Abstract
The incidence of extended-spectrum β-lactamases (ESBLs) has been increasing worldwide, but screening criteria for detection of ESBLs are not standardized for AmpC-producing Enterobacteriaceae such as Enterobacter species. In this study, we investigated the prevalence of ESBLs and/or AmpC β-lactamases in Japanese clinical isolates of Enterobacter spp. and the association of plasmid-mediated quinolone resistance (PMQR) determinants with ESBL producers. A total of 364 clinical isolates of Enterobacter spp. collected throughout Japan between November 2009 and January 2010 were studied. ESBL-producing strains were assessed by the CLSI confirmatory test and the boronic acid disk test. PCR and sequencing were performed to detect CTX-M, TEM, and SHV type ESBLs and PMQR determinants. For ESBL-producing Enterobacter spp., pulsed-field gel electrophoresis (PFGE) was performed using XbaI restriction enzyme. Of the 364 isolates, 22 (6.0%) were ESBL producers. Seven isolates of Enterobacter cloacae produced CTX-M-3, followed by two isolates producing SHV-12. Two isolates of Enterobacter aerogenes produced CTX-M-2. Of the 22 ESBL producers, 21 had the AmpC enzyme, and six met the criteria for ESBL production in the boronic acid test. We found a significant association of qnrS with CTX-M-3-producing E. cloacae. The 11 ESBL-producing Enterobacter spp. possessing bla CTX-M, bla SHV, or bla TEM were divided into six unique PFGE types. This is the first report about the prevalence of qnr determinants among ESBL-producing Enterobacter spp. from Japan. Our results suggest that ESBL-producing Enterobacter spp. with qnr determinants are spreading in Japan.
Introduction
Enterobacter species are an important opportunistic pathogen that can cause nosocomial outbreaks and invasive infections such as bloodstream infections [1]–[3]. The prevalence of Enterobacteriaceae with extended-spectrum β-lactamases (ESBLs) has been increasing worldwide [4]. The prevalence of ESBLs and/or AmpC β-lactamases in clinical isolates of Enterobacter spp. from Japan is unknown. The Clinical and Laboratory Standards Institute (CLSI) has established guidelines for detection of ESBLs in Escherichia coli, Klebsiella spp., and Proteus mirabilis [5]. However, there are no recommendations from the CLSI for detection of ESBLs in microorganisms with chromosomal AmpC β-lactamases since the presence of ESBLs can be masked by the AmpC β-lactamases. It has been reported that 3-aminophenylboronic acid (BA) is an inhibitor of AmpC [6], and it is possible to detect ESBLs in AmpC-producing strains by the boronic acid disk test [7].
Quinolone resistance was mainly caused by chromosomal mutations of the quinolone-resistance determining regions in DNA gyrase and DNA topoisomerase IV [8], [9]. Recently, plasmid-mediated quinolone resistance (PMQR) determinants such as qnr, aac(6′)-Ib-cr, and qepA, have been identified in clinical isolates of Enterobacteriaceae worldwide [8], [9]. In addition, the emergence of PMQR in ESBL-producing Enterobacteriaceae is raising public health concerns since the inappropriate use of antimicrobial agents can transfer PMQR genes on the same plasmid as β-lactamase genes [8].
In this study, we investigated the prevalence of ESBLs and/or AmpC β-lactamases in clinical isolates of Enterobacter spp. from Japan and the association of PMQR determinants with ESBL-producing isolates. We also evaluated the usefulness of the boronic acid disk test for detecting production of ESBLs by Enterobacter spp. This work was presented in part at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), Boston, 2010.
Materials and Methods
Bacterial Strains
We studied a total of 364 consecutive and nonduplicate clinical isolates of Enterobacter spp., consisting of 228 isolates of Enterobacter cloacae and 136 isolates of Enterobacter aerogenes. These isolates were collected between November 2009 and January 2010 from 10 regions of Japan, including 38 isolates from Hokkaido, 13 from Miyagi, 19 from Niigata, 95 from Tokyo, 7 from Aichi, 50 from Osaka, 15 from Hiroshima, 10 from Ehime, 99 from Fukuoka, and 18 from Okinawa. Enterobacter spp. were identified by the Vitek-2 System (Sysmex-bioMérieux Japan, Tokyo). The isolates included 115 from urine (31.6%), 95 from sputum (26.1%), 77 from throat swabs (21.2%), 26 from nasal (7.1%), and 51 from other specimens (14.0%). Ethical approval was not needed according to the ethical guidelines for epidemiological research by the Japanese government because this study focused on bacterial aspects.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing of ESBL-producing isolates was performed with a frozen plate (Eiken Chemical Co., Ltd., Tokyo, Japan) and the broth microdilution method described by the CLSI [10]. The following antimicrobial agents were tested: ampicillin (range: 1–32 mg/L), piperacillin (4–128 mg/L), piperacillin-tazobactam (4/4–128/4 mg/L), cefoxitin (1–32 mg/L), cefpodoxime (2–64 mg/L), cefotaxime (2–64 mg/L), ceftazidime (2–64 mg/L), cefepime (1–32 mg/L), meropenem (0.5–16 mg/L), aztreonam (1–32 mg/L), ciprofloxacin (0.5–16 mg/L), levofloxacin (0.5–16 mg/L), gentamicin (1–16 mg/L), amikacin (8–64 mg/L), and trimethoprim-sulfamethoxazole (2/38–4/76 mg/L). The CLSI interpretive criteria were employed for each antibacterial agent [5]. Quality control was conducted by using E. coli ATCC 25922 and E. coli ATCC 35218.
Phenotypic Detection of AmpC β-lactamases and Extended-spectrum β-lactamases
According to the characteristics of β-lactamase production, AmpC-producing stains with a zone diameter ≤14 mm for cefoxitin were classified as derepressed AmpC mutants when the zone diameter was ≤17 mm for cefpodoxime/clavulanic acid, while inducible AmpC-producing strains had a zone ≥18 mm for cefpodoxime/clavulanic acid and a positive cefoxitin/cefpodoxime antagonism test [7], [11]. Possible ESBL-producing strains were screened by a zone diameter ≤20 mm for cefpodoxime, followed by the CLSI confirmatory test for ESBLs using disks of ceftazidime (30 µg), cefotaxime (30 µg), and cefpodoxime (10 µg) with or without clavulanic acid (10 µg) [5]. The boronic acid disk test was also performed to detect ESBL production using disks with 400 µg of boronic acid, as described previously [11]. A ≥5 mm increase of the zone diameter was considered to be a positive result that indicated the presence of ESBLs.
ESBL and PMQR Gene Detection and Sequencing
PCR was performed to identify various resistance genes, including β-lactamase genes (bla TEM, bla SHV and bla CTX-M), PMQR genes (qnrA, qnrB, qnrC, qnrS, and qepA), and aac(6')-Ib [12], [13], [14]. For CTX-M positive strains, the CTX-M group was determined by PCR using CTX-M-1, CTX-M-2, and CTX-M-9 group-specific primers [15], [16], [17]. For positive controls, we used DNA extracts from clinical isolates which were confirmed to possess β-lactamase or PMQR genes by sequencing. PCR products were purified by using a QIA quick PCR Purification Kit (Qiagen K. K., Tokyo, Japan), and then were sequenced by using specific primers [14], [15], [16], [17] with an ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on an ABI 3730×l Analyzer (Applied Biosystems). Sequencing data were analyzed with BLAST version 2.2.24 (http://blast.ddbj.nig.ac.jp/top-j.html).
Pulsed-field Gel Electrophoresis
For 11 ESBL-producing isolates (nine isolates of E. cloacae and two isolates of E. aerogenes) in which bla CTX-M, bla SHV, or bla TEM were identified by PCR, evaluation of chromosomal polymorphisms was done by pulsed-field gel electrophoresis (PFGE) using the XbaI restriction enzyme (Takara Bio Inc., Otsu, Japan), as described previously [18]. Electrophoresis was performed on 1% PFGE agarose gel with a CHEF-DR III system (Bio-Rad Laboratories, Richmond, CA, USA). Electrophoretic patterns were analyzed with GelCompar II version 3.0 (Applied Maths, Kortrijik, Belgium). Similarity between two tracks was calculated by using the coefficient of Jaccard and the band positions. Cluster analysis was performed by the unweighted pair-group method using arithmetic averages, and a dendrogram was generated by the software. Isolates with ≥80% similarity were considered to reside within a single cluster.
Statistical Analysis
Statistical analysis was conducted with the chi-square test or Fisher’s test. A P value of less than 0.05 was considered significant.
Results and Discussion
A total of 206 (90.4%) of the 228 isolates of E. cloacae and 130 (95.6%) of the 136 isolates of E. aerogenes were phenotypically confirmed to be AmpC producers. Among the 228 E. cloacae isolates, there were 60 derepressed AmpC mutants (26.3%) and 146 inducible AmpC producers (64%), while the 136 E. aerogenes isolates included 21 derepressed AmpC mutants (15.4%) and 109 inducible AmpC producers (80.1%). We found that 18 (7.9%) of the 228 isolates of E. cloacae and 4 (2.9%) of the 136 isolates of E. aerogenes were positive in the ESBL confirmatory test. All of the ESBL-producing isolates, except one E. cloacae isolate, also produced AmpC β-lactamases. In Osaka prefecture, the prevalence of ESBL-producing E. cloacae (8/35) was significantly higher than that for all of Japan (18/228) (P<0.05). The prevalence of ESBL-producing Enterobacter spp. varies among countries and regions, as well as between detection methods. It was previously reported that ESBL-producing E. cloacae had a prevalence of 17.7% in Algeria [19], 28% in Taiwan [20], and 35.4% in Korea [21]. Our data suggest that although the prevalence of ESBL-producing E. cloacae in Japan is still lower than in those countries, nevertheless the spread of ESBL-producers is certainly occurring at large in our country.
Of the 22 ESBL producers, 16 isolates (72.7%) were only detected by the CLSI confirmatory test, while six isolates (27.3%) met the criteria for ESBL production in the boronic acid test. Jeong et al. reported that the CLSI confirmatory test only detected 72.1% of ESBL producers, while the boronic acid disk test detected 98.4% and showed no false-positive results [7]. In the present study, the boronic acid disk test was useful for detecting ESBL producers among chromosomal AmpC-producing Enterobacter spp.
All of the ESBL-producing Enterobacter spp. were resistant to ampicillin and piperacillin. Nineteen isolates (86.4%) and 22 isolates (100%) were resistant to piperacillin-tazobactam and cefoxitin, respectively, suggesting a high prevalence of AmpC β-lactamases among ESBL producers. Resistance to cefpodoxime, cefotaxime, ceftazidime, cefepime, and meropenem was shown by 22 (100%), 22 (100%), 20 (90.9%), 13 (59.1%), and 0 of these isolates, respectively. Thus, carbapenem may be the most effective treatment for ESBL-producing Enterobacter spp. Thirteen isolates (59.1%) and 21 isolates (95.5%) were susceptible to gentamicin and amikacin, respectively. Of the 10 qnr-positive isolates, 8 (80%) were resistant to both ciprofloxacin and levofloxacin, while only one (8.3%) of the 12 qnr-negative isolates was resistant to both ciprofloxacin and levofloxacin (P<0.01). qnr determinants can confer weak quinolone resistance [22]. There is concern that Enterobacteriaceae possessing qnr and lacking sufficient chromosomal quinolone resistance could be classified as susceptible to fluoroquinolones according to the CLSI criteria.
The distribution of antimicrobial resistance genes among ESBL-producing Enterobacter spp. is summarized in Table 1. Of the 22 ESBL-producing strains, nine were positive for CTX-M, two for SHV, and 10 for TEM by PCR. Sequencing revealed that seven of the 20 isolates of E. cloacae had CTX-M-3, suggesting that it is predominant among E. cloacae in Japan. An outbreak of CTX-M-3-producing E. cloacae infection arising from a patient with immature teratoma in the pediatric ward of a university hospital in Osaka prefecture has been described previously [2]. Two of the four isolates of E. aerogenes produced CTX-M-2. To our knowledge, this is the first report of CTX-M-2 among E. aerogenes in Japan. Both of the two SHV-positive isolates and all 10 TEM-positive isolates had SHV-12 and TEM-1, respectively.
Table 1. Distribution of antimicrobial resistance genes among ESBL-producing Enterobacter spp.
| β-lactamases | PMQR determinants | |||||||||||
| ESBL-producingEnterobacter spp. (n) | Inducible AmpC | Derepressed AmpC | CTX-M-2 | CTX-M-3 | TEM-1 | SHV-12 | qnrA * | qnrB | qnrC | qnrS | qepA | aac(6')-Ib-cr |
| E. cloacae (18) | 1 | 16 | 0 | 7 | 9 | 2 | 2 | 0 | 0 | 7 | 0 | 0 |
| E. aerogenes (4) | 1 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
All qnrA were qnrA1.
Of the 22 ESBL-producing strains, 10 were positive for PMQR determinants (45.5%). Of the 10 isolates possessing PMQR genes, eight and two were positive for qnrS and qnrA, respectively. This is also the first report about qnr determinants among ESBL-producing Enterobacter spp. from Japan. The profile of Enterobacter spp. carrying ESBL genes is shown in Table 2. All seven of the CTX-M-3-producing E. cloacae strains had qnrS, and qnrS positivity was significantly more common among CTX-M-3-producing E. cloacae (7/7) compared with other ESBL producers (7/7 vs. 1/4, P<0.05). Both of the two SHV-12-producing E. cloacae possessed qnrA1. None of the 22 ESBL-producing Enterobacter spp. had qnrB, qnrC, qepA, and aac(6')-Ib-cr. An association of PMQR determinants with ESBLs has been reported by several authors. Park et al. reported a high prevalence of qnrA and qnrB among ESBL-producing E. cloacae in Korea [23], while there was a close association of qnrA, qnrB, or qnrS with SHV-12 production by E. cloacae in France, Algeria, and Taiwan [19], [20], [24]. An association of qnrB or qnrS with CTX-M-15 production by E. cloacae was also reported in Algeria [19].
Table 2. Profile of Enterobacter spp. carrying ESBL genes and results of the boronic acid disk test.
| StrainNo. | Sample | Organism | AmpC | ESBL | PMQR | PFGEtype | MIC (mg/L) | ESBL confirmatory test | |||||||
| CTX | CAZ | FOX | FEP | MEM | LVX | AMK | without BA | with BA | |||||||
| A33 | pus | E. aerogenes | derepressed | CTX-M-2 | notdetected | EA1 | ≥128 | ≥128 | ≥64 | 32 | 2 | 1 | ≤8 | positive | positive |
| D35 | urine | E. cloacae | derepressed | SHV-12 | qnrA1 | EC3 | ≥128 | ≥128 | ≥64 | ≥64 | 8 | 16 | 16 | positive | positive |
| F8 | pus | E. aerogenes | inducible | CTX-M-2 | qnrS | EA2 | ≥128 | 8 | ≥64 | ≥64 | 1 | 1 | ≤8 | positive | positive |
| F10 | sputum | E. cloacae | inducible | CTX-M-3 | qnrS | EC4 | ≥128 | 8 | ≥64 | ≥64 | ≤0.5 | 8 | ≤8 | positive | positive |
| F22 | sputum | E. cloacae | derepressed | CTX-M-3 | qnrS | EC1 | ≥128 | ≥128 | ≥64 | ≥64 | 2 | 16 | ≤8 | negative | positive |
| F25 | sputum | E. cloacae | derepressed | CTX-M-3 | qnrS | EC1 | ≥128 | ≥128 | ≥64 | ≥64 | 2 | 16 | ≤8 | negative | positive |
| F33 | sputum | E. cloacae | derepressed | CTX-M-3 | qnrS | EC1 | ≥128 | ≥128 | ≥64 | ≥64 | 1 | 16 | ≤8 | negative | positive |
| F38 | sputum | E. cloacae | derepressed | CTX-M-3 | qnrS | EC1 | ≥128 | ≥128 | ≥64 | ≥64 | 2 | 16 | ≤8 | negative | positive |
| F43 | sputum | E. cloacae | derepressed | CTX-M-3 | qnrS | EC2 | ≥128 | 64 | ≥64 | ≥64 | ≤0.5 | 16 | ≤8 | negative | positive |
| F46 | pus | E. cloacae | derepressed | CTX-M-3 | qnrS | EC1 | ≥128 | ≥128 | ≥64 | ≥64 | 1 | 8 | ≤8 | Negative | positive |
| I62 | urine | E. cloacae | derepressed | SHV-12 | qnrA1 | EC3 | ≥128 | ≥128 | ≥64 | 32 | 2 | 2 | ≤8 | Positive | positive |
CTX: cefotaxime, CAZ: ceftazidime, FOX: cefoxitin, FEP: cefepime, MEM: meropenem, LVX: levofloxacin, AMK: amikacin, BA: 3-aminophenylboronic acid. EC: Enterobacter cloacae, EA: Enterobacter aerogenes.
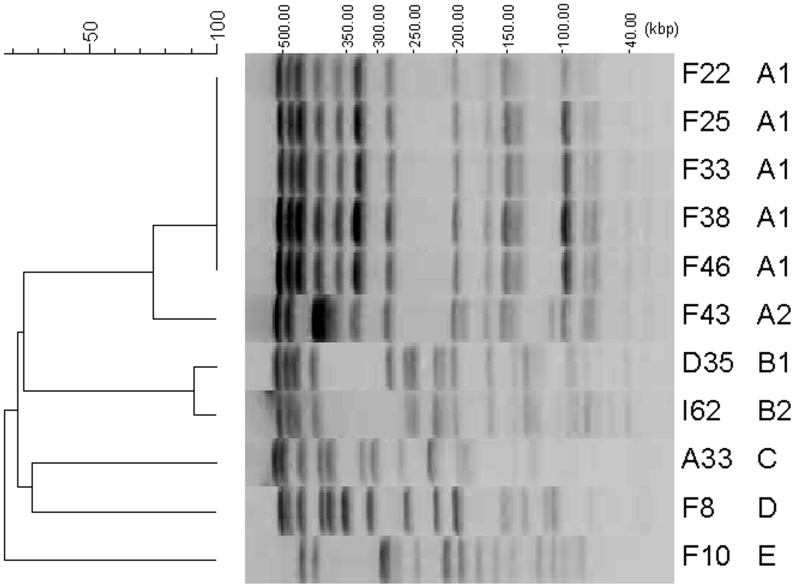
Of the 22 ESBL-producing isolates which were phenotipically identified by disk tests, 11 isolates (nine for E. cloacae and two for E. aerogenes) possessed ESBL genes (bla CTX-M-2, bla CTX-M-3, or bla SHV-12), but the remaining 11 isolates did not. Dissociation between disk tests and PCR detection results can occur since screening criteria for detection of ESBLs are not standardized for Enterobacter spp. Therefore, we focused on 11 isolates in which ESBL genes were identified by PCR and performed PFGE for them. Figure 1 shows dendrogram and PFGE of XbaI-digested genomic DNAs from 11 ESBL-producing Enterobacter spp. The 11 ESBL-producing isolates possessing bla CTX-M, bla SHV, or bla TEM were divided into six unique PFGE types. Of seven isolates possessing bla CTX-M-3 and qnrS from Osaka, five had identical PFGE type (EC1), while two had different types (EC2 and EC4). Two isolates possessing both bla SHV-12 and qnrA1 from Tokyo and Fukuoka were identical cluster (EC3). These results suggest that E. cloacae harbouring both bla CTX-M-3 and qnrS or both bla SHV-12 and qnrA1 are spreading via plasmids or transposons in Japan, although a limitation of our study is the relatively small sample size. Careful and continuous monitoring of antimicrobial resistance among Enterobacter spp. is needed.
Figure 1. Dendrogram and PFGE of XbaI-digested genomic DNAs from ESBL-producing Enterobacter species.
EC: Enterobacter cloacae, EA: Enterobacter aerogenes. Strain No. and PFGE type correspond to those in Table 2.
Acknowledgments
We thank clinical laboratory technologists for their assistance in this study.
Footnotes
Competing Interests: The authors have declared that Sadahiro Ichimura, Miho Ogawa, and Masahiro Shimojima belong to Department of Bacteriology, BML, Inc., but the affiliation to this company or this study is not related to employment, consultancy, patents, products in development or marketed products. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The authors have no support or funding to report.
References
- 1.Dalben M, Varkulja G, Basso M, Krebs VL, Gibelli MA. Investigation of an outbreak of Enterobacter cloacae in a neonatal unit and review of the literature. J Hosp Infect. 2008;70:14. doi: 10.1016/j.jhin.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Moriguchi N, Itahashi Y, Tabata N, Yamazumi T, Furuta I. Outbreak of CTX-M-3-type extended-spectrum β-lactamase-producing Enterobacter cloacae in a pediatric ward. J Infect Chemother. 2007;13:266. doi: 10.1007/s10156-007-0526-7. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi ZA, Paterson DL, Pakstis DL, Adams-Haduch JM, Sandkovsky G. Risk factors and outcome of extended-spectrum β-lactamase-producing Enterobacter cloacae bloodstream infections. Int J Antimicrob Agents. 2011;37:32. doi: 10.1016/j.ijantimicag.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Pitout JD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. Performance standards for antimicrobial susceptibility testing; Twentieth informational supplement M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA, USA. 2010.
- 6.Beesley T, Gascoyne N, Knott-Hunziker V, Petursson S, Waley SG. The inhibition of class C β-lactamases by boronic acids. Biochem J. 1983;209:233. doi: 10.1042/bj2090229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeong SH, Song W, Park MJ, Kim JS, Kim HS. Boronic acid disk tests for identification of extended-spectrum β-lactamase production in clinical isolates of Enterobacteriaceae producing chromosomal AmpC β-lactamases. Int J Antimicrob Agents. 2008;31:471. doi: 10.1016/j.ijantimicag.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 9.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22:689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Eighth Edition M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA, USA. 2009.
- 11.Kanamori H, Yano H, Hirakata Y, Endo S, Arai K. High prevalence of extended-spectrum β-lactamases and qnr determinants in Citrobacter species from Japan: dissemination of CTX-M-2. J Antimicrob Chemother. 2011;66:2262. doi: 10.1093/jac/dkr283. [DOI] [PubMed] [Google Scholar]
- 12.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47:3732. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melano R, Corso A, Petroni A, Centrón D, Orman B. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J Antimicrob Chemother. 2003;52:42. doi: 10.1093/jac/dkg281. [DOI] [PubMed] [Google Scholar]
- 14.Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother. 2009;53:645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G. Spread of bla CTX-M-type and bla PER-2 β-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother. 2006;57:978. doi: 10.1093/jac/dkl055. [DOI] [PubMed] [Google Scholar]
- 16.Moubareck C, Daoud Z, Hakimé NI, Hamzé M, Mangeney N. Countrywide spread of community- and hospital-acquired extended-spectrum β-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J Clin Microbiol. 2005;43:3313. doi: 10.1128/JCM.43.7.3309-3313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabaté M, Tarragó R, Navarro F, Miró E, Vergés C. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother. 2000;44:1973. doi: 10.1128/aac.44.7.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iabadene H, Messai Y, Ammari H, Ramdani-Bouguessa N, Lounes S. Dissemination of ESBL and Qnr determinants in Enterobacter cloacae in Algeria. J Antimicrob Chemother. 2008;62:136. doi: 10.1093/jac/dkn145. [DOI] [PubMed] [Google Scholar]
- 20.Wu JJ, Ko WC, Tsai SH, Yan JJ. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother. 2007;51:1227. doi: 10.1128/AAC.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park YJ, Park SY, Oh EJ, Park JJ, Lee KY. Occurrence of extended-spectrum β-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn Microbiol Infect Dis. 2005;51:269. doi: 10.1016/j.diagmicrobio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 23.Park YJ, Yu JK, Lee S, Oh EJ, Woo GJ. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J Antimicrob Chemother. 2007;60:871. doi: 10.1093/jac/dkm266. [DOI] [PubMed] [Google Scholar]
- 24.Cattoir V, Poirel L, Nordmann P. Plasmid-mediated quinolone resistance determinant QnrB4 identified in France in an Enterobacter cloacae clinical isolate coexpressing a QnrS1 determinant. Antimicrob Agents Chemother. 2007;51:2653. doi: 10.1128/AAC.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]