Abstract
To characterize protein phosphorylation in developing seed, a large-scale, mass spectrometry-based phosphoproteomic study was performed on whole seeds at five sequential stages of development in soybean (Glycine max), rapeseed (Brassica napus), and Arabidopsis (Arabidopsis thaliana). Phosphopeptides were enriched from 0.5 mg of total peptides using a combined strategy of immobilized metal affinity and metal oxide affinity chromatography. Enriched phosphopeptides were analyzed by Orbitrap tandem mass spectrometry and mass spectra mined against cognate genome or cDNA databases in both forward and randomized orientations, the latter to calculate false discovery rate. We identified a total of 2,001 phosphopeptides containing 1,026 unambiguous phosphorylation sites from 956 proteins, with an average false discovery rate of 0.78% for the entire study. The entire data set was uploaded into the Plant Protein Phosphorylation Database (www.p3db.org), including all meta-data and annotated spectra. The Plant Protein Phosphorylation Database is a portal for all plant phosphorylation data and allows for homology-based querying of experimentally determined phosphosites. Comparisons with other large-scale phosphoproteomic studies determined that 652 of the phosphoproteins are novel to this study. The unique proteins fall into several Gene Ontology categories, some of which are overrepresented in our study as well as other large-scale phosphoproteomic studies, including metabolic process and RNA binding; other categories are only overrepresented in our study, like embryonic development. This investigation shows the importance of analyzing multiple plants and plant organs to comprehensively map the complete plant phosphoproteome.
During seed development, plants accumulate starch, oil, and proteins at differing amounts, among the diversity of plant species (Weber et al., 2005). These nutrients serve a crucial role in human and animal diets as well as in feedstocks for the chemical industry (Thelen and Ohlrogge, 2002). A better understanding of the biology and biochemistry of seed development can provide opportunities for the rational engineering of seed content. In this regard, the recent use of transcriptomics and proteomics approaches has begun to provide global views of the compendium of regulatory, structural, and enzymatic proteins involved in carbon assimilation and reserve deposition in the diversity of model and agronomic oilseeds (Ruuska et al., 2004; Hajduch et al., 2005; Katavic et al., 2006; Agrawal et al., 2008; Houston et al., 2009). Although the metabolic pathways involved in seed maturation have been largely predicted using both “omics” (Hajduch et al., 2010) and in vivo labeling approaches (Schwender et al., 2003, 2004; for review, see Baud and Lepiniec, 2009), the regulation of the activities in these pathways is only beginning to be discovered.
A major form of metabolic control in higher eukaryotes occurs at the posttranslational level and most notably includes reversible protein phosphorylation (Huber, 2007). An increasing number of metabolic enzymes and enzyme complexes have been shown to be regulated by reversible phosphorylation; however, given the labile nature of protein phosphorylation and the detection difficulty, it is likely that there are as yet undiscovered enzymes regulated in this manner. Using modern approaches, recent global phosphoproteomic studies in developing Arabidopsis (Arabidopsis thaliana) and Brassica seed have revealed over 200 putative phosphoproteins (Agrawal and Thelen, 2006; Irar et al., 2006), including many enzymes. More targeted studies have identified several protein kinases involved in seed development. A receptor-like kinase, RPK1, has been shown to affect abscisic acid (ABA) sensitivity in plants. ABA is a key hormonal regulator of seed development, having roles in maturation and dormancy (Finch-Savage and Leubner-Metzger, 2006; Gutierrez et al., 2007). Another kinase, Suc nonfermenting kinase-related protein kinase 2.6, has been shown to affect ABA signaling and influence seed oil content (Zheng et al., 2010). The fatty acid composition of Arabidopsis seed is also influenced by a casein kinase I through the phosphorylation of transcription factors for the microsomal oleic acid desaturase (fad2) gene (Kim et al., 2010). The receptor-like kinases, somatic embryogenesis receptor-like kinases, and Crinkly4 have also been determined to be involved in seed development, although their substrate clients have yet to be elucidated (Wang et al., 2007).
The coupling of phosphopeptide enrichment strategies to high-resolution mass spectrometry has led to a renaissance in phosphorylation site mapping, and several recent large-scale studies have expanded the knowledge of phosphorylation in plants (Sugiyama et al., 2008; Reiland et al., 2009; Grimsrud et al., 2010; Nakagami et al., 2010). These studies discovered phosphorylation sites for hundreds of phosphorylated proteins in roots, shoots, and cell culture and suggested roles for phosphorylation in chloroplasts and root nodulation (Reiland et al., 2009; Grimsrud et al., 2010). Among other findings, the study performed by Nakagami et al. (2010) emphasized the importance of analyzing multiple plant species to enhance coverage of the phosphoproteome. This is likely due to the near singular use of trypsin as a protease in “bottom-up” phosphoproteomic investigations and the inherent bias associated with its cleavage specificity. Collectively, these large-scale studies represent a data set of over 10,000 redundant phosphorylation sites within Arabidopsis, barrel medic (Medicago truncatula), and rice (Oryza sativa).
In addition to the importance of a broader taxonomic representation for global phosphoproteomic studies, it is equally important that a broader spatiotemporal representation of plant organs becomes accessible. One example is developing seed. To better characterize the extent of protein phosphorylation during seed maturation, we performed a large-scale phosphorylation study on developing seeds from three species: Arabidopsis, rapeseed (Brassica napus), and soybean (Glycine max). A total of 956 proteins were identified containing 1,026 unambiguous phosphorylation sites involved in seed development. Of these proteins, 652 are novel to this study, showing the importance of analyzing the phosphoproteome of various plants and tissues to further expand knowledge of the plant phosphoproteome.
RESULTS
Phosphopeptide Identification from Developing Seeds
Phosphopeptide enrichment using both Fe-immobilized metal affinity chromatography (IMAC) and Ti-hydroxy acid-modified metal oxide chromatography in series was performed on three species, Arabidopsis, rapeseed, and soybean, at five stages of seed development in biological triplicate. The enriched phosphopeptides were separated by reverse-phase liquid chromatography and analyzed on an LTQ-Orbitrap mass spectrometer. The resulting RAW files were searched in parallel using Mascot, SEQUEST, and InsPecT against organism-specific databases concatenated to a randomized version of the forward database as a decoy. Filtering thresholds were compared for each search algorithm and verified to be below 1% false discovery rate (FDR; Elias and Gygi, 2007). Unambiguous phosphorylation sites were determined using PhosCalc (MacLean et al., 2008). A total of 2,414 phosphopeptides were identified spanning all three species, representing 956 proteins and 1,026 unambiguous phosphosites (Table I).
Table I. Number of phosphoproteins, phosphopeptides, and phosphorylation sites identified within developing seed of soybean, rapeseed, and Arabidopsis.
| Species | Proteins | Peptides | Sites | Mono | Di | Tri+ |
| Soybean | 459 | 1,007 | 1,195 | 737 (73.2%) | 215 (21.4%) | 55 (5.5%) |
| Rapeseed | 325 | 586 | 818 | 447 (76.3%) | 61 (10.4%) | 78 (13.3%) |
| Arabidopsis | 172 | 408 | 401 | 357 (87.5%) | 50 (12.5%) | 1 (0.2%) |
The distribution of unambiguous Ser, Thr, and Tyr phosphorylation sites from Arabidopsis seed is comparable to that found by Sugiyama et al. (2008; Table II). Both Thr and Tyr phosphorylation events in rapeseed and soybean are more prevalent than in Arabidopsis. There were a total of 92 unambiguous Tyr phosphorylation sites identified in the three species (Supplemental Table S1). Twenty-five of these phosphopeptides do not contain Ser or Thr residues within the peptide. Of the remaining 67 sites, 12 were assigned to mitogen-activated protein kinase (MAPK) proteins that are known to be Tyr phosphorylated, providing biological support for correct site assignment (Huang et al., 2000; Nühseet al., 2000).
Table II. Classification of phosphorylation sites according to matched ions assigned by PhosCalc version 1.2.
Phosphorylation sites that had at least one more matched ion than any other potential site were classified as unambiguous. Ambiguous class I sites are indeterminable and have more than one with equal numbers of matched ions. The remaining phosphorylation sites that had fewer matched ions than at least one other site were classified as ambiguous class II.
| Species | Class | Sites | Ser | Thr | Tyr |
| Soybean | Unambiguous | 534 | 408 (76.4%) | 79 (14.8%) | 47 (8.8%) |
| Ambiguous class I | 281 | 183 (65.1%) | 68 (24.2%) | 30 (10.7%) | |
| Ambiguous class II | 380 | 234 (61.6%) | 90 (23.7%) | 56 (14.7%) | |
| Rapeseed | Unambiguous | 320 | 234 (73.1%) | 49 (15.3%) | 37 (11.6%) |
| Ambiguous class I | 186 | 103 (55.4%) | 62 (33.3%) | 21 (11.3%) | |
| Ambiguous class II | 312 | 147 (47.1%) | 107 (34.3%) | 58 (18.6%) | |
| Arabidopsis | Unambiguous | 172 | 145 (84.3%) | 19 (11.0%) | 8 (4.7%) |
| Ambiguous class I | 78 | 46 (59.0%) | 20 (25.6%) | 12 (15.4%) | |
| Ambiguous class II | 151 | 83 (55.0%) | 49 (32.5%) | 19 (12.6%) |
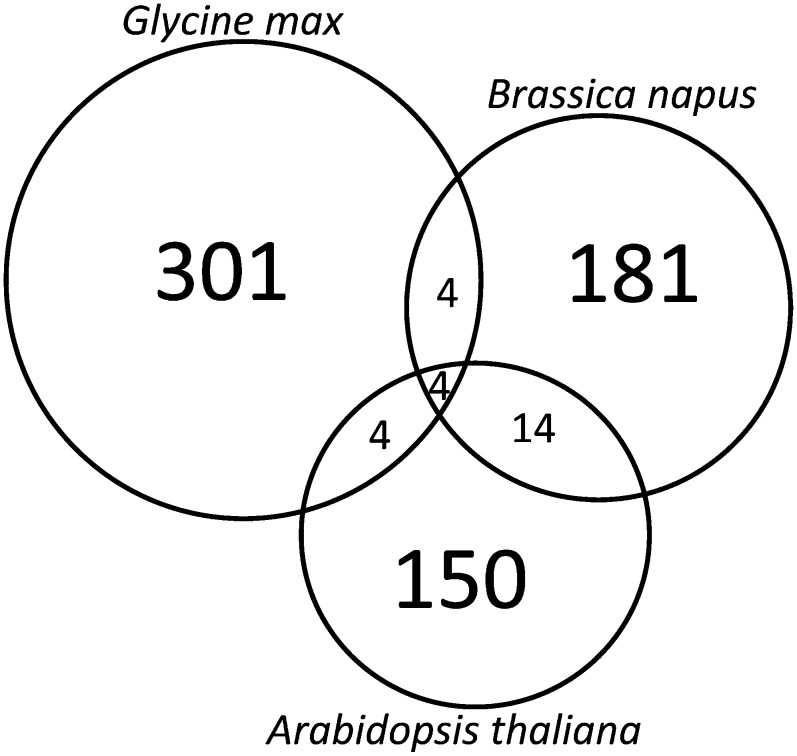
Conserved phosphopeptides among Arabidopsis, rapeseed, and soybean were identified by clustering the identified peptides using CD-HIT (Li and Godzik, 2006). Default parameters were used, including a 60% similarity cutoff and a bandwidth of 5. A total of 658 clusters were produced from the 2,001 identified peptides. Four clusters contained peptides from all three species, another four clusters contained soybean and Arabidopsis peptides, four clusters were also identified that contained soybean and rapeseed peptides, whereas Arabidopsis and rapeseed had the most represented clusters at 14 (Fig. 1). The peptides identified in each cluster along with the associated protein accessions and the annotation as determined by Blast2GO can be found in Supplemental Table S2.
Figure 1.
Conserved seed phosphopeptide clusters among seeds of soybean, rapeseed, and Arabidopsis as determined using CD-HIT. Identified phosphopeptides were clustered according to a 60% sequence identity cutoff and a bandwidth of 5. All other settings were default. The peptides within each cluster are shown in Supplemental Table S2.
Comparison of Identified Phosphoproteins from Developing Seed
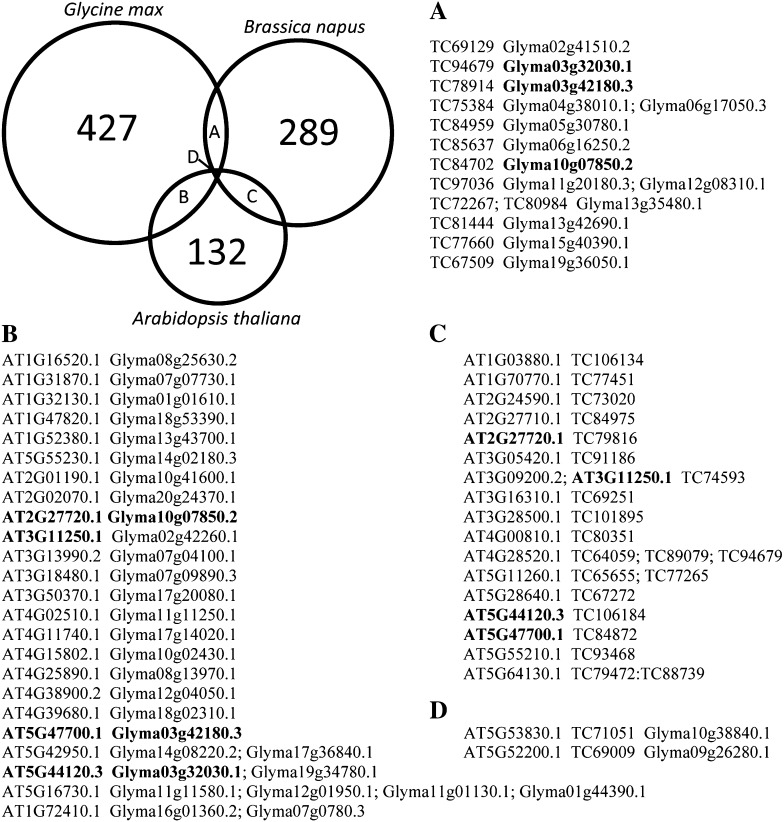
Orthologous phosphorylated proteins were identified among the three organisms analyzed using a reciprocal BLAST search (Altenhoff and Dessimoz, 2009). In some cases, paralogs matched to the same protein with the same E-value, so they could not be distinguished from one another. When one best match could not be determined based upon E-value, all proteins at that E-value were listed (Fig. 2; Supplemental Table S2). A developing seed nonredundant protein list consisting of 902 proteins was generated. This list contains all proteins with no identified orthologs, along with one representative protein for those where orthologs were identified.
Figure 2.
Identification of orthologous phosphorylated proteins during seed development. Pairwise reciprocal BLAST searches were performed between the three species analyzed. Proteins containing the highest E-value were considered orthologous. In some instances, multiple proteins were identified with the highest E-value, so all matches were used. Orthologous proteins identified within each pairwise comparison are listed. Proteins with different orthologs among the three species are shown in boldface.
The majority of the phosphoproteins identified were unique to each organism: 93.0%, 88.9%, and 76.6% for soybean, rapeseed, and Arabidopsis, respectively. There were only two protein groups that had phosphorylated orthologs among all three species. One of these proteins (AT5G52200.1 in Arabidopsis) is a putative peptidyl-prolyl cis-trans-isomerase, whereas the other protein (AT5G43830.1 in Arabidopsis) has an unknown function. The orthologous proteins between Arabidopsis and rapeseed contain a total of 48 unambiguous sites, 25 from Arabidopsis and 23 from rapeseed. Fifteen of the sites within the orthologous proteins overlapped between the two species, leaving 10 sites from Arabidopsis and eight sites in rapeseed as nonoverlapping. An additional eight nonambiguous sites in rapeseed overlapped with their orthologs in soybean out of a possible 14 sites in rapeseed and 23 in soybean. Arabidopsis and soybean have the most identified phosphorylation sites within the orthologous proteins, 33 in Arabidopsis and 42 in soybean. Only seven of the phosphorylation sites were conserved in both species.
Congruence with Other Large-Scale Phosphoproteomic Studies
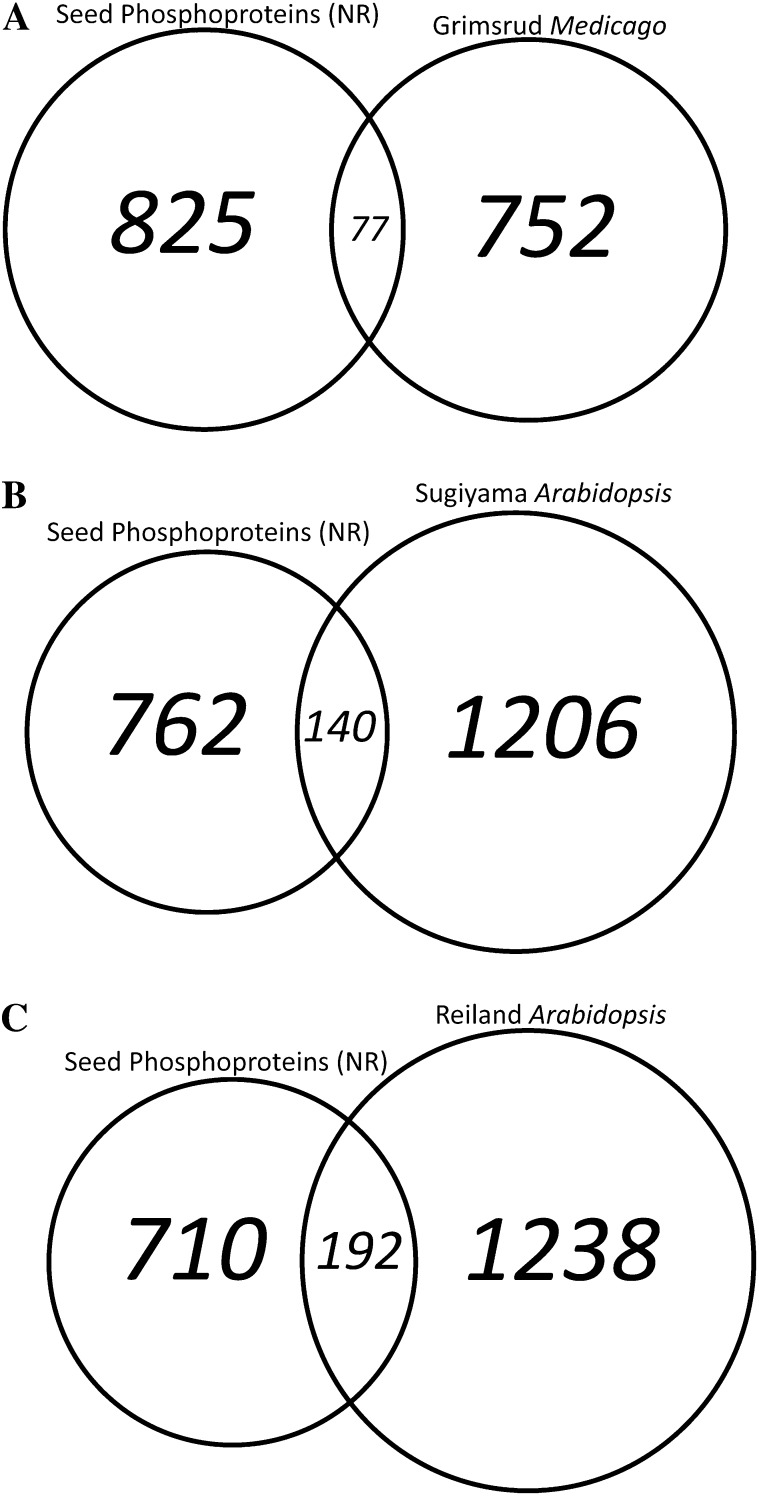
Our data were compared with other large-scale phosphoproteome studies to determine the uniqueness of the developing seed phosphoproteome. A reciprocal BLAST search was performed using the 902 nonredundant developing seed phosphoproteins against the proteins identified in M. truncatula roots (Grimsrud et al., 2010), Arabidopsis shoots (Reiland et al., 2009), and Arabidopsis cell culture (Sugiyama et al., 2008; Fig. 3). The nonredundant seed phosphoproteins had the least amount of overlap with M. truncatula root phosphoproteins (Grimsrud et al., 2010), including 77 orthologs representing 8.5% of the identified phosphoproteins in developing seeds. The Arabidopsis seedling phosphoprotein data set (Reiland et al., 2009) contained the highest percentage of overlap with our developing seed phosphoproteomic screen, including 192 orthologous proteins, 21.3% of the seed data set. Arabidopsis cell culture phosphoproteins (Sugiyama et al., 2008) had 140 orthologs (15.5% of nonredundant seed phosphoproteins) compared with developing Arabidopsis seed. The majority of the proteins in our developing seed data set, 72.3% (652 phosphoproteins), did not have any phosphorylated ortholog in the other studies.
Figure 3.
Comparison of phosphoproteins identified in developing seeds with prior phosphoproteomic studies. The 902 nonredundant phosphoproteins identified in developing seeds were compared with three other large-scale phosphoproteomic studies. Orthologous proteins were identified using a reciprocal BLAST search. This approach identified 652 phosphoproteins unique to this study.
Functional Comparison with Other Large-Scale Phosphoproteomic Studies
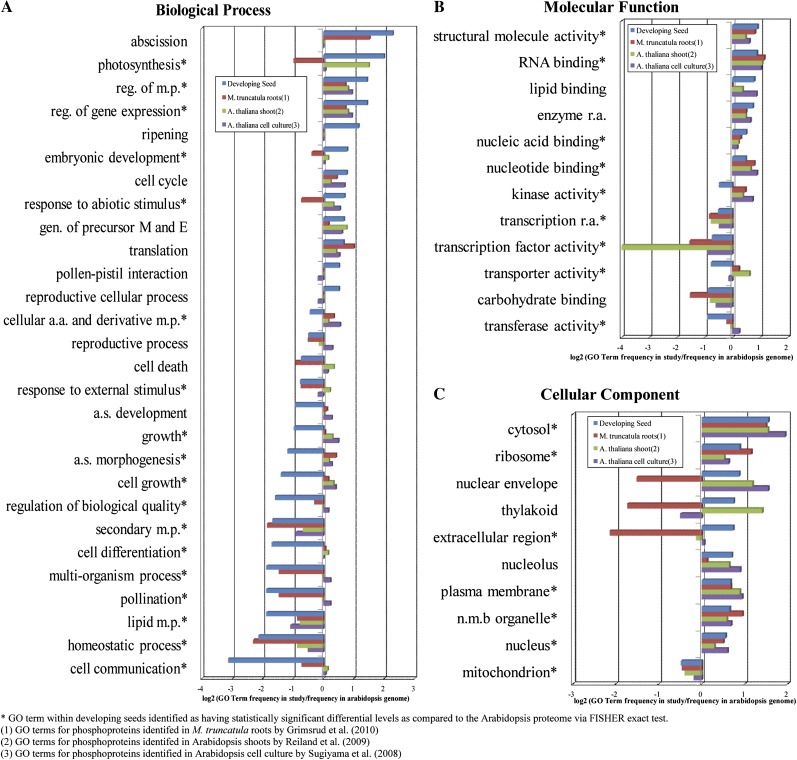
For a functional context of the seed phosphoproteomic results, functional classification using Gene Ontology (GO) terms of the nonredundant seed proteins was performed using Blast2GO (Götzet al., 2008). Overrepresented or underrepresented GO terms within our data set were determined by first obtaining GO terms for the Arabidopsis proteome (The Arabidopsis Information Resource [TAIR] 9) using Blast2GO. The frequency of each GO term in our data set was compared with the frequency in the Arabidopsis proteome, and the log2 ratio was graphed (Fig. 4; Supplemental Fig. S2). The abundance of the nonredundant seed GO terms was statistically compared with the TAIR9 results using Fisher’s exact test (asterisks indicate significant differences; Fig. 4). A majority of the GO terms with a 0.5-fold or greater change in developing seeds were also found to be statistically significantly overrepresented or underrepresented according to Fisher’s exact test. The proteins for those GO terms with a 0.5-fold or greater increase in our data set are listed in Supplemental Table S3.
Figure 4.
Comparison of identified phosphoproteins found in this study with three previous studies in plants. Frequency of GO terms for phosphoproteins identified from this study and three other large-scale studies as well as the entire Arabidopsis proteome were obtained using Blast2GO. The distribution of GO terms for each study was normalized to the frequencies in the Arabidopsis proteome and plotted on a log2 scale (Supplemental Fig. S2). A selection of the GO terms with the largest fold changes are plotted: biological process (A), molecular function (B), and cellular component (C). a.s., Anatomical structure; E, energy; M, metabolites; m.p., metabolic process; n.m.b., nonmembrane-bounded organelle; r.a., regulator activity. [See online article for color version of this figure.]
GO terms were also acquired for three other large-scale phosphoproteomic studies using Blast2GO for comparison (Sugiyama et al., 2008; Reiland et al., 2009; Grimsrud et al., 2010). The log2 ratios of the GO term frequency in the three large-scale studies as compared with the frequency within the Arabidopsis proteome are graphed in Figure 4 and Supplemental Figure S2.
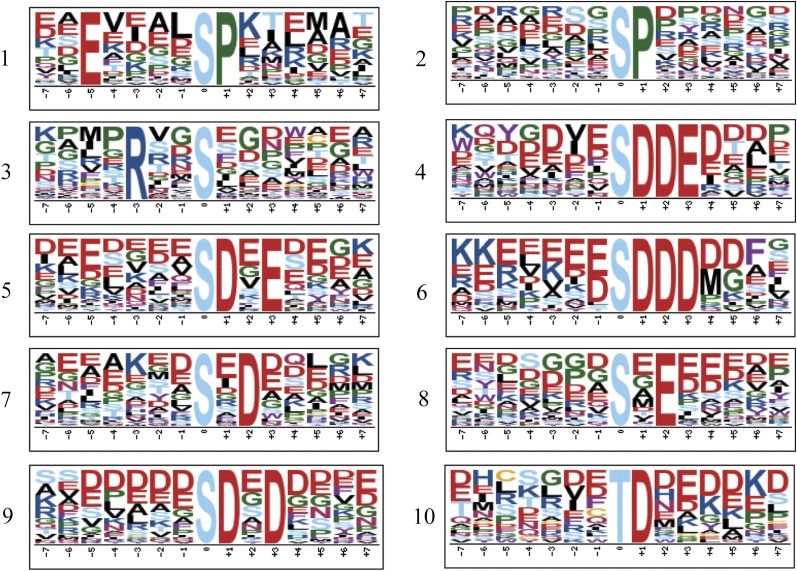
Phosphorylation Motifs in Seed Development
Consensus sequences flanking the sites of phosphorylation in developing seeds were identified using Motif-X version 1.2 (Schwartz and Gygi, 2005). Analysis was only performed on unambiguous sites with a minimum occurrence of 20 and a significance level of 10−6, identifying a total of 10 motifs (Fig. 5). Two Pro-targeted motifs (1 and 2), one calmodulin-dependent protein kinase II domain (3), and seven casein kinase II domains (4–10) were identified. In total, these motifs represent 36.8% of all unambiguous sites identified.
Figure 5.
Sequence motif analysis of developing seed phosphorylation sites. Motif-X analysis was performed with a 15-amino acid window, a minimum occurrence of 20, a significance of 10−6, and the Arabidopsis proteome as background to the unambiguous phosphorylation sites in developing seeds. This identified 10 motifs, of which 1 and 10 are unique to this study (Supplemental Table S4). [See online article for color version of this figure.]
To determine if any of these motifs are unique to developing seeds, parallel analyses were performed on unambiguous phosphorylation sites from the previous phosphoproteomic studies on Arabidopsis seedlings (Reiland et al., 2009) and Arabidopsis cell cultures (Sugiyama et al., 2008). The data were also compared with the motifs identified in M. truncatula roots (Grimsrud et al., 2010). One of the Pro-targeted motifs (motif 1; Fig. 5) and the Thr motif (motif 10; Fig. 5) are unique to this study.
The Plant Protein Phosphorylation Database
The identified phosphoproteins/peptides/sites and annotated spectra were deposited into the Plant Protein Phosphorylation Database (P3DB; http://www.p3db.org/), a Web resource we developed for hosting protein phosphorylation data in plants (Gao et al., 2009; Fig. 6). Phosphorylation data from previous large-scale plant phosphoproteomic studies were also collected and integrated into P3DB. Unlike other phosphorylation databases, this resource includes all plant phosphorylation data, associated meta-data, links to other major databases, and, when provided, individual annotated spectra in graphics. Currently, P3DB version 1.6 hosts phosphoproteomics data from 15 experimental studies, containing more than 10,000 phosphoproteins harboring more than 30,000 phosphorylation sites in five plant species: Arabidopsis, soybean, rapeseed, M. truncatula, and rice. With the user-friendly PHP-based Web interface, a user can easily access the data, which were stored in a MySQL relational database. P3DB supports seamless navigation among phosphoproteins, peptides, sites, and spectra. The search and BLAST utilities make it easy for users to find phosphoproteins and peptides of interest. Figure 6 illustrates the phosphopeptide BLAST utility in P3DB. All phosphorylation data in P3DB can also be downloaded in tab-delimited text files.
Figure 6.
Screenshot of the P3DB showing the phosphopeptide BLAST search function. As an example, the protein transcription factor BIM1 in rice was queried against all identified phosphopeptides from this study. A, Submission form for entering a query sequence and selecting organism, reference, and E-value threshold. B, BLAST result (partial). Phosphopeptide hits from P3DB (in angle brackets) were aligned with the query sequence and rendered in different colors according to the E-values as indicated in the color scheme legend. Phosphopeptides and phosphosites were hyperlinked to corresponding pages containing detailed information. Each Ser, Thr, or Tyr in the query sequence was rendered in blue if it aligned to any phosphosite. [See online article for color version of this figure.]
DISCUSSION
The compendium of experimentally determined plant phosphorylation sites has rapidly grown in the last few years, due to methodological advancements coupling phosphopeptide enrichment methods with high-resolution mass spectrometry. Prior investigations have focused on vegetative organs and tissues, primarily from Arabidopsis. Here, we extend prior studies by analyzing the phosphoproteome during five stages of seed maturation in soybean, rapeseed, and Arabidopsis, identifying a total of 956 phosphoproteins consisting of 1,026 unambiguous phosphorylation sites on 2,001 phosphopeptides (Table I).
Tyr Phosphorylation May Be More Prevalent in Crop Oilseeds
The extent of Tyr phosphorylation in plants has been recently debated with the publication of several mass spectrometry-based studies (Sugiyama et al., 2008; de la Fuente van Bentem and Hirt, 2009). In our study, a total of 92 unambiguous Tyr sites were identified. Of those 92 Tyr sites, 26 contained a Ser or Thr within two amino acids, allowing for potential misassignment. In an unlikely worst-case scenario, if each of these sites were incorrectly assigned, it would result in a Tyr phosphorylation occurrence at 7.1%, 7.2%, and 3.5% for soybean, rapeseed, and Arabidopsis, respectively. For Arabidopsis, this value is within the range of reported Tyr phosphorylation. However, the frequency of Tyr phosphorylation in soybean and rapeseed, even after removing potentially misassigned sites, is 2-fold higher than detected in Arabidopsis. This suggests that Tyr phosphorylation may be more prevalent in polyploid crops such as soy and canola and emphasizes a need to extend phosphoproteomic studies beyond nonmodel organisms.
Fewer Phosphoproteins Were Observed at Later Stages of Seed Maturation
Soybean seeds were collected from early maturation (27–42 mg fresh weight) to the end of mid maturation (green and more than 300 mg fresh weight), after which the seeds enter late maturation, begin to desiccate, and turn yellow. A majority of the phosphopeptides identified from soybean were within the first two stages of development, with only 37 (9.9%) unambiguous phosphopeptides being unique to the last three stages of development. The lower number of identified phosphopeptides at later stages of development may be due to an increasing abundance of storage proteins causing a decrease in the proportion of phosphoproteins. Of the few phosphoproteins identified during late seed development, three were putative transcription factors (Glyma08g15350.1, Glyma13g00200.1, and Glyma14g10830.1). Glyma08g15350.1 is homologous to ABR1 in Arabidopsis, which is involved in ABA signaling and affects seed germination (Pandey et al., 2005). The other two putative transcription factors are homologous to Arabidopsis GATA22 but contain different phosphorylation sites. GATA22 is a light-regulated transcription factor and part of a larger family of GATA-binding transcription factors (Manfield et al., 2007), which are involved in seed germination and sugar metabolism (Bi et al., 2005; Liu et al., 2005).
Arabidopsis staging was analogous to soybean, with seeds harvested from early maturation (5 d after flowering) to mid maturation (13 d after flowering; Baud and Lepiniec, 2008; Jones et al., 2010; Hajduch et al., 2011). Phosphopeptides identified from Arabidopsis had a comparable profile to soybean phosphopeptides, with a majority of the phosphopeptides identified early in development. Many of these phosphoproteins are involved in transcriptional regulation, although their precise function has yet to be determined. One phosphoprotein identified is FPA (AT2G43410.1), which is involved in transcription termination and 3′ processing (Sonmez et al., 2011). Another interesting phosphoprotein identified early in seed development, MINI ZINC FINGER2 (AT3G28917.1), may play a role in integrating signals from multiple hormones (Hu and Ma, 2006).
Unlike soybean and Arabidopsis, rapeseed staging represented all stages of seed development, including late maturation (beginning around 5 weeks after flowering), at which point photosynthesis stops and desiccation occurs. A majority (74.5%) of the rapeseed unambiguous phosphopeptides were identified within late maturation. A large fraction (26.0%) of the late-maturation-unique phosphopeptides mapped back to proteins annotated as late-embryogenesis-abundant proteins. There is little known about late-embryogenesis-abundant proteins beyond that they accumulate late in seed development and play a role in dehydration tolerance (Hundertmark and Hincha, 2008). In addition to the late-embryogenesis-abundant proteins, there were several proteins (4.2%) identified with roles in desiccation. During late maturation, proteins involved in cell wall structure are also phosphorylated to a higher degree, with 9.4% of late maturation phosphopeptides involved in secretion or cell wall polysaccharide synthesis. This suggests that phosphorylation may play a regulation role in the seed transition to a quiescent state.
Comparison of Protein Phosphorylation among Three Oilseeds
To identify conserved phosphorylation events in developing oilseeds, the identified phosphopeptides were clustered using CD-HIT with a 60% similarity cutoff (Fig. 1). There were four clusters containing peptides from Arabidopsis, soybean, and rapeseed; one of these clusters contains the Arabidopsis protein AN3 (AT5G28640.1), a transcriptional coactivator involved in cell proliferation, which may play an important role in seed structure and morphology. Another peptide cluster consisting of peptides from Arabidopsis and rapeseed is within the acyl-CoA (AT3G05420.1) binding protein 4, involved in fatty acid transport. Another interesting cluster contains Arabidopsis and soybean Tyr phosphopeptides that mapped back to MAPKs: the Arabidopsis MPK10 protein, seven soybean proteins with homology to MPK4 in Arabidopsis, two soybean proteins homologous to MPK6, and two additional soybean proteins homologous to MPK13. This may represent an expanded role for MAPKs in soybeans. The few proteins of known function found in these clusters indicate conservation in seed development, metabolism, and signaling.
Although several phosphorylation events of interest were identified by cluster analysis, the overlap between the three species was low. This may be due to a couple of reasons. One is that phosphorylation sites tend to occur in regions of disorder (Iakoucheva et al., 2004; Gao et al., 2010), which could mean the amino acids flanking the phosphorylation site are less constrained and may have a lower degree of conservation. Another possible reason for the low degree of overlap could be due to the phosphorylation data set being only a small sampling of all phosphorylation sites in developing seeds.
There was also a small degree of overlap among the three species determined by the phosphoprotein reciprocal BLAST, with 93.0%, 88.9%, and 76.6% of the soybean, rapeseed, and Arabidopsis phosphoproteins, respectively, having no phosphorylated ortholog in our study. This suggests that the current phosphoproteomics data set represents only a fraction of the phosphorylation events in developing seeds. To increase coverage of the developing seed phosphoproteome, it may be necessary to increase the quantity of starting material, perform further prefractionation, and potentially sample different environmental conditions (i.e. light and dark conditions).
The overlapping phosphoproteins among the three species are represented by 55 orthologous protein groups containing 111 phosphorylation sites. Of these sites, 82.0% were conserved, with 51.6% of conserved sites being identified as phosphorylated in multiple species. This indicates that although the data set is not comprehensive, there is a strong correlation among the phosphorylation sites within the orthologous proteins. Further analysis of the orthologous proteins determined that only nine of them were not identified as phosphorylated in other large-scale studies. Two of the novel orthologs are involved in transcription (AT5G28640.1/TC67272 and AT4G38900.2/Glyma12g04050.1) and may represent proteins important for regulating expression in seed development. Another two proteins have unknown function and will require further analysis to elucidate their role in seed development. The remaining novel orthologs are seed storage proteins and have been identified in other smaller scale phosphoproteomic studies (Agrawal and Thelen, 2006). An additional 11 orthologous proteins were identified as phosphorylated in other studies that contained phosphorylation sites unique to developing seeds. This may indicate altered regulation of these proteins in developing seeds as compared with other large-scale studies. Two of these proteins are involved with transcription, one is a protein phosphatase inhibitor, whereas the remaining eight have unknown functions.
Comprehensive Coverage of the Experimental Plant Phosphoproteome Requires Deeper Spatiotemporal and Phytological Analysis
Our analysis determined that less than 30% of identified phosphoproteins had phosphorylated orthologs in the M. truncatula and other Arabidopsis phosphoproteomic studies (Fig. 3). This is significantly lower than the over 50% overlap found between the phosphoproteins identified in rice and Arabidopsis (Nakagami et al., 2010). This discrepancy can be largely attributed to the method used for identifying orthologs. Nakagami et al. (2010) used OrthoMCL to cluster the proteins into closely related groups. This method identifies more orthologs but does not distinguish orthologs from paralogs and has a higher false-positive rate (Altenhoff and Dessimoz, 2009). The reciprocal BLAST method used in this study has a lower false-positive rate but is slightly less sensitive, producing fewer, but more accurate, orthologs.
The small degree of overlap between our study and other large-scale phosphoproteomic studies performed on different tissues might suggest that regulation by phosphorylation varies greatly among plant organs. This means that in order to obtain a better understanding of phosphorylation in plants, we need to begin sampling not only different organisms but also expand on the organs and tissues being analyzed.
Comparison of this data set and other large-scale phosphoproteomic studies from different organs at the protein function level, through the use of GO terminology, extrapolates on the variation among organs. As noted in other studies (Reiland et al., 2009; Nakagami et al., 2010), several GO categories were identified as overrepresented in the phosphoproteome, including regulation of metabolic process (GO:0019222), structural molecule activity (GO:0005198), RNA binding (GO:0003723), ribosome (GO:0005840), and plasma membrane (GO:0005886) as shown in Fig. 4. These categories represent overall targets for phosphorylation within plants, although the proteins represented within these categories can vary. There are also GO categories that are only overrepresented in this study, such as ripening (GO:0009835), embryonic development (GO:0009790), reproductive cellular process (GO:0048610), and extracellular region (GO:0005576). However, collectively, these GO categories represent only 17% of the 652 proteins unique to this study. Moreover, like most proteomic methodologies, experimental phosphoproteomics is not comprehensive in scope. This is due to a broad dynamic range in protein expression and the dearth of proteolytic peptides for some extreme proteins (e.g. low mass, membrane proteins). As regulatory proteins (e.g. protein kinases, transcription factors) tend to be low in abundance, it is likely that GO “preference” for phosphorylation will need to be continually revised as the experimental plant phosphoproteome matures.
Phosphorylation Motifs Unique to Seed and Seed Storage Proteins
There were two unique phosphorylation motifs identified in this study, one of which is the Pro-directed motif E-X-X-X-X-S-P (Fig. 5). Nucleic acid binding represents approximately 50% of the proteins containing the E-X-X-X-X-S-P motif. This motif might represent a recognition site for protein kinases involved in regulating transcription for seed development. The other unique motif identified in this study, X-T-D-X, is a nonspecific acidophilic motif and is similar to the Ser motif, X-S-D-X, identified in M. truncatula. About 25% of the proteins containing this motif in our study are seed storage proteins. Nucleic acid binding and methylation are also highly represented among the proteins with the X-T-D-X motif, at 17% and 14%, respectively. The large representation of seed storage proteins indicates that the X-T-D-X motif might be a unique motif for developing seed.
Our analysis identified a largely unique phosphorylation data set from developing seeds of soybean, rapeseed, and Arabidopsis. Functional comparison of our data with other large-scale phosphoproteomic studies identified the differences among these studies and emphasized the need to expand both the organisms and organs/tissues for global phosphoproteomic analysis.
MATERIALS AND METHODS
Materials
Sequencing-grade modified trypsin was from Promega. POROS 20 chromatographic material was purchased from Applied Biosystems. HPLC-grade acetonitrile and methanol were from Fisher Scientific. EDTA-free protease inhibitor cocktail was from Roche (catalog no. 11836170001). Unless otherwise stated, all other reagents used in this study were of analytical grade, including FeCl3 and TiO2.
Plant Materials and Growth Conditions
Rapeseed (Brassica napus var. Reston) seeds were sown in soil (Promix) in a greenhouse (light/dark cycles of 16 h [23°C]/8 h [20°C], 48% humidity, and supplemental light intensity of 112.8 µmol m−2 s−1) in Columbia, Missouri. Plants were fertilized (15:30:15, nitrogen:phosphorus:potassium [Miracle-Gro; product no. 1001232]) at 2-week intervals. Flowers were tagged immediately after opening of buds, and developing pods were collected at 2, 3, 4, 5, and 6 weeks after flowering. Seeds were excised from collected pods, frozen immediately with liquid nitrogen, and stored at −80°C until use. Three biological sample pools were collected at 2, 3, 4, 5, and 6 weeks after flowering.
Arabidopsis (Arabidopsis thaliana Columbia-0 ecotype) seeds were sown in soil (Promix) in a growth chamber (light/dark cycles of 14 h [23°C]/10 h [20°C], 50% humidity, and light intensity of 112.8 µmol m−2 s−1). Plants were fertilized (15:30:15, nitrogen:phosphorus:potassium) after first flower emergence. Flowers were tagged at petal emergence prior to opening and harvested at 5, 7, 9, 11, and 13 d after flowering. Seeds were excised from siliques, frozen with liquid nitrogen, and stored at −80°C until use. Three biological sample pools (each containing more than 100 seeds) were collected at 5, 7, 9, 11, and 13 d after flowering.
Soybean (Glycine max var. Jack) seeds were imbibed at 4°C for 2 d and then transferred to room temperature for 3 d. Germinated seeds were planted in soil (SOGEMIX) in a greenhouse (light/dark cycles of 14 h [28°C]/10 h [21°C]). Plants were fertilized (15:30:15, nitrogen:phosphorus:potassium) at 1-week intervals. Seeds were extracted from developing pods, weighed for staging, and frozen with liquid nitrogen. Five seed stages were used: stage 2 (27–42 mg per seed), stage 3 (70–95 mg per seed), stage 4 (115–150 mg per seed), stage 5 (200–250 mg per seed), and stage 6 (green and more than 300 mg per seed).
Isolation of Total Protein and In-Solution Digestion
Total seed protein was isolated according to a modified phenol-based procedure as described previously (Agrawal and Thelen, 2006). Briefly, 5 mm sodium vanadate, 5 mm sodium fluoride, and 25 mm glycerophosphate disodium salt pentahydrate were added to the phenol extraction buffer containing an EDTA-free protease inhibitor cocktail to prevent dephosphorylation by phosphoprotein phosphatases. Protein pellets were resuspended in 7 m urea, 50 mm Tris-HCl, pH 8.0, followed by centrifugation at 20,800g for 10 min to remove insoluble material. Supernatants were kept and protein concentration was assayed in quadruplicate using a Coomassie Protein Assay (Bio-Rad). Total proteins (500 µg) were subjected to in-solution digestion with trypsin (1:50, w/v), and after 20 h, digestions were stopped by adding formic acid to a final concentration of 5%. Samples were then lyophilized.
Phosphopeptide Enrichment
Lyophilized peptides were suspended in 0.1% trifluoroacetic acid and 50% acetonitrile and subjected to phosphopeptide enrichment using Fe(III)-IMAC columns of POROS 20 mass spectrometry resin packed in a 200-µL tip containing a C8 frit (Ishihama et al., 2006; Ndassa et al., 2006; Thingholm et al., 2008). Fe(III)-IMAC-enriched phosphopeptides were lyophilized and stored at −80°C until further use. The flow through and washes were combined and lyophilized for subsequent aliphatic hydroxy acid-modified metal oxide enrichment using lactic acid and TiO2 (Sugiyama et al., 2007). The eluted phosphopeptides were lyophilized and stored at −80°C until further use.
Mass Spectrometry
All samples were reconstituted in 15 µL of 0.1% formic acid in water. Peptides (10 µL) were analyzed on an LTQ Orbitrap (Thermo-Fisher) using collision-induced dissociation. Peptides were loaded onto peptide traps (C18, 5 × 1 mm; Michrom) for concentrating and desalting prior to analytical separation and ionization with a fused-silica capillary emitter (10.5 cm, 360 µm o.d., 150 µm i.d.) packed with BioBasic MAGIC C18 (300 Å, 5 µm) resin. Ion spraying was 1.4 kV with a 300 nL min−1 flow rate using an acetonitrile gradient (0%–35% solvent B for 120 min; solvent A = 0.1% formic acid in mass spectrometry-grade water, solvent B = 0.1% formic acid in acetonitrile). Nine data-dependent tandem mass spectrometry scans of the nine most intense parent ions were acquired in positive acquisition mode after each full scan (mass-to-charge ratio [m/z] 200–2,000). A minimal signal intensity of 1,000 along with monoisotopic precursor selection and rejection of ions with an unrecognized charge state and a charge state of 1 was used. Dynamic exclusion was enabled with a repeat count of 1, duration of 30 s, exclusion list size of 300, and exclusion duration of 45 s.
Peptide and Protein Identification
Acquired tandem mass spectrometry spectra were searched against organism-specific databases concatenated to a randomized form of the same database as a decoy (Elias and Gygi, 2007): TAIR9 for Arabidopsis (www.arabidopsis.com), Glyma1_highConfidence for soybean (http://www.phytozome.net), and BNGI.release_3-1 for rapeseed (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=oilseed_rape). Database randomizing and concatenation were performed using Decoy DB Creator (www.p3db.org). Three search algorithms were used for peptide identification: Mascot, SEQUEST, and InsPecT (Tanner et al., 2005). Peak lists were generated for Mascot and SEQUEST searches by the extract_msn.exe program (Thermo) with the following parameters: m/z of 200 to 2,000; threshold of 500; precursor ion tolerance of 10 ppm; and minimum ion count of 35. Prior to InsPecT searches, the raw files were converted to mzXML format using ReAdW. All other parameters were identical to those used for Mascot and SEQUEST searches. Two different modification searches were performed with each search engine. The first search contained a static modification of +57 D (carboxyamidomethylation) on Cys and variable modifications of +16 D (oxidation) on Met and +80 D on Ser, Thr, and Tyr residues. The second search allowed only variable +80-D modifications on Ser, Thr, and Tyr residues.
Bioinformatics Analysis
The in-house-developed PhosSite program was used to autoprocess the output result files for extraction, filtering (based upon FDR), and integration of phosphorylation results. All files were filtered first by ppm followed by ion score versus charge state for the Mascot files. Charge versus score and ppm filters were used to filter the InsPecT files. To determine the ideal filter values to use and obtain an FDR below 1%, the score frequencies at a given charge state for the target and decoy databases were plotted independently (Supplemental Fig. S1). The filter value was then set at the point where the frequency of target hits is higher than that of the decoy and resulted in an overall FDR below 1%. After filtering, all files had an FDR below 1%, estimated based on target-decoy strategy (Elias and Gygi, 2007). The identified phosphopeptides were deposited in P3DB (www.p3db.org; Gao et al., 2009).
Phosphorylation site localization was performed using PhosCalc version 1.2 for all identified phosphopeptides (MacLean et al., 2008). The default values of PhosCalc were used, matching the four most intense ions per 100 m/z with a tolerance of 0.4 to a list of potential ions including dehydro ions for all possible phosphorylation sites. The presence of additional matched ions to a phosphorylation site compared with all other candidate sites in the peptide is due to the presence of site-determining ions in the spectra and was labeled as unambiguous. If multiple phosphorylation sites contained an equal number of matched ions, these sites were classified as ambiguous class I. The remaining identified sites that had fewer matched ions than at least one alternative site were labeled as ambiguous class II. This procedure is based upon the method established by Nakagami et al. (2010).
CD-HIT and Reciprocal BLAST Search
Phosphopeptides among Arabidopsis, rapeseed, and soybean were clustered using the CD-HIT Web server (Li and Godzik, 2006; http://www.bioinformatics.org/cd-hit/). All phosphopeptides were merged into one Fasta file and uploaded into CD-HIT. Default parameters were used, including a 60% similarity cutoff and a bandwidth of 5.
Reciprocal BLAST search (Tatusov et al., 2000) was performed by comparing the identified phosphoproteins in one species against the entire proteome of the remaining two species separately using default parameters. The matched proteins with the lowest E-value were retained and compared with the matches of the BLAST queries performed in the opposite direction. For example, the soybean phosphoproteins were searched against the Arabidopsis proteome and compared with the BLAST results of the Arabidopsis phosphoproteins searched against the soybean proteome. If a pair of phosphoproteins had the lowest E-values when mined in both directions, they were determined to be tentative orthologs.
GO Classification
Blast2GO was used to obtain annotations and GO terms for all identified seed phosphoproteins using the default parameters, including ANNEX and plant GOslim augmentation (Götz et al., 2008). The distribution of the GO terms was statistically compared with that of the TAIR9 proteome (obtained with the same method as the seed phosphoproteins) using the built-in Fisher’s exact test. Blast2GO was also used to obtain GO terms for three other large-scale phosphoproteomic studies: Medicago truncatula roots (Grimsrud et al., 2010), Arabidopsis seedlings (Reiland et al., 2009), and Arabidopsis cell culture (Sugiyama et al., 2008).
Motif Analysis
Motif-X was used to identify the phosphorylation motifs from our data set (Schwartz and Gygi, 2005). The seven amino acids upstream and downstream of the unambiguous phosphorylation sites were extracted from the protein sequences using in-house software. Sequences were entered into Motif-X and executed using a 15-amino acid window, a minimum occurrence of 20, a significance of 10−6, and the Arabidopsis proteome as background.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Example of charge state 2 phosphopeptide filtering by Xcorr.
Supplemental Figure S2. Comparison of GO-classified proteins from four large-scale phosphoproteomic studies.
Supplemental Table S1. Identified phosphopeptides from developing seed.
Supplemental Table S2. Homologous phosphopeptides clustered using CD-HIT.
Supplemental Table S3. Orthologous seed phosphoproteins identified by reciprocal BLAST searches.
Supplemental Table S4. Phosphorylation sites assigned to the 10 motifs identified by sequence logo analysis.
Supplementary Material
Glossary
- ABA
abscisic acid
- FDR
false discovery rate
- MAPK
mitogen-activated protein kinase
- GO
Gene Ontology
- TAIR
The Arabidopsis Information Resource
- P3DB
Plant Protein Phosphorylation Database
- m/z
mass-to-charge ratio
References
- Agrawal GK, Hajduch M, Graham K, Thelen JJ. (2008) In-depth investigation of the soybean seed-filling proteome and comparison with a parallel study of rapeseed. Plant Physiol 148: 504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Thelen JJ. (2006) Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol Cell Proteomics 5: 2044–2059 [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Dessimoz C. (2009) Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol 5: e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S. (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Hirt H. (2009) Protein tyrosine phosphorylation in plants: more abundant than expected? Trends Plant Sci 14: 71–76 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Gao J, Agrawal GK, Thelen JJ, Xu D. (2009) P3DB: a plant protein phosphorylation database. Nucleic Acids Res 37: D960–D962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Thelen JJ, Dunker AK, Xu D. (2010) Musite, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol Cell Proteomics 9: 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, Ané JM, Coon JJ. (2010) Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol 152: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ. (2005) A systematic proteomic study of seed filling in soybean: establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M, Hearne LB, Miernyk JA, Casteel JE, Joshi T, Agrawal GK, Song Z, Zhou M, Xu D, Thelen JJ. (2010) Systems analysis of seed filling in Arabidopsis: using general linear modeling to assess concordance of transcript and protein expression. Plant Physiol 152: 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M, Matusova R, Houston NL, Thelen JJ. (2011) Comparative proteomics of seed maturation in oilseeds reveals differences in intermediary metabolism. Proteomics 11: 1619–1629 [DOI] [PubMed] [Google Scholar]
- Houston NL, Hajduch M, Thelen JJ. (2009) Quantitative proteomics of seed filling in castor: comparison with soybean and rapeseed reveals differences between photosynthetic and nonphotosynthetic seed metabolism. Plant Physiol 151: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Ma H. (2006) Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J 45: 399–422 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Gupta R, Morris PC, Luan S, Kieber JJ. (2000) ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol 122: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC. (2007) Exploring the role of protein phosphorylation in plants: from signaling to metabolism. Biochem Soc Trans 35: 23–32 [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irar S, Oliveira E, Pagès M, Goday A. (2006) Towards the identification of late-embryogenic-abundant phosphoproteome in Arabidopsis by 2-DE and MS. Proteomics (Suppl 1) 6: S175–S185 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M. (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J Proteome Res 5: 988–994 [DOI] [PubMed] [Google Scholar]
- Jones SI, Gonzalez DO, Vodkin LO. (2010) Flux of transcript patterns during soybean seed development. BMC Genomics 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ. (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Go YS, Lee SB, Kim YS, Shin JS, Min MK, Hwang I, Suh MC. (2010) Seed-expressed casein kinase I acts as a positive regulator of the SeFAD2 promoter via phosphorylation of the SebHLH transcription factor. Plant Mol Biol 73: 425–437 [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Liu PP, Koizuka N, Martin RC, Nonogaki H. (2005) The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J 44: 960–971 [DOI] [PubMed] [Google Scholar]
- MacLean D, Burrell MA, Studholme DJ, Jones AM. (2008) PhosCalc: a tool for evaluating the sites of peptide phosphorylation from mass spectrometer data. BMC Res Notes 1: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Devlin PF, Jen CH, Westhead DR, Gilmartin PM. (2007) Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol 143: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndassa YM, Orsi C, Marto JA, Chen S, Ross MM. (2006) Improved immobilized metal affinity chromatography for large-scale phosphoproteomics applications. J Proteome Res 5: 2789–2799 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Peck SC, Hirt H, Boller T. (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23: 1391–1398 [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y. (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278: 29442–29453 [DOI] [PubMed] [Google Scholar]
- Sonmez C, Bäurle I, Magusin A, Dreos R, Laubinger S, Weigel D, Dean C. (2011) RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc Natl Acad Sci USA 108: 8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics 6: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4: 12–21 [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics 7: 661–671 [DOI] [PubMed] [Google Scholar]
- Wang H, Guo J, Lambert KN, Lin Y. (2007) Developmental control of Arabidopsis seed oil biosynthesis. Planta 226: 773–783 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 253–279 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Xu X, Crosley RA, Greenwalt SA, Sun Y, Blakeslee B, Wang L, Ni W, Sopko MS, Yao C, et al. (2010) The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol 153: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.