Abstract
Epithelial cells (ECs) lining the secretory cavities of Citrus peel have been hypothesized to be responsible for the synthesis of essential oil, but direct evidence for such a role is currently sparse. We used laser-capture microdissection and pressure catapulting to isolate ECs and parenchyma cells (as controls not synthesizing oil) from the peel of young grapefruit (Citrus × paradisi ‘Duncan’), isolated RNA, and evaluated transcript patterns based on oligonucleotide microarrays. A Gene Ontology analysis of these data sets indicated an enrichment of genes involved in the biosynthesis of volatile terpenoids and nonvolatile phenylpropanoids in ECs (when compared with parenchyma cells), thus indicating a significant metabolic specialization in this cell type. The gene expression patterns in ECs were consistent with the accumulation of the major essential oil constituents (monoterpenes, prenylated coumarins, and polymethoxylated flavonoids). Morphometric analyses demonstrated that secretory cavities are formed early during fruit development, whereas the expansion of cavities, and thus oil accumulation, correlates with later stages of fruit expansion. Our studies have laid the methodological and experimental groundwork for a vastly improved knowledge of the as yet poorly understood processes controlling essential oil biosynthesis in Citrus peel.
Members of the genus Citrus (Rutaceae) produce some of the commercially most important tree fruit crops, which are grown in over 100 countries worldwide, most prominently in Brazil, the Mediterranean basin, the United States, and China. The two major markets in the Citrus sector are fresh fruit for direct consumption and fruit juice. Roughly 50% of the fruit weight consists of pulp, seeds, and peel, which are further processed into value-added by-products such as molasses, pectins, fiber, seed oils, fermentation products, and essential oils (Laufenberg et al., 2003). Citrus essential oils are obtained on an industrial scale by cold extraction and generally contain two classes of constituents: a volatile fraction consisting of monoterpenoids and small amounts of sesquiterpenoids (totaling 94%–98% of the oil) and a nonvolatile residue containing fatty acids, sterols, carotenoids, waxes, coumarins, and polymethoxylated flavonoids (2%–6% of the oil; Mondello et al., 2002). These oils are processed into various formulations for industrial cleaning applications and as sustainable alternatives to traditional solvents. The volatile fraction, which is gathered by steam or vacuum distillation, has important uses in the flavor, fragrance, aromatherapy, and agrochemical industries (Carson and Hammer, 2011).
Collaborative projects to generate genomic resources for Citrus, including EST libraries from various organs and cDNA microarrays (Forment et al., 2005; Reis et al., 2007), have yielded data sets that have been mined by several groups to discover genes with potential relevance to essential oil biosynthesis in the peel. For example, transcripts related to terpenoid synthases, which catalyze the first committed step in the biosynthesis of terpenoid essential oil constituents, were highly expressed in Citrus fruit peel when compared with other tissues and organs (Berger et al., 2007; Dornelas and Mazzafera, 2007; Takita et al., 2007). More recently, genome-wide oligonucleotide microarray analyses have been conducted with Citrus peel tissue (Maul et al., 2008; González-Candelas et al., 2010; Matas et al., 2010; Ballester et al., 2011; Hershkovitz et al., 2012). However, these studies were aimed at evaluating specific developmental processes or stress/defense responses of Citrus and did not provide direct insights into peel essential oil biosynthesis. The few functionally characterized genes involved in Citrus essential oil biosynthesis encode monoterpene and sesquiterpene synthases with activities for the synthesis of (+)-limonene, (−)-β-pinene, γ-terpinene, (E)-β-ocimene, 1,8-cineole, and (E)-β-farnesene (Maruyama et al., 2001; Lücker et al., 2002; Shimada et al., 2004, 2005).
In Citrus fruit, the pigmented region of the pericarp is called the flavedo and contains numerous oil glands consisting of secretory cavities that are lined by several layers of specialized epithelial cells (ECs; Fahn, 1979). Various authors have hypothesized that the ECs are responsible for essential oil biosynthesis (Esau, 1965; Schnepf, 1974; Turner et al., 1998; Lücker et al., 2002). It was also shown that plastid preparations obtained from the outer peel of Citrus could convert isopentenyl diphosphate into monoterpenes (Gleizes et al., 1983; Pauly et al., 1986). Because of the enrichment of this preparation in leucoplasts likely originating from ECs, the authors hypothesized that the ECs were the main location for essential oil biosynthesis. However, the only direct evidence for the site of essential oil biosynthesis comes from the in situ localization of transcripts for putative monoterpene synthases to ECs of rough lemon (Citrus jambhiri; Yamasaki and Akimitsu, 2007).
This study was initiated as a first step to comprehensively characterize essential oil biosynthesis and its regulation in Citrus, using peel from grapefruit (Citrus × paradisi ‘Duncan’) as an experimental model system. We employed an integrative approach to evaluate, throughout fruit development, the numbers, volume, and volume distribution of secretory cavities (microscopy and morphometrics), essential oil contents (gas chromatography [GC]-mass spectrometry [MS] and HPLC-MS), and global transcript profiles in isolated ECs (laser-capture microdissection and pressure catapulting followed by oligonucleotide microarray analysis). These data sets are invaluable resources for correlating relevant processes at the microscopic level (metabolic dynamics in ECs) with quantifiable outcomes at the macroscopic level (essential oil quantity and composition).
RESULTS
Correlation of the Distribution of Secretory Cavities and Essential Oil Quantities in the Peel of Developing Grapefruit
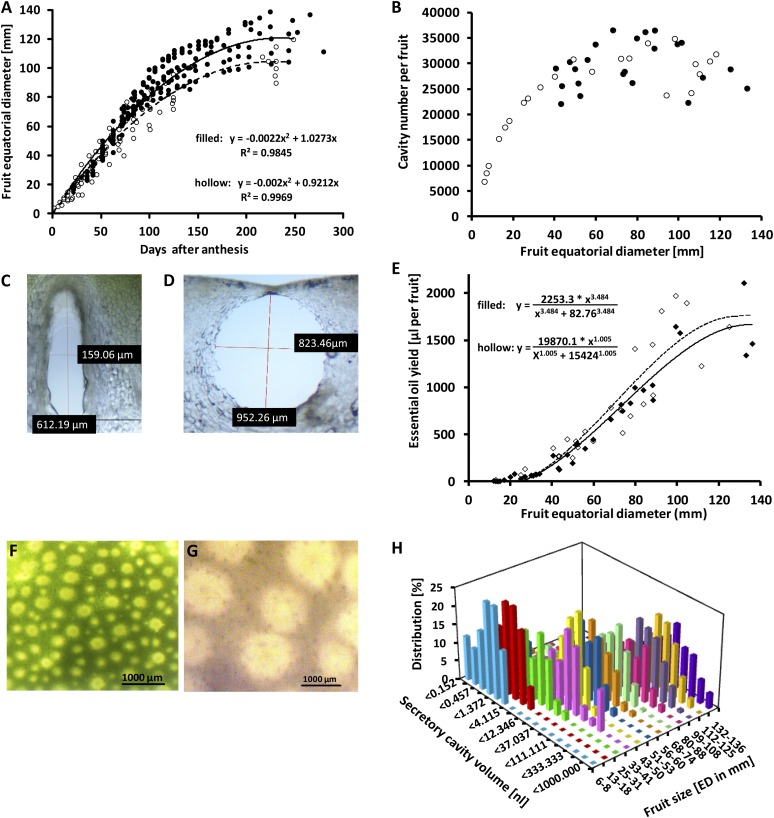
The grapefruit tree for this investigation was maintained under controlled greenhouse conditions. The fruit growth curve, monitored for two growth seasons (2009 and 2010) as equatorial diameter (ED), resembled the shape of a logistic function, with fairly rapid growth for the first 100 d post anthesis and a subsequently slower growth rate toward mature size (Fig. 1A). The average final size of fruit was about 15% larger in 2010 compared with 2009, but the growth trends were the same (Fig. 1A). Fruit from smaller clusters (one to three fruits per cluster) were generally of bigger size compared with those from larger clusters (four or more fruits). The transition from green to yellow peel color began between 80 and 110 d post anthesis and was completed within 90 d (170–200 d post anthesis). Secretory cavities were counted in analogy to Knight et al. (2001) from 6 to 133 mm ED (for details, see “Materials and Methods”). The cavity numbers increased rapidly from 6,840 (ED 6 mm) to 25,000 (ED 43 mm) and then much slower toward fruit maturity (Fig. 1B). A natural logarithmic function reflected these trends (r2 = 0.66). The variation in the number of cavities in mature fruit (29,040 ± 4,159 cavities per fruit [14.3%]) was larger than the variation in final fruit size (113 ± 10.1 mm ED [8.9%]).
Figure 1.
Developmental patterns of essential oil accumulation in grapefruit peel. A, Fruit growth in 2009 (white circles) and 2010 (black circles). B, Oil gland formation in 2009 (white circles) and 2010 (black circles). C, Prolate spheroid shape of secretory cavities in peel of young fruit (28 mm ED). D, Spheroid shape of secretory cavities in peel of mature fruit (100 mm ED). E, Oil yield as a function of fruit development (estimated based on cavity volume measurements as white diamonds and measured by chemoanalytical means as black diamonds). F, Peel surface in young fruit (28 mm ED). Mostly small and medium-sized cavities are visible. G, Peel surface in mature fruit (100 mm ED). Almost all cavities are fairly large. H, Volume distribution of cavities as a function of fruit development.
In addition to counting the total number of cavities per fruit, we also determined the volumes of cavities. To the best of our knowledge, such a survey has not been performed previously; thus, we will briefly outline our experimental strategy. Specimens were hand sectioned, rapidly placed under immersion oil to ensure optimal preservation of cavity shapes, and images were taken immediately thereafter. All immature cavities had prolate spheroid shapes, whereas mature cavities approached the shape of a regular sphere or were of an oblate spheroid shape (Fig. 1, C and D). Volumes were calculated for all cavities in a defined area of the fruit flavedo. The total cavity volume in this area was then used to extrapolate the total amount of oil per fruit (Fig. 1E). To evaluate the accuracy of these estimations experimentally, we determined the quantities of volatile essential oil components by hydrodistillation of finely ground peel tissue, followed by GC coupled with flame-ionization detection (FID; Supplemental Table S1). We also determined the contribution of nonvolatile constituents to the oil (which ranged between 2.5% and 3.4% [w/v] of the total oil), as described in the next paragraph. The oil quantities estimated based on secretory cavity volumes were very similar to the measured oil yields throughout fruit development, with final oil volumes of 1,738 ± 319 µL per fruit (estimated) and 1,586 ± 356 µL per fruit (measured) at maturity (Fig. 1E). The developmental changes of estimated oil quantities were fitted to a logistic function, with an initial lag phase (less than 70 µL per fruit at 24 mm ED), a subsequent more rapid increase in fruit of 40 to 100 mm ED, and then slower growth toward maturity (127–135 mm ED; Fig. 1E).
To evaluate why the rapid initial increase in the number of cavities (Fig. 1B) did not result in an equally fast increase in the accumulation of essential oil (Fig. 1E), we tested if the developmental distribution of cavities of different volumes might play a role in determining oil quantities (Fig. 1, F and G). Cavities were divided into different volume categories, and the volume distribution was determined throughout fruit development (Supplemental Protocol S1). In very young fruit (6–8 mm ED), cavity volumes ranged from 0.15 to 2.4 nL per fruit (distribution apex at 1.3 nL per fruit); in medium sized fruit (51–53 mm ED), the range was from 0.15 to 111 nL per fruit (distribution apex at 12.3 nL per fruit); and in mature fruit (99–136 mm), volumes from 0.15 to 1,000 nL per fruit (distribution apex at 64.2 nL per fruit) were obtained (Fig. 1H). These data indicate that the volume distribution of secretory cavities does indeed change, generally from smaller to larger cavities throughout fruit development. Oil yields estimated based on both cavity numbers and volume distribution are very similar to those measured experimentally (Fig. 1E).
Chemical Analysis of Grapefruit Peel Essential Oil Collected Directly with Microcapillaries
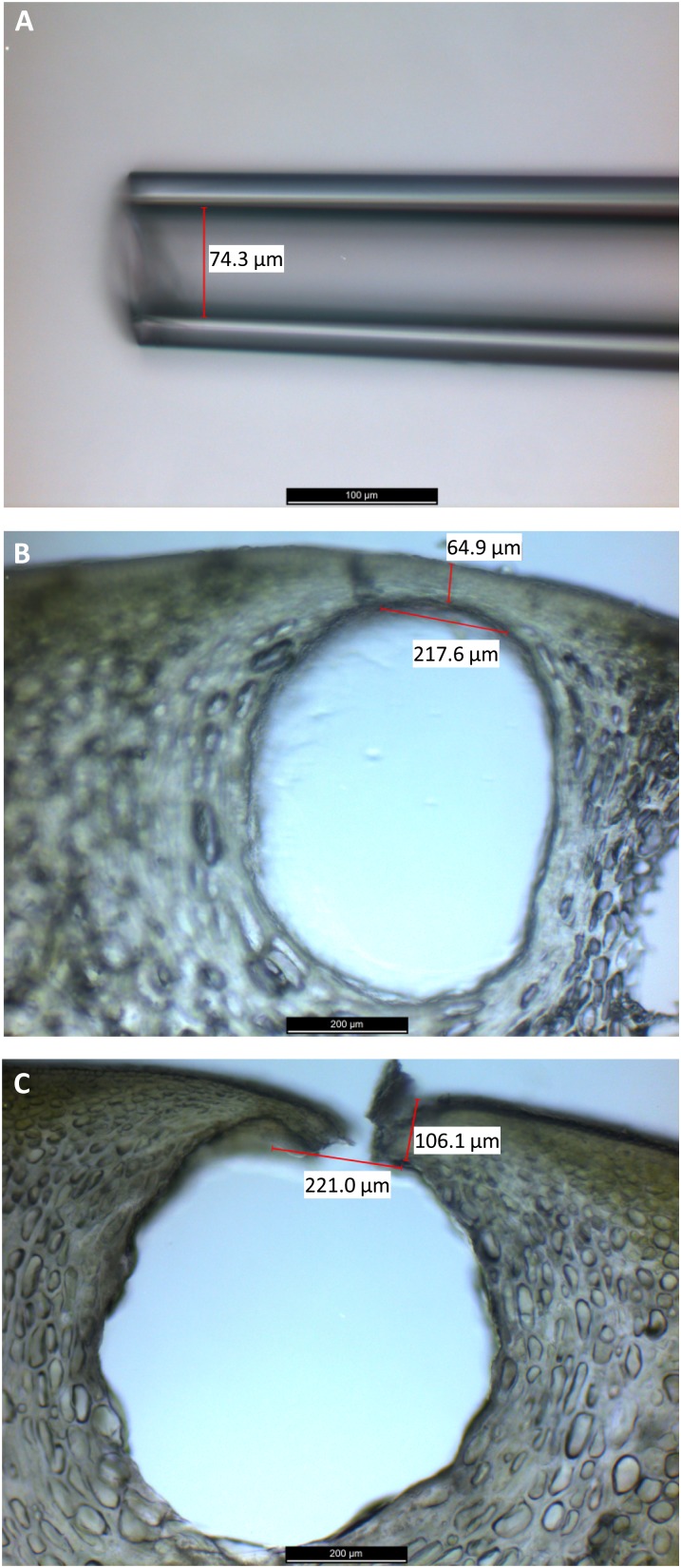
Studies on the nonvolatile constituents of Citrus essential oil began in the 1960s (for review, see Dugo et al., 2009), but reliable quantifications for grapefruit were not reported until much later (McHale and Sheridan, 1989; Frérot and Decorzant, 2004; Dugo et al., 2010). All of these studies used cold-pressed grapefruit oil for phytochemical and chemometric analyses. It is conceivable, however, that some components are not present in the essential oil itself but are instead dissolved from other peel tissues during the cold-pressing process. In the context of this work, which was aimed at characterizing the biosynthetic capabilities of ECs, it was critical to directly determine the oil composition in subepidermal cavities, which was achieved using microcapillaries (Fig. 2). We used this approach successfully before to analyze the contents of essential oil-accumulating glandular trichomes (Rios-Estepa et al., 2008). The main volatile terpenoids in the microcapillary-collected oil were (+)-limonene (93%–94%), myrcene (3%), and sabinene (0.7%–2.1%; Table I), which is in agreement with published data on the most abundant volatile constituents in cold-pressed oils (Kirbaslar et al., 2006; Espina et al., 2010). Sesquiterpenes occurred as minor components (Table I), which is also in accordance with previous reports (Flamini and Cioni, 2010). Oxygen-containing heterocyclic metabolites, consisting of coumarins, furanocoumarins, and polymethoxylated flavones, were present in the oil at 4.9% (w/v) before the peel started yellowing (70 mm ED) and then decreased slightly to 4.1% (w/v) in oil obtained from mature fruit (100 mm ED; Table I). This is consistent with prior studies (McHale and Sheridan, 1989; Frérot and Decorzant, 2004; Dugo et al., 2010). Carotenoids had been described as minor components of cold-pressed Citrus oils (Dugo et al., 2006) but were undetectable in our microcapillary-collected grapefruit oil samples (Table I). Sterols and fatty acids, which had been quantified in Citrus seed oils (El-Adawy et al., 1999), were present at very low levels in the grapefruit peel oil we collected with microcapillaries (0.38% or less and 0.003% or less, respectively; Table I).
Figure 2.
Collection of essential oil directly from secretory cavities of Citrus fruit peel using microcapillaries. A, Custom-made glass microcapillary (i.d. 75 ± 15 µm). B, Citrus peel section with secretory cavity. C, Citrus peel section after collection of secretory cavity contents with a microcapillary. Note that the cells surrounding the cavity are mostly undamaged.
Table I. Composition of grapefruit peel essential oil obtained directly from secretory cavities using microcapillaries.
| Constituents | Concentration |
|||||
|---|---|---|---|---|---|---|
| ED 70 mm | SD | ED 80 mm | SD | ED 100 mm | SD | |
| µg µL−1 oil | ||||||
| Monoterpenes | ||||||
| Limonene | 645.1 | 81.0 | 670.3 | 62.4 | 820.8 | 57.0 |
| Myrcene | 23.2 | 3.1 | 23.7 | 2.2 | 29.3 | 2.4 |
| Sabinene | 16.6 | 5.2 | 8.4 | 1.4 | 6.9 | 2.5 |
| γ-Terpinene | 2.7 | 0.7 | 2.0 | 0.6 | 7.1 | 1.8 |
| α-Terpinene | 1.6 | 0.4 | 1.2 | 0.5 | 5.9 | 2.0 |
| α-Pinene | 3.9 | 1.0 | 3.7 | 0.3 | 4.6 | 0.4 |
| β-Pinene | 1.3 | 0.4 | 0.6 | 0.2 | 0.6 | 0.1 |
| Terpinolene | 0.8 | 0.8 | 1.7 | 0.5 | 3.1 | 0.9 |
| Sum | 695.3 | 85.1 | 711.5 | 65.7 | 878.2 | 60.6 |
| Percentage (total) | 94.8 | 11.6 | 94.1 | 8.7 | 95.6 | 6.6 |
| Sesquiterpenes | ||||||
| β-Caryophyllene | 0.23 | 0.05 | 0.21 | 0.04 | 0.21 | 0.03 |
| β-Farnesene | 0.17 | 0.06 | 0.09 | 0.03 | 0.09 | 0.04 |
| Sum | 0.4 | 0.11 | 0.3 | 0.07 | 0.3 | 0.07 |
| Percentage (total) | 0.05 | 0.01 | 0.04 | 0.01 | 0.03 | 0.01 |
| Coumarins and furanocoumarins | ||||||
| Aurapten | 9.0 | 1.1 | 16.0 | 2.6 | 14.0 | 6.5 |
| Epoxyaurapten | 4.9 | 0.8 | 4.6 | 1.0 | 3.9 | 1.7 |
| Meranzin | 8.4 | 2.1 | 9.0 | 3.4 | 9.4 | 2.1 |
| Isomeranzin | 1.0 | 0.3 | 1.1 | 0.2 | 1.2 | 0.0 |
| Bergamottin | 0.6 | 0.2 | 1.3 | 0.1 | 1.3 | 0.8 |
| Epoxybergamottin | 9.9 | 3.2 | 8.5 | 0.2 | 7.0 | 2.9 |
| Osthol | 0.2 | 0.1 | 0.2 | 0.0 | 0.2 | 0.0 |
| Sum | 34.1 | 7.7 | 40.6 | 7.6 | 37.0 | 14.0 |
| Percentage (total) | 4.6 | 1.0 | 5.4 | 1.0 | 4.0 | 1.5 |
| Polymethoxylated flavones | ||||||
| Nobiletin | 1.1 | 0.3 | 0.6 | 0.1 | 0.5 | 0.2 |
| Tangeritin | 0.6 | 0.1 | 0.3 | 0.0 | 0.3 | 0.1 |
| Heptamethoxyflavone | 0.5 | 0.1 | 0.4 | 0.1 | 0.3 | 0.0 |
| Sum | 2.2 | 0.5 | 1.3 | 0.1 | 1.0 | 0.3 |
| Percentage (total) | 0.3 | 0.07 | 0.2 | 0.02 | 0.1 | 0.03 |
| Sterols | ||||||
| β-Sitosterol | 1.15 | 0.09 | 1.53 | 0.60 | 1.59 | 0.68 |
| Cycloartenol | 0.88 | 0.40 | 1.18 | 0.41 | 0.80 | 0.21 |
| Campesterol | 0.03 | 0.01 | 0.04 | 0.01 | 0.04 | 0.01 |
| Stigmasterol | 0.02 | 0.01 | 0.03 | 0.01 | 0.03 | 0.01 |
| Sum | 2.08 | 0.47 | 2.78 | 1.02 | 2.47 | 0.55 |
| Percentage (total) | 0.3 | 0.06 | 0.4 | 0.13 | 0.3 | 0.06 |
| Fatty acids | ||||||
| Palmitic acid (C16:0) | 0.003 | 0.001 | 0.005 | 0.000 | 0.006 | 0.001 |
| Stearic acid (C18:0) | 0.001 | 0.001 | 0.002 | 0.000 | 0.002 | 0.001 |
| Linoleic acid (C18:2) | 0.007 | 0.003 | 0.011 | 0.003 | 0.013 | 0.003 |
| α-Linolenic acid (C18:3) | 0.005 | 0.002 | 0.007 | 0.003 | 0.007 | 0.002 |
| Sum | 0.016 | 0.007 | 0.024 | 0.006 | 0.028 | 0.004 |
| Percentage (total) | 0.002 | 0.0009 | 0.003 | 0.0008 | 0.003 | 0.0004 |
Analysis of Global Transcript Expression Patterns in ECs Surrounding Secretory Cavities
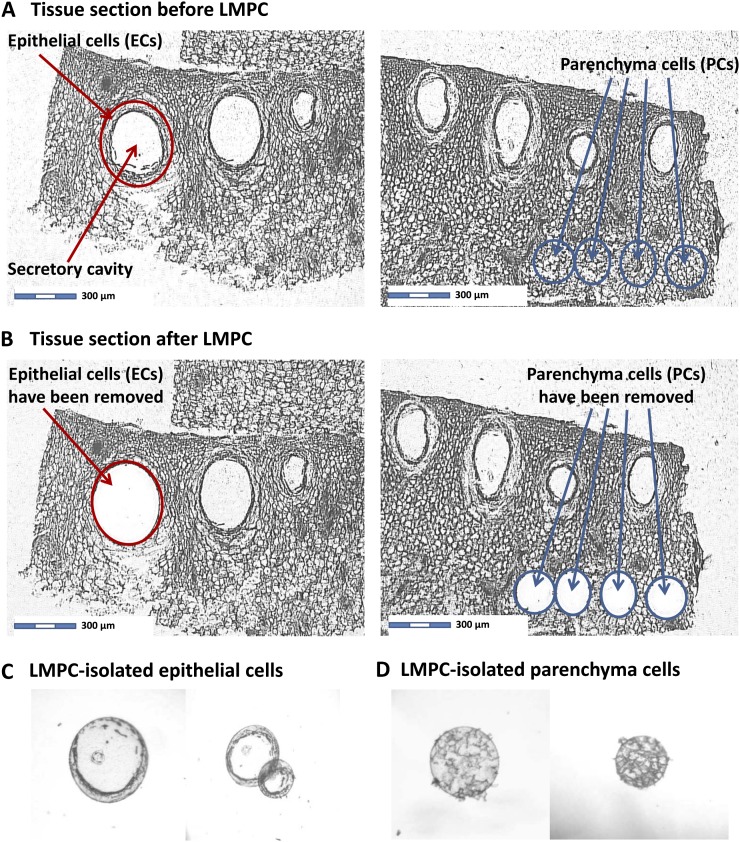
To test the hypothesis that ECs of peel glands are responsible for the biosynthesis of the oil accumulated in secretory cavities, it was crucial to study this cell type in isolation, without interference from neighboring cells. Previous transcriptome studies of Citrus fruit peel and peel sections were designed to investigate stress and defense responses but did not provide direct insights into essential oil biosynthesis (Maul et al., 2008; González-Candelas et al., 2010; Matas et al., 2010; Ballester et al., 2011; Hershkovitz et al., 2012). We optimized a protocol for the collection of intact ECs from the flavedo layer of grapefruit peel by laser microdissection and pressure catapulting, subsequent isolation of RNA, and linear amplification of transcripts for analysis on Affymetrix Citrus genome oligonucleotide microarrays. As negative controls, we collected parenchyma cells (PCs) of the albedo layer, which were processed in the same way (Fig. 3). For the microarray analysis, we selected fruit right before and during the highest oil accumulation (28 and 40 mm ED, respectively; Fig. 1E). Microarray data were uploaded into the Partek Genomics Suite software package for background correction and statistical processing. In 28 mm ED fruit, the expression levels of 4,079 transcripts were significantly different between ECs and PCs (Supplemental Table S2). The same comparison for transcripts from 40 mm ED fruit revealed 3,082 differentially expressed genes. This preliminary analysis provided evidence that there are substantial differences between gene expression patterns in ECs and PCs.
Figure 3.
To categorize the differentially expressed genes, gene lists were processed with the Web-based AgriGO tool (Du et al., 2010), which allows users to perform a singular enrichment analysis for Gene Ontology (GO) terms. The most positively enriched GO terms for metabolic genes in ECs (compared with PCs) for both fruit sizes were “phenylpropanoid metabolic process” (GO:0009698) and “isoprenoid metabolic process” (GO:0006720; Supplemental Table S3). A more comprehensive list of genes involved in these pathways is given in Table II and provides further evidence for the fairly high expression levels, in ECs, of genes involved in the biosynthesis of monoterpenes and sesquiterpenes, coumarins, and polymethoxylated flavones, the main components of Citrus peel essential oil. The list of metabolic genes with lower expression levels in ECs (when compared with PCs) was enriched in the GO categories “response to stimulus” (GO:0050896) and “polysaccharide metabolic process” (GO:0005976; Supplemental Table S3), which both contained genes related to cell wall biosynthesis. The expression levels of only 143 genes were different between ECs of fruits with EDs of 28 and 41 mm (26 higher and 117 lower in ECs of 28-mm compared with 41-mm fruit), and there was no significant enrichment in any GO categories. An analogous analysis of PCs revealed the differential expression of 356 genes in fruit with EDs of 28 and 41 mm (103 higher and 253 lower in PCs of 28-mm compared with 41-mm fruit), and there was also no enrichment of GO terms. These results indicate that there were no major differences between gene expression patterns at the two selected developmental stages.
Table II. Enrichment of genes involved in essential oil biosynthesis in ECs compared with PCs of grapefruit peel at two different stages of development (28 mm ED and 41 mm ED), expressed as fold change.
| Annotation | Citrus Affy Chip Identifier |
EC versus PC Enrichment |
|
|---|---|---|---|
| 28 mm | 41 mm | ||
| Terpenoid biosynthesis | |||
| MVA pathway | |||
| Acetyl-CoA thiolase | Cit.1287.1.S1_s_at | 6.941 | 4.117 |
| 3-Hydroxy-3-methylglutaryl-CoA synthase | Cit.29397.1.S1_s_at | 12.272 | 11.972 |
| Cit.1280.1.S1_s_at | 6.551 | 5.119 | |
| Cit.29249.1.S1_at | 4.446 | 5.080 | |
| 3-Hydroxy-3-methylglutaryl-CoA reductase | Cit.17889.1.S1_s_at | 14.403 | 8.488 |
| Cit.23097.1.S1_s_at | 10.603 | 6.856 | |
| MVA kinase | Cit.2723.1.S1_s_at | 5.617 | 3.106 |
| Phosphomevalonate kinase | Cit.37962.1.S1_at | 2.891 | 4.609 |
| Diphosphomevalonate decarboxylase | Cit.20947.1.S1_s_at | 3.849 | 2.764 |
| MEP pathway | |||
| 1-Deoxy-d-xylulose 5-phosphate synthase | Cit.10053.1.S1_at | 49.532 | 12.021 |
| Cit.10054.1.S1_s_at | 12.292 | 4.887 | |
| Cit.10056.1.S1_at | 11.740 | 8.216 | |
| 1-Deoxy-d-xylulose-5-phosphate reductoisomerase | Cit.23507.1.S1_at | 12.309 | 11.740 |
| Cit.4968.1.S1_s_at | 5.384 | 3.434 | |
| Cit.26985.1.S1_at | 3.538 | 2.404 | |
| 2-C-Methyl-d-erythritol 4-phosphate cytidylyltransferase | Cit.14478.1.S1_at | 5.452 | 4.375 |
| 4-(Cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase | Cit.29567.1.S1_s_at | 6.659 | 10.413 |
| Cit.3446.1.S1_s_at | 7.803 | 5.097 | |
| 2-C-Methyl-d-erythritol 2,4-cyclodiphosphate synthase | Cit.1430.1.S1_s_at | 5.645 | 4.046 |
| 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | Cit.985.1.S1_at | 8.931 | 4.878 |
| 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate reductase | Cit.17714.1.S1_at | 29.980 | 8.382 |
| Cit.9279.1.S1_at | 4.626 | 3.123 | |
| Cit.9280.1.S1_s_at | 4.483 | 2.612 | |
| Prenyltransferases and terpene synthases | |||
| Isopentenyl diphosphate isomerase (putative) | Cit.9842.1.S1_s_at | 3.430 | 2.800 |
| Cit.27203.1.S1_s_at | 2.875 | 2.350 | |
| Cit.9841.1.S1_s_at | 2.679 | 2.232 | |
| Farnesyl diphosphate synthase (putative) | Cit.1688.1.S1_s_at | 2.806 | 1.963 |
| Cit.35197.1.S1_at | 2.333 | 2.917 | |
| Geranylgeranyl pyrophosphate synthase (putative) | Cit.20724.1.S1_at | 25.436 | 7.288 |
| Cit.14050.1.S1_at | 9.553 | 3.298 | |
| Cit.16303.1.S1_at | 3.544 | 3.159 | |
| (+)-Limonene synthase (monoterpene synthase) | Cit.9964.1.S1_s_at | 9.540 | 3.194 |
| Cit.23560.1.S1_at | 9.116 | 4.338 | |
| β-Pinene synthase (monoterpene synthase) | Cit.2694.1.S1_at | 29.767 | 17.103 |
| (E)-β-Ocimene synthase (monoterpene synthase) | Cit.31295.1.S1_at | 3.119 | 1.821 |
| Monoterpene synthase (putative) | Cit.31559.1.S1_at | 2.158 | 1.416 |
| β-Farnesene synthase | Cit.2936.1.S1_at | 53.196 | 15.623 |
| Sesquiterpene synthase (putative) | Cit.17284.1.S1_at | 49.647 | 14.095 |
| Cit.30774.1.S1_at | 41.631 | 21.153 | |
| Cit.29748.1.S1_s_at | 27.134 | 17.274 | |
| Coumarin and flavonoid biosynthesis | |||
| Malonyl-CoA biosynthesis | |||
| ATP citrate lyase subunit B | Cit.11635.1.S1_at | 3.384 | 2.668 |
| Cit.30906.1.S1_s_at | 2.948 | 3.718 | |
| Cit.14414.1.S1_at | 2.796 | 3.078 | |
| Acetyl-CoA carboxylase (biotin carboxylase subunit) | Cit.26354.1.S1_s_at | 2.771 | 2.853 |
| Cit.26354.1.S1_s_at | 2.771 | 2.853 | |
| General phenylpropanoid pathway | |||
| Phe ammonia lyase | Cit.9590.1.S1_s_at | 4.452 | 5.469 |
| Cit.2241.1.S1_s_at | 2.810 | 2.654 | |
| 4-Coumarate CoA ligase | Cit.25648.1.S1_s_at | 3.473 | 4.061 |
| Coumarin and furanocoumarin pathway | |||
| CYP82C (hydroxylates psoralens) | Cit.5945.1.S1_at | 56.028 | 14.715 |
| Cit.29478.1.S1_s_at | 26.793 | 7.596 | |
| Cit.2373.1.S1_s_at | 17.277 | 4.347 | |
| Flavonoid pathway | |||
| Chalcone synthase | Cit.1966.1.S1_s_at | 14.402 | 1.299 |
| Cit.10216.1.S1_at | 3.008 | 2.146 | |
| Chalcone isomerase | Cit.5577.1.S1_at | 2.145 | 1.632 |
| Cit.29773.1.S1_s_at | 2.045 | 2.161 | |
| Flavanone 3-hydroxylase | Cit.2890.1.S1_s_at | 15.901 | 7.209 |
| Cit.2889.1.S1_at | 14.602 | 6.341 | |
| Cit.2890.1.S1_at | 8.796 | 5.203 | |
| Flavone synthase (CYP93D) | Cit.29489.1.S1_at | 26.434 | 13.133 |
| Cit.2475.1.S1_at | 21.786 | 13.355 | |
| Cit.29489.1.S1_s_at | 13.944 | 10.741 | |
| Flavonoid 3′-monooxygenase | Cit.11243.1.S1_at | 12.247 | 3.884 |
| Cit.30703.1.S1_at | 31.822 | 26.116 | |
| Cit.4610.1.S1_s_at | 49.621 | 12.614 | |
| Cit.4610.1.S1_at | 46.816 | 10.041 | |
| TT1 (glabra transcription factor) | Cit.2963.1.S1_s_at | 2.375 | 1.433 |
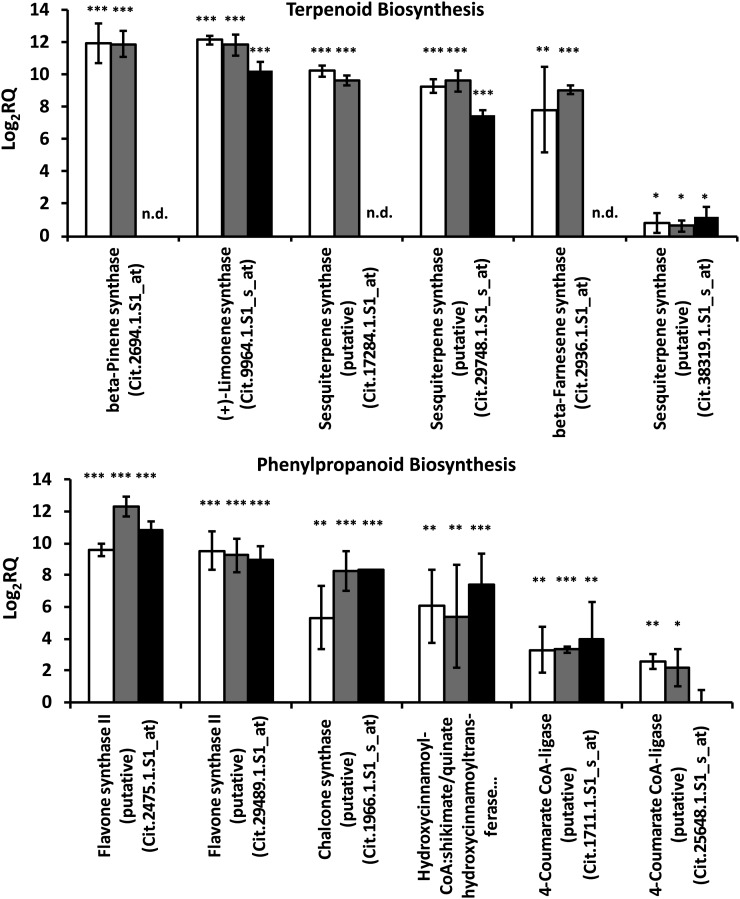
To further test the robustness of microarray data sets for transcripts highly expressed in ECs, real-time quantitative PCR (qPCR) analyses were performed for selected genes with relevance to Citrus essential oil biosynthesis. Because the laser microdissection and pressure catapulting of thousands of ECs had already been an enormously time-consuming task, we decided to compare gene expression patterns in thin sections (roughly 600–700 µm) of flavedo (exocarp layer containing ECs and secretory cavities) and upper albedo (mesocarp layer devoid of oil glands), which should provide similar insights into the apparent specialization of ECs. We selected three different developmental stages of grapefruit (34, 43, and 120 mm ED), which encompassed both early time points used in our microarray study and an additional time point close to maturity. Primers were designed to amplify fragments of genes representing the most highly enriched transcripts of ECs as indicated by our microarray data (Supplemental Table S4). Three monoterpene synthase genes [encoding (+)-limonene synthase, myrcene synthase, and β-pinene synthase], one characterized sesquiterpene synthase [(E)-β-farnesene synthase], and two putative sesquiterpene synthases (Citrus Chip identifiers Cit.29748.1.S1_s_at [orange1.1g015245m] and Cit.17284.1.S1_at [orange1.1g039366m]) were highly enriched in 34- and 43-mm fruit peel flavedo (Fig. 4), which is in full accordance with the microarray data. Two of these genes were also enriched in 120-mm fruit [(+)-limonene synthase and a putative sesquiterpene synthase of as yet unknown function (Cit.29748.1.S1_s_at)]. As a negative control, we evaluated the transcript levels of a putative terpene synthase (Cit.38319.1.S1_s_at) that, based on microarray results, was not found to be preferentially expressed in ECs, and we observed that its expression levels, as determined by qPCR, also did not differ significantly in the albedo and flavedo layers. We also selected six genes involved in phenylpropanoid/flavonoid biosynthesis (4-coumarate:CoA ligase [two genes], hydroxycinnamoyl-CoA:quinate transferase, chalcone synthase, and flavone synthase [two genes]) that were highly expressed in ECs (as determined by microarray analysis). All of these genes were found to be expressed at elevated levels in flavedo (compared with albedo) in qPCR assays (Fig. 4). Interestingly, transcript levels stayed at high levels throughout fruit development for five of the six selected phenylpropanoid/flavonoid pathway genes.
Figure 4.
Gene expression patterns of genes involved in grapefruit peel essential oil biosynthesis, as determined by real-time qPCR (n = 3), using the β-actin transcript (corresponding Chip identifier Cit.16435.1.S1_at) as an endogenous control. The log2 (relative quantification [RQ]) values are indicators of the fold change between the expression levels of a gene in the essential oil-producing flavedo and nonproducing albedo layers of the peel. Transcript abundance was quantified in peel of fruit at three developmental stages: 34 mm ED (white bars), 43 mm ED (gray bars), and 120 mm ED (black bars). Asterisks indicate statistical significance: * P < 0.05, ** P < 0.01, *** P < 0.001. n.d., Not determined.
DISCUSSION
Citrus Fruit Peel Oil Glands Develop Early, But the Filling of Secretory Cavities Correlates with Later Stages of Development
Our experiments showed that the increase in cavity numbers (Fig. 1B) in young grapefruit was much faster than the growth rate of the entire fruit (Fig. 1A), indicating that most cavities are formed early during fruit development. This is in agreement with previous data obtained with orange (Citrus sinensis) fruit (Knight et al., 2001). We also attempted to learn more about the process of cavity filling, which could not be discerned based on previously published data. In the youngest fruit (6 mm ED), we counted roughly 6,000 cavities, but their volumes were not uniform. The calculated volumes ranged from 0.15 to 2.4 nL, corresponding to a 16-fold difference from the lowest to the highest volume (Fig. 1, C and H). Interestingly, we found a distribution of vastly different cavity volumes at different stages of fruit development, most notably during the intermediate growth phase (50–90 mm ED), when volumes of 0.15 to 192 nL per fruit were observed (1,280-fold difference from lowest to highest volume). As expected, the distribution was shifted toward larger cavities at maturity, but there was still a considerable distribution of different volumes (1.4–1,000 nL per fruit; 714-fold difference; Fig. 1, D and H). These data indicate that, although the total number of cavities began to level off at approximately 60 mm ED (average of 29,040 cavities per fruit), the filling continued as long as the fruit was still growing.
While determining cavity diameters for the calculation of oil accumulated in all cavities of a fruit, we noticed that younger fruit, when compared with mature fruit, had a substantially higher proportion of cavities with fairly narrow, prolate spheroid shapes (Fig. 1C). As the fruit expanded, the cavities continued to fill until reaching the shape of a regular sphere or an oblate spheroid (Fig. 1H). It is generally accepted that the secretory cavities in Citrus are formed schizogenously (Thomson et al., 1976; Turner et al., 1998), that is, by a separation of gland cells at an early stage, which results in the formation of a storage space lined by ECs. It is thus tempting to speculate about the mechanism that leads to the observed cavity volume increases. We noted that the expansion of the peel during fruit growth was marked by the formation of air spaces between the exocarp PCs (Supplemental Fig. S1). Air spaces were most prominent in the albedo, which consists of an extensive aerenchyma. Flavedo air spaces were not apparent in small fruit (up to about 50 mm ED) but constituted more than half of the volume of the flavedo in mature fruit (Supplemental Fig. S1). No air spaces formed between the thick-walled sheath cells of the secretory cavities. Their expansion was apparently directed against each other, tangentially to the cavity surface, thereby causing the sheath cells to bow outward, whereas the secretory cavity assumed a more spherical shape. Whether the final cavity shape is determined by the expansion of sheath cells, the accumulation of secretion within the cavity, or both remains an open question that we are currently investigating.
Our estimations of the oil volume per fruit, based on the experimentally determined volume distribution of secretory cavities at different developmental stages, were in very good agreement with the experimentally determined oil yields (Fig. 1D). Similar observations were made in previous work with peppermint (Mentha × piperita [Lamiaceae]; Turner et al., 2000; Rios-Estepa et al., 2010), which demonstrated that the accumulation of leaf essential oil correlated with the developmental distribution of glandular trichomes. These anatomical structures harbor secretory cells that are hypothesized to be functionally related to ECs in Citrus glands (Fahn, 1988). Our study here provides, to our knowledge, the first direct evidence that the control of essential oil accumulation might follow similar patterns in the Lamiaceae and Rutaceae. It is also interesting that secretory cells in the Lamiaceae (Lange et al., 2000; Gang et al., 2001; Lane et al., 2010) and Citrus ECs (this work) are both highly enriched in transcripts related to essential oil biosynthesis. We are now expanding our analyses to further assess the evidence for generalizable patterns of metabolic specialization in cells involved in terpenoid oil and resin biosynthesis across different plant phyla.
Gene Expression Patterns in Citrus Fruit Peel ECs Generally Correlate with Essential Oil Composition
Although ECs surrounding secretory cavities have been hypothesized to be responsible for oil biosynthesis in Citrus peel (Turner et al., 1998; Lücker et al., 2002; Evert, 2006; Yamasaki and Akimitsu, 2007), our study has provided, to our knowledge, the first direct evidence for the biosynthetic capabilities of these highly specialized cells. It is important to note that the cavity filling in Citrus peel appears to be a relatively slow process when compared with the accumulation of essential oil in peppermint glandular trichomes (mean filling time of 20 h; Turner et al., 2000). The genes encoding all previously characterized terpene synthases of Citrus peel were expressed at high levels in ECs (when compared with PCs, which do not produce essential oil). Interestingly, although most terpene synthases are not expressed at appreciable levels in maturing fruit, the transcript for (+)-limonene synthase, which catalyzes the reaction to the major constituent of the essential oil of mature fruit [(+)-limonene; Supplemental Table S1], stays at high levels throughout fruit development (Fig. 4).
To our surprise, although grapefruit essential oil obtained from fruit of a greenhouse-grown tree contained less than 0.05% sesquiterpenes (Table I), two putative sesquiterpene synthases were enriched in the EC data set (Supplemental Table S2). In this context, it is important to note that the absolute expression values for the corresponding probe sets on the Affymetrix microarray were fairly low (15.2 for Cit.17284.1.S1_at and 14.7 for Cit.2936.1.S1_at) or very low (2.0 for Cit.29748.1.S1_s_at) in PCs, which were used as reference cells. A 10-fold increase of sesquiterpene synthase expression levels in ECs (compared with PCs), as determined by qPCR (Fig. 4), still means that the absolute expression levels were much lower than those for the most abundant genes, such as (+)-limonene synthase (expression level of 2,192.9 for Cit.9964.1.S1_s_at in ECs). Be that as it may, the enrichment of transcripts putatively related to sesquiterpene biosynthesis is interesting, and a possible explanation might be the contribution of ECs to terpenoid volatile formation. The complex terpenoid mixture emitted by grapefruit prior to maturity is significantly different from that of the oil accumulated in secretory cavities, with a fairly high proportion of sesquiterpenes (24% of emitted terpenoids as opposed to less than 2% in the oil; Flamini and Cioni, 2010).
A significant enrichment in ECs was also found for genes involved in terpenoid precursor biosynthesis via the mevalonate (MVA) and methylerythritol 4-phosphate (MEP) pathways. Both pathways are expressed in all cells, as they are involved in the biosynthesis of essential terpenoid end products (Phillips at al., 2008). In most plants, monoterpenes (the major constituents of Citrus oil) are derived primarily from the MEP pathway, with only little contribution of the MVA pathway (Phillips at al., 2008). Although the relative contributions of the MVA and MEP pathways for Citrus essential oil biosynthesis remain to be determined, it is possible that the MVA pathway, which is relevant for sesquiterpene biosynthesis in many plant species (Phillips at al., 2008), may contribute more to the sesquiterpene-rich terpenoid emissions.
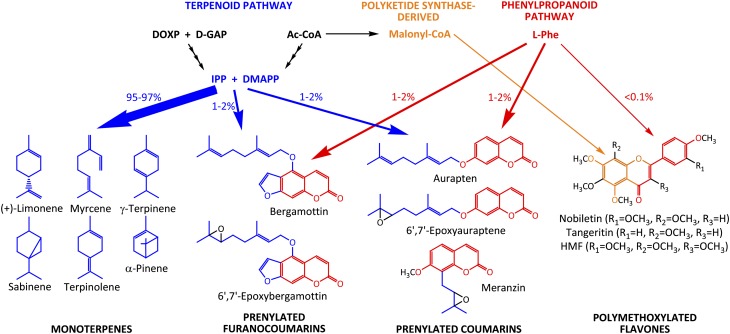
Citrus oil also contains prenylated coumarins and polymethoxylated flavones (Fig. 5). The core skeletons of both metabolite classes are derived from the phenylpropanoid pathway, and the corresponding genes were also expressed at high levels in ECs (Table II). Very few coumarin-specific genes have been characterized thus far (Bourgaud et al., 2006). A gene of the CYP82C subfamily, members of which have been demonstrated to be involved in the biosynthesis of 5-hydroxylated coumarins (Kruse et al., 2008), was found to be highly enriched in ECs (Table II). Polymethoxylated flavonoids are formed by successive hydroxylations and O-methylations (Willits et al., 2004; Schmidt et al., 2011), and candidate genes for enzymes involved in these steps were also found to be highly enriched in ECs (Supplemental Table S5). These observations indicate that our gene expression data from Citrus glandular ECs reflect their specialization for essential oil biosynthesis and present an excellent resource for gene discovery related to the biosynthesis of oil constituents.
Figure 5.
Contribution of the terpenoid and phenylpropanoid biosynthetic pathways to the accumulation of the major components of grapefruit peel essential oil. A color code is used to indicate the origin of structural moieties in oil constituents: blue, terpenoid pathway; red, phenylpropanoid pathway; orange, derived from a polyketide synthase. The thickness of the reaction arrows and the percentages next to them indicate the relative distribution of carbon flux through the different pathway branches. Ac, Acetyl; DMAPP, dimethyl allyl diphosphate; DOXP, 1-deoxy-D-xylulose-5-phosphate; D-GAP, D-glyceraldehyde 3-phosphate; HMF, heptamethoxyflavone; IPP, isopentenyl diphosphate.
MATERIALS AND METHODS
Plant Growth Conditions
Grapefruit (Citrus × paradisi ‘Duncan’) were harvested from a 30-year-old tree (height, 2.8 m; stem circumference, 12.5 cm) grown in the greenhouse of the Institute of Biological Chemistry on the Pullman, Washington, campus. The mean temperature was 26°C, and the humidity was set to 60%. Natural lighting was supplemented with sodium vapor lights that turned on at light intensities of 200 µmol m−2 s−1 or less, with a 14-h-day/10-h-night cycle. The tree was watered daily and fertilized twice a week with Peters 20/20/20. Fruits were labeled, and growth patterns (polar diameter [PD] and ED) were measured weekly starting in early June to late April of the following year for two growth seasons (2009/2010 and 2010/2011). The fruit surface area (S) was calculated based on the approximation for a specific shape, which depended on the developmental stage as follows.
For the surface area of prolate spheroid:
where
For the surface area of oblate spheroid:
Morphometric Measurements
To determine the number of secretory cavities in grapefruit peel, a defined area (seven to 10 patches of 300 mm2 each) of the flavedo layer (s) was hand sectioned horizontally, and images were taken with a Jenoptik ProgRes C12Plus camera mounted on a dissecting microscope. All cavities were counted for the smallest fruit. In this context, it is important to note that the density of cavities for a fruit of a defined developmental stage did not vary significantly in different positions of the fruit peel (assessed by counting cavities of fruit of five different developmental stages). The electronic image files were uploaded into GIMP 2 (an open-source image-processing program), and the total number of secretory cavities (n) per defined area was counted. The total number of cavities per fruit (N) was then calculated based on the surface area of the entire fruit (S):
To measure the volumes of individual secretory cavities, images of transversely hand-sectioned grapefruit peel were taken with a Leica DFC425C camera, and the polar diameter (PD) and ED of each cavity were determined using the Leica Application Suite version 3 software. Assuming that the shape of the cavities resembles that of a spheroid (of any kind), the cavity volume (V) can be estimated as follows:
The total volume of oil per fruit was then extrapolated by a series of calculations outlined in Supplemental Protocol S1.
For an evaluation of the volume distribution patterns, secretory cavities were divided into 19 classes by volume (ranging from 0.15 to 1,000 nL) and the number of cavities in each class was determined for fruit at various developmental stages (stage differentiation by size; 12 different sizes ranging from 6 to 136 mm ED).
Metabolite Analysis
Hydrodistillation of the Volatile Essential Oil Fraction
Three randomly taken sections of flavedo layer tissue (300 mm2 each) were obtained from freshly harvested grapefruit using a razor blade, placed in a mortar filled with liquid nitrogen, and ground to a fine powder. Sample material was then transferred to a glass flask for subsequent hydrodistillation using a modified Likens-Nickerson apparatus (Ringer et al., 2003). An aliquot of the n-hexane fraction, which contained the volatile oil constituents and camphor as an internal standard (final concentration at 5 ng µL−1), was transferred to a 2-mL glass vial for GC-FID analysis.
Collection of Citrus Essential Oil Using Microcapillaries
Microcapillaries of a defined size (internal diameter of 70–100 µm) were made using a custom-built capillary puller. Oil was collected by penetrating through the thin epidermal/subepidermal cell layers above each secretory cavity and allowing the liquid contents to enter the microcapillary. Roughly 4,000 cavities per fruit were accessed from fruit representing three developmental stages (EDs of 70, 80, and 100 mm; three biological replicates each). The microcapillary contents were emptied into a glass vial and spun at 4,000g for 2 min. A small volume of aqueous material was visible at the bottom of the vial, which was most likely attributable to small amounts of cellular content from ruptured cells. It is important to note, however, that most of the cells surrounding secretory cavities remained unaffected by the oil collection procedure (Supplemental Fig. S1).
Terpenoids
The essential oil was diluted 100-fold with n-hexane and transferred to 2-mL glass vials for GC-FID analysis. The separation of terpenoids was achieved by GC-FID as described by Ringer et al. (2003). Individual components were quantified based on calibration curves with known amounts of authentic standards and normalization to the peak area of camphor as an internal standard. The sum of all components was used to determine the total oil amount injected onto the GC device, and by taking into account all dilutions and losses, the total amount of oil recovered from each fruit was calculated.
Oxygen Heterocyclic Metabolites
Sample analysis was carried out using an ultra-high-pressure liquid chromatograph with diode array detector (1200 series; Agilent Technologies) coupled to a quadrupole time-of-flight mass spectrometer (6520 series; Agilent Technologies). The gradient separation was performed on an Eclipse Plus C-18 column (2.1 × 50 mm, 1.8 µm; Agilent Technologies) with a flow rate of 0.6 mL min−1. Solvent A was 0.2% (v/v) acetic acid in water and solvent B was 0.2% (v/v) acetic acid in acetonitrile. Analytes were separated isocratically at 70% B and 30% A (held for 16 min after injection). This was followed by a wash step at 98% B and 2% A (6 min) and a reequilibration at the initial conditions (5 min) between runs. Authentic standards were purchased from Enzo Life Sciences (aurapten, greater than 95% purity), Selleckchem (nobiletin, greater than 99% purity; osthole, greater than 99% purity; tangeretin, greater than 99% purity), and Sigma-Aldrich (bergamottin, greater than 98% purity). Calibration curves generated with authentic standards were used for the absolute quantification of nonvolatile essential oil constituents by diode array detection at 310 nm. Mass spectrometric data were used to confirm peak annotations. Additional minor nonvolatile oil components were meranzin (mass-to-charge ratio [m/z] 261.113), isomeranzin (m/z 261.119), epoxyaurapten (m/z 315.160), epoxybergamottin (m/z 355.155), and heptamethoxylated flavones (m/z 433.151), the concentrations of which were estimated based on the published literature (McHale and Sheridan, 1989; Frérot and Decorzant, 2004; Dugo et al., 2010).
Sterols
After addition of the internal standard (epi-cholesterol), sterol constituents of the oil collected with microcapillaries were saponified at 90°C for 60 min in 2 mL of 6% (w/v) KOH in methanol. Upon cooling to room temperature, 2 mL of n-hexanes and 2 mL of water were added, and the mixture was shaken vigorously for 20 s. After centrifugation (3,000g for 2 min) to separate the phases, the hexane phase was transferred to a 2-mL glass vial. The hexane phase was then evaporated to dryness using a SpeedVac concentrator, 50 µL of N-methyl-N-trimethylsilyltrifluoroacetamide was added to the residue, the sample was shaken vigorously for 20 s, and the mixture was transferred to a 2-mL autosampler glass vial with a 100-µL conical glass insert. After capping the vial, the reaction mixture was incubated at 23°C for at least 5 min. GC-MS analyses were performed on an Agilent 6890N apparatus coupled to an Agilent 5973 inert mass selective detector (MSD) detector. Samples were loaded (injection volume of 1 µL) with a CombiPAL autosampler (LEAP Technologies) onto an HP-5MS fused silica column (30 m × 250 µm, 0.25 µm film thickness; J&W Scientific). The temperatures of the injector and MSD interface were both set to 280°C. Analytes were separated at a flow rate of 1 mL min−1 using helium as the carrier gas and using a thermal gradient starting at 170°C (1.5 min), which was ramped first to 280°C at 37°C min−1 and then to 300°C at 1.5°C min−1, where it was held for 5.0 min. Analytes were fragmented in electron-impact mode with an ionization voltage of 70 eV. Data were acquired using the MSD ChemStation (revision D.01.02.SP1) software. Background was subtracted and peaks were deconvoluted using the automated mass spectral deconvolution and identification system (National Institute of Standards and Technology). Analytes were identified based on their mass fragmentation patterns by comparison with those of authentic standards. Quantification was achieved based on calibration curves acquired with authentic standards.
Fatty Acids and Carotenoids
Fatty acids were separated and quantified as methyl ester derivatives based on the protocol developed by Dyer et al. (2002). Carotenoid analysis was performed according to Fraser et al. (2000).
Gene Expression Analysis
Sample Processing for Laser Capture Microdissection and Pressure Catapulting
Grapefruit peel flavedo was cut into 3- × 3-mm sections and immediately dipped into ethanol:acetic acid (5:1, v/v) fixative (Deeken et al., 2008). Sample specimens were incubated in the fixative for 1 h under reduced pressure (23°C, 0.78 bar) in a vacuum oven. This fixation step was repeated twice with fresh fixative before incubation for 12 h at 4°C. The fixative was then replaced with 100% ethanol, and specimens were incubated for 30 min at 23°C. This step was also repeated twice with fresh ethanol. Samples were then incubated in a series of ethanol:xylene in different ratios (3:1, 1:1, and 1:3 [v/v]), each step for 1 h at 23°C. For tissue embedding, samples were vacuum infiltrated in 100% xylene for 1 h, followed by a series of xylene and liquid paraffin (melting temperature of 45°C; Sigma) mixtures (3:1, 1:1, and 1:3 [v/v]), each step for 1 h at 23°C. Lastly, specimens were infiltrated in 100% liquid paraffin for 1 h at 23°C. This step was repeated twice with fresh paraffin. Paraffin blocks were stored at 4°C in a sealed bag filled with silica gel desiccant.
Laser Capture Microdissection and Pressure Catapulting of Epithelial and PCs
Ribbons of paraffin sections (15 µm thickness) were cut with a rotary microtome, expanded by floatation on a bath of diethyl pyrocarbonate-treated water at 45°C, and placed onto polyethylene naphthalate (PEN)-membrane coated slides (Zeiss Microimaging). PEN slides were irradiated with UV light (254 nm) for 30 min and handled according to the manufacturer’s instructions. Slides were air dried for 20 min at 23°C, followed by melting of the paraffin on a slide warmer at 45°C for 30 min. This resulted in a better adherence of sections to PEN slides while further drying them. At this point, representative sections were scraped off from the slides for an assessment of RNA quality (RNA extraction using the method as described below). The slides were then stored in the dark at −80°C in a plastic box filled with desiccant until further use. Prior to microdissection, sample slides were deparaffinized in 100% xylene for 3 min at 23°C. This step was repeated three times with fresh xylene. Slides were then air dried in a fume hood for 15 min at 23°C. Laser capture microdissection and pressure catapulting (LMPC) was carried out using the PALM MicroBeam system (Zeiss Microimaging). The laser energy was set to 74 (scale from 0 to 100) and the laser focus to 57 (scale from 0 to 100). The excised target cells were catapulted and collected into caps of 0.5-mL Eppendorf vials containing lysis buffer (RLT buffer; Qiagen Micro RNeasy kit). Approximately 100 target cells were catapulted into one vial cap. The cell material was collected at the bottom of the tube by brief centrifugation (15,000g) and immediately stored in a −80°C freezer until enough cells had been collected for RNA extraction (roughly 2,500 cells per sample).
RNA Extraction from Cells Isolated by LMPC, cDNA Synthesis, Amplification, and Microarray Hybridization
RNA extractions were conducted using the RNeasy Micro kit (Qiagen). LMPC-derived cell material was incubated at 37°C for 5 min, followed by vigorous mixing for 1 min at 23°C. The suspension was centrifuged at 15,000g for 1 min, the supernatant was transferred into a new vial, and RNA was extracted according to the manufacturer’s protocol. The RNA yield and quality were assessed using a NanoDrop 2000c spectrophotometer (Thermo Scientific). RNA samples were stored at −80°C. The Whole-Transcriptome Ovation Pico RNA Amplification system version 1.0 (NuGEN Technologies) was used for cDNA synthesis and simultaneous amplification. The protocol involved a three-step process: (1) generation of first-strand cDNA; (2) generation of a DNA/RNA heteroduplex double-strand cDNA; and (3) linear isothermal DNA amplification. Prior to microarray hybridization, cDNA products were fragmented and biotin labeled using the FL-Ovation cDNA Biotin Module V2 kit (NuGEN Technologies). Once biotin labeled, cDNAs were hybridized to GeneChip Citrus Genome Arrays (Affymetrix).
Microarray Data Analysis
Microarray data analysis was performed with the Genomics Suite software (Partek). Affymetrix .cel files were imported, probe intensities were adjusted, and a background correction was carried out using the robust multiarray average algorithm, which included a correction on perfect match values, quantile normalization across all of the chips in the experiment, median polish summarization, and log2 transformation (Irizarry et al., 2006). Probe sets labeled as absent or marginal for any of the three replicate arrays for each time point and cell type were removed, leaving only those that were scored as present among all three biological replicates. A multiple testing correction was performed for assessing the false discovery rate (Benjamini and Hochberg, 1995), and only probe sets below a false discovery rate threshold of 0.01 were kept. Probe sets were filtered for a greater than 2.5-fold expression level difference in a two-way comparison between appropriate samples. Gene annotation was carried out based on similarity scores in BLASTX comparisons against sequences contained in the HarvEST (Close et al., 2007), The Arabidopsis Information Resource (Rhee et al., 2003), and Citrus (http://www.citrusgenomedb.org/) genome databases. All raw data files (.cel and .chp) were submitted to the National Center for Biotechnology Information Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE33964).
RNA Extraction of Fruit Peel Flavedo and Albedo and Subsequent qPCR
Flavedo and albedo layers of grapefruit peel were excised with a scalpel and processed separately. Samples were placed in a mortar filled with liquid nitrogen and ground to a fine powder. RNA was extracted using the Concert Plant RNA Reagent (Invitrogen) and further purified using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Isolated RNA (500–700 ng) was treated with RNase-free DNase (Fermentas Life Science), and first-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen). RNA isolation and cDNA synthesis were carried out with three different grapefruit sizes (ED 34, 43, and 120 mm). Three independent biological replicates were processed for each fruit size. In a 10-μL qPCR, concentrations were adjusted to 150 nm (primers), 1× SYBR Green PCR Master Mix, and 100× diluted first-strand cDNA as template. Reactions were performed on a 96-well optical plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60° for 10 min in a 7500 Real-Time PCR system (Applied Biosystems). Fluorescence intensities of three independent measurements (technical replicates) were normalized against the ROX reference dye (Roche Applied Science). For each sample, the amounts of target and endogenous control (β-actin gene selected from Citrus microarray: Cit.16435.1.S1_at) were determined using the comparative cycle threshold method according to the manufacturer’s instructions (Applied Biosystems).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Microscopic evaluation of secretory cavity expansion in grapefruit peel at different stages of fruit development.
Supplemental Table S1. Analysis of volatile essential oil components of grapefruit peel by hydrodistillation.
Supplemental Table S2. Microarray analysis of transcript patterns in grapefruit peel ECs and PCs at different stages of fruit development (28 mm and 41 mm ED of fruit).
Supplemental Table S3. GO analysis of microarray data obtained with grapefruit peel ECs and PCs at different stages of fruit development (28 mm and 41 mm ED of fruit).
Supplemental Table S4. Primers used for quantitative real-time PCR analyses of gene expression patterns in grapefruit peel flavedo and albedo tissues.
Supplemental Table S5. Cytochrome P450 and O-methyltransferase genes of unknown function enriched in grapefruit peel ECs compared with PCs.
Supplemental Protocol S1. Determining the volume distribution of secretory cavities in grapefruit peel.
Supplementary Material
Acknowledgments
We thank Dr. Daniel Cuthbertson and Dr. Glenn Turner (Institute of Biological Chemistry) for technical assistance and editorial comments regarding the manuscript and Mr. Derek Pouchnik (School of Molecular Biosciences) for technical assistance with microarray hybridization. We also thank Mr. Craig Whitney and Ms. Amy Hetrick (Institute of Biological Chemistry) for maintaining the plants used in this study. We are grateful for the technical support of the Franceschi Microscopy and Imaging Center.
Glossary
- ECs
epithelial cells
- PCs
parenchyma cells
- GC
gas chromatography
- MS
mass spectrometry
- ED
equatorial diameter
- FID
flame-ionization detection
- GO
Gene Ontology
- qPCR
real-time quantitative PCR
- MVA
mevalonate
- MEP
methylerythritol 4-phosphate
- m/z
mass-to-charge ratio
- LMPC
laser capture microdissection and pressure catapulting
- PEN
polyethylene naphthalate
References
- Ballester AR, Lafuente MT, Forment J, Gadea J, De Vos RCH, Bovy AG, González-Candelas L. (2011) Transcriptomic profiling of Citrus fruit peel tissues reveals fundamental effects of phenylpropanoids and ethylene on induced resistance. Mol Plant Pathol 12: 879–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Berger IJ, Freitas-Astúa J, Reis MS, Targon MLPN. (2007) In silico prediction of gene expression patterns in Citrus flavedo. Genet Mol Biol 30: 752–760 [Google Scholar]
- Bourgaud F, Hehn A, Larbat R, Perper S, Gontier E, Kellner S, Matern U. (2006) Biosynthesis of coumarins in plants: a major pathway still to be unraveled for cytochrome P450 enzymes. Phytochem Rev 5: 293–308 [Google Scholar]
- Carson CF, Hammer KA (2011) Chemistry and bioactivity of essential oils. In H Thormar, ed, Lipids and Essential Oils as Antimicrobial Agents. John Wiley & Sons, Chichester, UK, pp 203–238
- Close TJ, Wanamaker S, Roose ML, Lyon M. (2007) HarvEST. Methods Mol Biol 406: 161–177 [DOI] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Mazzafera P. (2007) A genomic approach to characterization of the Citrus terpene synthase gene family. Genet Mol Biol 30: 832–840 [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. (2010) AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugo P, Piperno A, Romeo R, Cambria M, Russo M, Carnovale C, Mondello L. (2009) Determination of oxygen heterocyclic components in Citrus products by HPLC with UV detection. J Agric Food Chem 57: 6543–6551 [DOI] [PubMed] [Google Scholar]
- Dugo P, Ragonese C, Russo M, Sciarrone D, Santi L, Cotroneo A, Mondello L. (2010) Sicilian lemon oil: composition of volatile and oxygen heterocyclic fractions and enantiomeric distribution of volatile components. J Sep Sci 33: 3374–3385 [DOI] [PubMed] [Google Scholar]
- Dugo P, Skeriková V, Kumm T, Trozzi A, Jandera P, Mondello L. (2006) Elucidation of carotenoid patterns in citrus products by means of comprehensive normal-phase × reversed-phase liquid chromatography. Anal Chem 78: 7743–7750 [DOI] [PubMed] [Google Scholar]
- Dyer JM, Chapital DC, Kuan JW, Mullen RT, Pepperman AB. (2002) Metabolic engineering of Saccharomyces cerevisiae for production of novel lipid compounds. Appl Microbiol Biotechnol 59: 224–230 [DOI] [PubMed] [Google Scholar]
- El-Adawy TA, Rahma EH, El-Bedawy AA, Gafar AM. (1999) Properties of some Citrus seeds. Part 3. Evaluation as a new source of protein and oil. Nahrung 6: 385–391 [Google Scholar]
- Esau K (1965) Plant Anatomy. John Wiley & Sons, New York
- Espina MS, Lorán S, Conchello P, García D, Pagán R. (2010) Chemical composition of commercial Citrus fruit essential oils and evaluation of their antimicrobial activity acting alone or in combined processes. Food Control 22: 896–902 [Google Scholar]
- Evert RF (2006) Esau’s Plant Anatomy. Meristems, Cells and Tissues of the Plant Body: Their Structure, Function and Development. John Wiley & Sons, Hoboken, NJ
- Fahn A (1979) Secretory Tissues in Plants. Academic Press, New York
- Fahn A. (1988) Secretory tissues in vascular plants. New Phytol 198: 229–257 [DOI] [PubMed] [Google Scholar]
- Flamini G, Cioni PL. (2010) Odour gradients and patterns in volatile emission of different plant parts and developing fruits of grapefruit (Citrus paradise L.). Food Chem 120: 984–992 [Google Scholar]
- Forment J, Gadea J, Huerta L, Abizanda L, Agusti J, Alamar S, Alos E, Andres F, Arribas R, Beltran JP, et al. (2005) Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol Biol 57: 375–391 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Pinto ME, Holloway DE, Bramley PM. (2000) Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Frérot E, Decorzant E. (2004) Quantification of total furocoumarins in citrus oils by HPLC coupled with UV, fluorescence, and mass detection. J Agric Food Chem 52: 6879–6886 [DOI] [PubMed] [Google Scholar]
- Gang DR, Wang JH, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E. (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125: 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes M, Pauly G, Carde JP, Marpeu A, Bernard-Dagan C. (1983) Monoterpene hydrocarbon biosynthesis by isolated leucoplasts of Citrofortunella mitis. Planta 159: 373–381 [DOI] [PubMed] [Google Scholar]
- González-Candelas L, Alamar S, Sánchez-Torres P, Zacarías L, Marcos JF. (2010) A transcriptomic approach highlights induction of secondary metabolism in Citrus fruit in response to Penicillium digitatum infection. BMC Plant Biol 10: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkovitz V, Ben-Dayan C, Raphael G, Pasmanik-Chor M, Liu J, Belausov E, Aly R, Wisniewski M, Droby S. (2012) Global changes in gene expression of grapefruit peel tissue in response to the yeast biocontrol agent Metschnikowia fructicola. Mol Plant Pathol 13: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Wu Z, Jaffee HA. (2006) Comparison of Affymetrix GeneChip expression measures. Bioinformatics 22: 789–794 [DOI] [PubMed] [Google Scholar]
- Kirbaslar SI, Boz I, Kirbaslar FG. (2006) Composition of Turkish lemon and grapefruit peel oils. J Ess Oil Res 18: 525–543 [Google Scholar]
- Knight TG, Klieber A, Sedgley M. (2001) The relationship between oil gland and fruit development in Washington Navel orange (Citrus sinensis L. Osbeck). Ann Bot (Lond) 88: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Ho K, Yoo HD, Johnson T, Hippely M, Park JH, Flavell R, Bobzin S. (2008) In planta biocatalysis screen of P450s identifies 8-methoxypsoralen as a substrate for the CYP82C subfamily, yielding original chemical structures. Chem Biol 15: 149–156 [DOI] [PubMed] [Google Scholar]
- Lane A, Boecklemann A, Woronuk GN, Sarker L, Mahmoud SS. (2010) A genomics resource for investigating regulation of essential oil production in Lavandula angustifolia. Planta 231: 835–845 [DOI] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber E, Sanchez C, Pouchnik D, Croteau R. (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufenberg G, Kunz B, Nystroem M. (2003) Transformation of vegetable waste into value added products: (A) the upgrading concept; (B) practical implementations. Bioresour Technol 87: 167–198 [DOI] [PubMed] [Google Scholar]
- Lücker J, El Tamer MK, Schwab W, Verstappen FWA, van der Plas LH, Bouwmeester HJ, Verhoeven HA. (2002) Monoterpene biosynthesis in lemon (Citrus limon): cDNA isolation and functional analysis of four monoterpene synthases. Eur J Biochem 269: 3160–3171 [DOI] [PubMed] [Google Scholar]
- Maruyama T, Ito M, Honda G. (2001) Molecular cloning, functional expression and characterization of (E)-β farnesene synthase from Citrus junos. Biol Pharm Bull 24: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Matas AJ, Agustí J, Tadeo FR, Talón M, Rose JKC. (2010) Tissue-specific transcriptome profiling of the Citrus fruit epidermis and subepidermis using laser capture microdissection. J Exp Bot 61: 3321–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul P, McCollum GT, Popp M, Guy CL, Porat R. (2008) Transcriptome profiling of grapefruit flavedo following exposure to low temperature and conditioning treatments uncovers principal molecular components involved in chilling tolerance and susceptibility. Plant Cell Environ 31: 752–768 [DOI] [PubMed] [Google Scholar]
- McHale D, Sheridan JB. (1989) The oxygen heterocyclic compounds of Citrus peel oils. J Ess Oil Res 1: 139–149 [Google Scholar]
- Mondello L, Zappia G, Dugo P, Dugo G (2002) Advanced analytical techniques for the study of Citrus oils. In G Dugo, A DiGiacomo, eds, Citrus: The Genus Citrus. Taylor and Francis, New York, pp 179–200
- Pauly G, Belingheri L, Marpeu A, Gleizes M. (1986) Monoterpene formation by leucoplasts of Citrofortunella and Citrus unshiu. Plant Cell Rep 5: 19–22 [DOI] [PubMed] [Google Scholar]
- Phillips MA, León P, Boronat A, Rodríguez-Concepción M. (2008) The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci 13: 619–623 [DOI] [PubMed] [Google Scholar]
- Reis MS, Takita MA, Palmieri DA, Machado MA. (2007) Bioinformatics for the Citrus EST project (CitEST). Genet Mol Biol 3: 1024–1029 [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer KL, McConkey ME, Davis EM, Rushing GW, Croteau RB. (2003) Monoterpene double-bond reductases of the (−)-menthol biosynthetic pathway: isolation and characterization of cDNAs encoding (−)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch Biochem Biophys 418: 80–92 [DOI] [PubMed] [Google Scholar]
- Rios-Estepa R, Lange I, Lee JM, Lange BM. (2010) Mathematical modeling-guided evaluation of biochemical, developmental, environmental, and genotypic determinants of essential oil composition and yield in peppermint leaves. Plant Physiol 152: 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Estepa R, Turner GW, Lee JM, Croteau RB, Lange BM. (2008) A systems biology approach identifies the biochemical mechanisms regulating monoterpenoid essential oil composition in peppermint. Proc Natl Acad Sci USA 105: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Li C, Shi F, Jones AD, Pichersky E. (2011) Polymethylated myricetin in trichomes of the wild tomato species Solanum habrochaites and characterization of trichome-specific 3′/5′- and 7/4′-myricetin O-methyltransferases. Plant Physiol 155: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf E (1974) Gland cells. In AW Robards, ed, Dynamic Aspects of Plant Ultrastructure. McGraw-Hill, New York, pp 331–357
- Shimada T, Endo T, Fujii H, Hara M, Omura M. (2005) Isolation and characterization of (E)-beta-ocimene and 1,8 cineole synthases in Citrus unshiu Marc. Plant Sci 168: 987–995 [Google Scholar]
- Shimada T, Endo T, Fujii H, Hara M, Ueda T, Kita M, Omura M. (2004) Molecular and functional characterization of four monoterpene synthase genes from Citrus unshiu Marc. Plant Sci 166: 49–58 [Google Scholar]
- Takita MA, Berger IJ, Basilio-Palmieri AC, Borges KM, de Souza JM, Targon MLNP. (2007) Terpene production in the peel of sweet orange fruits. Genet Mol Biol 30: 841–847 [Google Scholar]
- Thomson WM, Platt-Aloia KA, Endress AG. (1976) Ultrastructure of oil gland development in the leaf of Citrus sinensis L. Bot Gaz 137: 330–340 [Google Scholar]
- Turner GW, Berry AM, Gifford EM. (1998) Schizogenous secretory cavities of Citrus limon (L.) Burm. F. and a reevaluation of the lysigenous gland concept. Int J Plant Sci 159: 75–88 [Google Scholar]
- Turner GW, Gershenzon J, Croteau RB. (2000) Development of peltate glandular trichomes of peppermint. Plant Physiol 124: 665–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willits MG, Giovanni M, Prata RTN, Kramer CM, De Luca V, Steffens JC, Graser G. (2004) Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites. Phytochemistry 65: 31–41 [DOI] [PubMed] [Google Scholar]
- Yamasaki Y, Akimitsu K. (2007) In situ localization of gene transcriptions for monoterpene synthesis in irregular parenchymic cells surrounding the secretory cavities in rough lemon (Citrus jambhiri). J Plant Physiol 164: 1436–1448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.