Abstract
The goal of many metabolomic studies is to identify the molecular structure of endogenous molecules that are differentially expressed among sampled or treatment groups. The identified compounds can then be used to gain an understanding of disease mechanisms. Unfortunately, despite recent advances in a variety of analytical techniques, small molecule (<1000 Da) identification remains difficult. Rarely can a chemical structure be determined from experimental “features” such as retention time, exact mass, and collision induced dissociation spectra. Thus, without knowing structure, biological significance remains obscure. In this study we explore an identification method in which the measured exact mass of an unknown is used to query available chemical databases to compile a list of candidate compounds. Predictions are made for the candidates using models of experimental features that have been measured for the unknown. The predicted values are used to filter the candidate list by eliminating compounds with predicted values substantially different from the unknown. The intent is to reduce the list of candidates to a reasonable number that can be obtained and measured for confirmation. To facilitate this exploration, we measured data and created models for two experimental features; MS Ecom50 (the energy in eV required to fragment 50% of a selected precursor ion) and HPLC retention index. Using a dataset of 52 compounds, Ecom50 models were developed based on both Molconn and CODESSA structural descriptors. These models gave r2 values of 0.89 to 0.94 depending on the number of inputs, the modeling algorithm chosen, and whether neutral or protonated structures were used. The retention index model was developed with 400 compounds using a back propagation artificial neural network and 33 Molconn structure descriptors. External validation gave a v2 = 0.86 and standard error of 38 retention index units. As a test of the validity of the filtering approach, the Ecom50 and retention index models, along with exact mass and collision induced dissociation spectra matching, were used to identify 1,3-dicyclohexylurea in human plasma. This compound was not previously known to exist in human biofluids and its elemental formula was identical to 315 other candidate compounds downloaded from PubChem. These results suggest that the use of Ecom50 and retention index predictive models can improve non-targeted metabolite structure identification using HPLC/MS derived structural features.
Keywords: metabolomics, retention index, Ecom50, mass spectrometry, HPLC, database searching, structure identification, molecular topology, Molconn, Codessa, artificial neural network
Introduction
Over the past decade, the field of metabolomics1 has expanded to encompass a wide range of applications in genetics, environmental sciences, human health, and pre-clinical toxicity studies. Improvements in mass spectrometry and liquid chromatography have enabled high-throughput detection of a very large number of experimental peaks from complex biological samples. Measurements can be made in a short period of time and metabolites can be detected even when present at low concentration. Likewise, improvements in chemometric tools have enabled the high throughput extraction and comparison of raw data to determine which experimental ‘features’ are significantly different between sample groups. In this context a ‘feature’ (or ‘target feature’) refers to a single observed experimental endpoint, or peak, which corresponds to a specific molecular structure. When a complex biologic sample is processed by HPLC and mass spectrometry, a very large number of features are observed in the selected ion chromatograms. The ultimate goal is to match a specific molecular structure to each observed feature. Unfortunately, the process of structure elucidation for these features has remained time consuming, costly, and is frequently unsuccessful2.
Currently, structure identification for mass spectrometry based metabolomics relies heavily on database searching. To facilitate the identification process, a number of metabolite databases have been compiled. These often contain detailed structural and chemical information of known metabolites3–5 (i.e. molecular weight, elemental formula, logP, mass spectral fragmentation patterns, retention index). For example, the combined use of exact mass, retention index, and MS fragmentation pattern has been previously demonstrated to be a robust technique for identifying metabolites using an in-house database of these descriptors3. Because these metabolite databases are incomplete, their use precludes identification of metabolites or metabolite classes not contained in the database. Therefore, there are clear advantages in using chemical databases that contain a much larger number and greater diversity of compounds, including compounds that have not been confirmed to exist in biological systems. One such database, PubChem, consists of molecules compiled from more than 125 smaller databases6,7 and currently contains approximately 30 million compounds with masses between 50 and 1000 Da. Obviously, it is not feasible for any single laboratory to measure experimental features (i.e. fragmentation patterns and retention indices) for such a large number of compounds. However, as previously described2, large databases such as PubChem can be annotated with chemical and structural information predicted by computational techniques, thus increasing the utility of the database for identification purposes. For example, computational predictions of logP are now included for most compounds in PubChem.
Previous reports have demonstrated the validity of identifying an unknown compound by matching its experimental collision induced dissociation (CID) spectra with predicted CID spectra of candidate compounds contained in databases8,9. In these cases, the CID spectra of the database-derived compounds were predicted using commercial and non-commercial software. The initial list of candidate compounds was obtained from PubChem based on their having the same exact mass as the unknown. All compounds with the exact mass of the unknown compound were extracted from the database and their CID spectra were predicted. The predicted CID spectra were compared to the observed spectra and database compounds are ranked based on number of matches. This combination of accurate mass and accurate mass fragmentation prediction was sufficient for identifying a majority of the 102 compounds tested. Some structures were not correctly identified due to the inability of the software to accurately predict their fragmentation mechanisms. In addition, Hill et al found no a priori way to determine which compounds or compound classes were poorly predicted by the software8. Current CID prediction software is not sufficiently accurate to discriminate unambiguously among all potential candidates contained in large databases. Thus, the prediction of additional orthogonal experimental features could be of tremendous value to aid in the identification of unknown compounds using HPLC/MS. Recently, Kumari et. al. combined exact mass, CID fragmentation prediction, and retention index prediction for identifying metabolites using accurate mass gas-chromatography/time-of-flight mass spectrometry10. Although analyte derivitization required for gas chromatography can complicate data analysis, their results also suggest that chemical feature prediction is a paradigm worth pursuing for non-targeted metabolomics.
In the present study, Ecom50 and retention index (RI) were evaluated as additional experimental features that are applicable to electrospray-based MS analysis and could augment exact mass and predicted CID spectra in structure identification. These features can be measured using HPLC/MS, and if the endpoints are successfully modeled using readily calculated structure descriptors, use of predicted values could improve database filtering efficiency. We reasoned that by comparing exact mass, along with experimental vs. predicted values for Ecom50, retention index, and CID spectra, we could better discriminate among candidate compounds from large databases such as PubChem and reduce the number of potential matches to a target unknown.
Materials and Methods
Chemicals
Ethisterone was purchased from Steroloids (Newport, RI). HPLC grade methanol (Chromasol®), trifluoroacetic acid, and 1,3-dicyclohexylurea (DCHU) were purchased from Sigma-Aldrich (St. Louis, MO). N-cyclohexyl-2-methyl-piperidine-1-carboxamide was purchased from Chembridge (San Diego, CA). HPLC grade acetonitrile was purchased from Fisher Scientific (Hampton, NH). Nitromethane, nitroethane, n-nitropropane, n-nitrobutane, n-nitropentane and n-nitrohexane were purchased from Aldrich (St. Louis, MO). n-nitroheptane, n-nitrooctane, n-nitrononane and n-nitrodecane were synthesized as described by Aderjan and Bogusz (1988). HPLC grade heptafluorobutyric acid was purchased from Thermo Scientific (Rockford, IL). Reagent grade water was generated by a Barnstead Diamond® reverse-osmosis water purification system. 1,3-bis(cyclopentylmethyl)urea was synthesized and characterized as described in supporting information SI10. The sources of all other chemical compounds for which Ecom50 and RI were measured are listed in supporting information SI1–SI3.
Biological Samples
Human serum and cerebral spinal fluid (CSF) were obtained from the Rocky Mountain Multiple Sclerosis Center (Englewood, CO). Samples were used in accordance with the University of Connecticut Institutional Review Board.
Detection of Unknown Compound in Human Serum Sample MSC205s
During an HPLC/MS analysis of human CSF taken from control patients and with multiple sclerosis, an unknown compound with a retention time of approximately 13 min and an apparent protonated mass of m/z 225 (exact protonated mass = 225.1958 Da) was detected in many of the control and multiple sclerosis samples. The m/z 225 unknown compound was targeted for additional investigation because it was a significant peak in these samples. More importantly, a search of the 2009 Metlin4, HMDB5, DrugBank11, CSF-metabolome12, and KEGG13 databases of known biological and bioactive compounds returned no matches at that exact mass. Thus, this unknown compound represented an ideal opportunity to test our approach. Further analysis of CSF samples was precluded by insufficient volume; however, a sample of human serum (labeled MSC205s) from a control patient was obtained and this sample was extracted and analyzed by HPLC/MS for presence of the m/z 225 compound. One hundred μL of the serum sample was pipetted into a 1.5 mL polypropylene centrifuge tube (Eppendrof, Hauppauge, NY). Cold methanol (400 μL) was added to the tube, vortex mixed for 1 min, and allowed to incubate at −20° C for 1 h. The sample was centrifuged at 14,000 rcf for 10 min and an aliquot (450 μL) of the supernatant phase was transferred to a clean 1.5 mL polypropylene tube. The solvent was evaporated to dryness under reduced pressure in a Vacufuge® (Eppendrof, Hauppauge, NY) evaporator at ambient temperature. The sample residue was dissolved in 25 μL of methanol by vortex mixing for 30s and then diluted with 25 μL of reagent grade water with vortex mixing for 5s. Eight μL of the sample was analyzed on the described HPLC/UV system. The Ecom50 value of a peak in the sample (detected at ~13 min with a nominal m/z value of 225) was determined by reanalyzing the sample extract on the HPLC system in the MS/MS mode, isolating the m/z 225 ion, and collecting the CID spectrum at 17.0, 19.0 and 21.0 eV at 0.5 second intervals. The CID mass spectral profile was obtained on the m/z 225 ion at 25 eV. The Ecom50 value of the m/z 225 compound was calculated as described by equation 2 and the retention index was calculated as described in the retention index measurement section. A mouse serum sample was processed by the same methods to ensure that the m/z 225 peak was not the result of a contaminant associated with our methodology. The mass of the m/z 225.1958 Da ion was assumed to be the protonated molecular ion of the unknown compound. The monoisotopic molecular weight (MIMW) of the unknown was therefore calculated as:
| Equation(1) |
The MIMW for the m/z 225 ion was determined to be 224.1885 Da. In order to determine if the structure of this unknown could be identified using our database filtering method, predictive models were created for the Ecom50 and RI experimental features and were used in conjunction with exact mass and CID fragment predictions.
Measurement of Ecom50 values
The Ecom50 experimental feature was chosen as a compliment to exact mass and CID fragment spectra matching because recent work has shown that isomeric structures can be differentiated using this experimental feature14. In addition, Ecom50 data is easily obtained during the collection of CID spectra data
In preparation for the development of an Ecom50 model, measurements were taken on a diverse set of 54 compounds listed in supporting information SI1. Collision energy at 50% survival yield (CE50) values were measured as described by Kertesz et al.15 and converted to center of mass energy at 50% survival yield (Ecom50) using the following formula:
| Equation(2) |
where Mrg is the natural isotope weighted mass of the reagent gas (argon in this case, 39.945) and Mx is the monoisotopic mass of the test compound’s molecular ion. Ecom50 values were used (rather than CE50 values) because Ecom50 values are independent of the type of collision gas used and are thus more compatible with data obtained in different laboratories. The range of experimental Ecom50 values was 0.70 to 7.82 eV. Structures of the neutral compounds for the Ecom50 dataset were downloaded from PubChem16 as computer-readable 3-dimensional structure data (SD) files and verified by cross-referencing with the Merck Index17.
Development of Ecom50 models
Ecom50 models were developed using multiple structure description and modeling methods for comparison. Version 1.4 of winMolconn18 was used to generate structure descriptors, which were used as input to SAS19 for the generation of both multiple linear regression (MLR) and partial least squares (PLS) models. Also, the CODESSA20 software package (in conjunction with AMPAC21) was used to generate both structure descriptors and MLR models based on the CODESSA input selection algorithm. The Ecom50 model data set and these two approaches are described in the following sections.
Ecom50 data set structure profile
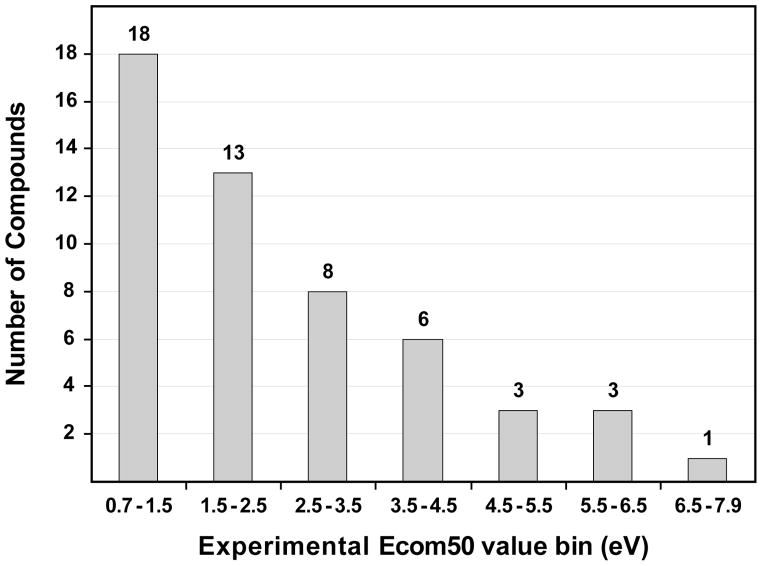
The compounds used to generate the Ecom50 model contain from one to five nitrogen atoms in several functional group types. The number of oxygen atoms in a given molecule ranges from zero to eleven. Forty-eight molecules contain at least one oxygen atom. Oxygen is present in several functional group types including alcohol, ether, ketone, ester, acid, amide, and phosphate. Four compounds have permanently charged quaternary nitrogens and one N-oxide is present. Molecular weight ranges from 88.2 Da to 607.7 Da with an average of 263.2 Da and standard deviation of 140.5 Da. Acyclic, cyclic, and multiply cyclic structures are present. The data set contains nitrogen atoms in 15 different chemical environments. This data set contains a useful overall variety of nitrogen atom types but the groups are heavily loaded as alkyl amines as well as pyridines and amides. Ureas are underrepresented and others such as imines, oximes, and nitriles are missing. In this sense the set is not well balanced in overall chemical space with respect to nitrogen functional groups. However, the data are sufficient to provide an initial evaluation for using Ecom50 as a filtering option for non-targeted metabolomics that is approximately orthogonal to retention index. Two compounds that were initially considered for inclusion in the models (gallamine, a tri-cation, and ATP, a triphosphate) were subsequently considered outside the chemical space of the 52 other structures and were removed from the model dataset. A histogram of the measured Ecom50 values for the 52 remaining compounds is given in Figure 1.
Figure 1.
Distribution of measured experimental values for 52 compound Ecom50 dataset.
Protonation Site Selection
Models of Ecom50 were generated from descriptors calculated for both the neutral and mono-protonated forms of the structures. Protonated structures were considered because positive ion electrospray produces mono-protonated ions. Protonation alters the internal energy distribution of a molecule, so it was reasonable to infer that it could be useful to calculate descriptors based on the mono-protonated form. The actual protonation site is not known with certainty. Indeed, evidence exists that protonation site can be influenced by prevailing conditions in an LC-MS experiment such as mobile phase pH, aqueous-organic ratio, and buffer concentration22. However, the most likely protonation site for molecules in this data set is a nitrogen atom. For molecules that contain a single nitrogen atom, the proton was assigned to that atom. For structures that contain more than one nitrogen atom, it was necessary to evaluate protonation site options. Two methods were examined in this study; one based on partial charge, and a second based on a combination of experimental and quantum chemically computed proton affinity.
To assign the protonation site according to partial charge, the SD file of the neutral structure was used to compute semi-empirical quantum chemical geometry optimizations using the AMPAC21 program and the AM1 (Austin Model 1) semi-empirical model, originally developed by Dewar et. al.23–26. Results were verified by frequency calculation to ensure that a stationary state geometry (i.e. a conformation with at least a local energy minimum) was achieved. The electrostatic charge distribution of the geometry-optimized neutral molecule was examined. The atom showing the highest partial negative charge was chosen as the site to attach the H+. Following protonation at that site, the geometry of the resulting cationic structure was re-optimized using the same frequency calculation criterion. Most structures in this test set contain an obvious single highly negatively charged atom. Several compounds show multiple atoms with negative partial electrostatic charges comparable in magnitude. The single site with highest partial negative charge was consistently chosen.
A second method assigned the site of protonation based on proton affinity. For those structures which contained more than one nitrogen atom, a combination of experimental and calculated proton affinity values was used to make the assignment. Proton affinity data available from NIST consists largely of molecules with a single nitrogen atom27. This data does, however, suggest the relative proton affinity of nitrogen in different chemical environments. For example, proton affinity values for alkyl amines are in the order tertiary > secondary > primary; also pyridine > pyrrole. However, when a nitrogen atom in a particular group occurs in varied electronic surroundings, the picture is less clear.
Quantum mechanical computations were carried out to compare computed proton affinities for the structures with multiple nitrogen atoms. Density functional computations at both the 6-31G* and the 6-311+G* level were used in addition to a semi-empirical method (RM1)28. All computations were done with Spartan-08 software29. Of the 52 compounds, 32 have nitrogen atoms in more than one type of electronic setting. For 22 of these 32 structures it was reasonable to assign a protonation site on the basis of simple rules derived from experimental proton affinity data. An examples is the protonation preference for alkyl amines in the order tertiary > secondary > primary and also for pyridines and ureas > pyrroles. Since there were combinations of nitrogen functional groups for which there was no experimental data, and confirmation of the simple rule assignments was useful, density functional theory DF(B3LYP 6-31G*) was used to compute the proton affinity for all 32 structures as:
| Equation(3) |
The energy of all possible mono-protonated nitrogen forms was calculated, along with the energy of the neutral form. For molecules which contain more than one nitrogen atom, the protonated species with the greatest proton affinity was taken to be the most stable. For nine compounds (isoniazid, allantoin, 3-methyl-L-histidine, N-methyl-L-histidine, 3-hydroxy-2-naphthone hydrazide, nalidixic acid, 8-hydroxy-2-deoxyguanosine, 4-dimethylaminopyridine, and ormetoprim) assignments were determined entirely on the basis of the greatest computed proton affinity. These computations confirmed the simple rule assignment for the other 22 structures, as the rule based assignment agreed with the calculated proton affinity in all cases. It is of note that, except for N-methyl-L-histidine and 4-aminohippuric acid, the semi-empirical method RM1 gave the same protonation site indication as the DF 6-31G* computations.
Molconn Modeling of Ecom50
Structure descriptors for the Ecom50 compounds were generated from an SD file using winMolconn18. The representations of molecular structure calculated by winMolconn are generally referred to as topological structure descriptors and are divided into two broad categories, electrotopological state (E-State indices) and molecular connectivity (chi indices). E-State descriptors represent the electron accessibility at each atom, a combination of electron distribution and local atom topology. In addition to individual atom descriptors, the basic atom-level E-State information is developed into several types of structure descriptors including atom types and bond types as well as functional group descriptors, and others such as the minimum and maximum atom-level values in a structure, and hydrogen bonding descriptors (including internal hydrogen bonding). The molecular connectivity (chi) descriptors represent features of molecular skeletal structure including degree of branching, ring structure, and overall skeletal ramification. For this modeling, the difference valence chi indices are used because they are independent of molecular size and represent skeletal variation, including the influence of heteroatoms. In contrast to the largely electronic information in E-State descriptors, chi indices primarily encode the topology of the molecular skeleton. This overall method of structure descriptor development has become known as Structure Information Representation (SIR)30–36.
Molconn structure descriptors most closely related to the training set structure features were used to create a pool of descriptors available to the modeling algorithms. Descriptors were additionally filtered so that no pair shows a pairwise correlation greater than 0.75. For modeling on protonated structures, descriptors included the protonated form of the E-State index for all protonated nitrogen and quaternary nitrogen N+ atoms. The names and descriptions of indices in the descriptor pools for Molconn modeling are given in supporting information SI4–SI7.
Model building and prediction were done with the SAS Statistical System 9.119. To facilitate variable selection for MLR models, the SAS RSQUARE procedure was used. This method determines models for all possible combinations of a specified number of descriptors. RSQUARE does not use a stepwise procedure for descriptor selection. SAS REG was used for creation of multiple linear regression (MLR) models and associated statistics. SAS was also used to develop partial least squares (PLS) models. In the PLS method37,38 the procedure constructs linear combinations (latent factors) of all the descriptors in the pool such that each successively created variable (latent factor) improves the correlation with the dependent variable. The resulting latent factors are created as an orthogonal (independent) set. Up to eight latent factors were considered for PLS models and up to 8 descriptors were considered for MLR models. The best MLR and PLS models for the protonated and non-protonated forms of the data set were kept for further analysis.
CODESSA modeling of Ecom50
Katritzky and coworkers explored structure-property and structure-activity relations using a multidimensional approach39,40. They introduced the CODESSA method, in which a large number of molecular descriptors can be calculated, based upon a quantum chemical optimization of a molecular geometry, and can be correlated with experimental results to find a structure–property correlation.
Model data for the CODESSA study was the same as for the Molconn study described above. SD files for both the neutral and protonated structures were used to compute semi-empirical quantum chemical geometry optimizations using AMPAC with AM1. Results were verified by frequency calculation to ensure that a stationary state geometry (a conformation with at least a local energy minimum) was achieved. While doing the geometry optimization of the neutral and protonated structures, AMPAC was set to generate output results for use by CODESSA20. Quantum mechanical calculation results were then used to calculate CODESSA models for Ecom50. CODESSA calculates upwards of 500 molecular descriptors for each structure submitted for consideration. Modeling then consists of systematically examining each of the descriptors available for all structures to find the best empirical model(s), which are based on multiple linear regression (MLR) of the selected descriptors. The best 8 variable MLR models for the protonated and non-protonated forms of the data set were kept for further analysis.
Development of a Retention Index (RI) Model
Predictive models for RI have been previously developed for specific classes of compounds41–47. A model suitable for non-targeted metabolite structure identification would require the ability to predict RI values for a diverse set of biologically relevant small molecules. In this regard, we previously developed a robust HPLC RI predictive model demonstrating the potential for predicting RI for a structurally diverse group of compounds based on chemical structure30. The mobile phase used to measure this previously published dataset is not appropriate for the currently proposed LC/MS metabolomic method because it is not compatible with electrospray ionization mass spectrometry. This necessitated the measurement of new data and the corresponding development of a new model.
RI Model Compounds and Source
In order to build a robust predictor of chromatograph RI for small molecules, it was first necessary to develop a training set of experimentally measured values. RI values were experimentally determined for a structurally diverse group of 411 small molecules consisting of endogenous compounds, endogenous metabolites, drugs, and drug metabolites. Compounds chosen for the study were limited to the set of biologic elements (C, H, N, O, S, and P) and contained at least one protonatable atom to facilitate detection by mass spectrometry in positive ion mode. For the majority of compounds, the protonatable atom was nitrogen. A total of 12 steroid-like compounds with alpha-beta unsaturated ketone groups were also measured because of the prevalence of this scaffold in biologic systems. The ketone group has been observed to protonate at electrospray pH levels allowing these compounds to be detected in positive ion mode30.
Retention Index Measurements
Retention index was determined for the 411 compounds on a Zorbax Stable Bond C-18, 1 mm × 150 mm (3.5 μm particle size) column (Agilent, Santa Clara, CA) with a mobile phase consisting of 0.01% (v/v) heptafluorobutyric acid (HFBA) in water (solvent A) and 0.01% HFBA in 90% acetonitrile (solvent B) using an Agilent 1100 capillary HPLC system (Agilent, Santa Clara, CA). A linear gradient from 0% solvent B to 100% solvent B at a flow rate of 75 μL/min was used to elute the compounds from the column. One μL of 0.34 mM mixtures of approximately 20 test compounds per solution were analyzed in duplicate using gradient times of 17 min and 35 min. At the beginning and end of each batch of sample analyses 1 μL of a homologous mixture of C1 through C10 n-nitroalkanes (1.12 mM in mobile solvent phase B) was analyzed with the effluent column tubing attached to a Agilent 1100 diode array detector (Agilent, Santa Clara, CA) monitoring 210 nm.
For the analysis of test samples the tubing from the end of the column was attached to the electrospray source of a Micromass Q-TOF-2 (Waters Associates, Beverly, MA) mass spectrometer operated in positive ion mode. The electrospray source of the mass spectrometer was operated at a cone potential of 20 V, a capillary potential of 3.0 kV and a source temperature of 120° C. Ethisterone (0.3 mM) was co-analyzed with the nitroalkanes and the test samples as a retention reference to determine the void time difference between the diode array detector and the mass spectrometer. The retention times of test compounds were determined from their respective reconstructed molecular ion chromatographic peak.
The retention times of the respective n-nitroalkanes analyzed before and after the batch analysis of a set of test samples were averaged and used as the calibration references for calculating the RI of the test compounds. The offset in the retention time between the diode array and the mass spectrometer was calculated as the difference between the average retention time of ethisterone determined using the diode array detector and the retention time of ethisterone obtained on the mass spectrometer during a respective set of test compound analyses. This value was added to the retention time of test compounds analyzed on the mass spectrometer to correct for the void time difference between the two detectors. During solvent gradient analysis the composition of the mobile phase remains constant at the starting composition until the mixing of the two mobile phases transverses the dwell volume of the system. During this period compounds are eluted under isocratic conditions.
We have determined that a linear relationship exists between the log of the retention times of the homologous series of n-nitroalkanes and the number of carbon atoms in the structure of each individual compound in an isocratic reverse phase HPLC system. The retention index of each n-nitroalkane was defined as 100 times the number of carbon atoms. Therefore the retention indices of test compounds that eluted during the isocratic portion of the mobile phase were calculated by the following formula:
| Equation(4) |
where, Tx is the corrected retention time of the analyte, Tz is the retention time of the nitroalkane eluting just before the analyte, Tz+1 is the retention time of the nitroalkane eluting just after the analyte and z is the number of carbons in the nitroalkane eluting just before the analyte.
The dwell time for the system used in this study was determined as the greatest retention time from the set of 411 compounds in which the retention time in the 17 min gradient and the 34 min gradient was the same value. For this system, 1-methylpyrrolidine had a 17 min solvent gradient retention time of 5.54 min and a 35 min solvent gradient retention time of 5.53 min. The respective retention times for the next highest retained compound, 2-aminopyrimidine, were 5.83 min and 6.01 min. Therefore a dwell time of 5.54 min was used as the end of the isocratic portion of the mobile phase and this time corresponded to a retention index of 221.8. The C3 through C10 n-nitroalkanes eluted during the gradient portion of the mobile phase. The trend function in an Excel spreadsheet was used to determine the 4th order polynomial equation between the retention time and the retention index of the dwell time and the C3 through C10 n-nitroalkanes. This equation was then used to calculate the retention index of each test compound from the corrected retention time. The assigned retention index for each compound used in the development of the retention model was the average value for duplicate analysis of each compound in the 17 min gradient system.
Experimental reproducibility of the RI measurements was calculated by performing 12 replicate measurements of 15 test compounds and generating a two-factors mixed effects model48 from the resulting data. Using this method, the total measurement error for a given compound was calculated to be 2.24 RIU. This suggests an experimental reproducibility of ± 6.7 RIU where, on a large number of replicate measurements on the same compound, 99.7% of measurements could be expected to fall within ± 6.7 RIU of the mean. Repeatability was calculated to be 1.07 and reproducibility 3.93. Total measurement error for a given compound is equal to repeatability + reproducibility, so it is calculated that reproducibility contributes 78.6% of the variability of measurement error.
Data Set Retention Index Profile
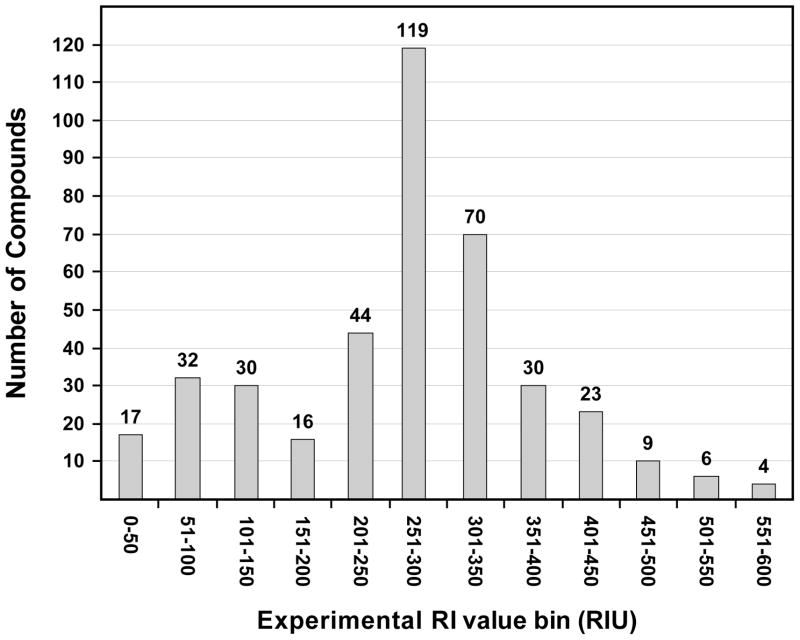
A histogram of the measured RI values is given in Figure 2. The measurement range was 578 RIU. Phosphoserine had the minimum value of 15 RIU, while carbazole had the maximum at 593 RIU. The mean is at 264 RIU with a standard deviation of 114 RIU. The distribution of measurement values is not normal, but the majority of the data points cluster around the mean. The largest RI value is 2.9 standard deviations above the mean and the smallest is 2.2 standard deviations below the mean. The median value of 268 RIU is very close to the mean. It was not necessary to remove compounds from the study as experimental outliers since no values were more than 3 standard deviations from the mean value.
Figure 2.
Distribution of measured experimental values for the retention index dataset. RIU = 100z where z = the number of carbons in the nitroalkane series (see equation 4).
Dataset Structural Profile and Diversity
Compounds measured for the RI study have an average molecular weight of 218 Da with a minimum of 79 Da, maximum of 609 Da, and standard deviation of 94 Da. More than 40 common organic functional groups are observed in the dataset. Additionally, there are many analogues of the common functional groups and a variety of carbon skeletal features such as phenyl, napthyl, cyclohexyl, and t-butyl groups (Table 1). There also exists in the data lower population functional subgraphs that are related to more highly populated functional groups such as amide. Some of these are “amide-like” features that contain at least one amide subgraph that is part of an extended conjugated system of additional carbon and nitrogen atoms. Examples in the dataset include urea, imide, guanine, xanthine, cytosine, uracil, barbiturate, uric acid, carbamte, hydantoin, pyrazolidinone, and flavin (Table 2).
Table 1.
HPLC-RI dataset high population features
| Feature | Counta | %b | Averagec |
|---|---|---|---|
| MWd | 400 | – | 218.2 |
| ringe | 319 | 79.8% | 1.7 |
| aromatic ring | 186 | 46.5% | 0.6 |
| heteroaromatic ring | 95 | 23.8% | 0.3 |
| non-aromatic ring | 180 | 45.0% | 0.77 |
| fused ring system | 128 | 32.0% | 0.33 |
| rotatable bonds | 381 | 95.3% | 6.1 |
| heteroatom | 400 | 100% | 4.4 |
| hydrogen bond acceptor | 396 | 99.0% | 4.1 |
| hydrogen bond donor | 343 | 85.8% | 1.9 |
| internal hydrogen bond | 232 | 58.0% | 1.8 |
| TPSAf | 399 | 99.8% | 68.2 |
| amine | 212 | 53.0% | 0.57 |
| anilline | 44 | 11.0% | 0.12 |
| pyridine | 40 | 10.0% | 0.11 |
| pyrole | 33 | 8.3% | 0.09 |
| ether | 80 | 20.0% | 0.32 |
| ester | 27 | 6.8% | 0.08 |
| ketone | 33 | 8.3% | 0.11 |
| alcohol | 107 | 26.8% | 0.42 |
| carboxylic acid | 113 | 28.3% | 0.32 |
| phenol | 41 | 10.3% | 0.12 |
| amide | 73 | 18.3% | 0.20 |
Number compounds with specified attribute.
Percent of compounds in the dataset with at least one example of the specified feature.
Average value in the dataset for the specified feature (rings average = 1.7 indicates that compounds in the dataset have an average or 1.7 rings).
MW = Molecular weight.
Compound contains a ring structure.
TPSA = static surface area of O, N, P and S along with associated hydrogen atoms calculated by the Etrl49 method.
Table 2.
HPLC-RI low population structure features
| Feature | Counta | %b | IGroupc |
|---|---|---|---|
| aldehyde | 3 | 0.8% | aliphatic O |
| diazole | 12 | 3.0% | pyridine, pyrrole |
| purine | 12 | 3.0% | pyridine, aniline, pyrrole |
| guanadine | 6 | 1.5% | aliphatic N |
| pyrimidine | 4 | 1.0% | pyridine, aniline |
| diphenylamine | 1 | 0.3% | pyrrole |
| carbamate | 8 | 2.0% | amide, aliphatic O |
| oxime | 1 | 0.3% | aliphatic N, aliphatic O |
| guanine | 9 | 2.3% | amide, pyridine, pyrrole |
| xanthine | 9 | 2.3% | amide, pyridine, pyrrole |
| cytosine | 8 | 2.0% | amide |
| uracil | 6 | 1.5% | amide |
| urea | 6 | 1.5% | amide |
| barbiturate | 4 | 1.0% | amide |
| uric acid | 4 | 1.0% | amide |
| imide | 3 | 0.8% | amide |
| hydantoin | 3 | 0.8% | amide |
| flavin | 2 | 0.5% | amide |
| pyrazolidinone | 2 | 0.5% | amide |
| phosphate | 16 | 4.0% | acid O |
| sulfonic acid | 7 | 1.8% | acid O |
| sulfinic acid | 1 | 0.3% | acid O |
| phosphonate | 1 | 0.3% | acid O |
| thioether | 9 | 2.3% | aliphatic S |
| thioanisole | 6 | 1.5% | aliphatic S |
| disulfide | 3 | 0.8% | aliphatic S |
| sulfoxide | 1 | 0.3% | aliphatic S, aliphatic O |
| vinyl | 13 | 3.3% | acidic CH |
| quaternary N+ | 9 | 2.3% | protonated N |
| pyridinium N+ | 5 | 1.3% | protonated N |
| thiazole | 3 | 0.8% | aromatic S, pyridine |
| thiophene | 2 | 0.5% | aromatic S |
Count of compounds with at least one example of the feature.
Percent of compounds with at least one example of the feature.
IGroup descriptor or descriptors where structure information for atoms in the group was assigned.
Structure descriptors for RI compounds
Based on the structure features of the compounds for which the RI was determined, a group of 33 structure information representation (SIR) descriptors35 was selected for use in creating a predictive model. All descriptors values were calculated using the winMolconn software, v2.150. An automated feature selection algorithm was not used to pick descriptors. Descriptors were chosen in an attempt to explicitly describe every feature of every molecule that would likely influence the retention time under the known measurement conditions.
Certain challenges exist when modeling a dataset that is small compared to the level of diversity. The most significant issue involves the attempt to encode chemical information relative to the modeled endpoint in a statistically acceptable number of inputs. The Interaction Group E-State (IGroup) family of descriptors was utilized as an alternative to counting each possible variation of every functional group as a separate descriptor, which would have resulted in a very large number of low population inputs. The IGroups methodology30 attempts to explicitly represent every atom in the molecule by combining atom level contributions from atoms that would be expected to undergo similar non-covalent interactions in solution. Functional sub-graphs are categorized into groups such as the amide-like features described above. There are several other groups including acids (carboxylate, phosphate, phosphonate, sulfonate, sulfinate), anilines (aniline, benzamidine), aromatic nitrogen (pyridine, quinoline, pyrimidine), permanent charge nitrogen, and others. For each group, a separate descriptor is created for each non-carbon atom in the group. The amide-like IGroup descriptors consist of three indices; one for double bond oxygen, one for nitrogen atoms, and one for hydrogen bonded to nitrogen. The separate indices consist of the sum of the atom-level E-State value for all atoms of the type in the molecule. A list of the IGroups indices and corresponding atom types is given in supporting information SI8.
This method allows for the description of all sub-graphs similar to the parent functional group in a small number of indices. Additionally, when a model is used for screening, sub-graphs similar to those in the training data can be accounted for in a reasonable manner, even if the exact sub-graph is not present in training data. Including amide, urea, imide, and other similar NC(=O) subgraphs, the amide-like group of features requires at least 100 different feature counts to enumerate all the possible combinations of nitrogen atom types that could be present in screening data. The IGroups method condenses this information into to three inputs that can be used to encode information for any subgraph of the amide-like type.
Along with the IGroup indices, a number of descriptors were added to encode information about size, shape, branch points, rings, flexibility, hydrophilicity, lipophilicity, and internal hydrogen bonding. These indices encode bulk structure features that likely influence partitioning characteristics during liquid chromatography. These descriptors and their definitions are listed in supporting information SI9. Two of these indices are new descriptors developed to characterize hydrophilicity and lipophilicity. The ratio valence hydrophilic index (rvalHyd) is the sum of the atom level E-State values of all hydrophilic atoms divided by the sum of atom level E-State values for every atom in the molecule. Since the atom level E-State value is a measure of the build up of valence electron density at an atom, this index describes the proportion of valence electron density that is associated with hydrophilic atoms in a size independent index. The sumLip index (total lipophilic estate) is the sum of the atom level EState value for all lipophilic atoms in the molecule and is size dependent.
Input normalization and scaling
In preparation for modeling, descriptor values for the 33 inputs were normalized and scaled from 0.1 to 0.6. The maximum value of 0.6 leaves headroom in the event that a compound to be predicted has a larger value than was observed in the model dataset. For feature descriptors, including the IGroups, a value of zero indicates the absence of the feature in the molecule. The 0.0 input values were preserved through scaling and normalization; also the mean and standard deviation used for normalization was taken on rows with a non-zero value for the input. After normalization, any compound with a normal value of 5 or greater for any input was excluded. These are compounds with a descriptor input value greater than 5 standard deviations above or below the mean value for other compounds with a non-zero value for the index. A total of 11 compounds were excluded on the basis of large normal values leaving a total of 400 compounds for modeling.
Dataset partitioning
The remaining 400 compounds were partitioned into a 4×10×10 ensemble configuration by creating four validation sub-sets, each containing approximately 25% of the compounds. This procedure results in four splits of the data, each with approximately 75% of the data for model fitting, and a unique 25% reserved for validation. The validation sub-sets were chosen so that the structural diversity and experimental activity were as similar as possible across all four sets. Each compound was used in exactly one validation set. The 75% fit set for each split was further subdivided into 10 train-test folds. Each training fold has a unique 10% of the 75% set aside for cross validation and the remainder used for neural net training.
Unlike the 1×10×10 ensemble described previously30, the use of a 4×10×10 ensemble allows each compound to be included in model training, validation, and cross validation. One of the four ensemble models gives a validation prediction for each compound, and each compound is used for model fitting in the other three models. Since the four validation sets are created by the same method, it is unlikely that any single set will yield a picture of model validation capability that is intrinsically more representative than the others. It is suggested that the four validation sets together give a more complete picture of prediction accuracy and limit the likelihood of bias associated with validation compound selection.
Neural Network Methodology
The Emergent ANN software package51,52 was used to create a separate ensemble model for each of the four data splits. For each split, a model was developed for each of the 10 individual train-test folds. A three layer back propagation net with 33 input neurons and 16 hidden neurons was used with online weight updates, 0.25 learning rate, and 0.9 momentum. For each fold, learning was conducted from a total of 50 different random initial weight starting points and the network was allowed to iterate through learning epochs until the mean absolute error (MAE) for the validation set reached a minimum. Training was stopped at this point and statistics were calculated for train, test, and validate subsets. After learning from all 50 initial weight sets was completed, the run that produced the smallest validation MAE was kept as the model for that fold. The final model used for database filtering consists of the average predicted value of the individual models from the 40 total training sets.
Filtering of candidate list of possible matches to the m/z 225 unknown
A search of possible matches for the m/z 225 unknown (candidates with similar chromatographic and mass spectral characteristics) was performed by retrieving all compounds from the PubChem database (July 2009) with a MIMW of 224.1885 Da ± 10 ppm. The search was limited to compounds composed of carbon, hydrogen, nitrogen, oxygen, sulfur, and phosphorous, with the additional stipulation that the structure contain at least one protonatable atom. Redundant compounds and multiple diasteriomers and enantiomers were eliminated. The RI values and the Ecom50 values of candidate compounds were predicted using the respective models described above. Additionally, the predicted protonated CID fragment ions for each candidate compound were calculated using Mass Frontier53 in the rules prediction mode as described previously8. The list of candidates was filtered using the Ecom50 and RI models with a filter range of ± three standard errors of prediction from the respective measured values. The ± 3 SE range corresponds to a 99.8% confidence interval and gives a high probability that a compound matching the unknown will survive the filter process if present in the candidate list. Based on experimental values for the m/z 225 unknown (Ecom50 = 2.76 eV, RI = 499 RIU), the resulting filter ranges were 2.76 ± 1.59 eV for Ecom50 and 499 ± 114 RIU for RI. Candidates with predicted values outside the specified ranges were removed from the list and surviving candidate compounds were rank ordered on the number of predicted fragment ions that matched the measured CID fragment ions of the unknown.
Results
Ecom50 modeling based on Molconn descriptors
For models based on Molconn descriptors, the use of PLS resulted in significantly better statistics than MLR (Table 3). The probable reason for this is that the set of structures in the training set covers a wide range of structure types and functional groups. Based on the typically accepted ratio of observations to statistical variables, the number of statistical variables should not be greater than eight for a data set of 52 compounds. Using eight descriptors in an MLR model places a limitation on the amount of structure information that can be included. On the other hand, PLS includes a larger fraction of the available structure information in the full pool of descriptors by making use of orthogonal linear combinations (latent factors) of all 75 descriptors in the pool. Our preliminary work indicates that up to seven latent factors could be used while maintaining statistical significance; beyond seven, little statistically significant improvement is observed.
Table 3.
Ecom50 Molconn PLS and MLR modeling results (protonated and neutral structures)
| Model | na | r2b | sc | Fd | q2e | spressf | inputsg |
|---|---|---|---|---|---|---|---|
| Protonated Structures | |||||||
| PLS | 52 | 0.931 | 0.46 | 84.3 | 0.910 | 0.53 | 7 |
| MLR | 52 | 0.862 | 0.65 | 34.3 | 0.812 | 0.76 | 8 |
| Neutral Structures | |||||||
| PLS | 52 | 0.905 | 0.54 | 59.9 | 0.855 | 0.67 | 7 |
| MLR | 52 | 0.848 | 0.68 | 30.7 | 0.762 | 0.90 | 8 |
Number of compounds.
Correlation coefficient for training set calculations.
Standard error (RMSE) for training set calculations.
Fisher F-test for training set calculations.
Correlation coefficient for leave-one-out cross validation.
Standard error for leave-one-out cross validation predications.
Number of descriptor inputs.
For MLR modeling the pool of descriptors was deceased to 68 by removing descriptors which are highly inter-correlated with other descriptors. The SAS RSQUARE procedure was run to select descriptors up to a maximum of 8. With these settings, the procedure performs MLR regressions on all possible combinations of 1 to 8 variables. The statistically best models are reported and the model with the highest r2 for eight variables was selected.
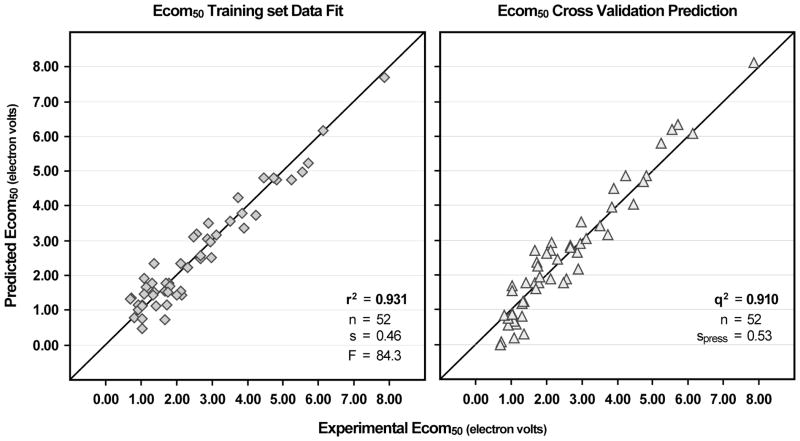
Based on these modeling results (Table 3), we focused primarily on the PLS model for protonated structures since the statistics for that model are significantly better than for the neutral structures. The direct statistics for the PLS model on protonated structures yields r2 = 0.931, s = 0.46, F = 84.3, n = 52. The leave-one-out method of cross-validation yields q2 = 0.910 and spress = 0.53. A plot of calculated versus measured Ecom50 values (Figure 3) reveals a reasonable spread of predicted values about the line that represents the measured values. In the statstical sense there are no outliers (residual > 3 sigma) resulting from the PLS model for protonated structures. Also this plot shows the sparseness of data with Ecom50 values above 3.50eV (25% of compounds covering 68% of data range) and the much higher density of values below 2.00eV (47% of compounds covering 18% of data range).
Figure 3.
Molconn modeling results for Ecom50 data giving experimental versus both calculated and predicted values from the PLS model (seven latent factors) using protonated structures. Model fitting calculation data points are given on the left and leave-one-out cross validation prediction data points are given on the right.
CODESSA modeling
We developed a preliminary Ecom50 model using CODESSA descriptors based on 3-D structure representations. CODESSA modeling calculations were initially limited to finding the best model using 5 molecular descriptors. For neutral structures, several models were found with the best 5-descriptor model giving an r2 correlation of 0.874. To obtain a model with an R2 greater than 0.9, and to be consistent with the Molconn modeling, the number of allowed descriptors was increased to 8. The best 8-descriptor CODESSA model obtained had an r2 = 0.920 (Table 4).
Table 4.
Ecom50 CODESSA MLR modeling results (protonated and neutral structures)
| Model | na | r2b | sc | Fd | q2e | inputsf |
|---|---|---|---|---|---|---|
| Protonated Structures | ||||||
| MLR | 52 | 0.943 | 0.38 | 89.6 | 0.917 | 8 |
| Neutral Structures | ||||||
| MLR | 52 | 0.920 | 0.45 | 63.2 | 0.882 | 8 |
Number of compounds.
Correlation coefficient for training set calculations.
Sigma for training set calculations.
Fisher F-test statistic for training set calculations.
Correlation coefficient for leave-one-out cross validation.
Number of descriptor inputs.
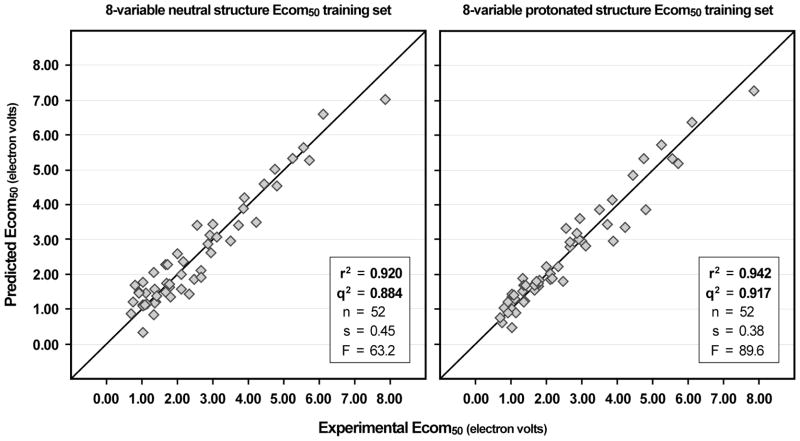
In addition to modeling neutral structures, we also used CODESSA for developing an Ecom50 model using protonated structures. Protonation sites based on both partial charge and proton affinity were assigned as described above. Appropriate structures files were created for compounds under the two criteria and CODESSA models built for Ecom50 with both sets of structures. For the criterion whereby a proton is attached to the most electrostatically negative site on the molecule, the best 8-descriptor CODESSA model had an r2 of 0.917 and q2 of 0.882. For protonation at a site assigned by highest proton affinity, the best 8-descriptor model gave an r2 of 0.943 and q2 of 0.917 (Table 4).
A comparison of the statistics of the various models suggests that the use of protonated structures was of marginal value for the CODESSA modeling procedure. The training set standard error of the model based on protonated structures is 15% smaller than for the model based on neutral structures and the correlations coefficients are different by 0.03. For the purpose of creating CODESSA based models, it is questionable whether the additional computational effort to compute a minimized structure, examine its partial negative charge distribution or assign a charge site by proton affinity, and then re-compute optimized geometries is warranted to obtain statistical improvements of that magnitude. Plots of the neutral and protonated models are given in Figure 4.
Figure 4.
CODESSA training set results for Ecom50 data. Plots show experimental versus calculated values from two 8 variable models. The left plot shows the model based on neutral structures and the right plot shows the model based on protonated structures. The q2 leave one out cross validation correlation coefficient is also given.
Retention Index Model
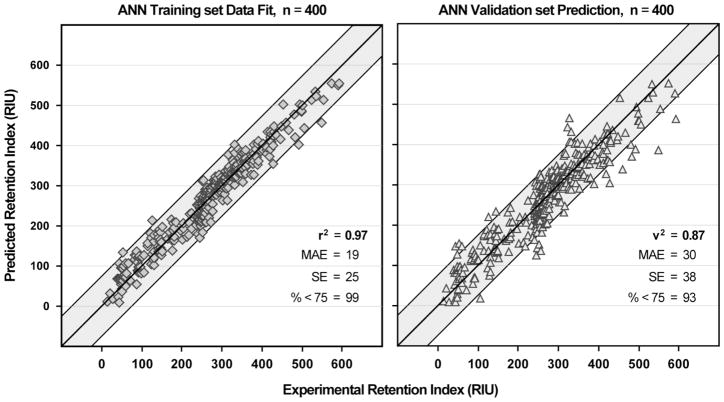
An artificial neural network (ANN) ensemble was used to generate the retention index model because previous work showed linear modeling by MLR to produce substantially inferior results30. Statistics were generated for training set fit, cross-validation prediction of the training data by 10-fold leave-10%-out, and external validation prediction on a set of compounds not used for modeling.
For each of the 4 data splits described in the methods section, each compound was used for training in 9 of the 10 train-test folds. This resulted in 36 training fit values for each compound, each calculated by a different neural net model. All 36 values were averaged to generate the calculated 4×10×10 ensemble RI value from training. Training set calculations show a high correlation with measured data with r2 = 0.95, mean absolute error (MAE) = 19 RIU, and standard error (SE) = 25 RIU. Each compound was left out of exactly one of the 4 data splits for use as validation data. Validation compounds were predicted by all 10 models from the split in which they were left out, and all 10 values were averaged to generate the predicted RI value from validation. External validation predictions are also well correlated with experimental values with v2 = 0.87, MAE = 30 RIU, and SE = 38 RIU. A total of 93% of validation predictions are within 75 RIU of the measured value (Table 5). The largest validation error was observed for bitolterol at 161 RIU.
Table 5.
Retention Index ANN modeling results
| Sub-set | na | r2b | MAEc | SEd | < 75RIUe |
|---|---|---|---|---|---|
| Train | 400 | 0.95 | 19 | 25 | 99% |
| External validation | 400 | 0.87 | 30 | 38 | 93% |
| Cross validation | 400 | 0.83 | 36 | 46 | 90% |
Number of compounds.
Square of the correlation coefficient.
Mean absolute error = 1/n * Σ(|RIpred − RIexp|).
standard error (root mean square error).
Percent of predicted values ± 75RIU of the measured value.
Each compound was used for cross validation in one of the 10 folds of each of the 3 splits where the compound was part of the fit set used for training. This resulted in three cross-validation predictions for each compound which were averaged to generate the predicted RI value from cross-validation. Being an average of only 3 predicted values, the cross validation predictions show the lowest correlation with q2 = 0.83, MAE = 36 RIU, and SE = 46 RIU. A total of 90% of cross-validation predictions are within 75 RIU of the measured value. These statistics are still quite reasonable given the small number of predictions that are averaged for this value.
Examination of the corresponding plots (Figure 5) shows that performance is similar across the RI range for both training and validation. Validation performance, however, is somewhat diminished above 400 RIU (MAE of 40 RIU as compared to 30 RIU for the overall data set) and the majority of predictions above 500 RIU are low. This could be the result of limited data in this experimental range, insufficient non-linearity in the neural net architecture, or a deficit in the description of lipophilic features likely responsible for longer retention times.
Figure 5.
Artificial Neural Network results for training set and validation set are shown with the squared correlation coefficient, mean absolute error, standard error and percent of compounds with residuals less than 75 RIU. The shaded region on the validation plot represents ± 75 RIU from the experimental value.
Given the large number of features of low population, an analysis was performed to tabulate the performance of the model predicting compounds with at least one low population feature (Table 6). The MAE for compounds with at least one low population feature is 32 RIU, which is only slightly higher than the overall MAE. The number of low population features in a compound does not appear to be a significant factor as predictions for compounds with multiple low population features appear to be of similar quality to the rest of the data. The largest MAE values occur in subsets where only one or two compounds with the feature are present in the data. These values are still well within two standard errors. The overall performance of the model in predicting compounds with low population features implies that the IGroup method of combining features with similar solution interaction characteristics into groups is at least partially successful in mitigating the issue of low population features.
Table 6.
Mean absolute error of prediction for compounds with low population features
| Group | Counta | MAEb |
|---|---|---|
| overall dataset | 400 | 30.4 |
| 1–4 low population features | 165 | 32.3 |
| 1 low population feature | 140 | 32.2 |
| 2 low population features | 23 | 33.4 |
| 3–4 low population features | 2 | 26.5 |
| Feature | Count | MAE |
|---|---|---|
| phosphate | 16 | 30.7 |
| vinyl | 13 | 22.9 |
| diazole | 12 | 48.9 |
| purine | 12 | 36.3 |
| xanthine | 9 | 20.9 |
| thioether | 9 | 40.7 |
| quaternary N+ | 9 | 33.6 |
| cytosine | 8 | 22.3 |
| carbamate | 8 | 33.6 |
| sulfonic acid | 7 | 33.4 |
| guanosine | 6 | 31.9 |
| guanadine | 6 | 33.7 |
| uracil | 6 | 28.9 |
| urea | 6 | 52.9 |
| thioanisole | 6 | 24.9 |
| pyridinium N+ | 5 | 25.2 |
| barbiturate | 4 | 25.4 |
| pyrimidine | 4 | 35.9 |
| methyluric acid | 4 | 24.0 |
| aldehyde | 3 | 15.4 |
| disulfide | 3 | 21.9 |
| guanine | 3 | 37.8 |
| hydantoin | 3 | 38.0 |
| imide | 3 | 19.8 |
| thiazole | 3 | 31.3 |
| flavin | 2 | 26.6 |
| pyrazolidine | 2 | 56.5 |
| thiophene | 2 | 35.2 |
| phosphonate | 1 | 19.0 |
| diphenylamine | 1 | 40.1 |
| oxime | 1 | 65.1 |
| sulfinic acid | 1 | 49.3 |
| sulfoxide | 1 | 8.4 |
Count of compounds with at least one example of the feature.
Mean absolute error for all compounds with at least one example of the indicated feature.
Filtering list of possible matches to unknown m/z 225 in human serum sample MSC205s
As stated previously, a compound was detected in human serum sample MSC205s with a measured protonated molecular mass of 225.1958 Da (MIMW 224.1885 Da), Ecom50 value of 2.76 eV, and RI value of 499. The Ecom50 and RI models developed in the study were used to filter 315 compounds with MIMW within ± 10 ppm of 224.1885 Da that were retrieved from the PubChem database as possible matches to the unknown. The goal of filtering was to eliminate structures that were unlikely to be a match because their predicted Ecom50 and RI values were greater than three standard errors of prediction from the measured values of the unknown.
Both the CODESSA and Molconn models were evaluated for Ecom50 filtering. The Molconn PLS model based on protonated structures was chosen for final filtering because it eliminated a larger number of the 315 candidate structures as compared to the CODESSA models, or Molconn model based on neutral structures. The improved filtering efficiency was likely a consequence of the Molconn protonated model predicting a larger range of Ecom50 values for the 315 compounds compared to the best CODESSA model (5.7 eV compared to 3.12 eV). Since the model statistics and corresponding 3 standard error filter ranges were similar, the smaller range of predicted values for the CODESSA model resulted in fewer compounds outside the boundaries of the filter range. At this point, however, the size of the Ecom50 data set is too small to draw meaningful conclusions about the potential suitability of either method as they relate to predictive models of the Ecom50 experimental feature.
Of the initial 315 compound candidate set, a total of 280 compounds passed the Ecom50 filter, having predicted values within ± 1.59 eV of the 2.76 eV measured value for the unknown (Table 7). Of these 280 compounds, 65 had predicted RI values within ± 114 RIU of the measured RI value for the unknown. These final 65 compounds were rank ordered based on the number of matching predicted CID fragments. A match was defined as a predicted CID fragment ion mass within ± 10 ppm of the mass of an experimentally observed fragment ion from the unknown. Of these 65 compounds, 11 were found to have 3 predicted fragments matches to the unknown spectrum. The same three matching predicted fragments were found in all 11 final candidates.
Table 7.
Model filtering results on 315 PubChem candidates for match to m/z 225 unknown
| Filter | rangea | passb |
|---|---|---|
| Exact mass (from PubChem) | 224.1885 Da ± 10 ppm | 315 |
| Ecom50 filter | 2.76 eV ± 1.59 eV | 280 |
| RI filter | 499 RIU ± 114 RIU | 65 |
| Predicted CID fragment | 3 matches | 11 |
range of values used for filter, compounds outside of range do not pass.
Count of compounds passing filter.
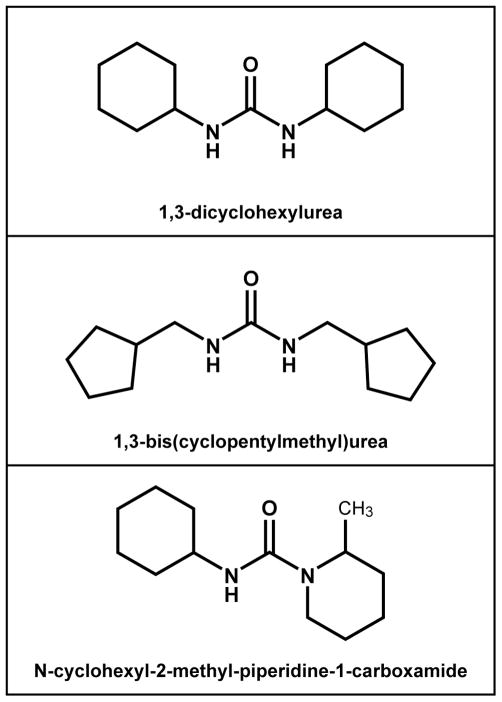
The final list of 11 compounds was rank ordered on the absolute value of the difference between the predicted RI value and the m/z 255 unknown measured RI value. The predicted RI was chosen for final candidate ranking because of the larger dataset for the RI model as compared to the Ecom50 model. The rank order of the 11 final candidate compounds is given in Table 8 along with the PubChem CID number, name and filter predicted values. None of the top four final candidates was available for commercial purchase. The top ranking final candidate, 1,3-bis(cyclopentylmethyl)urea, was synthesized according to the method described in supplemental material SI10. Samples of both 1,3-dicyclohexylurea and N-cyclohexyl-2-methyl-piperidine-1-carboxamide were commercially available and were also obtained. These three compounds (Figure 6) were analyzed experimentally to determine their Ecom50 values, HPLC retention times, and CID mass spectral profiles for direct comparison with those of the unknown compound.
Table 8.
PubChem compounds of mass 224.1885 ± 10 ppm with the closest match of predicted Ecom50, predicted RI, and predicted CID fragments to the experimental data for the unknown compound m/z 225 found in human serum
| PubChem CIDa | Compound Name | Predictedb | Predictedd | predicted fragmentsf | fragment matchesg | ||
|---|---|---|---|---|---|---|---|
| RI | a-resc | Ecom50 | a-rese | ||||
| 19071309 | 1,3-bis(cyclopentylmethyl)urea | 492 | 7 | 2.77 | 0.01 | 20 | 3 |
| 23592100 | 5,6-ditert-butyl-2-methyl-2,3-dihydro-1H-pyrimidin-4-one | 437 | 62 | 3.11 | 0.35 | 58 | 3 |
| 18384398 | 4-butyl-1-(2-ethylbutyl)-3H-imidazol-2-one | 428 | 71 | 2.72 | 0.04 | 65 | 3 |
| 18384329 | 4-butyl-1-(3,3-dimethylbutyl)-3H-imidazol-2-one | 423 | 76 | 2.73 | 0.03 | 56 | 3 |
| 4277 | 1,3-dicyclohexylurea | 413 | 86 | 3.29 | 0.53 | 24 | 3 |
| 19906651 | 1,1-dicyclohexylurea | 411 | 88 | 3.11 | 0.35 | 31 | 3 |
| 4207227 | N-cyclohexyl-4-methyl-piperidine-1-carboxamide | 410 | 89 | 3.46 | 0.70 | 25 | 3 |
| 2903296 | N-cyclohexyl-2-methyl-piperidine-1-carboxamide | 410 | 89 | 3.48 | 0.72 | 46 | 3 |
| 896432 | (3S)-N-cyclohexyl-3-methyl-piperidine-1-carboxamide | 409 | 90 | 3.48 | 0.72 | 31 | 3 |
| 21027155 | 1-cyclohexyl-3-[(E)-4-methylpent-2-enyl]urea | 396 | 103 | 2.63 | 0.13 | 26 | 3 |
| 20497885 | 1-allyl-1-cyclohexyl-3-propylurea | 385 | 114 | 2.77 | 0.01 | 66 | 3 |
All compounds have the same molecular formula C13H24N2O. PubChem CID number.
RI model prediction.
Absolute residual, absolute value of difference between RI prediction and measured RI value for unknown.
Ecom50 model prediction.
Absolute residual, absolute value of difference between Ecom50 prediction and measured Ecom50 value for unknown
Number of fragment ions predicted by Mass Frontier software.
Number of predicted CID fragment ions matching measure fragments from unknown. The following matching fragments were predicted in all 11 candidates [predicted(experimental)], 83.0855(83.0860), 100.1121(100.1117), 143.1179(143.1173).
Figure 6.
Three C13H24N2O isomer candidate compounds resulting from the PubChem database filtering process.
Confirmation of 1, 3-dicyclohexylurea in human serum
The MSC205s sample extract containing the unknown m/z 225 ion peak and solutions of standards for the three candidate compounds were sequentially analyzed on the HPLC system in MS/MS mode isolating the m/z 225 as a precurson ion and using a collision energy of 25 eV. For Ecom50 determinations, samples and reference standards were analyzed in duplicate on the same system collecting the CID spectrum at 17.0, 19.0 and 21.0 eV at 0.5 second intervals. The retention times, calculated Ecom50 values, and CID fragmentation profile of the unknown were compared to those of each standard. A linear regression correlation coefficient between the unknown CID spectrum and that of each standard spectrum was calculated and used as a measure of the similarity CID spectral profiles. The HPLC retention and mass spectral data for the unknown compound in sample MSC205s and for candidate compounds 1,3-bis(cyclopentylmethyl)urea, 1,3-dicyclohexylurea, and N-cyclohexyl-2-methyl-piperidine-1-carboxamide are shown in Table 9 and Figure 7.
Table 9.
HPLC retention, Ecom50, and molecular ion mass for m/z 225 ion in test sample MSC205s and candidate compounds
| Sample ID | RTa | Ecom50b | (M+H+)c |
|---|---|---|---|
| MSC205s | 12.947 ± 0.008 | 2.68 ± 0.00 | 225.1958 |
| 4277 | 12.946 ± 0.000 | 2.67 ± 0.01 | 225.1953 |
| 19071309 | 13.480 ± 0.008 | 2.75 ± 0.03 | 225.1952 |
| 2903296 | 13.111 ± 0.003 | 1.99 ± 0.03 | 225.1966 |
MSC205s = human serum sample with m/z 225 unknown
4277 = 1,3-dicyclohexylurea
19071309 = 1,3-bis(cyclopentylmethyl)urea
2903296 = N-cyclohexyl-2-methyl-piperidine-1-carboxamide
Measured HPLC retention time in minutes.
Measured Ecom50 value in eV.
Molecular ion mass (for mono-protonated ion).
Figure 7.
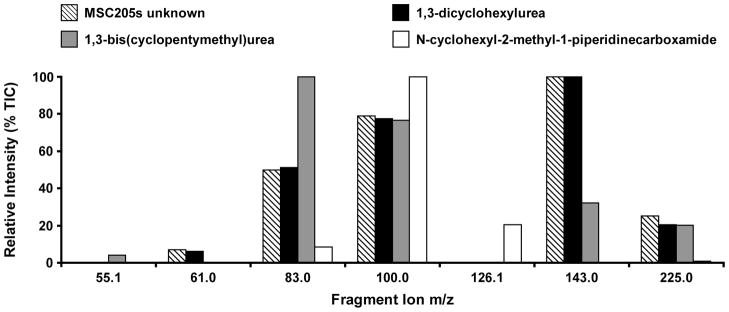
Comparison of the CID fragmentation profile of unknown m/z 255 compound in MSC205s to those of the 1,3-bis(cyclopentymethyl)urea, 1,3-dicyclohexylurea, and N-cyclohexyl-2-methyl-piperidine-1-carboxamide standards.
All three candidate compounds are isomers and thus have the same theoretical protonated MIMW of 225.1961 Da. During the confirmation analysis the measured protonated MIMW of 1,3 dicyclohexylurea was 6 ppm of the theoretical value; whereas, the measured protonated MIMW of 1,3-bis(cyclopentylmethyl)urea and N-cyclohexyl-2-methyl-piperidine-1-carboxamide differered from this value by 2 and 11 ppm, respectively. The measure protonated MIMW of the unknown compound differed from the theoretical mass of the reference compounds by 2 ppm. The standard deviation of the mass accuracy of the mass spectrometer used in this study was determined in our laboratory to be −4.4 ± 8.9 ppm.
Table 9 lists the average retention times obtained for the unknown and 3 reference compounds. A T-test comparison of the average retention time of the unkown to that of the standards indicated a significant (p<0.005) difference between the retention times of the unknown and 1,3- bis(cyclopenylymethyl)urea and N-cyclohexyl-2-methyl-piperidine-1-carboxamide but no significant (p<0.005) difference between the retention time of the unknown and 1,3-dicyclohexylurea.
Figure 7 shows that the molecular ion peak and 4 fragment ion peaks in the 25 eV CID spectrum of the unknown are also present in the spectra of 1,3-dicyclohexylurea. The CID spectrum of 1,3-bis(cyclopentylmethyl)urea had 3 fragment ion peaks and the moleclar ion peak in common with that of the unknown. This compound also had an additional low intenisty fragment ion peak at m/z 55.1 that was not detected in the unknown spectrum. The CID spectrum of N-cyclohexyl-2-methyl-piperidine-1-carboxamide shows a very low intensity ion peak for the moleuclar ion, two ion peaks in common with the CID spectrum of the unknown and one ion peak that is not present in the other 2 standards or the unknown CID spectra. The measured masses of all common fragment ion peaks between spectra differed by less than 17 ppm. For a complete qualitative comparison of the mass spectral profiles, the correlation coefficient of the relative intensities of the matching ion peaks between the CID spectrum of the unknown and those of the three standards was calculated. The cross correlation value approaches 1 as the relative intensities of matched ion peaks between two spectra approach the same values, thus indicating a close match between the two profiles. The cross correlation between the unknown and 1,3-dicyclohexylurea is 0.9977, the cross correlation between the unknown and 1,3-bis(cyclopentylmethyl)urea is 0.4033 and the cross correlation between N-cyclohexyl-2-methyl-piperidine-1-carboxamide is 0.1563.
The Ecom50 value for the unknown (Table 9) was not significantly (T-test, p<0.005) different from that of 1,3 dicyclohexylurea or 1,3 bis(cyclopenylmethyl)urea, but was significantly (T-test, p<0.005) different from the Ecom50 value of N-cyclohexyl-2-methyl-piperidine-1-carboxamide.
These data suggest that the overall chromatographic and mass spectral characteristics of the unknown are consistent with those of 1,3-dicyclohexylurea but not with 1,3 bis(cyclopenylmethyl)urea or N-cyclohexyl-2-methyl-piperidine-1-carboxamide.
Discussion
In this study we obtained new experimental data and developed two new models designed to filter large chemical databases for possible matches to an experimentally observed unknown. This work also served as a test of the overall filtering methodology in the identification and confirmation of 1,3-dicyclohexylurea as an unknown compound in a human serum sample observed during HPLC/MS analysis.
A significant aspect of the Ecom50 modeling described here is the use of input variables based largely on molecular structure but not specifically designed to describe bond breaking. Both the Molconn and CODESSA structure descriptors are representations of molecular structure information, rather than quantities related to bond energy. The encoded information includes atom and bond arrangement, electron distribution, skeletal ramification, and geometric quantities, encoded at both atom and bond levels. Both MLR and PLS methods were used to develop models with correlation coefficients > 0.93 for a truly eclectic data set. This outcome could be considered surprising given the absence of descriptor information specifically related to bond energy. The modeling results suggest that use of the molecular structure features described herein is a workable basis for model development of the novel molecular characteristic, Ecom50. Furthermore, because the modeling descriptors are a representation of molecular structure, future models based on a much larger data set with better distribution in chemical data space may permit elucidation of structure features most strongly related to fragmentation. Molconn structure descriptors, based on molecular topology, do not require geometry optimization and CODESSA descriptors use a semi-empirical level quantum chemical determination of geometry. Because the statistics of the CODESSA models were not significantly different from the Molconn PLS model, and the CODESSA models were not as effective in filtering the candidate database, the use of semi-empirical optimized geometry did not appear to provide a significant advantage for modeling this dataset.
Molconn descriptor calculations are not computationally intensive. This permits the development of high speed methods for filtering and compound identification as demonstrated in this work. However, since the best Molconn model made use of protonated input structures, additional work is required to more completely develop a computationally expedient algorithm for assigning the site of protonation. The CODESSA model does not have the limitation of requiring an assigned protonation site, but is computationally more intensive and did not prove to be as effective in filtering. Of the 315 structures taken from the PubChem database with an identical monoisotopic molecular weight to the unknown compound, the Molconn PLS protonated Ecom50 model was able to filter out 35, or ~11%. This is not an especially large percentage, but given that this is a linear model based on very limited data, there is reason to believe that a non-linear model based on a much larger dataset will provide significantly better filtering. Neither the Molconn or CODESSA method provided optimum results, but we assert that, with limited data available for modeling, it is far too early in the investigation to draw any meaningful conclusions about which method might finally prove more effective. It remains to be seen if the findings of this study hold true when modeling a larger database that is more normally populated in both structure and activity space.
The model of retention index was found to show reasonable proficiency in differentiating among a group of structures with identical monoisotopic molecular weight and molecular formula. Of the 280 structures from the PubChem database that pass the Ecom50 filter, the RI model was able to filter out an additional 212 compounds (based on a 99.8% confidence interval). Used in combination with the Ecom50 model, approximately 80% of the candidate structures from the PubChem database were eliminated.
As was found in our previous RI modeling study30, good validation statistics were obtained despite an 8 to 1 ratio between the number of training rows and input descriptors. A 10 to 1 ratio is one of the least conservative ratios commonly deemed acceptable. The question arises as to whether or not maintaining a small number of descriptors relative to the number of training rows is actually necessary in order for a model to have acceptable validation statistics or if some other effect is involved. It is possible that a ratio of less than 10 to 1 is sufficient in some cases, but it is also possible that the population of input descriptors should be considered along with the number of descriptors. Looking at the non-zero population, on average, compounds have a non-zero input value for only 18.6 of the 33 descriptors (σ = 2.98, min = 10, max = 26). A ratio of much closer to 15 to 1 is maintained if only descriptors with a non-zero values are counted. This 15 to 1 ratio would pass the much more conservative (√n)−1 rule. Eriksson suggests that models do not have a significant number of irrelevant inputs when the difference between train and validation correlation coefficients does not exceed 0.254, which is well satisfied in this case. The quality validation results of the RI study suggest that the population of input descriptors should be taken into account when evaluating the ratio of inputs to training rows or if the traditionally accepted ratios are overly conservative.
The data used for the RI model is still lacking in structures with long retention times. Retention times of over 1000 RIU have been observed for some endogenous metabolites, yet the largest retention index in the data used was 593 RIU. The addition of compounds with longer retention times would likely give access to a larger domain of applicability for the final model. The data set is also lacking a significant number of simple structures which may provide useful information to the model about more complex structures, which can be considered as various combinations of simple compounds. As stated in the methods section, there are a large number of features of low population in the dataset. Although we suggest that the good validation statistics of the RI model indicate that the IGroups method was reasonably successful in mitigating the commonly seen detrimental effects of under populated features, it is also likely that increasing the population of these features would enhance the predictive performance of the model. Measurement of additional compounds will also provide invaluable independent validation data for the current model. Such data would help define the applicability domain by unambiguously identifying compounds the model makes accurate predictions for. Revealing the strengths and weakness of current models will assist in tuning both the input descriptors and neural net modeling process. Though we believe the validation criteria used in this study were well constructed, validation with data on which the model is partially based will never enumerate predictive capability in the way that newly measured data can.
As mentioned previously, 1,3-dicyclohexylurea has not been previously detected in human serum, so our finding is rather surprising. 1,3-dicyclohexylurea is a known by-product formed during automated solid-phase peptide synthesis and thus the potential for contamination in our samples should be considered. However, we feel this is unlikely for several reasons. First, this compound was not found in all samples, even those coming from the same source. Second, in samples where it was found, the amounts were highly variable, ranging from barely detectible to approximately 10 μg/L. Third, we found this compound in human serum and CSF but not in mouse serum using the same methodology. Interestingly, 1,3-dicyclohexylurea is a potent inhibitor of soluble epoxide hydrolase with an Ki of approximately 30 nM55. Soluble epoxide hydrolase has been implicated in cardiovascular disease56 and inflammation in mammals57, and thus soluble epoxide hydroase inhibitors have been considered for various cardiovascular indications58. On the other hand, soluble epoxide hydrolase inhibitors have recently been shown to dramatically enhance tumor metastasis in rodent models59. We estimate that in the human samples used in our study, the concentration of 1,3-dicyclohexylurea was as high as 10 μg/L or 45 nM, which is approximately equivalent to its Ki for soluble epoxide hydrolase. Although our findings will require verification in larger studies, it is temping to speculate that by regulating soluble epoxide hydrolase activity in vivo, endogenously produced 1,3-dicyclohexylurea may be physiologically significant. The endogenous source of 1,3-dicyclohexylurea remains unknown. However, it has been previously identified in two species of plants, Portulaca oleracea60 and Toddalia asiatica61, suggesting the existence of a biochemical pathway for its synthesis in vivo.
This study illustrates the use of predictive algorithms for Ecom50, retention index, and mass spectral fragmentation in discriminating among compounds retrieved from structural databases to aid in the identification of compounds detected in biological samples. A unique feature of these predictive algorithms is that they can be applied to any chemical database that provides structure files. Therefore, structure identification is not limited to the contents of small biological databases containing premeasured experimental variables. Thus, as demonstrated here, unknown endogenous compounds can be successfully identified. This would not be possible using experimental libraries of endogenous compounds or searching databases comprised of known biological compounds.
Although this approach is promising, there remain several limitations. The three predictive models used (Ecom50, Retention index, and CID fragmentation) will require considerable improvements; Ecom50 by using larger numbers of compounds and neural network modeling, RI by increasing the number and diversity of compounds used for developing the neural net model, and CID fragmentation by including bond energy considerations. Our goal is to increase model predictive accuracy so that the 3 standard error filter ranges are approximately ± 0.75 eV for the Ecom50 model and ± 75 RIU for the RI model. In addition, this approach still relies on the assumption that the unknown compound is contained in the chosen database. Although the presence of matches to an unknown seems likely for many mammalian metabolites, it is clearly not true for metabolomic studies of plants or other phyla for which potential metabolite lists are far from complete. Thus, the incorporation of structure prediction software that uses an elemental formula as a starting point will allow this procedure to become database-enabled rather than database-dependent.
Clearly, the addition of models for other orthogonal experimental features would be advantageous in this regard. There are two reasons for this. First, no single experimental feature, or even combination of featuers, is unique to a single compound; multiple compound candidates will have the same exact mass and RI value, for example. Second, since models are created using data sets with a limited selection of structures, compared to the databases that are screened with them, it is unlikely that predicted values will be as accurate as the corresponding experimental measurement. Increasing the number of orthogonal chemical descriptors could allow for enhanced discrimination between candidates without the need for models based on an exhaustive and impractical number of compounds. However, we emphasize that even in their current state, the models described here reduced the number of potential matches for the unknown by approximately 80%, and by including CID spectra matching, approximately 96% of the 315 candidate compounds were eliminated. Additionally, the combination of three experimental features used for final verification of the unknown was successful in unambiguously differentiating among three structurally similar isomers in the final candidate list without the need for purifying the unknown compound. Taken together, these results suggest promise of this approach for identifying the structures of unknown compounds in non-targeted metabolomics research.
Supplementary Material
Acknowledgments
Funding for this work was provided by NIH (1R01GM087714); Pfizer Inc., Groton, CT; The University of Connecticut Foundation and by an American Foundation for Pharmaceutical Education Predoctoral Fellowship to Tziporah M. Kertesz.
Footnotes
Supporting Information Available
SI1: Measured Ecom50 data, material sources, and model values for neutral structures. SI2: Measured Ecom50 data, material sources, and model values for protonated structures. SI3: Measured retention index data, material sources and model values. SI4-7: Tables of Molconn descriptors used in Ecom50 PLS model. SI8-9: Tables of Molconn descriptors used in RI model. SI10: Description of the procedure used to synthesize and confirm 1,3-bis(cyclopentylmethyl)urea. SI11: Structures of 8 final candidates that were not obtained and measured. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Kertesz TM, Hill DW, Albaugh DR, Hall LH, Hall LM, Grant DF. Database searching for structural identification of metabolites in complex biofluids for mass spectrometry-based metabonomics. Bioanalysis. 2009;1(9):1627–1643. doi: 10.4155/bio.09.145. [DOI] [PubMed] [Google Scholar]
- 3.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81(16):6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 4.Smith CA, O’Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 5.Wishart DS, Tzur D, Knox C, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35(Database issue):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kind T, Scholz M, Fiehn O. How large is the metabolome? A critical analysis of data exchange practices in chemistry. PLoS One. 2009;4(5):e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill DW, Kertesz TM, Fontaine D, Friedman R, Grant DF. Mass spectral metabonomics beyond elemental formula: chemical database querying by matching experimental with computational fragmentation spectra. Anal Chem. 2008;80(14):5574–5582. doi: 10.1021/ac800548g. [DOI] [PubMed] [Google Scholar]
- 9.Wolf S, Schmidt S, Müller-Hannemann M, Neumann S. In silico fragmentation for computer assisted identification of metabolite mass spectra. Bioinformatics. 2010;11:148–160. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumari S, Stevens D, Kind T, Denkert C, Fiehn O. Applying in-silico retention retention index mass spectra matching for identification of unknown metabolites in accurate mass GC-TOF mass spectrometry. Analytical Chemistry. 2011;83:5895–5902. doi: 10.1021/ac2006137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y, Cheng D, Eisner R, Gautam B, Tzur D, Sawhney S, Bamforth F, Greiner R, Li L. The human cerebrospinal fluid metabolome. J Chromatogr B Analyt Technol Biomed Life Sci. 2008 Aug 15;871(2):164–73. doi: 10.1016/j.jchromb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memboeuf A, Jullien L, Lartia R, Brasme B, Gimbert Y. Tandem mass spectrometric analysis of a mixture of isobars using the survival yield technique. Journal of the American Society for Mass Spectrometry. 2011;22:1744–1752. doi: 10.1007/s13361-011-0195-8. [DOI] [PubMed] [Google Scholar]
- 15.Kertesz TM, Hall LH, Hill DW, Grant DF. CE50: quantifying collision induced dissociation energy for small molecule characterization and identification. J Am Soc Mass Spectrom. 2009;20(9):1759–1767. doi: 10.1016/j.jasms.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 16. [accessed 2011];The PubChem Project, Compounds Database. http://pubchem.ncbi.nlm.nih.gov/
- 17.O’Neil Maryadele J., editor. The Merck Index. 14. Whitehouse Station, NJ: Merck & Co; 2006. [Google Scholar]
- 18.winMolconn, version 1.1.14. Hall Associates Consulting; Quincy, MA: 2008. [Google Scholar]
- 19.SAS, 9.1. SAS Institute; Cary, NC: [Google Scholar]
- 20.CODESSA 2.7.16, © 1995–2011 Semichem, Inc., 12456 W 62nd Terrace Suite D, Shawnee, KS 66216. http://semichem.com.
- 21.AMPAC 9, © 1992–2011 Semichem, Inc., 12456 W 62nd Terrace Suite D, Shawnee, KS 66216. http://semichem.com.
- 22.Wang J, Aubry A, Bolgar MS, Gu H, Olah TV, Arnold M, Jemal M. Effect of mobile phase pH, aqueous-organic ratio, and buffer concentration on electrospray ionization tandem mass spectrometric fragmentation patterns: implications in liquid chromatography/tandem mass spectrometric bioanalysis. Rapid Communications in Mass Spectrometry. 2010;24:3221–3229. doi: 10.1002/rcm.4748. [DOI] [PubMed] [Google Scholar]
- 23.Dewar MJS, Zoebisch EG, Healey EF, Stewart JJP. AM1: a new general-purpose quantum mechanical molecular model. J Am Chem Soc. 1985;107:3902–3909. [Google Scholar]
- 24.Dewar MJS, Zoebisch EG. Extension of AM1 to the halogens. J Mol Struct (Theochem) 1988;180:1–21. [Google Scholar]
- 25.Dewar MJS, Jie C. AM1 parameters for phosphorus. J Mol Struct (Theochem) 1989;187:1–13. [Google Scholar]
- 26.Dewar MJS, Yuan YC. AM1 parameters for sulfur. Inorg Chem. 1990;29:3881–3890. [Google Scholar]
- 27. [Accessed 8-02-11]; http://webbook.nist.gov/chemistry/pa-ser.html.
- 28.Rocha GB, Freire RO, Simas AM, Stewart JJP. RM1: A reparameterization of AM1 for H, C, N, O. P, S F, Cl, Br, and I. J Comput Chem. 2006;27:1101–1111. doi: 10.1002/jcc.20425. [DOI] [PubMed] [Google Scholar]
- 29.Spartan ‘08. WAVEFUNCTION, Inc; Irvine, CA: [Google Scholar]
- 30.Albaugh DR, Hall LM, Hill DW, et al. Prediction of HPLC retention index using artificial neural networks and IGroup E-State indices. J Chem Inf Model. 2009;49(4):788–799. doi: 10.1021/ci9000162. [DOI] [PubMed] [Google Scholar]
- 31.Hall LH, Kier LB, Hall LM. ADME-Tox Approaches, Electrotopological State indices to Assess Molecular Absorption, Distribution, Metabolism, Excretion, and Toxicity. In: Triggle David J, Taylor John B., editors. Comprehensive Medicinal Chemistry II. Vol. 5. Elsevier; Oxford, UK: 2007. pp. 555–576. [Google Scholar]
- 32.Hall LH, Kier LB, Hall LM. Computer Assisted Drug Design, Topological Quantitative Structure-Activity Relationship Applications: Structure Information in Drug Discovery. In: Triggle David J, Taylor John B., editors. Comprehensive Medicinal Chemistry II. Vol. 4. Elsevier; Oxford, UK: 2007. pp. 537–574. [Google Scholar]
- 33.Hall LH, Kier LB. Molecular Structure Description: The Electrotopological State. Academic Press; San Diego, CA: 1999. [Google Scholar]
- 34.Kier LB, Hall LH. Topological Indices and Related Descriptors in QSAR and QSPR. In: James Devillers James, Balaban Alexandru T., editors. The Kappa Indices for Modeling Molecular Shape and Flexibility. Gordon and Breach; Reading, UK: 1999. pp. 455–489. [Google Scholar]
- 35.Hall LH. A Structure-Information Approach to Prediction of Biological Activities and Properties. Chem Biodiversity. 2004;1:183–201. doi: 10.1002/cbdv.200490010. [DOI] [PubMed] [Google Scholar]
- 36.Hall LH, Hall LM, Kier LB, Parham ME, Votano JR. Interpretation of the role of the Electrotopological State and Molecular Connectivity Indices in the prediction of physical properties and ADME-Tox behavior. Case study: Human Plasma Protein Binding. In: Testa Bernard, Turski Les., editors. Virtual ADMET Assessment in Target Selection and Maturation; Proceedings of the Solvay Conference; Luzerne, Switzerland. IOS Press; 2006. pp. 67–100. Proceedings of the Solvay Conference, Luzerne, Switzerland (2006) [Google Scholar]
- 37.Ränner S, Lindgren F, Geladi P, Wold S. A PLS Kernel Algorithm for Data Sets with Many Variables and Fewer Objects. Journal of Chemometrics. 1994;8:111–125. [Google Scholar]
- 38.Tobias R. An Introduction to Partial Least Squares Regression. Proceedings of the Twentieth Annual SAS Users Group International Conference; Cary, NC: SAS Institute Inc; 1995. pp. 1250–1257. [Google Scholar]
- 39.Katritzky AR, Karelson M, Lobanov VS. SPR as a means of predicting and understanding chemical and physical properties in terms of structure. Pure Appl Chem. 1997;69:245–248. [Google Scholar]
- 40.Katritzky AR, Oliferenko AA, Oliferenko PV, Petrukhin R, Tatham DB. A general treatment of solubility: 1. The QSPR correlation of solvation free energies of single solutes in series of solvents. J Chem Inf Comput Sci. 2003;43:1794–1805. doi: 10.1021/ci034120c. [DOI] [PubMed] [Google Scholar]
- 41.Acevedo-Martinez J, Escalona-Arranz JC, Villar-Rojas A, et al. Quantitative study of the structure-retention index relationship in the imine family. J Chromatogr A. 2006;1102(1–2):238–244. doi: 10.1016/j.chroma.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Guo W, Lu Y, Zheng XM. The predicting study for chromatographic retention index of saturated alcohols by MLR and ANN. Talanta. 2000;51(3):479–488. doi: 10.1016/s0039-9140(99)00301-x. [DOI] [PubMed] [Google Scholar]
- 43.Hadjmohammadi MR, Fatemi MH, Kamel K. Quantitative structure-property relationship study of retention time of some pesticides in gas chromatography. J Chromatogr Sci. 2007;45(7):400–404. doi: 10.1093/chromsci/45.7.400. [DOI] [PubMed] [Google Scholar]
- 44.Jalali-Heravi M, Kyani A. Use of computer-assisted methods for the modeling of the retention time of a variety of volatile organic compounds: a PCA-MLR-ANN approach. J Chem Inf Comput Sci. 2004;44(4):1328–1335. doi: 10.1021/ci0342270. [DOI] [PubMed] [Google Scholar]
- 45.Konoz E, Fatemi MH, Faraji R. Prediction of Kovats retention indices of some aliphatic aldehydes and ketones on some stationary phases at different temperatures using artificial neural network. J Chromatogr Sci. 2008;46(5):406–412. doi: 10.1093/chromsci/46.5.406. [DOI] [PubMed] [Google Scholar]
- 46.Quiming NS, Denola NL, Saito Y, Jinno K. Multiple linear regression and artificial neural network retention prediction models for ginsenosides on a polyamine-bonded stationary phase in hydrophilic interaction chromatography. J Sep Sci. 2008;31(9):1550–1563. doi: 10.1002/jssc.200800077. [DOI] [PubMed] [Google Scholar]
- 47.Rouhollahi A, Shafieyan H, Ghasemi JB. A QSPR study on the GC retention times of a series of fatty, dicarboxylic and amino acids by MLR and ANN. Ann Chim. 2007;97(9):925–933. doi: 10.1002/adic.200790077. [DOI] [PubMed] [Google Scholar]
- 48.Montgomery DC. Design and Analysis of Experiments. 5. John Wiley &. Sons, Inc; New York: 2008. Experiments with Random Factors; pp. 511–556. [Google Scholar]
- 49.Ertl P, Rohde B, Selzer P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J Med Chem. 2000;43 (20):3714–371. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 50.winMolconn, version 1.2.2.1. Hall Associates Consulting; Quincy, MA: 2011. [Google Scholar]
- 51.Emergent, Version 5.2. University of Colorado; Bolder, CO: [Google Scholar]
- 52.Aisa B, Mingus B, O’Reilly R. The Emergent Neural Modeling System. Neural Networks. 2008;21:1045–1212. doi: 10.1016/j.neunet.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 53.HighChem. HighChem Mass Frontier 5.0. 2007 http://www.highchem.com.
- 54.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi- and Megavariate Data Analysis, Principles and Applications. Umetrics AB; Umeå Sweeden: 2001. p. 107. [Google Scholar]
- 55.Morisseau C, Goodrow MH, Dowdy D, Zheng J, Greene JF, Sanborn JR, Hammock BD. Potent urea and carbamate inhibitors of soluble epoxide hydrolases. Proc Natl Acad Sci USA. 1999;96(16):8849–8854. doi: 10.1073/pnas.96.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8(10):794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elmarakby AA, Faulkner J, Al-Shabrawey M, Wang MH, Maddipati KR, Imig JD. Deletion of the soluble epoxide hydrolase gene improves renal endothelial function and reduces renal inflammation and injury in streptozotocin-induced type 1 diabetes. Am J Physiol Regul Integr Comp Physiol. 2011;301(5):R1307–17. doi: 10.1152/ajpregu.00759.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imig JD. Epoxides and soluble epoxide hydrolase in cardiovascular physiology. Physiol Rev. 2012;92(1):101–30. doi: 10.1152/physrev.00021.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panigrahy D, Edin ML, Lee CR, Huang S, Bielenberg DR, Butterfield CE, Barnés CM, Mammoto A, Mammoto T, Luria A, Benny O, Chaponis DM, Dudley AC, Greene ER, Vergilio JA, Pietramaggiori G, Scherer-Pietramaggiori SS, Short SM, Seth M, Lih FB, Tomer KB, Yang J, Schwendener RA, Hammock BD, Falck JR, Manthati VL, Ingber DE, Kaipainen A, D’Amore PA, Kieran MW, Zeldin DC. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J Clin Invest. 2012;122(1):178–91. doi: 10.1172/JCI58128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rashed AN, Afifi FU, Shaedah M, Taha MO. Investigation of the active constituents of Portulaca oleraceae L. (Portulacaceae) growing in Jordan. Pakistan J Pharm Sci. 2004;17(1):37–45. [PubMed] [Google Scholar]
- 61.Ian-Lih Tsai IL, Song-Chwan Fang SC, Tsutomu Ishikawa T, Chang CT, Chen I-S. N-cyclohexyl amides and a dimeric coumarin from formosanToddalia asiatica. Phytochemistry. 1997;44(7):1383–1386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.