||||
| 3 | At1g19800 | Ps023468d | TGD1 | 14 | 8 | 10 | 28 | 6 | 4 | 6 | 24.0 | 17.7 | 24.0 |
| At5g46110 | Ps006793e | APE2 | 11 | 44.63 | 5 | 9.6 | |||||||
| At2g37220 | Ps002377c | Ribonucleoprotein,putative | 4 | 5 | 30.72 | 3 | 2 | 14.8 | 14.4 | ||||
| At5g24650 | Ps010239c | TIM17/22/23 family protein | 4 | 9 | 11 | 27.77 | 2 | 3 | 4 | 10.0 | 16.6 | 20.1 | |
| At3g25920 | Ps010043a | Ribosomal protein L15 | 5 | 4 | 29.71 | 4 | 2 | 21.1 | 10.8 | ||||
| At2g43030 | Ps002441d | Ribosomal L3 family protein | 3 | 14 | 29.36 | 2 | 7 | 12.6 | 43.9 | ||||
| At5g43070 | Ps015737a | WPP domain protein 1 | 2 | 16.63 | 1 | 16.5 | |||||||
| Atcg00490 | Ps021077b | Small subunit of RUBISCO | 2 | 26.91 | 2 | 4.2 | |||||||
| Atcg00160 | Ps069182c | Ribosomal protein S2 | 2 | 52.96 | 2 | 12.3 | |||||||
| At3g27160 | Ps001242c | Ribosomal protein S21 | 5 | 20.91 | 2 | 15.4 | |||||||
| At1g49970 | Ps029444f | CLP protease subunit 1 | 4 | 42.63 | 2 | 6.6 | |||||||
| At3g20320 | Ps000114b | TGD2 | 3 | 2 | 35 | 1 | 1 | 3.6 | 3.6 | ||||
| At3g13470 | Ps009941b | Chaperone, CPN60b-2 | 1 | 63.34 | 1 | 2.4 | |||||||
Band 1 had the highest spectral counts of a predicted potassium efflux antiporter (KEA), for which there are two, 76% identical Arabidopsis homologs: KEA1 (At1g01790) and KEA2 (At4g00630). Only TOC159 was also found in each biological replicate, and it is unlikely to be relevant for two reasons. First, it is known to be in its own large complex, which has been described previously and does not include KEA2 or TGDs (30). Second, TOC159 was not co-precipitated by TGD1HA or TGD3HA proteins (Fig. 1E). Therefore, only KEA1 and -2 were considered from band 1 as possible TGD complex components.
Band 2 consistently contained spectral counts of TGD2 and TGD3 as well as a pea homolog of malate dehydrogenase. Malate dehydrogenase has been characterized as part of a 96-kDa stromal complex (43) and is believed to be active as a homodimer (44). For these reasons, it is probably only a contaminant rather than a possible complex component.
Band 3 had consistent spectral counts for TGD1 and a small translocase of the inner mitochondrial membrane (TIM) family protein. The TIM family protein has two 83% identical homologs in Arabidopsis: At5g24650 and At3g49560. Each of these was shown to be targeted to the chloroplast, but they have no known roles there (45). Because nothing was known about their complex formation or function, we considered the possibility that the TIMs were associated with the TGD complex.
Investigation of KEAs and TIMs
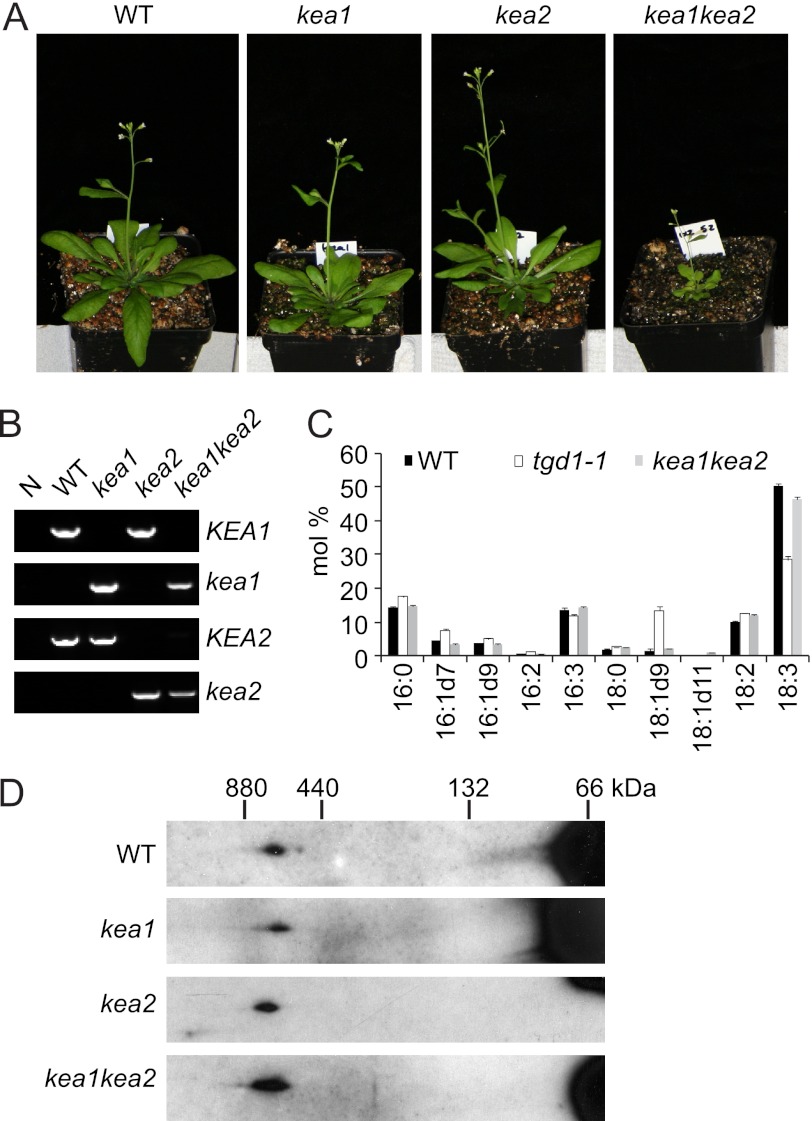
The possible presence of KEA1 or -2 in the TGD complex was investigated through the use of homozygous T-DNA lines carrying insertions into the coding regions of KEA1 or KEA2, respectively. These were crossed, and double homozygous lines were selected among the offspring (Fig. 4A). Genotypes of the lines were confirmed by PCR of genomic DNA (Fig. 4B) and then tested for lipid defects similar to tgd mutant lines. In tgd mutant total fatty acid profiles, there is a strong reduction of 18:3 fatty acids and an increase in 18:1 and 16:0 fatty acids compared with wild type, indicative of a reduction in ER to chloroplast lipid transport. However, this was not seen for the kea1kea2 double mutant lipid extracts (Fig. 4C). If KEA1 and -2 were members of the complex, the complex size should be smaller in the double mutant. However, the TGD-containing complex remained the same size (Fig. 4D). These data support the probability that KEA1 and -2 form a separate complex from that of the TGDs and are not involved in lipid trafficking from the ER to the chloroplast. The function of KEA1 and -2 must be important to plant growth and perhaps chloroplast function because the double mutant plants were reduced in growth and slightly yellow. However, the precise molecular function of these proteins remains to be determined.
FIGURE 4.
Potassium efflux antiporters are not part of the TGD complex. A, 35-day-old Arabidopsis are shown with genotypes labeled above. B, genomic DNA extracted from genotypes listed above (N, no-DNA control) was genotyped using PCR with primers recognizing alleles indicated at the right (KEA1 is the wild-type KEA1 allele, and kea1 is the T-DNA insertion into KEA1; KEA2 and kea2 for the KEA2 allele are designated accordingly). C, total fatty acids of lipids converted to fatty acid methylesters and quantified by gas chromatography for genotypes listed above. The S.D. value of three biological replicates is shown (error bars). D, dodecylmaltoside-solubilized leaf proteins from Arabidopsis genotypes listed at the left were separated in the first dimension by 4–14% HDN-PAGE and in the second dimension by 12% SDS-PAGE and then immunoblotted and detected with TGD2 antiserum.
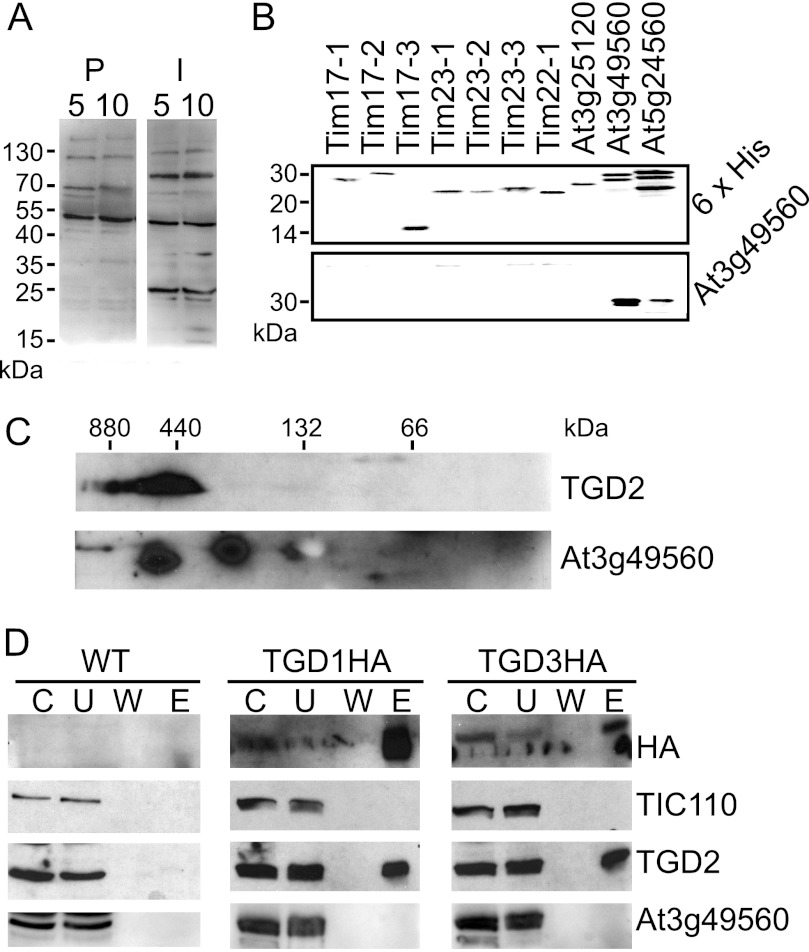
To investigate whether TIM family proteins, At3g49560 and At5g24650, were associated with the TGD complex, antiserum was raised against At3g49560. The antiserum specifically recognized two bands of a similar size (∼25 kDa) in isolated chloroplasts when compared with preimmune serum (Fig. 5A). Moreover, it recognized heterologously produced proteins for both TIMs of interest but not other, less related TIM proteins (Fig. 5B). Two-dimensional analysis with HDN- and SDS-PAGEs showed that At3g49560 antiserum does recognize a ∼25-kDa protein in a high molecular mass complex that overlaps with the TGD2 complex (Fig. 5C); however, this protein was not co-immunoprecipitated by HA in TGD1HA- and TGD3HA-producing plants (Fig. 5D). Thus, it is likely that the identified TIM proteins, like the KEAs, are forming their own large complexes rather than forming parts of the TGD complex.
FIGURE 5.
TIM family proteins are not part of the TGD complex. A, immunoblots of isolated chloroplast protein equivalent to 5 or 10 μg of chlorophyll were probed with At3g49560 antiserum (I) or its preimmune serum (P), as indicated at the top. B, antiserum raised against proteins encoded by At3g49560 was tested for specificity against the preprotein and amino acid transporter family. Full-length recombinant proteins for Tim17-1 (At1g20350), Tim17-2 (At2g37410), Tim17-3 (At5g11690), Tim23-1 (At1g17530), Tim23-2 (At1g72750), Tim23-3 (At3g04800), Tim22-1 (At1g18320/At3g10110), At3g25120, At3g49560, and At5g24650 (indicated at the top) were detected with antiserum recognizing proteins indicated at the right. C, chloroplasts isolated from wild-type Arabidopsis were separated by 4–14% HDN-PAGE in the first dimension and 12% SDS-PAGE in the second and then immunoblotted and detected with antisera recognizing proteins indicated at the right. D, immunoprecipitations of HA-tagged proteins, similar to those in Fig. 1E, were immunoblotted and probed with antisera recognizing proteins indicated at the right. C, chloroplast starting material; U, unbound fraction; W, final wash; E, eluate.
There Are Multiple TGD2s per Functional TGD Complex
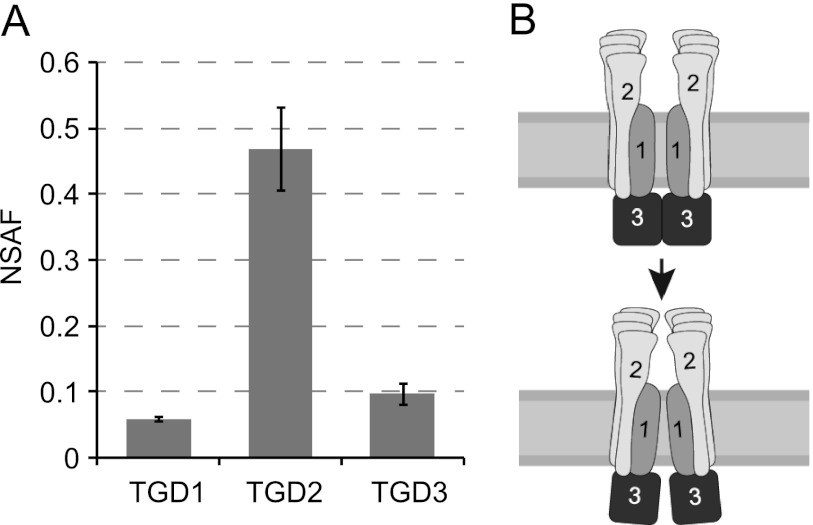
Because other complex components were not identified, TGD1, -2, and -3 must themselves make up the bulk of the large TGD complex. To quantify levels of each, spectral counts (Table 1) were normalized based on molecular mass. This method of quantification, known as spectral counting, has been shown to be a reliable method when compared with other methods of labeled and label-free quantification (46, 47). Normalized spectral abundance factors (NSAF) for TGD1, -2, and -3 show that 4–6 copies of TGD2 are present for every copy of TGD1 and -3 (Fig. 6A). Assuming that TGD1 and -3 remain similar to other ABC transporters and form a tetramer (dimer of homodimers), there would be an overall subunit stoichiometry of 2 TGD1, 2 TGD3, 8–12 TGD2, yielding an overall complex with a size of 416–560 kDa.
FIGURE 6.
There are 8–10 copies of TGD2 per functional transporter. A, calculated normalized spectral abundance factors (NSAF) are displayed with S.D. (error bars) for TGD1, -2, and -3. B, model of the TGD1, -2, and -3 complex suggests that changes in TGD1 geometry due to ATPase activity by TGD3 could be shared by multiple TGD2 polypeptides.
DISCUSSION
We have demonstrated that TGD1 and -3 associate with TGD2 in vivo to form a complex of greater than 500 kDa. This complex was shown to be stable and to be protected from protease digestion by the outer but not the inner chloroplast envelope membrane. Although other chloroplast envelope proteins in similarly sized complexes were discovered in this study, no additional components of the TGD complex were identified. Instead, we conclude that TGD1, -2, and -3 form an ABC transporter in which the substrate-binding domain (TGD2) is associated at 4–6 times the levels of the permease domain (TGD1) and the nucleotide-binding domain (TGD3).
Normalized spectral abundance factors of TGD1, -2, and -3 were used to quantify their relative levels (Fig. 6A), although the precise number of TGD2s associating per complex remains unclear. Part of the uncertainty is because levels of TGD1 and TGD3 were not the same. Although it is possible that there is more TGD3 than TGD1 in the complex, a large amount of data concerning structures, mechanism, and function of other ABC transporters indicates that two permease and two ATPase domains are required for transport (2). To our knowledge, no ABC transporter varies from this pattern; therefore, we consider it unlikely. Further, if the levels of TGD2 were calculated directly from the levels of TGD1, there would be 16 copies per complex, for a total size of 700 kDa, which is higher than that observed by native gel (Fig. 3A). More probable is that TGD1 is underrepresented because it is a smaller protein and also more hydrophobic than TGD3. Thus, we have calculated the levels of TGD2 based on the levels of TGD3 alone, assuming that the true levels of TGD1 match those of TGD3. The second source of uncertainty is variation between biological replicates. By isolating the complex without affinity purification, we ensured that individual components would not be overrepresented; however, this also increased the number of steps and therefore sample-to-sample variation. The first sample suggests a 5:1 ratio, the second a 6:1 ratio, and the third a 4:1 ratio of TGD2 to TGD3 (Table 1). It is likely that the average 5:1 ratio is correct; however, we can neither confirm nor rule out this possibility. A future structural study of the complex by electron microscope tomography or x-ray crystallography will resolve the precise number of TGD2 (i.e. SBD-containing) proteins in the complex.
Why does the TGD ABC transporter have so many SBD-containing proteins? We assume that they are biologically relevant, for two reasons. First, the size of the complex has been conserved across species (compare Figs. 2A and 3A). Second, when the size of the complex is reduced, as in the TGD2 point mutant tgd2-1 (G234R), the plant shows a lipid transport defect (18). In the case of the ABC transporter OpuA from L. lactis, it was shown that binding of the substrate was cooperative when two SBD-containing proteins were present (48). Binding of TGD2 to its proposed lipid substrate, phosphatidic acid, has similarly been shown to be cooperative (17). As a possible rationale, cooperative binding can increase sensitivity to small amounts of substrate, and levels of phosphatidic acid are low in the chloroplast envelope membranes (49). Therefore, it seems likely that TGD2, like the SBD-containing protein of OpuA, is abundant to increase cooperativity, allowing substrate sensitivity.
Although there are additional TGD2s in the functional TGD complex, this does not necessarily mean there is any change in the generally accepted ABC transporter mechanism. On the contrary, because TGD2 subunits are anchored firmly to the complex, it is tempting to hypothesize that conformational changes originating from ATP binding and hydrolysis by TGD3 are transmitted through the entire complex and confer further changes in the substrate binding capability of TGD2 (Fig. 6B). Increased amounts and firm association of TGD2 with the complex could be consistent with overcoming the large energy needed to remove phosphatidic acid from a bilayer. If true, this would imply that other lipid ABC transporters may also have abundant SBD-containing proteins. Analogous bacterial lipid transport systems (MlaD-F and LptB, -C, -F, and -G) are each implicated in removing lipids from a membrane and would also need to overcome a large energy barrier (50, 51). Similar to the TGD system and disparate from other Gram-negative SBD-containing proteins, the SBD-containing proteins of the Mla and Lpt systems are membrane-anchored by an α-helical transmembrane domain. To our knowledge, the abundance of system components has not yet been verified for either complex.
Finally, it is noteworthy that chloroplastic TIM family proteins and KEAs each form large complexes in the chloroplast envelopes. Little is known of the proteins' function in the chloroplast, and the complexes described here may be useful tools for understanding their function in the future.
Acknowledgments
We thank Doug Whitten of the Michigan State University Proteomics Facility for assistance with mass spectrometry and excellent discussion, Masato Nakai and John Froehlich for antiserum contributions, and Zhen Wang for discussion and critical comments.
This work was supported by National Science Foundation Grant MCB0741395 and the US Department of Energy, Basic Energy Sciences, DE-FG02–98ER20305 (to C. B.).
- ABC
- ATP binding cassette
- DDM
- dodecylmaltoside
- HDN-PAGE
- histidine deoxycholate native PAGE
- KEA
- potassium efflux antiporter
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- TGD
- TRIGALACTOSYLDIACYLGLYCEROL protein
- TGDG
- trigalactosyldiacylglycerol lipid
- TIC and TOC
- translocon at the inner and outer envelope membrane of chloroplasts, respectively
- TIM
- translocase of the inner mitochondrial membrane
- ER
- endoplasmic reticulum
- ACN
- acetonitrile
- contig
- group of overlapping clones
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Albers S. V., Koning S. M., Konings W. N., Driessen A. J. (2004) Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 36, 5–15 [DOI] [PubMed] [Google Scholar]
- 2. Higgins C. F. (1992) ABC transporters. From microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 [DOI] [PubMed] [Google Scholar]
- 3. Saier M. H., Jr. (2000) A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64, 354–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hollenstein K., Dawson R. J., Locher K. P. (2007) Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 [DOI] [PubMed] [Google Scholar]
- 5. van der Heide T., Poolman B. (2002) ABC transporters. One, two, or four extracytoplasmic substrate-binding sites? EMBO Rep. 3, 938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee S. J., Böhm A., Krug M., Boos W. (2007) The ABC of binding protein-dependent transport in Archaea. Trends Microbiol. 15, 389–397 [DOI] [PubMed] [Google Scholar]
- 7. Schuurman-Wolters G. K., Poolman B. (2005) Substrate specificity and ionic regulation of GlnPQ from Lactococcus lactis. An ATP-binding cassette transporter with four extracytoplasmic substrate-binding domains. J. Biol. Chem. 280, 23785–23790 [DOI] [PubMed] [Google Scholar]
- 8. Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71–91 [DOI] [PubMed] [Google Scholar]
- 9. Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., Klein M., Kolukisaoglu U., Lee Y., Martinoia E., Murphy A., Rea P. A., Samuels L., Schulz B., Spalding E. J., Yazaki K., Theodoulou F. L. (2008) Plant ABC proteins. A unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159 [DOI] [PubMed] [Google Scholar]
- 10. Roughan P. G., Holland R., Slack C. R. (1980) The role of chloroplasts and microsomal fractions in polar-lipid synthesis from [1-14C]acetate by cell-free preparations from spinach (Spinacia oleracea) leaves. Biochem. J. 188, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jarvis P., Dörmann P., Peto C. A., Lutes J., Benning C., Chory J. (2000) Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. U.S.A. 97, 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunst L., Browse J., Somerville C. (1988) Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 85, 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heinz E., Roughan P. G. (1983) Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 72, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Awai K., Xu C., Tamot B., Benning C. (2006) A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. U.S.A. 103, 10817–10822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu C., Fan J., Riekhof W., Froehlich J. E., Benning C. (2003) A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 22, 2370–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu B., Xu C., Awai K., Jones A. D., Benning C. (2007) A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J. Biol. Chem. 282, 35945–35953 [DOI] [PubMed] [Google Scholar]
- 17. Lu B., Benning C. (2009) A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284, 17420–17427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roston R., Gao J., Xu C., Benning C. (2011) Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex. Plant J. 66, 759–769 [DOI] [PubMed] [Google Scholar]
- 19. Xu C., Fan J., Froehlich J. E., Awai K., Benning C. (2005) Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17, 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson D. T., Froehlich J. E., Keegstra K. (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273, 16583–16588 [DOI] [PubMed] [Google Scholar]
- 21. Bruce B. D., Perry S., Froehlich J., Keegstra K. (1994) In vitro import of protein into chloroplasts. in Plant Molecular Biology Manual (Gelvin S. B., Schilperoort R. A., eds) pp. 1–15, Kluwer Academic Publishers, Boston [Google Scholar]
- 22. Keegstra K., Yousif A. E. (1986) Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 118, 316–325 [Google Scholar]
- 23. Cline K., Andrews J., Mersey B., Newcomb E. H., Keegstra K. (1981) Separation and characterization of inner and outer envelope membranes of pea chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 78, 3595–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 25. McElver J., Tzafrir I., Aux G., Rogers R., Ashby C., Smith K., Thomas C., Schetter A., Zhou Q., Cushman M. A., Tossberg J., Nickle T., Levin J. Z., Law M., Meinke D., Patton D. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clough S. J., Bent A. F. (1998) Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 27. Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7, 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z., Benning C. (2011) Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp. 49, e2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kikuchi S., Hirohashi T., Nakai M. (2006) Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol. 47, 363–371 [DOI] [PubMed] [Google Scholar]
- 30. Ladig R., Sommer M. S., Hahn A., Leisegang M. S., Papasotiriou D. G., Ibrahim M., Elkehal R., Karas M., Zickermann V., Gutensohn M., Brandt U., Klösgen R. B., Schleiff E. (2011) A high-definition native polyacrylamide gel electrophoresis system for the analysis of membrane complexes. Plant J. 67, 181–194 [DOI] [PubMed] [Google Scholar]
- 31. Akita M., Nielsen E., Keegstra K. (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cooper H. M., Paterson Y. (2009) Production of polyclonal antisera. Curr. Protoc. Neurosci. Chapter 5, Unit 5.5 [DOI] [PubMed] [Google Scholar]
- 33. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 34. Bräutigam A., Shrestha R. P., Whitten D., Wilkerson C. G., Carr K. M., Froehlich J. E., Weber A. P. (2008) Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species. Comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J. Biotechnol. 136, 44–53 [DOI] [PubMed] [Google Scholar]
- 35. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 36. Eubel H., Braun H. P., Millar A. H. (2005) Blue-native PAGE in plants. A tool in analysis of protein-protein interactions. Plant Methods 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uteng M., Hauge H. H., Markwick P. R., Fimland G., Mantzilas D., Nissen-Meyer J., Muhle-Goll C. (2003) Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and a sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 42, 11417–11426 [DOI] [PubMed] [Google Scholar]
- 38. Buck M. (1998) Trifluoroethanol and colleagues. Cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31, 297–355 [DOI] [PubMed] [Google Scholar]
- 39. Zou Q., Habermann-Rottinghaus S. M., Murphy K. P. (1998) Urea effects on protein stability. Hydrogen bonding and the hydrophobic effect. Proteins 31, 107–115 [PubMed] [Google Scholar]
- 40. Kikuchi S., Oishi M., Hirabayashi Y., Lee D. W., Hwang I., Nakai M. (2009) A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 21, 1781–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cline K., Werner-Washburne M., Andrews J., Keegstra K. (1984) Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75, 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Franssen S. U., Shrestha R. P., Bräutigam A., Bornberg-Bauer E., Weber A. P. (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peltier J. B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A. J., Rutschow H., van Wijk K. J. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5, 114–133 [DOI] [PubMed] [Google Scholar]
- 44. Krimm I., Goyer A., Issakidis-Bourguet E., Miginiac-Maslow M., Lancelin J. M. (1999) Direct NMR observation of the thioredoxin-mediated reduction of the chloroplast NADP-malate dehydrogenase provides a structural basis for the relief of autoinhibition. J. Biol. Chem. 274, 34539–34542 [DOI] [PubMed] [Google Scholar]
- 45. Murcha M. W., Elhafez D., Lister R., Tonti-Filippini J., Baumgartner M., Philippar K., Carrie C., Mokranjac D., Soll J., Whelan J. (2007) Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 143, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu W., Smith J. W., Huang C. M. (2010) Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U.S.A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Biemans-Oldehinkel E., Poolman B. (2003) On the role of the two extracytoplasmic substrate-binding domains in the ABC transporter OpuA. EMBO J. 22, 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siebertz H. P., Heinz E., Linscheid M., Joyard J., Douce R. (1979) Characterization of lipids from chloroplast envelopes. Eur. J. Biochem. 101, 429–438 [DOI] [PubMed] [Google Scholar]
- 50. Malinverni J. C., Silhavy T. J. (2009) An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chng S. S., Ruiz N., Chimalakonda G., Silhavy T. J., Kahne D. (2010) Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U.S.A. 107, 5363–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]