Abstract
Background
A century ago, Chestnut Blight Disease (CBD) devastated the American chestnut. Backcross breeding has been underway to introgress resistance from Chinese chestnut into surviving American chestnut genotypes. Development of genomic resources for the family Fagaceae, has focused in this project on Castanea mollissima Blume (Chinese chestnut) and Castanea dentata (Marsh.) Borkh (American chestnut) to aid in the backcross breeding effort and in the eventual identification of blight resistance genes through genomic sequencing and map based cloning. A previous study reported partial characterization of the transcriptomes from these two species. Here, further analyses of a larger dataset and assemblies including both 454 and capillary sequences were performed and defense related genes with differential transcript abundance (GDTA) in canker versus healthy stem tissues were identified.
Results
Over one and a half million cDNA reads were assembled into 34,800 transcript contigs from American chestnut and 48,335 transcript contigs from Chinese chestnut. Chestnut cDNA showed higher coding sequence similarity to genes in other woody plants than in herbaceous species. The number of genes tagged, the length of coding sequences, and the numbers of tagged members within gene families showed that the cDNA dataset provides a good resource for studying the American and Chinese chestnut transcriptomes. In silico analysis of transcript abundance identified hundreds of GDTA in canker versus healthy stem tissues. A significant number of additional DTA genes involved in the defense-response not reported in a previous study were identified here. These DTA genes belong to various pathways involving cell wall biosynthesis, reactive oxygen species (ROS), salicylic acid (SA), ethylene, jasmonic acid (JA), abscissic acid (ABA), and hormone signalling. DTA genes were also identified in the hypersensitive response and programmed cell death (PCD) pathways. These DTA genes are candidates for host resistance to the chestnut blight fungus, Cryphonectria parasitica.
Conclusions
Our data allowed the identification of many genes and gene network candidates for host resistance to the chestnut blight fungus, Cryphonectria parasitica. The similar set of GDTAs in American chestnut and Chinese chestnut suggests that the variation in sensitivity to this pathogen between these species may be the result of different timing and amplitude of the response of the two to the pathogen infection. Resources developed in this study are useful for functional genomics, comparative genomics, resistance breeding and phylogenetics in the Fagaceae.
Background
The family Fagaceae includes more than 900 species in nine genera [1] found in temperate, subtropical, and tropical regions of the world [2]. In the Western Hemisphere, the Fagaceae range from southern Canada to Colombia [1] where they grow as tall trees or, occasionally, as shrubs. The Fagaceae include major species such as chestnuts, oaks, and beeches [2]. The Fagaceae have significant ecological and economic value. They are predominant species of most hardwood forests in the Northern Hemisphere. Nuts of Fagus (beeches), Castanea (chestnuts), Quercus (oaks) and most Castanopsis species are important sources of food for forest animals and produce a high quality extractable oil [3]. The Fagaceae are important as a timber resource for construction, telephone poles, floors, furniture, cabinets, and other applications. American chestnut played an especially important economic and ecological role by providing food to various insects, birds, and mammals, and food, fiber, and wood for rural communities, prior to the introduction of the Chestnut Blight Disease (CBD) early in the 20th century [4].
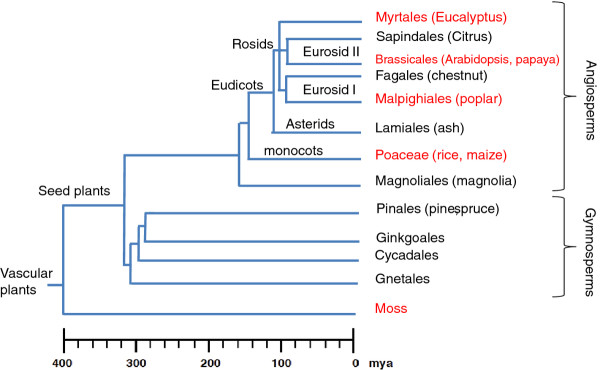
Fossil dating reveals that species of the order Fagales existed 84 million years ago (Mya) [5]. The time of divergence of the families of the Fagales has been estimated to be as early as 103 mya [6]. Fagaceae species hold a key position in the phylogeny of angiosperms (Figure 1); they cluster closely to Cucurbitales in the Eurosid I group, which includes several model plant species, such as Medicago truncatula, Glycine max, Populus trichocarpa, and Eucalyptus grandis for which complete or nearly complete whole-genome sequences exist. The close phylogenetic relationships of several model plants should make members of the Fagaceae good models for comparative genomics between woody and herbaceous species.
Figure 1.
Simplified phylogeny of the major groups of woody plants. Species for which genome sequencing has been completed are indicated in red. (Modified from [7].
Genomic resources for the Fagaceae also have practical applications. CBD is a stem canker disease caused by the fungus Cryphonectria parasitica. The pathogen infects stem tissues and kills the above ground portions of trees by girdling the cambium. Today, American chestnut exists primarily as an understory shrub, repeatedly sprouting from the root collar of blight-topped trees [8,9]. Programs to breed timber-type blight resistant chestnut were initiated after appearance of the blight [8], but failed to produce the desired timber-type tree with acceptable levels of resistance. In 1983, a program was initiated by the American Chestnut Foundation [10] to introgress genes from blight-resistant Asian chestnut species into American chestnut via backcross breeding [11]. The American Chestnut Cooperators Foundation, has made crosses between pure American chestnut trees that have limited resistance [8] for future selection. Introductions of hypovirulent fungal genotypes of C. parasitica [12], have successfully controlled the severity of blight in Europe, but have had only limited success in North America. Understanding the molecular basis of resistance to the CBD could enable the eventual restoration of American chestnut to the eastern North American forests [13].
The Genomic Tool Development for the Fagaceae (GTDF) project [14] was organized to develop genomic resources for several North American Fagaceae species to help address major forest health problems of American chestnut and other trees within this botanical family. The over 1.5 million cDNA sequences generated in this study for American and Chinese chestnut have been used to advance knowledge about the genetics of resistance to the CBD. A preliminary comparison of transcriptome sequences of healthy and blight infected American chestnut and Chinese chestnut resulted in the identification of potential candidate disease resistance genes [15]. However, the 2009 report was based on only 13,000 and 15,000 contigs for American chestnut and Chinese chestnut, respectively, available at the time. We now report on over 83,000 contigs in total for the two species, which includes 9000 Sanger sequences from Chinese chestnut in addition to 454 sequence reads. Moreover, the assembly of transcript contigs was performed using the SeqMan™ NGen™ v1.2 software (DNAStar, Inc) which provided better assembly of the 454 reads than the version of the Newbler program available in 2008. The orthologs used for comparison of DTA genes between American and Chinese chestnut were better defined in this study through the use of a reciprocal best hits analysis. The DEGseq software package [16] used for assessing transcript abundance (TA) includes statistical analyses that make it a more powerful tool than the one used the previous analysis.
This paper reports on further analysis of the transcriptome of chestnut and the identification of candidate genes involved in the defense against CBD. The coverage of the transcriptome was analyzed for number of genes identified, sequence length, gene family size, and similarity to the transcriptome of other woody and herbaceous species. The transcript contigs include full length coding sequences from American and Chinese chestnut respectively which represents a valuable resource for genomic studies in the Fagaceae. The analysis of the differential transcript abundance (DTA) of chestnut genes using DEGseq [16] allowed the identification of hundreds of defense related candidate genes with DTA in canker versus healthy stem tissues. A small set of candidate genes for resistance to CBD were verified for their DTA using real time quantitative RT-PCR.
Results
Sequencing summary
More than 1.5 million (1,526,670) reads were generated corresponding to 400 million nucleotides of cDNA sequences from the two studied species. Transcript contigs were assembled from the pyrosequencing reads using Newbler software (Roche 454) and designated version 1 on the Fagaceae website [14]. A second assembly, performed on sequences generated from both pyrosequencing and capillary sequencing reads using SeqMan™ NGen™ v1.2 software (DNAStar, Inc), was designated version 2. The version 2 contig set included longer contigs and more sequences were integrated into the contigs when compared to the original 454 Newbler assemblies. The combination of Sanger sequences and 454 sequences also resulted in slightly fewer but longer contigs. General information about the sequences and contigs identified from each species are summarized (Tables 1 and 2).
Table 1.
Sources of tissues and 454 sequencing results per tissue for the American and Chinese chestnut EST Datasets.
| Library | Tissues Sampled | Genotype | # 454 plates | Ave. 1 Read Length, bp | Total # Reads | Total bp Sequence |
|---|---|---|---|---|---|---|
| ACCanker | American chestnut cankers (infected stems) | TACF2, BA69 | 1 | 101 | 129,508 | 13,080,308 |
| ACHS1 | American chestnut healthy stems | CAES3, Wisneiwski | 3/4 | 247 | 222,939 | 29,828,910 |
| ACHS2 | American chestnut healthy stems | CAES, Watertown | 3/4 | 246 | 254,810 | 38,165,054 |
| ACWP1 | American chestnut whole plant tissues, pooled | CAES, Wisneiwski | 1/4 | 239 | 47,653 | 11,380,607 |
| ACWP2 | American chestnut whole plant tissues, pooled | CAES, Watertown | 1/4 | 221 | 33,288 | 7,364,484 |
| AC Totals | 5 cDNA libraries | 3 genotypes | 3 | 210.8 | 688,198 | 99,819,363 |
| CCCanker | Chinese chestnut var Nanking, cankers | TACF, VA37 | 1 | 101 | 235,635 | 23,799,135 |
| CCMHS | Chinese chestnut var Mahogany, healthy stems | TACF, BX316 | 3/4 | 246 | 228,594 | 56,051,191 |
| CCNHS | ChinesecChestnut var Nanking, healthy stems | TACF, GR119 | 3/4 | 247 | 259,859 | 64,271,926 |
| CCWP1 | Chinese chestnut var Nanking, whole plant tissues | TACF, GR119, | 1/4 | 242 | 60,445 | 14,643,040 |
| CCWP2 | Chinese chestnut var Mahogany whole plant tissues | TACF, BX316; | 1/4 | 246 | 53,939 | 13,249,397 |
| CC Total | 5 cDNA libraries | 3 genotypes | 3 | 205 | 838,472 | 172,014,689 |
1, Ave, average; 2, TACF, The American Chestnut Foundation's Meadowview VA Farm; 3, CAES, Connecticut Agricultural Research Station. (NB: Canker tissue RNAs were sequenced with the GS20 while other RNAs were sequenced using the FLX version of the 454 system)
Table 2.
Results of mass assembly of sequence reads from all libraries for each of American chestnut and Chinese chestnut into contigs1.
| Reads | Assembly V1 | Assembly V2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | # 454 plates | # 454 reads | # of bp | AL2 of reads | #contigs | AL. of contigs | % of reads in contigs | # contigs | AL of contigs | % reads in contigs |
| ACa3 | 2 | 398,783 | 78,183,046 | 197 bp | 22,714 | 374 | 59.4% | NA | NA | NA |
| ACb4 | 1 | 688,198 | 146,479,551 | 200 | NA | NA | NA | 34,800 | 412 | 80.9% |
| CCa5 | 3 | 838,472 | 171,849,098 | 207 | 32,738 | 501 | 66.8% | 48,501 | 526 | 87.4% |
| CCb6 | 3 | 838,472 | 171,849,098 | 207 | NA | NA | NA | 48,335 | 543 | 87.7% |
1, All of the assemblies are available for download at the http://www.fagaceae.org.website; 2, AL, Average length; 3, American chestnut, 2 initial 454 runs; 4, ACb, American chestnut, 3 454 runs; 5, CCa, Chinese chestnut 454 reads only; 6, CCb, Chinese chestnut 454 and Sanger reads combined; 7, NA, Not appropriate. (NB: Version 1 assemblies were conducted 1-31-2008 with 454 Newbler software; Version 2 assemblies were conducted 8-15-2008 with SeqManPro software)
Analysis of the American and Chinese chestnut transcriptomes
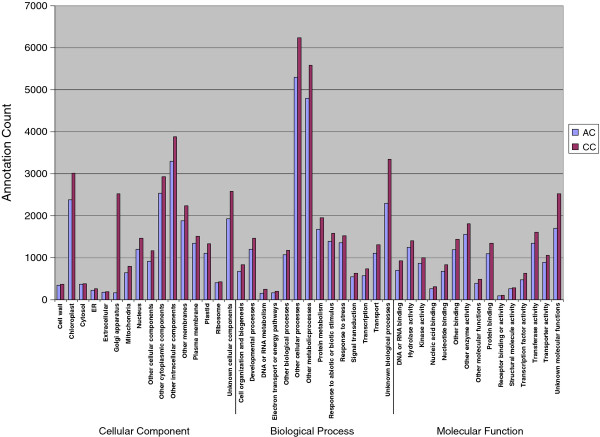
For the two Fagaceae species in this study, over one and half million sequencing reads were generated and yielded a total of 93,018 contigs in total for the separate assemblies of the cDNA libraries for the 10 tissues sampled. A small fraction of contigs matched mitochondrial (1.3%) and chloroplast (3%) genes. Similarly, ~2.5% of American and Chinese chestnut sequences obtained from canker tissues had best BLASTX alignments to the Cryphonectria parasitica proteome. Transcriptome assembly, version 2 (using all of the reads combined across all tissues), led to the identification of 34,800 and 48,501 contigs from American and Chinese chestnut respectively, from pyrosequencing alone, and 34,800 and 48,335 contigs, with the addition of the Sanger sequences for American chestnut and Chinese chestnut, respectively (Table 2). GO annotation using the Arabidopsis thaliana proteome as reference showed that the transcriptome of these species covers a wide range of biological processes (Figure 2) suggesting that the cDNA libraries were unbiased and well-suited for studies of development and physiology. The distribution of biological processes of the identified contigs from American and Chinese chestnut (Figure 2) did not show any statistically significant differences (p-value > 0.05) [17]. BLASTX alignments to model system proteomes showed that ~ 60% of the transcript contig sequences from the chestnut species studied have strong similarity to predicted proteins in Arabidopsis thaliana or Populus trichocarpa. Of the contig sequences that did not have any significant matches to Arabidopsis thaliana genes, 5 to 6% had a match to Populus trichocarpa genes. The remaining contigs (~30%) did not match any sequence in either the Arabidopsis thaliana or Populus trichocarpa proteomes. We observed a bias toward longer sequences in the contigs with BLASTX alignments to the model proteomes. The distribution of contig length showed that ~85% of sequences without BLASTX alignments to the proteomes of the two model species were short (< 250 nt). In contrast, only about 50% of contigs with good BLASTX hits on the model species proteomes were shorter than 250 nt.
Figure 2.
Histogram presentation of Gene Ontology classification of putative biological processes of contigs from American chestnut (AC), and Chinese chestnut (CC). The Y axis indicates the annotation count corresponding to each biological process indicated on the X axis.
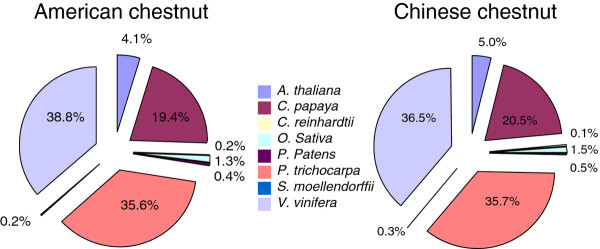
BLASTX searches were then conducted against the proteomes of all of the plant species for which the whole genome sequence were available at the time of this study, including Vitis vinifera, Carica papaya, Medicago truncatula, Oryza sativa, Populus trichocarpa, Physcomitrella patens, and Selaginella moellendorffii. A large fraction of chestnut contigs had better BLAST alignment scores to woody species than to the herbaceous species (Figure 3). For instance, over 35% of contigs from American and Chinese chestnut had best alignments to Vitis vinifera and Populus trichocarpa. Only ~5% of the contigs had best alignments to Arabidopsis thaliana. This bias cannot be attributed to a GC content difference between contigs from woody versus herbaceous species as their GC content have similar distributions (data not shown).
Figure 3.
Comparison of the percentage of American and Chinese chestnut contigs that have best hits on the proteome of each of the model plants.
Coverage of the transcriptomes
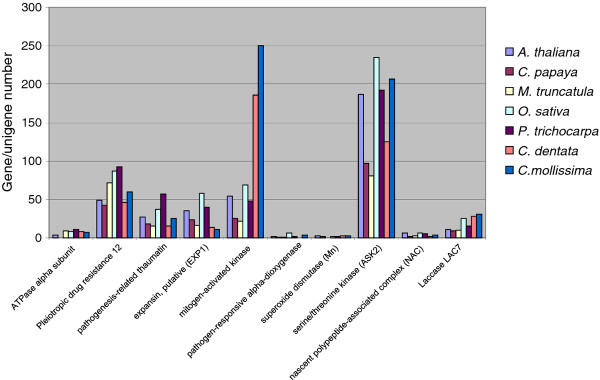
From the 34,800 and 48,335 contigs, a total of 11,431 and 10,016 large transcript contigs (more than 800 nucleotides) were identified from American chestnut and Chinese chestnut, respectively. The size of the transcript contigs assembled including large ones ranged from 258 bp to 1038 bp with approx. 4-12% of the American and Chinese chestnut contigs covering at least 70% of the length of the coding sequences relative to the respective genes in Arabidopsis thaliana. Analyses of the length of contigs showed that 344 (6.0%) and 874 (6.7%) contigs were full length in American chestnut and Chinese chestnut, respectively. Analysis of gene family size, both in American and Chinese chestnut as well as in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa, showed that the numbers of genes per family identified in chestnuts is similar to their counterpart in the model plant species suggesting a good coverage of the transcriptome in these two Fagaceae species (Figure 4). For instance, the number of members per gene family correlates well between American chestnut, Chinese chestnut, and the other model plant species (Additional file 1: Figure S1), with correlations coefficients of R = 0.8 for Chinese chestnut versus Arabidopsis and R = 0.74 for Chinese chestnut versus Populus.
Figure 4.
Size of some defense related gene families in model species: Arabidopsis thaliana, Populus trichocarpa, Oryza sativa, Carica papaya, Medicago truncatula, Castanea molissima, and Castanea dentate.
Defense related genes in American and Chinese chestnut
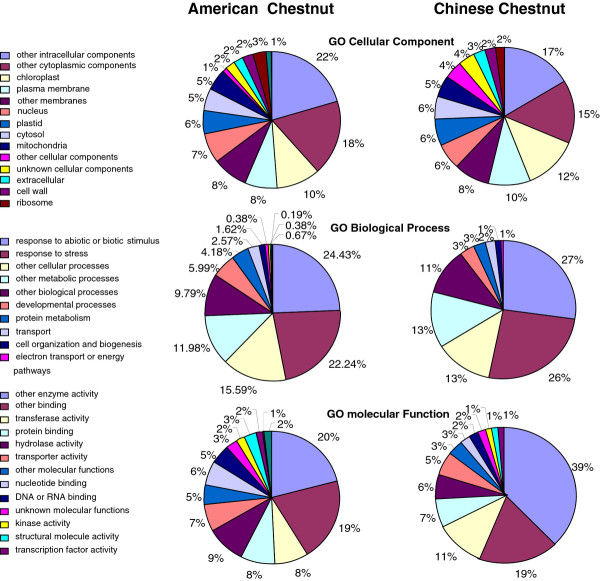
In silico analysis of transcript abundance using the DEGseq approach [16] identified 1715 GDTA in canker tissues versus healthy stems in American chestnut and 720 G DTA in Chinese chestnut (Figure 5, Additional file 2: Table S1, and Additional file 3: Table S2). The number of reads per transcript contig ranged between 7 and 6388 in canker and between 0 and 756 in healthy stem tissues. GO annotation distribution (Figure 5) showed that 177 and 86 of the identified genes from American and Chinese chestnut, respectively, were involved in response to abiotic or biotic stimuli. Twenty two percent and twenty three percent of American chestnut and Chinese chestnut genes from this functional category were involved in defense against biotic stresses. Most of the gene transcripts were highly abundant in canker tissues of both species (Additional file 4: Table S3) and thus represent good candidates for defense against the CBD fungus. GO annotation distribution (Figure 5) showed that the most frequent molecular functions of the identified defense-related genes were hydrolases, protein binding, transferases, and transporters. Several annotation categories including "secondary metabolic process", "oxidoreductase", "cellulose and pectin-containing cell wall", "hydrolases", and "lyases activity" were significantly over-represented in Chinese than American chestnut. A statistical analysis using the GOstat program [18] confirmed the enrichment of Chinese chestnut transcritpome in these functional categories (p-value < 0.01). On the opposite, several functional categories including mainly house-keeping genes such "structural constituent of the ribosome", "translation", "ribosome biogenesis and assembly", and "protein metabolic process" were over-represented in American chestnut than Chinese chestnut (p-value < 0.01). The over-representation of house-keeping GDTAs in American chestnut could be associated with the increase in protein synthesis at the infection site for defense against the pathogen.
Figure 5.
Pie chart representation of Gene Ontology classification of putative molecular functions of American and Chinese chestnut predicted genes involved in response to biotic stimuli as well as biological processes in which they are involved.
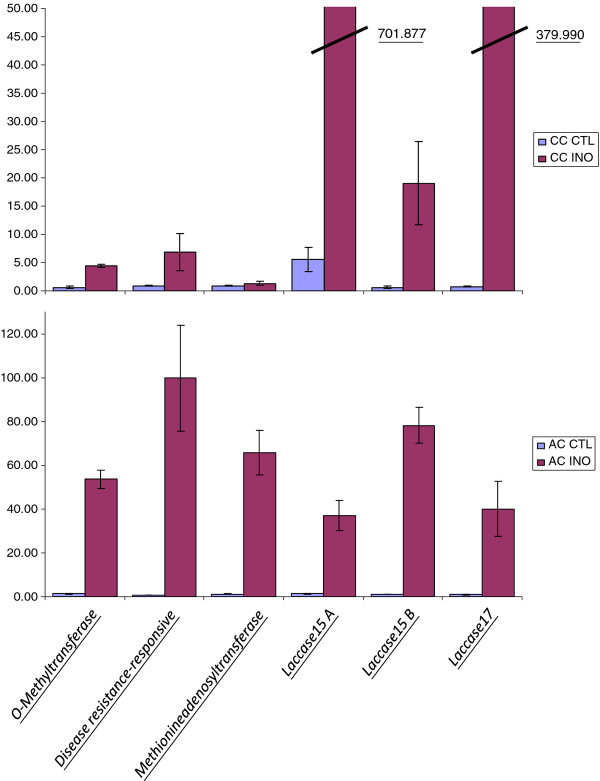
The list of the identified defense-related genes showing DTA involves several related pathways (Tables 3 and 4, Additional file 2: Table S1, Additional file 3: Table S2, and Additional file 4: Table S3). The first category includes genes involved in the biosynthesis of lignin and other cell wall components such as 4-coumarate:CoA ligase (4CL), Cinnamyl-Alcohol Dehydrogenase (CAD), cinnamoyl CoA reductase (CCR), peroxidase, Myb transcription factor, and UDP-glucose:thiohydroximate S-glucosyltransferase. Genes involved in programmed cell death and hypersensitivity such as Myo-inositol-1-phosphate, ATPase transporter, voltage-dependent anion channel, 2-deoxy-D-arabino-heptulosonate 7-phosphate, and cysteine proteinase precursor-like protein were also identified in canker tissues. However, one of the highly represented categories was phytohormone signaling including ethylene, jasmonic acid (JA), salicyc acid (SA), and abscisic acid (ABA) (Tables 3 and 4, Additional file 1: Figure S1, Additional file 2: Table S1). For example, transcripts of 12 genes involved in JA response were differentially abundant in Chinese chestnut. These include allene oxide cyclase, JAZ1, lipoxygenase, 12-oxophytodienoate reductase, 3-ketoacyl-CoA thiolase, chitinase, plastidic fatty acid desaturase, and others. Lipooxygenase, chitinase, and ACC oxidase are among genes with the most DTA in canker versus healthy stem (Additional file 3: Table S3). Genes involved in the response to SA include alpha-dioxygenase, mitochondrial chaperonin HSP, senescence-associated gene, and others. Genes related to the ABA response include ABA 8'-hydroxylase, 26S proteasome regulatory subunit, protein phosphatase 2 C, hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBPP) synthase and others. Several other genes involved were identified such as MYB transcription factors, proteases, and kinases. Comparison of GDTAs from American and Chinese chestnut showed that similar set of genes were induced in American and Chinese chestnut (Additional file 4: Table S3). Few genes were induced in American versus Chinese and vice versa (Additional file 4: Table S3). Analyses of a small set of these candidate genes using quantitative RT-PCR in healthy stem tissue versus Cryphonectria parasitica inoculated stem tissues from American and Chinese chestnut confirmed the differential expression of several of these candidates (Figure 6).
Table 3.
List of some defense related GDTA in canker versus healthy stem in American chestnut.
| Contig name | Ath. BH1 | # ACC Reads2 | # ACHS Reads3 | log2norm | Function |
|---|---|---|---|---|---|
| AC454_contig77_v3 | AT1G15950.1 | 58 | 34 | 3.19 | cinnamoyl CoA reductase. |
| AC454_contig559_v3 | AT1G65060.1 | 17 | 0 | 7.50 | isoform of 4-coumarate:CoA ligase (4 cl) |
| AC454_contig968_v3 | AT2G30490.1 | 11 | 5 | 3.55 | cinnamate-4-hydroxylase. |
| AC454_contig15349_v3 | AT3G12500.1 | 49 | 8 | 5.03 | basic chitinase involved |
| AC454_contig1339_v3 | AT1G74100.1 | 6 | 1 | 5.00 | desulfoglucosinolate sulfotransferase |
| AC454_contig17741_v3 | AT3G10920.1 | 15 | 25 | 1.68 | manganese superoxide dismutase (MSD1) |
| AC454_contig14459_v3 | AT2G35690.1 | 59 | 72 | 2.13 | acyl-CoA oxidase. Involved in jasmonate biosynthesis. |
| AC454_contig2314_v3 | AT1G15520.1 | 10 | 1 | 5.74 | abc transporter family |
| AC454_contig15056_v3 | AT2G41430.1 | 25 | 69 | 0.95 | hydrophilic protein lacking Cys residues |
| AC454_contig8521_v3 | AT3G60160.1 | 6 | 3 | 3.42 | member of MRP subfamily |
| AC454_contig38515_v3 | AT1G80600.1 | 6 | 0 | 6.00 | HopW1-1-Interacting protein 1 (WIN1). |
| AC454_contig1398_v3 | AT2G23620.1 | 9 | 3 | 4.00 | carboxylesterase/methyl jasmonate esterase |
| AC454_contig27747_v3 | AT5G60600.1 | 30 | 65 | 1.30 | hydroxy-2-methyl-2-(E)- butenyl 4-diphosphate synthase |
| AC454_contig4243_v3 | AT4G11260.1 | 43 | 142 | 0.69 | SCF(TIR1) mediated degradation of Aux/IAA proteins |
| AC454_contig117_v3 | AT4G37980.1 | 63 | 0 | 9.39 | oxidoreductase activity |
| AC454_contig12110_v3 | AT3G60190.1 | 8 | 5 | 3.09 | Arabidopsis dynamin-related protein 1E, DRP1E |
| AC454_contig1301_v3 | AT2G26560.1 | 6 | 6 | 2.42 | lipid acyl hydrolase with wide substrate specificity |
| AC454_contig2362_v3 | AT3G04720.1 | 6 | 0 | 6.00 | similar to the antifungal chitin-binding protein hevein |
| AC454_contig575_v3 | AT1G73260.1 | 6 | 0 | 6.00 | trypsin inhibitor |
| AC454_contig33210_v3 | AT3G20600.1 | 6 | 4 | 3.00 | non-race specific resistance to bacterial and fungal pathogens |
| AC454_contig22127_v3 | AT4G37870.1 | 39 | 101 | 1.04 | putative phosphoenolpyruvate carboxykinase |
| AC454_contig9688_v3 | AT4G23130.1 | 22 | 25 | 2.23 | receptor-like protein kinase. |
| AC454_contig18720_v3 | AT4G09320.1 | 39 | 32 | 2.70 | nucleoside diphosphate kinase type 1 (NDPK1) gene |
| AC454_contig7385_v3 | AT4G29040.1 | 24 | 28 | 2.19 | 26S proteasome AAA- ATPase subunit RPT2a (RPT2a) |
| AC454_contig5037_v3 | AT2G18960.1 | 8 | 5 | 3.09 | plasma membrane proton ATPase |
| AC454_contig38744_v3 | ATCG00120.1 | 5 | 2 | 3.74 | ATPase alpha subunit |
| AC454_contig16366_v3 | AT1G33970.2 | 6 | 4 | 3.00 | avirulence-responsive protein |
| AC454_contig14337_v3 | AT1G35720.1 | 31 | 78 | 1.08 | a member of the annexin gene family |
| AC454_contig28989_v3 | AT4G11600.1 | 43 | 23 | 3.32 | glutathione peroxidase. |
| AC454_contig1677_v3 | AT1G45249.1 | 5 | 1 | 4.74 | Leucine zipper transcription factor |
| AC454_contig1141_v3 | AT3G11410.1 | 4 | 1 | 4.42 | protein phosphatase 2 C |
| AC454_contig6419_v3 | AT1G69530.1 | 56 | 35 | 3.09 | Member of Alpha-expansin Gene Family |
| AC454_contig1070_v3 | AT4G37990.1 | 5 | 1 | 4.74 | alcohol:NADP + oxidoreduct ase |
| AC454_contig12900_v3 | AT1G04410.1 | 48 | 59 | 2.12 | malate dehydrogenase, cytosolic, putative. |
| AC454_contig2256_v3 | AT4G16260.1 | 6 | 4 | 3.00 | catalytic/cation binding /hydrolase |
| AC454_contig1025_v3 | AT3G57240.1 | 24 | 0 | 8.00 | glycosyl hydrolase family 17 |
| AC454_contig4431_v3 | AT3G25070.1 | 4 | 2 | 3.42 | R protein complex |
| AC454_contig2340_v3 | AT5G48485.1 | 20 | 31 | 1.78 | putative apoplastic lipid transfer protein |
| AC454_contig6410_v3 | AT1G20030.1 | 27 | 10 | 3.85 | pathogenesis-related thaumatin family protein |
| AC454_contig18835_v3 | AT4G38660.2 | 5 | 3 | 3.15 | thaumatin, putative |
| AC454_contig3796_v3 | AT5G47390.1 | 19 | 48 | 1.08 | myb family transcription factor |
| AC454_contig23403_v3 | AT5G35620.1 | 14 | 15 | 2.32 | Cap-binding protein |
1, Ath. BH: Arabidopsis best hit; 2, # ACC Reads: number of reads in American chestnut canker libraries; 3, # ACHS Reads: number of reads in American chestnut healthy stem libraries
Table 4.
List of some defense related GDTA in canker versus healthy stem tissues in Chinese chestnut.
| Contig Name | Ath. BH1 | # CCC Reads2 | # CCHS Reads3 | log2 norm4 | Function |
|---|---|---|---|---|---|
| CCall_contig18_v2 | AT1G02500 | 68 | 31 | 2.33 | S-adenosylmethionine synthetase |
| CCall_contig24687_v2 | AT1G02800 | 29 | 1 | 6.05 | endo-1,4-beta glucanase (CEL2) |
| CCall_contig45501_v2 | AT1G04410 | 111 | 155 | 0.71 | malate dehydrogenase |
| CCall_contig26701_v2 | AT1G05010 | 260 | 12 | 5.63 | 1-aminocyclopropane-1- carboxylate oxidase |
| CCall_contig35157_v2 | AT1G06180 | 8 | 0 | 5.20 | MYB3R- and R2R3- type MYB- encoding genes |
| CCall_contig28632_v2 | AT1G13280 | 33 | 9 | 3.07 | allene oxide cyclase |
| CCall_contig39585_v2 | AT1G13440 | 294 | 211 | 1.67 | Glyceraldehyde-3-phosphate dehydrogenase C2 |
| CCall_contig27982_v2 | AT1G15520 | 26 | 8 | 2.90 | ABC transporter family |
| CCall_contig42826_v2 | AT1G15950 | 66 | 56 | 1.43 | cinnamoyl CoA reductase |
| CCall_contig13029_v2 | AT1G19180 | 45 | 30 | 1.78 | JAZ1 is a nuclear-localized protein |
| CCall_contig9319_v2 | AT1G22450 | 35 | 37 | 1.12 | subunit 6b of cytochrome c oxidase |
| CCall_contig48116_v2 | AT1G24020 | 9 | 0 | 5.37 | MLP-LIKE PROTEIN 423 (MLP423) |
| CCall_contig22194_v2 | AT1G47128 | 94 | 87 | 1.31 | cysteine proteinase precursor- like protein |
| CCall_contig8876_v2 | AT1G51470 | 8 | 0 | 5.20 | myrosinase. |
| CCall_contig35286_v2 | AT1G52340 | 23 | 7 | 2.91 | cytosolic short-chain dehydrogenase/reductase |
| CCall_contig38717_v2 | AT1G55020 | 19 | 0 | 6.44 | lipoxygenase |
| CCall_contig12135_v2 | AT1G58440 | 15 | 5 | 2.78 | squalene monooxygenase activity. |
| CCall_contig30860_v2 | AT1G59960 | 133 | 155 | 0.97 | aldo/keto reductase |
| CCall_contig42753_v2 | AT1G64520 | 26 | 20 | 1.57 | Regulatory Particle non-ATPase 12a (RPN12a) |
| CCall_contig1090_v2 | AT1G65930 | 80 | 109 | 0.75 | NADP +- isocitrate dehydrogenase |
| CCall_contig24182_v2 | AT2G06050 | 25 | 16 | 1.84 | 12-oxophytodienoate reductase |
| CCall_contig41807_v2 | AT2G16500 | 84 | 50 | 1.94 | arginine decarboxylase (ADC) |
| CCall_contig12372_v2 | AT2G18950 | 19 | 13 | 1.74 | homogentisate phytyltransferase |
| CCall_contig11468_v2 | AT2G22240 | 171 | 74 | 2.40 | Myo-inositol-1-phosphate synthase isoform 2 |
| CCall_contig19221_v2 | AT2G26710 | 8 | 0 | 5.20 | cytochrome p450 family |
| CCall_contig3763_v2 | AT2G30080 | 10 | 3 | 2.93 | member of Fe(II) transporter isolog family |
| CCall_contig43865_v2 | AT2G30490 | 28 | 16 | 2.00 | cinnamate-4-hydroxylase |
| CCall_contig3045_v2 | AT2G33150 | 56 | 71 | 0.85 | organellar 3-ketoacyl-CoA thiolase |
| CCall_contig28249_v2 | AT2G37170 | 140 | 194 | 0.73 | plasma membrane intrinsic protein (PIP2) |
| CCall_contig45019_v2 | AT2G38710 | 14 | 8 | 2.00 | AMMECR1 family |
| CCall_contig22956_v2 | AT2G38870 | 39 | 0 | 7.48 | PR (pathogenesis-related) peptide |
| CCall_contig28937_v2 | AT2G47510 | 10 | 2 | 3.52 | FUM1: fumarase |
| CCall_contig3196_v2 | AT3G01280 | 47 | 56 | 0.94 | voltage-dependent anion channel |
| CCall_contig46205_v2 | AT3G01420 | 7 | 0 | 5.00 | alpha-dioxygenase |
| CCall_contig28792_v2 | AT3G04120 | 12 | 1 | 4.78 | cytosolic GADPH (C subunit) |
| CCall_contig8907_v2 | AT3G04720 | 17 | 4 | 3.28 | similar to the antifungal chitin- binding |
| CCall_contig41513_v2 | AT3G10920 | 42 | 26 | 1.89 | manganese superoxide dismutase (MSD1) |
| CCall_contig3523_v2 | AT3G10985 | 36 | 34 | 1.28 | A senescence-associated gene |
| CCall_contig27333_v2 | AT3G12360 | 39 | 34 | 1.39 | protein with an ankyrin motif |
| CCall_contig39986_v2 | AT3G12490 | 39 | 33 | 1.44 | cysteine proteinase inhibitor activity |
| CCall_contig42078_v2 | AT3G12500 | 73 | 13 | 3.69 | basic chitinase |
| CCall_contig46415_v2 | AT3G23600 | 72 | 6 | 4.78 | dienelactone hydrolase family protein |
| CCall_contig11692_v2 | AT3G45140 | 103 | 63 | 1.90 | Chloroplast lipoxygenase |
| CCall_contig8863_v2 | AT4G01070 | 8 | 1 | 4.20 | glycosyltransferase (UGT72B1) |
| CCall_contig11269_v2 | AT4G15480 | 217 | 99 | 2.33 | sinapic acid:UDP-glucose glucosyltransferase |
| CCall_contig38534_v2 | AT4G34135 | 11 | 2 | 3.66 | flavonol 7-O-glucosyltransferase (EC 2.4.1.237) |
| CCall_contig18339_v2 | AT4G37530 | 7 | 1 | 4.00 | peroxidase, putative |
| CCall_contig4088_v2 | AT4G37980 | 150 | 15 | 4.52 | ELICITOR-ACTIVATED GENE 3-1 (ELI3-1) |
| CCall_contig24556_v2 | AT4G37990 | 29 | 0 | 7.05 | aromatic alcohol:NADP + oxidoreductase |
| CCall_contig16847_v2 | AT4G39980 | 8 | 0 | 5.20 | 2-deoxy-D-arabino-heptulosonate 7-phosphate synthase |
| CCall_contig14020_v2 | AT5G06950 | 17 | 11 | 1.82 | Transcription factor of the B-ZIP family |
| CCall_contig45136_v2 | AT5G14740 | 147 | 19 | 4.15 | beta carbonic anhydrase |
| CCall_contig41707_v2 | AT5G42650 | 184 | 0 | 9.72 | cytochrome p450 CYP74 gene family |
| CCall_contig46027_v2 | AT5G45340 | 16 | 0 | 6.20 | protein with ABA 8'-hydroxylase activity |
| CCall_contig9507_v2 | AT5G52640 | 22 | 15 | 1.75 | cytosolic heat shock protein AtHSP90.1 |
| CCall_contig42347_v2 | AT5G64250 | 13 | 4 | 2.90 | 2-nitropropane dioxygenase family /NPD family |
1, Ath. BH: Arabidopsis best hit; 2, # CCC Reads: number of reads in Chinese chestnut canker libraries; 3, # CCHS Reads: number of reads in Chinese chestnut healthy stem libraries; 4, log2 norm, Expression ratio normalized
Figure 6.
Expression profiles of defense related genes in Cryphonectria parasitica inoculated and control plants in American and Chinese chestnut. The names of genes studied are indicated on the bottom of diagrams. The tissues analyzed are indicated on the right of the figure. CC CTL, Chinese chestnut control; CC INO, Chinese chestnut inoculated plants; AC CTL, American chestnut control; AC INO, American chestnut inoculated.
Discussion
In this study, over 1.5 million sequencing reads from various tissues were generated and assembled into 34,800 and 48,501 contigs, for American and Chinese chestnut respectively. The low level of contamination with organelle DNA, the fraction of complete or nearly complete full length cDNA sequences, and the depth coverage of genes involved in various biological processes indicate that pyrosequencing is an excellent tool for gene discovery, EST sequencing, and transcriptome analysis in non-model tree species. Combining 454 and Sanger sequences can improve contig construction as the assembly using both data sets resulted in more reads being integrated into contigs than by assembly of pyrosequences alone, resulting in slightly fewer but longer contig sequences. Using a tool developed recently [19], that estimates the amount of sequencing needed to cover the transcriptome of a given species, that takes into consideration the sequencing platform and the number of contigs generated from each species, we determined that as few as two or three additional plates of 454 sequence from different developmental stages and physiological conditions should allow for 100% coverage of the transcriptomes of Chinese and American chestnut.
The sequencing effort of the GTDF increased the number of Fagaceae cDNAs available in GenBank from a few sequences to hundreds of thousands of sequences for each of the species studied. The cDNA sequences and contig sequences, which are publicly accessible at the website [14] hosted at Clemson University, provides tools for Fagaceae species-specific BLAST searches. The cDNA sequences have been used to select molecular markers such as Simple Sequence Repeats (SSRs), and Single Nucleotide Polymorphisms (SNPs) for genetic mapping in Chinese and American chestnut (Kubisiak et al., in preparation), for ordering the physical map of Chinese chestnut (Fang et al., in preparation), and for characterizing microRNAs (Barakat et al., unpublished data). The sequences were also used to identify candidate genes involved in resistance to CBD [15], some of which are being functionally characterized (Powell et al., unpublished data). The genes with DTA were also being examined for colocalization with QTLs for resistance to CBD.
BLASTX alignments to the proteomes of two model systems (Arabidopsis or Populus) showed that ~ 60% of the transcript contig sequences from the Fagaceae species studied have strong similarity to predicted proteins. The remaining contigs did not match any sequence in the Arabidopsis or Populus proteomes. A large fraction of these are short sequences that may originate from 3' or 5' untranslated regions, which tend to be highly divergent between species (Additional file 1: Figure S1). Part of these sequences may also correspond to non-coding RNAs, or potential chestnut-specific genes. BLAST searches against the proteomes of eight plant species with complete genome sequences (Vitis vinifera, Carica papaya, Medicago truncatula, Oryza sativa, Populus trichocarpa, Selaginella moellendorffii, Physcomitrella patens, and Chlamydomonas reinhardtii) showed that a large fraction of the Fagaceae contigs have better alignments with genes in woody species such as Vitis vinifera, Populus trichocarpa, and Carica papaya versus the non-woody species angiosperm species. Similar results have been reported for other tree species such as Liriodendron tulipifera and Carica papaya [20,21]. Many long-lived woody plants, including Fagaceae species, exhibit extended phases of juvenile development before they reach flowering age, whereas most herbaceous plants reach flowering age in a single growing season. Thus, these observations could be associated with a slower evolutionary rate of genes in woody species versus herbaceous species.
In silico analysis of gene expression identified over two fold GDTAs in American than Chinese chestnut. However, most of the difference between the two DGTA sets lies in the number of induced housekeeping genes associated with the increase in resource utilization for plant defense at the infection site. The difference in the number of house-keeping genes induced in American and Chinese is maybe due to the amplitude of response of the two species to C. parasitica infection. The number of genes belonging to the category "response to biotic and abiotic stimuli" in American and Chinese chestnut from this is over 14 and 6 times higher than the ones reported previously using a partial dataset [22]. Also, several genes identified previously were not confirmed in this analysis. However, many new genes with DTA were identified in this study. The discrepancy between these results is linked to the larger dataset, which includes both 454 and capillary sequences, the better contig assembly, and the use of a more powerful tool for DTA analysis.
Genes with DTA identified in this study belong to well known plant pathways such as phenylpropanoid metabolism, phytohormone (JA, ABA, ethylene and SA) signaling, cell wall biosynthesis, proteolysis, and others. These genes and pathways function at different times in the plant response to pathogens. The category of genes involved in phenylpropanoid metabolism act early in plant defense, serving to inhibit or to block the penetration and the progression of the plant pathogen. This category includes genes for biosynthesis of monolignol and other phenolic compounds. Previous studies [23-27] showed that lignin biosynthesis is crucial for cell wall apposition, one of the first lines of plant defense against invading fungi. Besides lignin, the biosynthesis of other polymers such as callose seems to follow infection as suggested by the increased transcript abundance of UDP-glucose:thiohydroximate S-glucosyltransferase [28]. Other phenolic products that are involved in plant defense against pest and pathogens seem to be produced as well, as deduced by the presence of transcripts encoding genes such as flavanone 3-hydroxylase and flavonol 7-O-glucosyltransferase known to regulate flavonoid biosynthesis [29].
The second most important category of genes detected in response to the blight infection includes genes from phytohormone signaling pathways including JA, SA, and ethylene. These hormones trigger the activation of induced systemic resistance and systemic acquired resistance (SAR) to nectrotrophic pathogens [30,31]. The SAR is an effective defense mechanism against a broad range of pathogens and insects. Several genes from the JA response pathway such as methyl jasmonate esterase (MES1), acyl-CoA oxidase, a phyB pathway, and ATPase transporter were identified [32]. Genes involved in SA response such as hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate, HopW1-1-Interacting protein 1 (WIN1) were identified [33]. The SA pathway, which is considered one of the major pathways involved in defense against nectrotrophic pathogens, regulates the expression of defense effector genes and systemic acquired resistance through the repression of the auxin signaling pathway [16,33-36]. Another hormone that seems to play a role in the resistance of chestnut to CBD is abscissic acid (ABA). While ABA was described as a susceptibility factor, other studies [37,38] showed that it activates plant defense by priming for callose deposition or by restricting the progression of the fungus Cochliobolus miyabeanus in the mesophyll of rice [38]. Other signaling genes involved in SAR that induce numerous defense genes include apoplastic lipid transfer protein, and basic chitinase, etc [39].
The third category of genes with DTA in canker tissues includes genes involved in early response as part of the HR. Among these are transcripts encoding proteins such as ATPase transporter, kinases, carbonic anhydrase, AMMECR1, MIPS1, voltage-dependent anion channel, 2-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, and glutathione peroxidase that were reported previously to be involved in the hypersensitivity resistance (HR) and cell death in plants under pathogenic attack [18,31,40-43]. Reactive oxygen species (ROS) seem to be induced following C. parasitica infection as several genes involved in oxidative stress (alpha-dioxygenase, fumarase, cytosolic GADPH (C subunit), cytosolic ascorbate peroxidase APX1) had more abundant transcripts. Furthermore, several pathogenesis related (PR) genes such as elicitor-activated gene 3-1 (ELI3-1), aromatic alcohol:NADP + oxidoreductase, thaumatin, pathogenesis-related, and antifungal chitin-binding protein had differentially abundant transcripts in canker versus healthy stem tissues. PR proteins, of which some have antimicrobial functions [44], are mainly induced in localized pathogen attack around HR lesions. It is unknown what roles the HR and cell death play for chestnut defense against a necrotrophic pathogen such as C. parasitica. Alternatively, some of the genes involved in the HR may activate a systemic response of the plant or the pathogen may trigger HR to facilitate its colonization of the plant as reported for other pathogens [45]. Several other genes involved in defense such as eIF(iso)4E [46] were also implicated.
The candidate genes identified in this study represent a valuable resource for studying the genetic basis underlying resistance to CBD and the isolation of the fungal pathogen resistance genes. Comparative mapping of the blight resistance quantitative trait loci (QTL) of chestnut with peach disease resistance QTL is revealing that the genes for several of the differentially abundant chestnut transcripts in canker versus healthy stem tissues map to disease resistance QTL regions in both species (Fang and collaborators, in preparation). This suggests that some of the genes identified in this study may play a major role in plant defense against the CBD. Because most of the defense genes and gene networks were induced in both American and Chinese chestnut canker tissues, the question that remains is what then accounts for one of these species being susceptible and the other resistant to CBD. The timing of the response to the pathogen infection, and the amplitude of the response, could result in Chinese chestnut resistance to the blight. More information from transcript profiling during the time course of infection in these two species is required to address this question.
Conclusions
This project has generated over 83,000 transcript contigs from American and Chinese chestnuts and identified hundreds of genes and regulation pathways which may be involved in chestnut resistance to the pathogen C. parasitica. These resources have also been used for SNP and SSR marker development, genetic mapping, physical mapping, and genome organization comparison among chestnut species and between chestnut and other closely related species. Several of the candidate genes identified in this study are in the process of being analyzed for their function using transformation in planta. The cDNA database generated in this project is also being used to map expressed genes and to annotate the proteome in the Chinese chestnut genome that is being assembled during submission of this report (John Carlson, personal communication).
Methods
Plant materials
Ten cDNA libraries, representing a range of tissues from American and Chinese chestnut were prepared for EST sequencing by 454 technology [47] (Table 1). Tissue samples were submerged in liquid nitrogen immediately upon collection and stored at - 80°C until use. To create cankers, the stems of chestnut trees were inoculated with the hypervirulent C. parasitica strain EP155 as described by Hebard and collaborators [15,48]. For American chestnut and Chinese chestnut, canker tissue was collected at 5 and 14 days post-inoculation as previously described [15,48]. These times correspond to early and late stages of interaction between the plant and the pathogen [48].
RNA preparation, cDNA library synthesis, and 454 sequencing
Total RNA was prepared as described previously [49], and assessed with a 2100 Bioanalyzer (Agilent Technologies). cDNA and 454 libraries were constructed as described [15]. The 454 libraries were sequenced using the model GS20, for the American chestnut canker and Chinese chestnut canker libraries, and an FLX model 454 DNA sequencer (Roche Diagnostics), as previously described [15], for the remaining libraries. The sequence data was deposited into the Short Read Archive at the National Center for Biotechnology Information (study accession SRP000395).
Sanger sequencing
To analyze the transcriptomes of American and Chinese chestnut, this project generated Sanger sequences for about ~9000 cDNA clones from a subtractive library enriched in genes highly expressed in canker tissues in Chinese versus American chestnut using capillary sequencing. A total of 8,101 cDNA sequences were obtained after filtering reads for quality. RNA was prepared using the method described previously [49] and reverse transcribed using the SMART PCR cDNA Synthesis Kit (Clontech, Mountain View, CA). Substrative libraries were constructed using Chinese chestnut as the tester and American chestnut as the driver following the manufacturer recommendation (Clontech). Sequencing of the substractive libraries was conducted at the Clemson University Genomics Institute by an automated Sanger sequencing protocol.
Transcript assembly and contig annotation
The 454 sequence reads were assembled into contigs using 454 Newbler (Roche Diagnostics) or SeqMan™ NGen™ v1.2 software (DNAStar, Inc), optimized for 454 next generation data. The new assemblies are available on the Fagaceae website [14]. cDNA libraries were constructed using random priming which results in low poly A/T tail contamination and therefore no filtering was performed. Also, SeqMan removes low quality ends including homopolymer runs of poly(A/T) that have lower qualities in 454 sequencing. Contamination with mitochondrial and chloroplast genes was assessed by running a BLASTX search against Arabidopsis mitochondrion and chloroplast proteomes. An assembly using 454 and 9000 Sanger sequences was performed and compared the one that used 454 sequences only. Full-length contigs were identified by running a BLASTX search against the Arabidopsis thaliana proteome and comparing the lengths of the aligned portion of each contig and the putative proteins [50]. The annotation of contigs was performed by BLASTX [51] against the Arabidopsis thaliana proteome (e-value = e-5) and the Gene Ontology (GO) [52] system [15]. Comparison of GO annotation distribution between species was conducted using the GOstat program [17] set to the following parameters: GO-DB: tair; Min Sub-GO length: 3; P-Value Cutoff: 0.01; GO-Cluster Cutoff: -1; with no correction for multiple testing because the high dependence between GO terms will cause the test to be overly conservative. To determine which model species with most best hits to Fagaceae transcript contigs, BLAST alignments were conducted by querying the Fagaceae contigs against the proteomes of algal, moss and higher plant species with fully sequenced genomes (Chlamydomonas reinhardtii, Physcomitrella patens, Selaginella moellendorffii, Oryza sativa, Vitis vinifera, Populus trichocarpa, Carica papaya, and Arabidopsis thaliana) and the e-values of the best hits from each species were compared.
Identification of DTA in canker tissues
DEGseq [16] was used to identify gene specific differences in transcript abundance. The DEGseq package was chosen because it integrates several statistical methods, can estimate a theoretical replicate when an experimental one is not provided, and has been used routinely to identify DTA [16,53,54]. The number of 454 reads per contig for each gene was compared between canker and healthy stem tissues in American and Chinese chestnut separately. Similar analyses were performed for gene orthologs from both species. Orthologs were identified using a reciprocal best hit approach. DEGseq employs a random sampling model based on the read count in canker and healthy stem tissue libraries and performs a hypothesis test based on that model. Two theoretical four-fold local standard deviation lines can be drawn on the expression MA-plot to estimate the noise level of genes with different intensities and identify gene expression differences in different libraries. Genes passing the threshold are identified as exhibiting DTA. GO enrichment analyses were performed using Blast2Go software [55].
Validation tests of GDTA by real-time quantitative RT-PCR
Real-time quantitative RT-PCR tests were conducted to determine the extent to which the number of EST reads per gene obtained by shotgun sequencing accurately reflected transcript levels in the source tissues. RT-PCR estimates of transcript abundance were conducted on RNA from healthy and canker stem tissues from American chestnut and Chinese chestnut. RT-PCRs were performed as described previously [56,57]. Quantitative real time PCRs (qRT-PCRs) were prepared using the SYBR Green Master Mix kit (Applied Biosystems) and run in an Applied Biosystems 7500 Fast Real-Time PCR system with default parameters. Primers were designed using Primer Express® software (Applied Biosystems). A gene encoding 18S rRNA was used as an endogenous standard to normalize template quantity. We used only one standard because we did not observe any tissue specific differences in expression of 18S rRNA gene in our study. In addition, RT-PCR analyses were performed to confirm the expression of GDTA already identified using in silico expression analysis. For each gene, three biological replicates (three different trees) and three technical replicates were performed. Statistical analyses used Statistica 6.0 software (StatSoft Poland Inc., Tulsa, OH, USA), to estimate the significance of the differences.
Abbreviations
CBD: Chestnut blight disease; cDNA: Complementary DNA; EST: Expressed sequence tag; DTA: Differential transcript abundance; GTDF: Genomic tool development for the Fagaceae; Mya: Million years ago; PCD: Programmed cell death; QTL: Quantitative trait locus; SNP: Single nucleotide polymorphism; SSR: Simple sequence repeat; JA: Jasmonic acid; SA: Salicylic acid; ABA: Abscisic acid.
Competing interests
The authors declare that they have no competing interests
Authors' contributions
AB conceived and performed the transcriptome study, analyzed the DEG results, performed the RT-PCR experiments, wrote the manuscript, and supervised the work of NBMY, JP, and CY. NBMY, JP and CY contributed to the RT-PCR analyses and helped prepare the figures and Tables. MS, SF, and CC assembled the 454 cDNA reads and run the DEG analysis. FH performed the chestnut inoculation experiments and provided canker and healthy stem tissues. BP and KB constructed the cDNA subtractive libraries. SS conducted 454 library sequencing. NW, BP, JEC, AA, and RS designed the 454 library construction, provided advice and oversight on sequencing, and edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Correlation between the number of genes per gene family in American chestnut, Chinese chestnut, Arabidopsis, and Populus.
List of genes with differential accumulated transcripts in canker tissue versus healthy stem in American chestnut.
List of genes with differential accumulated transcripts in canker versus healthy stem tissues in Chinese chestnut.
List of genes with differential accumulated transcripts in infected tissues of Chinese chestnut (CC) and American chestnut (AC), or in one species but not in the other.
Contributor Information
Abdelali Barakat, Email: abaraka@clemson.edu.
Meg Staton, Email: mestato@yahoo.com.
Chun-Huai Cheng, Email: chunhuaicheng@wsu.edu.
Joseph Park, Email: enceunki@gmail.com.
Norzawani Buang M Yassin, Email: norzawanibuang@gmail.com.
Stephen Ficklin, Email: spficklin@gmail.com.
Chia-Chun Yeh, Email: yeh0104@gmail.com.
Fred Hebard, Email: fred@acf.org.
Kathleen Baier, Email: kbaier@esf.edu.
William Powell, Email: wapowell@esf.edu.
Stephan C Schuster, Email: scs@bx.psu.edu.
Nicholas Wheeler, Email: nickwheeler@scattercreek.com.
Albert Abbott, Email: aalbert@clemson.edu.
John E Carlson, Email: jec16@psu.edu.
Ronald Sederoff, Email: ron_sederoff@ncsu.edu.
Acknowledgements
We would like to thank our colleague Kerr Wall for his help with the GO annotation analysis and the Joint Genome Institute (Department of Energy) for providing us with the Cryphonectria parasitica proteome and EST sequences in advance of publication. This work was supported by grant DBI-PGPR-TRPGR 0605135 from The National Science Foundation (to RS), The Schatz Center for Tree Molecular Genetics (JEC), the World Class University Project R31-2009-000-20025-0 funded by the Ministry of Education, Science and Technology of Korea (to JEC), and The American Chestnut Foundation (FH).
References
- Flora of North America. http://www.efloras.org
- Oh S, Manos PS. Molecular phylogenetics and cupule evolution in Fagaceae as inferred from nuclear CRABS CLAW sequences. Taxon. 2008;57:434–451. [Google Scholar]
- Anagnostakis S. The effect of multiple importations of pests and pathogens on a native tree. Biological Invasions. 2001;3:245–254. doi: 10.1023/A:1015205005751. [DOI] [Google Scholar]
- Hill J. Proceedings of the International Chestnut Conference: 1994; Morgantown, West. Virginia. West Virginia University Press; 1994. Wildlife value of Castanea dentata past and present, the historical decline of the chestnut and its future use in restoration of natural areas; pp. 186–193. [Google Scholar]
- Herendeen PS, Magallon-Puebla S, Lupia R, Crane PR, Kobylinska J. A preliminary conspectus of the Allon Flora from the Late Cretaceous (Late Santonian) of central Georgia, U.S.A. Ann Missouri Bot Garden. 1999;86:407–471. doi: 10.2307/2666182. [DOI] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA. 2009;106(10):3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederoff R, Myburg A, Kirst M. Genomics, domestication, and evolution of forest trees. Cold Spring Harb Symp Quant Biol. 2009;74:303–317. doi: 10.1101/sqb.2009.74.040. [DOI] [PubMed] [Google Scholar]
- Griffin GJ. Blight Control and Resoration of the American Chestnut. J Forestry. 2000;99:22–27. [Google Scholar]
- Andrade GM, Merkle SA. Enhancement of American chestnut somatic seedling production. Plant Cell Rep. 2005;24(6):326–334. doi: 10.1007/s00299-005-0941-0. [DOI] [PubMed] [Google Scholar]
- The American Chestnut Foundation. http://www.acf.org
- Hebard FV. Restoration of American chestnut to forest lands--Proc of a conference and workshop: 2006; The North Carolina Arboretum. National Park Service; 2006. The backcross breeding program of the American chestnut foundation. Natural Res Rep NPS/NCR/CUE/NRR-2006/001. [Google Scholar]
- Milgroom MG, Cortesi P. Biological control of chestnut blight with hypovirulence: a critical analysis. Annu Rev Phytopathol. 2004;42:311–338. doi: 10.1146/annurev.phyto.42.040803.140325. [DOI] [PubMed] [Google Scholar]
- Wheeler N, Sederoff R. Role of genomics in the potential restoration of the American chestnut. Tree Genetics Genomes. 2008;5:181–187. [Google Scholar]
- The Fagaceae Genomic Tool Project. http://www.fagaceae.org
- Barakat A, DiLoreto D, Zhang Y, Smith C, Baier K, Powell W, Wheeler N, Sederoff R, Carlson J. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biology. 2009;9(1):51. doi: 10.1186/1471-2229-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20(9):1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bovie C, Ongena M, Thonart P, Dommes J. Cloning and expression analysis of cDNAs corresponding to genes activated in cucumber showing systemic acquired resistance after BTH treatment. BMC Plant Biol. 2004;4:15. doi: 10.1186/1471-2229-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Muller KF, Field D, Altman NS, de Pamphilis CW. PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Res. 2008. pp. D970–D976. [DOI] [PMC free article] [PubMed]
- Liang H, Ayyampalayam S, Wickett N, Barakat A, Xu Y, Landherr L, Ralph P, Jiao Y, Xu T, Schlarbaum S. et al. Generation of a large-scale genomic resource for functional and comparative genomics in Liriodendron tulipifera L. Tree Genetics & Genomes. 2011;7(5):941–954. doi: 10.1007/s11295-011-0386-2. [DOI] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL. et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452(7190):991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, DiLoreto DS, Zhang Y, Smith C, Baier K, Powell WA, Wheeler N, Sederoff R, Carlson JE. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol. 2009;9:51. doi: 10.1186/1471-2229-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell. 2005;17(7):2059–2076. doi: 10.1105/tpc.105.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichhart D, Schaller H, Lesot A. et al. A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc Natl Acad Sci USA. 2006;103(49):18848–18853. doi: 10.1073/pnas.0607849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Hisako K, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, Umemura K, Umezawa T, Shimamoto K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. PNAS. 2006;103:230–235. doi: 10.1073/pnas.0509875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalleck P, Plesch G, Hahlbrock K, Somssich IE. Induction by fungal elicitor of S-adenosyl-L-methionine synthetase and S-adenosyl-L-homocysteine hydrolase mRNAs in cultured cells and leaves of Petroselinum crispum. Proc Natl Acad Sci USA. 1992;89(10):4713–4717. doi: 10.1073/pnas.89.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ding X, Yuan M, Qiu D, Li X, Xu C, Wang S. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation. Plant Mol Biol. 2006;60(3):437–449. doi: 10.1007/s11103-005-4770-x. [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323(5910):95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RL, Hammerschmidt R. Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol. 1992;30:369–389. doi: 10.1146/annurev.py.30.090192.002101. [DOI] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16(10):449–455. doi: 10.1016/S0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Liu PP, Cameron RK, Park SW, Yang Y, Kumar D, Zhou F, Padukkavidana T, Gustafsson C, Pichersky E. et al. Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 2008;56(3):445–456. doi: 10.1111/j.1365-313X.2008.03618.x. [DOI] [PubMed] [Google Scholar]
- Lee MW, Jelenska J, Greenberg JT. Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J. 2008;54(3):452–465. doi: 10.1111/j.1365-313X.2008.03439.x. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Belghazi M, Huguet R, Robin C, Moreau A, Job C, Job D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 2006;141(3):910–923. doi: 10.1104/pp.106.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba A, Nishihara M, Tsukatani N, Nakatsuka T, Kato Y, Yamamura S. A peroxiredoxin Q homolog from gentians is involved in both resistance against fungal disease and oxidative stress. Plant Cell Physiol. 2005;46(6):1007–1015. doi: 10.1093/pcp/pci109. [DOI] [PubMed] [Google Scholar]
- Gil MJ, Coego A, Mauch-Mani B, Jorda L, Vera P. The Arabidopsis csb3 mutant reveals a regulatory link between salicylic acid-mediated disease resistance and the methyl-erythritol 4-phosphate pathway. Plant J. 2005;44(1):155–166. doi: 10.1111/j.1365-313X.2005.02517.x. [DOI] [PubMed] [Google Scholar]
- Wiese J, Kranz T, Schubert S. Induction of pathogen resistance in barley by abiotic stress. Plant Biol (Stuttg) 2004;6(5):529–536. doi: 10.1055/s-2004-821176. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D, Yang Y, Cruz CV, Hofte M. Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 2010;152(4):2036–2052. doi: 10.1104/pp.109.152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, La Camera S, Lamotte O, Metraux JP, Gatz C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010;61(2):200–210. doi: 10.1111/j.1365-313X.2009.04044.x. [DOI] [PubMed] [Google Scholar]
- La Camera S, Balague C, Gobel C, Geoffroy P, Legrand M, Feussner I, Roby D, Heitz T. The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol Plant Microbe Interact. 2009;22(4):469–481. doi: 10.1094/MPMI-22-4-0469. [DOI] [PubMed] [Google Scholar]
- Chen K, Du L, Chen Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol. 2003;53(1-2):61–74. doi: 10.1023/B:PLAN.0000009265.72567.58. [DOI] [PubMed] [Google Scholar]
- Li Y, Zou J, Li M, Bilgin DD, Vodkin LO, Hartman GL, Clough SJ. Soybean defense responses to the soybean aphid. New Phytol. 2008;179(1):185–195. doi: 10.1111/j.1469-8137.2008.02443.x. [DOI] [PubMed] [Google Scholar]
- Pennell RI, Lamb C. Programmed Cell Death in Plants. Plant Cell. 1997;9(7):1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem. 2008;46(11):941–950. doi: 10.1016/j.plaphy.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Govrin EM, Levine A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol. 2000;10(13):751–757. doi: 10.1016/S0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Okade H, Muto S, Suehiro N, Nakashima H, Tomoo K, Natsuaki T. Turnip mosaic virus VPg interacts with Arabidopsis thaliana eIF(iso)4E and inhibits in vitro translation. Biochimie. 2008;90(10):1427–1434. doi: 10.1016/j.biochi.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebard F, Griffin G, Elkins J. Developmental histopathology of cankers incited by virulent and hypovirulent Endothia parasitica on susceptible and resistant chestnut trees. Phytopathology. 1984;74:140–149. doi: 10.1094/Phyto-74-140. [DOI] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, Liang H, Landherr L, Tomsho LP, Hu Y, Carlson JE. et al. Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics. 2009;10:347. doi: 10.1186/1471-2164-10-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Consortium GO. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21(6):915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Matsumura K, Wang H, Arellano SM, Yan X, Alam I, Archer JA, Bajic VB, Qian PY. Toward an understanding of the molecular mechanisms of barnacle larval settlement: a comparative transcriptomic approach. PLoS One. 2011;6(7):e22913. doi: 10.1371/journal.pone.0022913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Frost CJ, Carlson JE. Phylogeny and expression profiling of CAD and CAD-like genes in hybrid Populus (P. deltoides × P. nigra): evidence from herbivore damage for subfunctionalization and functional divergence. BMC Plant Biol. 2010;10:100. doi: 10.1186/1471-2229-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto DS, Yellanki P, Carlson JE. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol. 2009;9:26. doi: 10.1186/1471-2229-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between the number of genes per gene family in American chestnut, Chinese chestnut, Arabidopsis, and Populus.
List of genes with differential accumulated transcripts in canker tissue versus healthy stem in American chestnut.
List of genes with differential accumulated transcripts in canker versus healthy stem tissues in Chinese chestnut.
List of genes with differential accumulated transcripts in infected tissues of Chinese chestnut (CC) and American chestnut (AC), or in one species but not in the other.