Abstract
Objectives
We aimed to define effects of PPARγ and PPARα agonist mono and combination therapy on adipose tissue and skeletal muscle gene expression in relation to insulin sensitivity. We further investigated the role of genetic polymorphisms in PPAR ligand-modulated genes in transcriptional regulation and glucose homeostasis.
Methods
Genome-wide transcript profiles of subcutaneous adipose and skeletal muscle and metabolic phenotypes were determined before and after 10 weeks of pioglitazone and fenofibrate mono or combination therapy in 26 subjects with impaired glucose tolerance. To establish the functional role of SNPs in genes modulated by pioglitazone alone or in combination with fenofibrate, we interrogated genome-wide association data of continuous glycemic phenotypes from the MAGIC study and adipose eQTL data from the MuTHER study.
Results
PPARγ, alone or in combination with PPARα agonists, mediated up-regulation of genes involved in the TCA cycle, branched chain amino acid metabolism, fatty acid metabolism, PPAR signaling, AMPK and cAMP signaling, and insulin signaling pathways, and downregulation of genes in antigen processing and presentation, immune and inflammatory response in adipose tissue. Remarkably few changes were found in muscle. Strong enrichment of genes involved in propanoate metabolism, fatty acid elongation in mitochondria, and acetyl-CoA metabolic process were observed only in adipose tissue of the combined pioglitazone and fenofibrate treatment group. After interrogating MAGIC data, SNPs in 22 genes modulated by PPAR ligands were associated with fasting plasma glucose and the expression of 28 transcripts modulated by PPAR ligands was under control of local genetic regulators (cis-eQTLs) in adipose tissue of MuTHER study twins.
Conclusions
We found differences in transcriptional mechanisms that may describe insulin sensitizing effects of PPARγ agonist monotherapy or in combination with PPARα agonist. The regulatory and glucose homeostasis trait-associated SNPs in PPAR agonist-modulated genes are important candidates for future studies that may explain the inter-individual variability in response to thiazolidinedione and fenofibrate treatment.
Keywords: insulin resistance, gene expression profile, muscle, adipose, pioglitazone, fenofibrate, eQTL
Introduction
Peroxisomal proliferator activated receptors (PPAR) are a subfamily of nuclear receptors that regulate gene expression in response to ligand binding. They consist of three major subtypes, PPARγ, PPARα, and PPARδ [1]. Thiazolidinediones (TZD) are PPARγ ligands which are used for treatment of type 2 diabetes (T2D) as insulin sensitizers [2]. PPARγ is highly expressed in adipose tissue and is involved in adipocyte differentiation, lipid storage, glucose homeostasis, and adipocytokine regulation [3]. TZDs such as pioglitazone improve insulin sensitivity by different mechanisms, including diverting fat from ectopic sites into subcutaneous depots [4] and decreasing macrophage number in adipose tissue [5]. Approximately 40–50% of individuals with impaired glucose tolerance (IGT) progress to T2D over their lifetimes [6]. Although recent studies have shown that pioglitazone can reduce the risk of this progression by 72% [7, 8], approximately 30% of patients do not respond to treatment with TZDs [9]. The precise mechanism by which PPARγ ligands improve insulin sensitivity, and the role of regulatory genetic polymorphisms that may modulate the PPARγ ligand-mediated gene expression response, are poorly understood.
A recent animal study suggested that combined PPARγ and PPAR α ligand therapy is more effective in improving glycemic control compared to monotherapy [10]. PPARα is expressed predominantly in muscle, liver, heart, and vessel wall, and regulates fatty acids metabolism. Fenofibrate is a PPARα activator and is used to treat hypertriglyceridemia, which is associated with insulin resistance and T2DM. Development of novel therapeutic agents, including dual PPARγ/α agonists and agents that selectively block cdk5-mediated PPARγ-Ser273 phosphorylation, is currently underway [11, 12]. Understanding the synergistic actions of PPARγ and PPARα combination therapy compared to PPARγ agonist monotherapy on adipose and muscle gene expression of patients with IGT will expedite the development of more efficacious therapy against insulin resistance with fewer side effects.
Based on these considerations, we designed a randomized clinical trial to determine the global transcript profiles of subcutaneous adipose and skeletal muscle before and after 10 weeks of pioglitazone and fenofibrate monotherapy or combination therapy in subjects with IGT. We selected individuals with IGT for this study to avoid variable degrees of hyperglycemia and different anti-diabetes medications as confounding factors. We tested four specific hypotheses: 1) treatment of pioglitazone alone or in combination with fenofibrate will improve insulin sensitivity by restoring gene expression pattern in adipose and muscle of IGT subjects; 2) compared to pioglitazone monotherapy, pioglitazone and fenofibrate combination therapy will cause a more significant activation of biological pathways and gene networks involved in lipid metabolism; 3) polymorphisms in a subset of PPAR ligand-modulated genes will be associated with insulin resistance; and 4) genes in key biological pathways modulated by PPAR are under control of local genetic regulators (cis-eQTLs).
Genome-wide expression analysis allowed us an unbiased assessment of the data. We utilized statistical and bioinformatics analyses to investigate the differences and similarities in biological pathways modulated by combination or monotherapy of PPAR agonists. Combining these data with our previous genome-wide study on insulin-sensitive versus insulin-resistant subjects [13], we could distinguish specific pathways involved in PPAR agonist-mediated improvement of insulin sensitivity. Further, to establish the role of SNPs in genes modulated by PPAR agonists, we interrogated genome-wide association data comprised of meta-analyses of continuous glycemic phenotypes from the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) [14, 15]. Finally, to develop a causal model of PPAR ligand pharmacogenomics, we analyzed the publicly available subcutaneous adipose eQTL data from the MuTHER (Multi Tissue Human Expression Resource) study [16] to explore the role of cis-regulatory variants in the expression of genes in pathways modulated by PPAR ligands.
Methods
Study Subjects
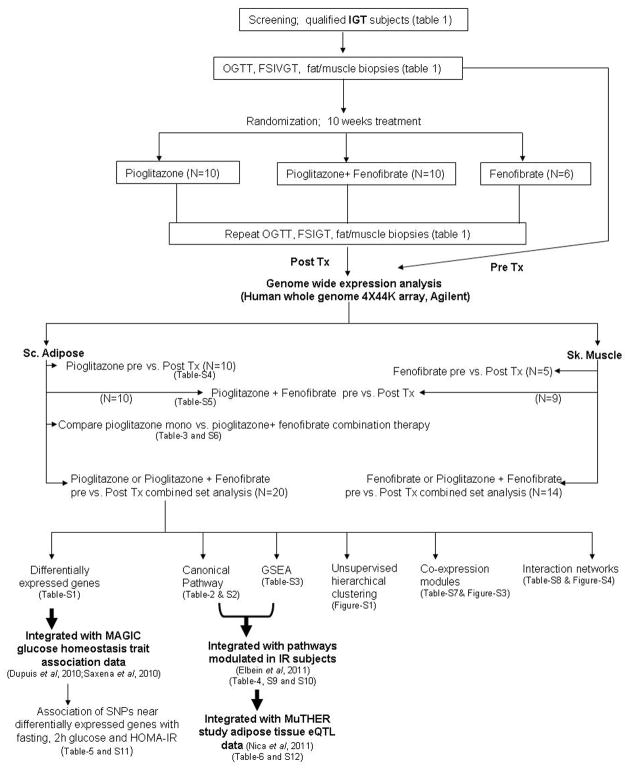
Men and women (age 18–65 yrs and BMI 28–38 kg/m2) with IGT (defined by fasting blood glucose <126 mg/dl and a two-hour glucose of 140–199 mg/dl after a 75 g oral glucose tolerance test [OGTT]) were recruited for this study as a part of randomized clinical trial (ClinicalTrials.gov identifier: NCT00470262). All subjects provided written, informed consent under a protocol that was approved by the local Institutional Review Board. Studies were conducted at the University of Arkansas for Medical Sciences/Central Arkansas Veterans Health Care System General Clinical Research Center. Subjects with a history of major health problem such as renal insufficiency (creatinine >1.4), liver disease (AST or ALT> 2x normal), congestive heart failure, and coronary artery disease were excluded. In addition, we excluded subjects who were taking medications known to affect adipose tissue inflammation or insulin resistance, such as HMG Co-A reductase inhibitors (statins), fibrates, anti-diabetes medications, or anti-inflammatory agents. During the first visit, subjects underwent OGTT, routine blood tests, and body composition measurement by dual-energy X-ray absorptiometry (DXA). Eligible participants underwent dietary stabilization for one week on a 35% fat, eucaloric diet and were maintained on this diet throughout the study. Insulin-modified, frequently sampled intravenous glucose tolerance tests (FSIGT) were performed at the second visit after an overnight fast, followed by subcutaneous adipose and skeletal muscle biopsies at their third visit. Subjects were then randomized to receive either pioglitazone (45 mg/day, N=10), fenofibrate (145 mg/day, N=6), or both medications (N=10) in an open-label design. Compliance and laboratory tests were monitored three times during follow-up. After 10 weeks of treatment, the OGTT, FSIGT, DXA, and biopsies were repeated. All biopsies were obtained in the fasting state as previously described [4]. Adipose biopsies were obtained under local anesthesia from abdominal subcutaneous adipose tissue by incision. Adipose tissue biopsy specimens were rinsed in saline, and frozen in liquid nitrogen. All muscle samples were obtained from the vastus lateralis by Bergstrom needle biopsy under local anesthesia and frozen in liquid nitrogen immediately. The study design is outlined in Figure 1.
Figure 1.
Study design and experimental plan.
Laboratory measurements
Insulin was measured by the University of Arkansas General Clinical Research Center core laboratory using an immuno-chemiluminometric assay (Invitron Limited, UK). Plasma glucose was measured by using a glucose oxidase method at LabCorp, Inc. (Burlington, NC).
RNA extraction
Total RNA was isolated from adipose tissue using the RNAeasy Lipid Tissue Mini kit (Qiagen Inc-USA, Valencia, CA), and from muscle (for 14 subjects treated with fenofibrate) using the Ultraspec RNA kit (Biotecx Laboratories, Inc, Houston, TX). The quantity and quality of the isolated RNA were determined by ultraviolet spectrophotometry and electrophoresis using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively. Similar to our published studies [13], high-quality RNA with RIN (RNA integrity number) > 8 was used for genome-wide transcriptome analysis.
Microarray studies
Genome-wide transcriptome analysis and initial array processing was performed by GenUs Biosystems (Northbrook, IL) using Human Whole Genome 4x44k arrays (Agilent Technologies) according to the vendor’s recommended standard protocol, similar to our published studies [13]. In brief, labeled cRNA was prepared by linear amplification of the Poly(A)+ RNA population within the total RNA sample. Total RNA (1 μg) was reverse transcribed after priming with a DNA oligonucleotide containing the T7 RNA polymerase promoter 5′ to a d(T)24 sequence. After second-strand cDNA synthesis and purification of double-stranded cDNA, in vitro transcription was performed using T7 RNA polymerase. The quantity and quality of the cRNA was assayed by spectrophotometry and on the Agilent Bioanalyzer. We fragmented 1 μg of purified cRNA to a uniform size and hybridized samples to Human Whole Genome 4x44k arrays (Agilent Technologies) at 37° C for 18 hrs in a rotating incubator. Arrays were washed and scanned with a G2565 Microarray Scanner (Agilent Technologies). Arrays were processed and background corrected with default settings of the Agilent Feature Extraction software v.9.5.3.1 (Agilent Technologies). The Agilent FE plug-in converts the complex set of 16 binary flag columns into three levels of GeneSpring flags: Absent (A), Marginal (M) or Present (P). Raw data were analyzed with GeneSpring GX v7.3.1 software (Agilent Technologies). To compare individual expression values across arrays, raw intensity data from each gene were quantile normalized to the 75th percentile intensity of each array. Only genes designated as present in at least 80% of samples from the baseline or post-treatment groups were included in further analyses.
Processed arrays were analyzed using two-class paired sample analysis on normalized data in Statistical Analysis for Microarray (SAM) V3.11 software [17]. In this analysis, baseline (−k) and post treatment (k) expression values of each study subject are considered as a pair (observation −k is paired with observation k); random exchanges are performed within each −k; k pair to calculate false discovery rate (FDR) by permutation. We considered results significant for a false discovery rate (q value) ≤ 10%, and average fold difference between baseline and post treatment samples of ≥1.2 (or ≤0.83), based on at least 500 permutations. Additional validation of SAM analysis was performed by paired sample t-test (p ≤ 0.05), and we report only on probes corresponding to transcripts with NCBI/Entrez identifiers. We excluded predicted and hypothetical genes (ORFs, FLJs and LOCs) from our significant gene lists. Genes represented by multiple probes were considered significant only if at least one probe met our stringent selection criteria and all probes were in the same direction of differential expression (formatted data will be available at: http://www.wakehealth.edu/research/research_default.aspx?id=29781 ).
Functional annotation of differentially expressed genes
Bioinformatic analysis was performed to investigate the enrichment of differentially expressed genes in known biological pathways or Gene Ontology (GO) categories. We performed canonical pathway analysis and interaction network analysis of differentially expressed genes using Ingenuity Pathway analysis (IPA ver9.0-3211; https://analysis.ingenuity.com). Pathways with corrected p-value (Benjamini and Hochberg p-value) ≤0.05 were considered significantly enriched in our gene lists. Additional annotations of differentially expressed genes were performed by singular enrichment analysis (SEA) using the DAVID v6.7 functional annotation tool (http://david.abcc.ncifcrf.gov/) [18]. Detailed analysis parameters for SEA using the functional annotation chart module of DAVID are described elsewhere [13]. In SEA analysis, a category with a gene count >5, an EASE score ≤0.05 (modified Fisher’s exact p-value), and FDR ≤15% was considered as a significance threshold. Gene set enrichment analysis (GSEA) using advanced GSEA methods was performed in GeneTrail software (http://genetrail.bioinf.uni-sb.de/) [19]. We used all probes corresponding to transcripts with NCBI/Entrez identifiers irrespective of fold change for this analysis. In the GSEA input file, we ranked our gene list based on SAM score (d, the T-statistic value). GeneTrail was run with median occurrence for genes with multiple probes; we considered the KEGG pathway and all GO categories with a minimum number of 6 genes in a category in our ranked expressed gene list. The p-value of ≤0.05 after correcting for FDR (Benjamini and Hochberg p-value) was considered as significance threshold for detecting an enriched category in this analysis. Microsoft Access queries were generated for data management and comparing genes differentially expressed and enriched pathways between data sets.
We recently showed enrichment of canonical pathways and GO categories among genes differentially expressed in insulin-resistant subjects (IS, n=31) compared to insulin-sensitive subjects (IR, n=31) matched for age, gender and BMI [13]. To understand the mechanisms involved in improved insulin sensitivity due to PPAR ligand treatment, we analyzed the overlap between pathways and GO categories enriched (in IPA and GSEA analysis) in our IR/IS data set and our current study involving pre- and post-PPAR ligand treatment.
Clustering analysis
We performed unsupervised hierarchical clustering using PermutMatrix version 1.9.3EN software to evaluate the predictive value of differentially expressed genes in identifying pre- and post-treatment transcript patterns [20]. A dissimilarity matrix based on Pearson’s correlation coefficient between each individual was generated using Z-score-normalized values for each differentially expressed probes and seriation under multiple fragment heuristics. The hierarchical clustering-based tree and heat map was generated using McQuitty’s criteria. We further performed modulated modularity clustering (MMC) analysis to identify coherent transcriptional modules among transcripts differentially expressed by PPAR ligands [21]. We utilized the MMC tool (http://mmc.gnets.ncsu.edu/) for this analysis and selected Spearman correlation coefficients to quantify pairwise relationships among transcripts to define modules. Modules with ≥25 transcript members elicited by MMC were further tested for their enrichment in known pathways or GO categories using DAVID v6.7.
Association of SNPs in PPAR ligand-modulated genes with glucose homeostasis traits
To establish whether the SNPs in genes modulated by pioglitazone were associated with continuous glycemic phenotypes, we obtained meta-analysis association data (effect allele, regression coefficient and p-values) from the MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium) investigators (http://www.magicinvestigators.org). In MAGIC, fasting glucose and insulin resistance (HOMA-IR) datasets were generated by performing a meta-analysis of up to 21 genome-wide association studies in up to 46,186 non-diabetic Caucasian participants. Two-hour glucose datasets were generated by a meta-analysis of nine genome-wide association studies in 15,234 non-diabetic Caucasian individuals [14, 15]. We interrogated the MAGIC data for association (p<0.001) of SNPs within and ±2 kb of 983 genes differentially modulated by pioglitazone treatment in subcutaneous adipose tissue.
eQTL analyses
We explored the role of local regulatory elements or cis-regulatory variants in modulating the expression of genes in pathways modulated by PPAR ligands to improve insulin sensitivity. We used Genevar 3.0.2 software (http://www.sanger.ac.uk/resources/software/genevar/) and analyzed subcutaneous adipose gene expression data from the MuTHER study [16, 22]. This study implemented a matched co-twin study design and includes 166 adipose tissue biopsies from Caucasian female twins (twin 1, N=79 and twin 2, N=87). We searched for cis-eQTLs within ±500 kb of genes in selected pathways by linear regression analysis. We considered an eQTL significant if the genotype of a SNP was associated (unadjusted p ≤ 0.01) in linear regression analysis with the expression of a transcript in both twin set analyses. Independent validation of selected cis-eQTLs was performed in adipose tissue of 89 Caucasian non–diabetic subjects from our laboratory.
Results
The baseline characteristics of the 26 subjects (10 who received pioglitazone, 10 pioglitazone plus fenofibrate, and 6 fenofibrate) did not differ by treatment group (Table 1). Significant improvement in insulin sensitivity (SI) was observed in subjects treated with pioglitazone alone (WSRT p-value = 0.011) or a combination of pioglitazone and fenofibrate (WSRT p-value = 0.022) for 10 weeks, but no significant improvement in insulin sensitivity was observed in subjects treated with fenofibrate alone (Table 1). Treatment with pioglitazone alone or pioglitazone and fenofibrate in our study mediated similar improvements in insulin sensitivity (SI). As expected, treatment with fenofibrate decreased serum triglyceride concentrations, but pioglitazone did not have a significant effect on lipid profiles. Interestingly, the combination of pioglitazone and fenofibrate had favorable effects on triglyceride, LDL cholesterol, and HDL cholesterol concentrations (Table 1). Pioglitazone is a ligand of nuclear transcription factor PPARγ, while fenofibrate is a ligand for PPARα. Since PPARγ is most abundantly expressed in adipose tissue while PPARα is highly expressed in muscle [2], we examined the effects of pioglitazone and fenofibrate treatment in modulating the transcript profile of subcutaneous adipose and skeletal muscle, respectively.
Table 1.
Baseline characteristics of subjects and changes in response to 10-week treatment with PPAR ligands
| Phenotype | Pioglitazone | Pioglitazone+Fenofibrate | Fenofibrate | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | PostTx | P-value | Baseline | PostTx | P-value | Baseline | PostTx | P-value | |
| N | 10 | 10 | 10 | 10 | 6 | 6 | |||
| age | 49.4±9.6 | 47.3±10.7 | 51.0±8.5 | ||||||
| Gender (M/F) | 2/8 | 2/8 | 4/2 | ||||||
| Ethnicity (EA/AA) | 7/3 | 7/3 | 6/0 | ||||||
| BMI, kg/m2 | 32.5±3.6 | 33.2±4.0 | 0.07 | 34.4±3.6 | 35.0±4.2 | 0.11 | 35.67±3.44 | 36.77±4.33 | 0.046 |
| Weight, Kg | 93±17.7 | 95.0±17.1 | 0.028 | 98.9±14.7 | 100.5±14.2 | 0.09 | 107.0±15.35 | 109.50±16.62 | 0.07 |
| Total Cholesterol, mg/dl | 228.5±61.0 | 204.6±56.0 | 0.09 | 202.8±32.7 | 171.9±29.4 | 0.008 | 169.67±53.05 | 156.83±39.38 | 0.11 |
| Triglyceride, mg/dl | 229.6±166.9 | 196±206.1 | 0.09 | 125.9±44.3 | 79.1±28.7 | 0.021 | 138.33±52.01 | 100.17±31.06 | 0.046 |
| HDL Cholesterol, mg/dl | 49.8±9.0 | 51.7±13.3 | 0.51 | 55.8±13.3 | 64.9±11.0 | 0.038 | 44.33±10.25 | 47.83±11.27 | 0.040 |
| LDL Cholesterol, mg/dl | 125.9±36.7 | 113.5±31.8 | 0.12 | 121.8±34.1 | 91.2±31.8 | 0.011 | 97.67±48.67 | 89.17±37.07 | 0.42 |
| HbA1c, % | 5.7±0.7 | 5.6±0.5 | 1.00 | 6.0±0.5 | 5.9±0.5 | 0.13 | 5.87±0.48 | 6.02±0.64 | 0.14 |
| % Body fat | 40.3±7.2 | 40.8±6.9 | 0.28 | 42.8±4.5 | 43.3±4.9 | 0.24 | 36.83±9.39 | 38.58±7.09 | 0.50 |
| SI (10−4min−1/μU/ml) | 1.6±0.2 | 2.7±0.7 | 0.011 | 1.8±0.7 | 3.0±1.4 | 0.022 | 1.60±0.86 | 2.22±1.07 | 0.25 |
Data are presented as means ± SD. BMI, body mass index; LDL and HDL, low- and high-density lipoprotein, respectively; HbA1c, glycosylated hemoglobin; SI, Insulin sensitivity from MINMOD analysis; M, Male; F, Female; EA, European American (Caucasian); AA, African American; P-value, statistical significance was determined from Wilcoxon’s signed rank test (WSRT); Post Tx, post treatment
Changes in adipose tissue expression profile in response to PPAR agonists
Comparison of the pre- and post-treatment adipose transcript profiles in treatment groups showing improved SI (combined set of 20 subjects treated either with pioglitazone or pioglitazone plus fenofibrate) showed differential expression of 983 genes at an average fold change of ≥1.2 fold and false discovery rate (q-value) of <10% (Supplementary Table 1). Large inter-individual variability was observed in gene expression response and only 39 genes showed differential expression when we used more stringent criteria (average fold change of ≥1.5 and q-value 0%). Unsupervised hierarchical clustering for transcripts differentially expressed by PPAR agonists grouped the pre- and post- treatment samples in two major clusters (Supplementary Figure 1); 17 of 19 samples in cluster 1 were from pretreatment subjects, whereas 18 of 21 samples in cluster 2 were from post-treatment subjects. As expected, the insulin sensitivity (SI) of samples in cluster 2 was significantly higher than in cluster 1 samples (p= 4.32 × 10−4). Nonetheless, despite this heterogeneity, we identified a set of genes that may characterize the transcriptional response of adipose tissue associated with PPAR agonist-mediated improvement in insulin sensitivity in most study subjects. However, the dissimilarity between pre- and post-treatment gene expression at mRNA level was low in five subjects; thus they were not classified into different clusters. Pre-treatment expression levels of three of these subjects were classified in a post-treatment cluster and indicate a high inter-individual variation in transcript expression. This incomplete clustering based on mRNA expression cannot be explained by differences in response to PPAR agonist as measured by changes in insulin sensitivity (SI). Thus, we speculate that PPAR agonist- mediated improvement in insulin sensitivity is at least partly contributed by its non-genomic effect and is independent of transcriptional modulation.
Ingenuity pathway analysis (IPA) among differentially expressed genes showed significant enrichment of genes in important biological pathways, including degradation of the branched chain amino acids (BCAA) valine, leucine, and isoleucine (p= 0.000068), propanoate metabolism (p= 0.0025), angiogenesis (P= 0.0091), PPARα/RXRα activation (p= 0.0095), and AMPK and cAMP signaling (p= 0.012 and p= 0.021; Table 2 and Supplementary Table 2). IPA also calculated the statistically significant overlap between differentially expressed genes and those regulated by a transcription factor. As expected, genes regulated by PPARγ were most significantly enriched (p= 2.77 × 10−6, 21 PPAR γ–modulated genes including ACSL1, SCD, INSIG1 and SORBS1) followed by the SREBF1 (p= 1.64 × 10−4). We also observed marginal enrichment for PPAR-alpha targets (p= 0.026). Supplementary Figure 2 shows the regulatory relationships among genes modulated by transcription factor PPARγ and SREBF1. Further, GSEA using the SAM score-ranked list of all transcripts expressed in adipose tissue also indicated PPAR agonist-mediated up-regulation of genes involved in the citrate (TCA) cycle; PPAR and insulin signaling; and fatty acid, butanoate and propanoate, and BCAA metabolism. By contrast, genes involved in antigen processing and presentation, and in immune and inflammatory responses, were downregulated (Supplementary Table 3).
Table 2.
Subcutaneous adipose tissue transcripts modulated by pioglitazone or pioglitazone + fenofibrate treatment in subjects with impaired glucose tolerance are enriched for important canonical pathways
| Ingenuity Canonical Pathways | p-value (B-H) | # Gene Downregulated | #Gene Upregulated |
|---|---|---|---|
| Valine, Leucine and Isoleucine Degradation | 6.76E-05 | 1 | 15 |
| Propanoate Metabolism | 2.51E-03 | 0 | 13 |
| Inhibition of Angiogenesis by TSP1 | 9.12E-03 | 1 | 8 |
| PPARα/RXRα Activation | 9.55E-03 | 6 | 16 |
| AMPK Signaling | 0.012 | 1 | 18 |
| Role of NFAT in Cardiac Hypertrophy | 0.012 | 6 | 17 |
| Relaxin Signaling | 0.012 | 2 | 17 |
| Renin-Angiotensin Signaling | 0.019 | 2 | 14 |
| Molecular Mechanisms of Cancer | 0.020 | 9 | 26 |
| RAR Activation | 0.020 | 2 | 19 |
| Glutamate Metabolism | 0.021 | 1 | 7 |
| Breast Cancer Regulation by Stathmin1 | 0.021 | 4 | 19 |
| GM-CSF Signaling | 0.021 | 1 | 10 |
| Citrate Cycle | 0.021 | 0 | 7 |
| p53 Signaling | 0.021 | 5 | 9 |
| Nitric Oxide Signaling in the Cardiovascular System | 0.021 | 0 | 12 |
| TR/RXR Activation | 0.021 | 1 | 12 |
| cAMP-mediated signaling | 0.021 | 4 | 20 |
| PTEN Signaling | 0.021 | 2 | 13 |
| RANK Signaling in Osteoclasts | 0.021 | 4 | 9 |
| Endothelin-1 Signaling | 0.021 | 6 | 14 |
| LXR/RXR Activation | 0.021 | 4 | 8 |
| B Cell Receptor Signaling | 0.022 | 5 | 13 |
| Glyoxylate and Dicarboxylate Metabolism | 0.024 | 0 | 5 |
| Amyloid Processing | 0.029 | 0 | 9 |
| Cardiac Hypertrophy Signaling | 0.030 | 7 | 17 |
| Glucocorticoid Receptor Signaling | 0.030 | 8 | 19 |
| IL-1 Signaling | 0.030 | 4 | 9 |
| Leptin Signaling in Obesity | 0.033 | 2 | 9 |
| Pancreatic Adenocarcinoma Signaling | 0.034 | 4 | 10 |
| Antiproliferative Role of TOB in T Cell Signaling | 0.034 | 2 | 4 |
| IL-2 Signaling | 0.034 | 1 | 8 |
| Role of Tissue Factor in Cancer | 0.034 | 4 | 10 |
| VEGF Signaling | 0.036 | 2 | 10 |
| Clathrin-mediated Endocytosis Signaling | 0.043 | 2 | 16 |
| Protein Kinase A Signaling | 0.043 | 9 | 20 |
| β-alanine Metabolism | 0.043 | 0 | 8 |
| Butanoate Metabolism | 0.043 | 0 | 9 |
| Fatty Acid Metabolism | 0.049 | 0 | 13 |
| Docosahexaenoic Acid (DHA) Signaling | 0.049 | 1 | 6 |
| Thrombin Signaling | 0.049 | 4 | 16 |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 0.049 | 4 | 12 |
| FLT3 Signaling in Hematopoietic Progenitor Cells | 0.050 | 0 | 10 |
Result from IPA canonical pathway analysis among genes modulated by pioglitazone in adipose tissue (in the combined set of 20 subjects treated with pioglitazone alone or pioglitazone and fenofibrate) is shown. Only pathways showing enrichment at p ≤ 0.05 after Benjamini and Hochberg multiple testing corrections (B-H P-Value) are presented.
To investigate the differences in transcriptional response between subjects treated with pioglitazone alone and pioglitazone plus fenofibrate, we further analyzed the adipose transcript profile of these two groups separately. Despite the same statistical power (n=10 in each group), 1839 genes showed differential expression (average fold change ≥1.2 and q-value <10%) in subjects treated with pioglitazone alone (Supplementary Table 4), while only 261 genes showed differential expression in subjects treated with pioglitazone plus fenofibrate (Supplementary Table 5). Ninety-four genes showed differential expression in both data sets (45 upregulated and 49 downregulated). Among the genes showing upregulation were cholesteryl ester transfer protein (CETP), phosphoenolpyruvate carboxykinase 1 (PCK1), serum glucocorticoid regulated kinase 2 (SGK2), protein phosphatase 1-regulatory subunit 1B (PPP1R1B), adiponectin (ADIPOQ), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (PFKFB1), and vascular endothelial growth factor A (VEGFA). Downregulated genes included hydroxysteroid-11-beta dehydrogenase 1 (HSD11B1), ectonucleotide pyrophosphatase/phosphodiesterase 4 (ENPP4), phospholipase C- beta 4 (PLCB4), integrin, alpha-V (ITGAV), and p53 apoptosis effector related to PMP22 (PERP). Interestingly, GSEA analysis showed a stronger upregulation of genes involved in fatty acid metabolism, the BCAA metabolic process, TCA cycle, oxidative phosphorylation, PPAR signaling pathway, and down-regulation of genes in antigen processing and presentation in adipose tissue from those taking pioglitazone plus fenofibrate compared to pioglitazone monotherapy (Table 3 and Supplementary Table 6). Strong enrichment of genes in propanoate metabolism, fatty acid elongation in mitochondria, and the acetyl-CoA metabolic process was observed in adipose samples of subjects treated with pioglitazone plus fenofibrate, but these pathways did not reach statistical significance in the pioglitazone monotherapy group.
Table 3.
KEGG pathways enriched in GSEA analysis in pioglitazone and pioglitazone + fenofibrate-treated subjects (N=10 per group)
| KEGG pathway | Pioglitazone | Pioglitazone + Fenofibrate | ||
|---|---|---|---|---|
| p-value (FDR) | enrichment | p-value (FDR) | enrichment | |
| Fatty acid metabolism | 0.03 | up | <1.0E-05 | up |
| Valine, leucine and isoleucine degradation | 0.01 | up | <1.0E-05 | up |
| Citrate cycle (TCA cycle) | 0.004 | up | <1.0E-05 | up |
| Oxidative phosphorylation | 0.004 | up | 3.63E-04 | up |
| PPAR signaling pathway | 0.02 | up | 6.89E-04 | up |
| Parkinson’s disease | 0.004 | up | 7.07E-04 | up |
| Butanoate metabolism | 0.01 | up | 0.007 | up |
| Arginine and proline metabolism | 0.04 | up | 0.007 | up |
| Lysine degradation | 0.03 | up | 0.010 | up |
| Insulin signaling pathway | 0.003 | up | 0.02 | up |
| Propanoate metabolism | NS | <1.0E-05 | up | |
| Glyoxylate and dicarboxylate metabolism | NS | 0.00069 | up | |
| Fatty acid elongation in mitochondria | NS | 0.00095 | up | |
| Ribosome | 0.03 | down | 5.14E-05 | down |
| Antigen processing and presentation | 0.004 | down | 3.78E-04 | down |
| Graft-versus-host disease | 0.004 | down | 0.002 | down |
| Autoimmune thyroid disease | 0.004 | down | 0.009 | down |
| Allograft rejection | 0.007 | down | 0.02 | down |
| Viral myocarditis | 0.03 | down | 0.03 | down |
| Type I diabetes mellitus | 0.004 | down | 0.03 | down |
| Proteasome | NS | 1.96E-06 | down | |
| Natural killer cell-mediated cytotoxicity | NS | 6.71E-06 | down | |
Result of advanced GSEA analysis (GeneTrail) for KEGG pathways showing enrichment p-value ≤0.05 after false discovery rate (FDR) correction is shown. Pathways highly enriched in Pioglitazone + Fenofibrate group are marked. NS, not significant.
PPAR agonists modulate regulatory networks of genes in lipid metabolism
To better understand the effect of PPAR agonists on gene regulatory networks, we utilized modulated modularity clustering analysis (MMC) and IPA knowledge-based biological interaction network analysis. The MMC analysis identified 54 coherent transcriptional modules among genes differentially expressed in adipose tissue in response to PPAR agonists (Supplementary Table 7 and Supplementary Figure 3). Considering the 983 differentially expressed genes as statistical background, we used singular enrichment analysis (SEA in DAVID) to access the degree to which biological processes and pathways were overrepresented in large transcriptional modules (modules with ≥ 25 gene members). Module 52 was enriched for genes in lipid metabolism (P= 0.0000684, FDR= 0.05). The IPA biological interaction network analysis within differentially expressed genes further revealed 25 highly significant interaction networks (Supplementary Table 8). Interestingly, two of these top 25 interaction networks showed an important role in lipid metabolism (networks 4 and 14) (Supplementary Figure 4). The network 4 (score= 34 with 28 differentially expressed genes) indicates a link between upregulation of lipid mobilization factor zinc-α2-glycoprotein (AZGP1) and adiponectin (ADIPOQ) [23] and downregulation of class A macrophage scavenger receptor (MSR1) [24]. This network also shows the positive regulatory interaction of adiponectin with protein kinase B (AKT1), AMP/cAMP kinases (PRKAG1 and PRKAR1A), and the IKK complex. The network 14 (score= 23 with 22 differentially expressed genes) showed a positive regulatory interaction with several nuclear receptors (NCOA4, NCOR1, NR1D2 and NR1H3), lipoprotein receptor (CD36), and lipid-metabolizing enzymes (ACACA, CETP, DGAT2, FASN and SCD). Thus, our study indicates an important role of PPAR agonists in modulating regulatory networks involved in lipid metabolism in adipose tissue.
Changes in skeletal muscle expression profile in response to PPAR agonists
Comparison of the pre- and post-treatment muscle transcript profile was performed in our combined set of 14 subjects treated either with fenofibrate alone or in combination with pioglitazone. Large inter-individual variability in transcriptional response was observed in muscle after PPAR agonist treatment. Only the phosphatidylinositol-4-phosphate 5-kinase-type II-gamma (PIP5K2C) gene showed significant differential expression in muscle of 14 subjects treated either with fenofibrate (N= 5) or fenofibrate and pioglitazone (N= 9). A total of 172 genes, including fatty acid binding protein 4 (FABP4) and diacylglycerol O-acyltransferase-2 (DGAT2), showed differential expression (average fold change ≥1.2 and p-value ≤0.05) but failed to meet our false discovery rate criterion (≤10%). Similar results were obtained when we considered only subjects treated with pioglitazone + fenofibrate (N=9). We further performed GSEA to identify enrichment of genes in biological pathways. GSEA considers all expressed genes by rank without a fold-change threshold, and thus may identify altered pathways in which no individual members meet criteria of differential expression. In contrast to adipose tissue, we found no enriched pathways that may be modulated by PPAR agonists in skeletal muscle.
PPAR agonists restore a normal insulin-sensitive gene expression pattern in adipose tissue
We recently showed significant enrichment of important biological pathways among genes differentially modulated in insulin-resistant compared to insulin-sensitive individuals [13]. In the present study, we investigated if the pathways dysregulated by insulin resistance overlap with those modulated by PPAR agonists in adipose tissue. As shown in Table 4 (and in Supplementary Table 9), GSEA indicates that the genes in the TCA cycle, BCAA metabolism (valine, leucine and isoleucine degradation), PPAR and insulin signaling, fatty acid, butanoate and propanoate metabolism, and biosynthesis of unsaturated fatty acids pathways – all of which were significantly downregulated in insulin-resistant subjects – were upregulated in adipose tissue after 10 weeks of treatment with the PPARγ ligand alone or in combination with the PPARα agonist. Similarly, genes in KEGG pathways related to immune and inflammatory function (antigen processing and presentation, proteasomes, cell adhesion molecules, and cytokine-cytokine receptor interaction) that were upregulated in insulin-resistant subjects were downregulated by the PPARγ therapy alone or combined with PPARα agonists. When we considered only the top differentially expressed gene, IPA analysis additionally showed the downregulation of AMPK pathway genes in insulin-resistant subjects and was upregulated by PPAR agonist treatment (Supplementary Table 10). Thus, our genome-wide analysis of the adipose tissue transcriptome successfully narrowed down the pathways involved in PPAR agonist-mediated improvement of insulin sensitivity.
Table 4.
KEGG pathways enriched in adipose tissue of insulin-resistant subjects overlaps with pathways modulated by PPAR agonist treatment
| KEGG pathway | in IR/IS analysis | in PostTx/baseline analysis | ||
|---|---|---|---|---|
| p-value (FDR) | enrichment | p-value (FDR) | enrichment | |
| Citrate cycle (TCA cycle) | 0.000693 | down | <1.0E-5 | up |
| Valine, leucine and isoleucine degradation | 5.4E-05 | down | 3.91E-05 | up |
| PPAR signaling pathway | 0.000567 | down | 0.000465 | up |
| Insulin signaling pathway | 0.000727 | down | 0.000693 | up |
| Fatty acid metabolism | 0.009271 | down | 0.000908 | up |
| Butanoate metabolism | 0.003297 | down | 0.001082 | up |
| Propanoate metabolism | 0.000281 | down | 0.00148 | up |
| Lysine degradation | 0.040107 | down | 0.002943 | up |
| Pyruvate metabolism | 0.026626 | down | 0.014727 | up |
| Biosynthesis of unsaturated fatty acids | 0.006467 | down | 0.04186 | up |
| ECM-receptor interaction | 0.043291 | up | 0.043069 | up |
| Antigen processing and presentation | 0.001375 | up | 7.34E-05 | down |
| Proteasome | 0.000194 | up | 0.008954 | down |
| Cell adhesion molecules (CAMs) | 6.37E-06 | up | 0.01419 | down |
| Cytokine-cytokine receptor interaction | 0.000194 | up | 0.042528 | down |
Result of advanced GSEA analysis (Genetrail) for KEGG pathways showing enrichment p-value ≤0.05 after false discovery rate (FDR) correction is shown. KEGG pathways enriched in insulin resistant subjects were detected by GSEA analysis of ranked list of all expressed transcripts in adipose of 31 insulin-resistant and 31 insulin-sensitive subjects (reanalyzed from our published data, Elbein et al, 2011). KEGG pathways enriched after PPAR agonist treatment was detected by GSEA analysis of a ranked list of all expressed transcripts in adipose of 20 subjects with impaired glucose tolerance, treated with either pioglitazone or pioglitazone and fenofibrate for 10 weeks.
Interestingly, our analysis showed upregulation of 18 transcripts in the AMPK signaling pathway in adipose tissue after PPAR agonist treatment. Published studies have demonstrated decreased AMPK activity (due to reduced phosphorylated AMPK) in different tissues, including subcutaneous adipose tissue of insulin-resistant subjects [25]. Several in vitro and animal studies also demonstrated rapid activation of AMPK in different cells and tissues after PPARγ agonist treatment independent of PPARγ–mediated gene transcription [26, 27]. Activation of AMPK by phosphorylation triggers downstream signaling events, including increased fatty acid oxidation and metabolism [28]. The mechanism of PPAR agonist-mediated AMPK activation is incompletely understood. However, our transcriptome analysis supports the concept that activation of AMPK signaling in human adipose tissue is one mechanism involved in PPAR agonist-mediated restoration of insulin sensitivity.
Polymorphisms in PPAR ligand-modulated genes are associated with glucose homeostasis traits
Improved insulin sensitivity with PPAR treatment resulted in a differential expression of 983 genes in subcutaneous adipose tissue. We investigated whether the SNPs within and ±2 kb of these 983 genes are associated with glucose homeostasis traits. Interrogation of large meta-analysis data from the MAGIC consortium [14, 15] showed significant associations (p <0.001) of 97 SNPs in 22 genes with fasting plasma glucose. Table 5 shows the most significant SNPs for each of these 22 genes. The most significant SNPs associated with fasting plasma glucose include rs11717195 (p= 1.11×10−9) in ADCY5 (adenylate cyclase 5) and rs4308429 (p= 9.15×10−7) in WFS1 (Wolfram syndrome 1). Similarly, the MAGIC data also showed significant association of SNPs with 2-hour glucose and HOMA-IR in 14 and 15 genes, respectively (Supplementary Table 11). Thus, a subset of genes modulated by PPAR ligands harbor polymorphisms associated with glucose homeostasis traits, and are genetic risk factors that may modulate insulin sensitivity.
Table 5.
SNPs in PPAR agonist-modulated genes are significantly associated with fasting plasma glucose values
| SNP | Chromosome | Gene | Effect allele | Other allele | MAF | Effect | Stderr | p-value |
|---|---|---|---|---|---|---|---|---|
| rs11717195 | 3 | ADCY5 | t | c | 0.23 | 0.029 | 0.0047 | 1.11E-09 |
| rs4308429 | 4 | WFS1 | a | g | 0.254 | −0.018 | 0.0037 | 9.15E-07 |
| rs1449627 | 11 | NR1H3-MADD | t | g | 0.31 | −0.018 | 0.0039 | 1.87E-06 |
| rs7120118 | 11 | NR1H3 | t | c | 0.301 | −0.019 | 0.004 | 1.88E-06 |
| rs7681334 | 4 | ACSL1 | a | g | 0.455 | 0.016 | 0.0037 | 8.37E-06 |
| rs11621188 | 14 | GPHN | a | g | 0.184 | −0.024 | 0.0056 | 1.86E-05 |
| rs194518 | 7 | STEAP2 | a | g | 0.482 | 0.015 | 0.0036 | 3.36E-05 |
| rs12601608 | 17 | TAOK1 | a | g | 0.232 | −0.018 | 0.0045 | 6.15E-05 |
| rs9516213 | 13 | GPC6 | c | g | 0.39 | −0.014 | 0.0037 | 1.83E-04 |
| rs2167079 | 11 | ACP2 | t | c | 0.309 | 0.018 | 0.0049 | 1.94E-04 |
| rs1564374 | 12 | GEFT | a | g | 0.385 | 0.014 | 0.0037 | 2.47E-04 |
| rs39284 | 7 | STEAP1 | t | c | 0.483 | −0.013 | 0.0037 | 3.36E-04 |
| rs11849129 | 14 | ZNF219 | c | g | 0.008 | 0.056 | 0.016 | 3.45E-04 |
| rs6557097 | 6 | PLEKHG1 | a | c | 0.371 | 0.015 | 0.0042 | 5.07E-04 |
| rs6724567 | 2 | GFPT1 | a | g | 0.376 | −0.013 | 0.0037 | 6.48E-04 |
| rs6959536 | 7 | MAGI2 | a | c | 0.345 | −0.013 | 0.0038 | 6.78E-04 |
| rs8054039 | 16 | DNAJA2 | t | c | 0.103 | 0.026 | 0.0077 | 7.01E-04 |
| rs3753490 | 1 | AGL | t | g | 0.144 | 0.017 | 0.005 | 7.57E-04 |
| rs2231328 | 13 | UFM1 | t | c | 0.018 | −0.054 | 0.016 | 8.21E-04 |
| rs7712111 | 5 | SLIT3 | a | g | 0.363 | −0.013 | 0.0038 | 8.74E-04 |
| rs6072269 | 20 | TOP1 | a | c | 0.155 | 0.016 | 0.0049 | 9.60E-04 |
| rs225710 | 6 | VTA1 | t | c | 0.496 | −0.012 | 0.0036 | 9.73E-04 |
The most significant SNPs within ±2 kb of the genes are shown. Additional data are presented in Supplementary Table 11. MAF, Minor allele frequency; Stderr, standard error; P-value, global meta-analysis p-values from MAGIC study.
Cis-regulatory SNPs modulate the expression of genes in pathways modulated by PPAR agonists
Our study identified several biological pathways in adipose tissue that are dysregulated in insulin-resistant subjects and restored by PPAR agonist treatment. We investigated whether local SNPs (± 500 kb) regulate transcript level expression of the genes in these key pathways in adipose tissue. Among the 189 selected genes annotated in four key pathways (BCAA metabolism, PPAR signaling, AMPK- and cAMP-mediated signaling, and propanoate metabolism), 28 genes were significantly regulated by local SNPs (cis-eQTLs) in subcutaneous adipose gene expression data from the MuTHER study [16] (Table 6 and Supplementary Table 12). Top cis-eQTLs included PRKAR1A (cAMP-dependent protein kinase regulatory-type I-alpha) regulated by SNP rs2302234 (p <1×10−12) and CD36 (lipoprotein receptor) regulated by SNP rs3211931 (p <1×10−12). Independent validation of these top cis-eQTLs was performed in adipose tissue of 89 Caucasian non–diabetic subjects utilizing microarray data followed by genotyping and real time q-PCR (see Supplementary Table 13 for genotyping and real-time q-PCR assay). Genome-wide gene expression data generated by microarray for these subjects were available from our previous study [29]. Genotyping of three selected SNPs (or their proxies) in this cohort replicated associations between PRKAR1A-rs6958 (additive model p-value = 3.2×10−27, in r2 =1 with rs2302234), CD36-rs7755 (additive model p-value = 1.38×10−11, in r2 =0.96 with rs3211931) and CD36- rs3211870 (additive model p-value = 0.0083). Regulatory SNPs for CD36 were further confirmed by gene expression data generated by real-time PCR in the same sample set. Individuals with the TC or TT genotype for SNP rs7755 showed higher expression of CD36 transcript in adipose tissue compared to individuals with the CC genotype (dominant model p-value =0.013). Thus, our study indicates that gene expression in key biological pathways modulated by PPAR agonists is under control of local genetic regulators.
Table 6.
Genotypic association of SNPs with expression of PPAR agonist-modulated genes in adipose tissue
| SNP ID | Gene Symbol | chr | SNP Position | Gene Position | Distance | twin1 | twin2 | ||
|---|---|---|---|---|---|---|---|---|---|
| r | p-value | r | p-value | ||||||
| rs2302234 | PRKAR1A | 17 | 64049834 | 64019705 | 30129 | 0.844 | 1.45E-22 | 0.678 | 5.75E-13 |
| rs3211931 | CD36 | 7 | 80136109 | 80069459 | 66650 | 0.701 | 6.31E-13 | 0.705 | 2.62E-14 |
| rs8074500 | ALDH3A2 | 17 | 19486426 | 19492641 | 6215 | −0.677 | 7.63E-12 | −0.52 | 2.38E-07 |
| rs17366568 | ADIPOQ | 3 | 188053147 | 188043157 | 9990 | 0.446 | 3.82E-05 | 0.538 | 7.65E-08 |
| rs17351864 | ALDH4A1 | 1 | 18696490 | 19101659 | 405169 | 0.439 | 5.19E-05 | 0.408 | 8.91E-05 |
| rs3856806 | PPARG | 3 | 12450557 | 12304070 | 146487 | 0.49 | 5.16E-06 | 0.397 | 0.0002 |
| rs4776832 | SMAD3 | 15 | 64861969 | 65145249 | 283280 | −0.468 | 1.39E-05 | −0.378 | 0.0003 |
| rs7584253 | LHCGR | 2 | 48780750 | 48836373 | 55623 | −0.401 | 0.0002 | −0.407 | 9.21E-05 |
| rs7590991 | HIBCH | 2 | 190821498 | 190917164 | 95666 | 0.419 | 0.0001 | 0.379 | 0.0003 |
| rs6989592 | DUSP4 | 8 | 28901570 | 29264104 | 362534 | 0.383 | 0.0005 | 0.418 | 5.70E-05 |
| rs1046108 | ACSL3 | 2 | 223515501 | 223433976 | 81525 | 0.471 | 1.17E-05 | 0.355 | 0.0008 |
| rs10747699 | PDE1B | 12 | 53241591 | 53229671 | 11920 | 0.344 | 0.0019 | 0.424 | 4.35E-05 |
| rs15401 | SLC27A1 | 19 | 17477747 | 17442300 | 35447 | 0.332 | 0.0028 | 0.314 | 0.0031 |
| rs851027 | MAPK14 | 6 | 36098853 | 36103551 | 4698 | −0.341 | 0.0021 | −0.303 | 0.0043 |
| rs2295885 | ACSS2 | 20 | 33177624 | 32926406 | 251218 | 0.396 | 0.0003 | 0.286 | 0.0072 |
| rs1897115 | HADHA | 2 | 26407567 | 26321098 | 86469 | 0.367 | 0.0009 | 0.284 | 0.0077 |
| rs11929529 | ACAA1 | 3 | 38149474 | 38153619 | 4145 | 0.294 | 0.0085 | 0.373 | 0.0004 |
| rs7223304 | ACOX1 | 17 | 71958685 | 71487039 | 471646 | 0.293 | 0.0088 | 0.36 | 0.0006 |
| rs4903245 | ALDH6A1 | 14 | 74117041 | 73620949 | 496092 | −0.3 | 0.0072 | −0.323 | 0.0022 |
| rs9859538 | P2RY12 | 3 | 152573653 | 152585229 | 11576 | 0.326 | 0.0035 | 0.287 | 0.0070 |
| rs2542532 | MCEE | 2 | 71349147 | 71210875 | 138272 | 0.325 | 0.0036 | 0.298 | 0.0073 |
| rs10030631 | SCD5 | 4 | 83815769 | 83939034 | 123265 | 0.312 | 0.0051 | 0.293 | 0.0059 |
| rs12021720 | DBT | 1 | 100444648 | 100487997 | 43349 | 0.344 | 0.0019 | 0.278 | 0.0092 |
| rs12451 | ILK | 11 | 6589026 | 6581540 | 7486 | −0.338 | 0.0023 | −0.278 | 0.0090 |
| rs4752973 | NR1H3 | 11 | 47225735 | 47227043 | 1308 | 0.306 | 0.0061 | 0.297 | 0.0052 |
| rs6849076 | ACOX3 | 4 | 8100613 | 8493352 | 392739 | −0.325 | 0.0035 | −0.276 | 0.0096 |
| rs1500988 | IRS1 | 2 | 227045601 | 227372719 | 327118 | −0.305 | 0.0062 | −0.283 | 0.0079 |
| rs11809495 | RXRG | 1 | 163859346 | 163681054 | 178292 | 0.298 | 0.0084 | 0.298 | 0.0058 |
Most significant SNPs within ±500 kb of PPAR modulated genes are shown. Additional data are presented in Supplementary Table 12. SNP and gene positions are based on NCBI build36 or hg18; r, Linear regression coefficient; p-value, unadjusted p-values from MuTHER study.
Discussion
Morbidity and mortality in T2D arises from long-term complications. Early detection and prevention of T2D is expected to have tremendous medical, economic, social, and human implications. Lifestyle modifications (weight loss and increased physical activity) can reduce the rate of conversion from impaired glucose tolerance to T2D, but it is difficult to achieve and maintain weight loss [30, 31]. In contrast, oral antidiabetic agents that improve insulin sensitivity do prevent progression of IGT to T2D [7, 8]. Utilizing a systematic genetic approach, we examined the mechanism of actions of pioglitazone (PPARγ agonist), fenofibrate (PPARα agonist), and their combination in IGT subjects in an unbiased and statistically robust manner. To our knowledge, this is the first study to compare effects of different PPAR ligands (pioglitazone, fenofibrate, or both) on adipose tissue and muscle transcripts to elucidate mechanistic differences at the genome-wide transcriptional network and biological pathway levels.
Our study was not designed to definitively show whether the combination or monotherapy is more effective in preventing the progression of IGT to T2D, but provided mechanistic information that could expedite the development of more efficacious therapy against insulin resistance. This study delineated the pathways involved in PPAR agonist-mediated improvement of insulin sensitivity in IGT subjects. Utilizing publicly available genomic databases, we explored the role of regulatory polymorphisms that may modulate the expression of genes in these pathways. These findings will form the basis of larger clinical trials to understand the role of SNPs in modulating the PPAR agonist-mediated gene expression response that might determine the outcome of therapeutic response.
Despite several advantages, our study cohort had limited statistical power to confidently detect smaller changes in PPAR agonist-mediated gene expression. Thus, we used moderately stringent statistical thresholds to avoid false-positive results. Our sample set also was not sufficiently powered to detect gender-specific effects of drug response, and we have not studied differential expression in response to PPAR agonists after stratifying for sex. We compared pre-and post-PPAR agonist treatment differences within an individual (two-class paired sample analysis) to minimize effects of gender-specific differences in expression response. Our study was focused on changes in expression at the mRNA level after PPAR agonist treatment. However, PPAR agonist-mediated improvement is insulin sensitivity may also occur via changes in other molecular phenotypes, including miRNA, protein and metabolite levels, and chromatin or epigenetic modifications. More inclusive studies will be required to expand our knowledge on molecular mechanisms of PPAR agonist-mediated improvement of insulin sensitivity.
A recent study in mice indicated that rosiglitazone (a PPARγ ligand) plus fenofibrate was more effective in improving glycemic control compared to monotherapy with either agent [10]. Treatment of IGT subjects with either pioglitazone alone or pioglitazone and fenofibrate in our study mediated similar improvement in insulin sensitivity (SI), as measured by the minimal model analysis of the FSIGT data. However, the combination of pioglitazone and fenofibrate had more favorable effects on lipid profiles compared to either PPARα or γ ligand monotherapy. Interestingly, enrichment analyses indicated that those taking the combined therapy had a stronger adipose tissue upregulation of genes involved in fatty acid metabolism, BCAA degradation, the TCA cycle, and oxidative phosphorylation, and downregulation of genes in antigen processing and presentation, when compared to pioglitazone monotherapy. Significant enrichment of genes involved in propanoate metabolism, fatty acid elongation in mitochondria, and the acetyl-CoA metabolic process was only observed in the pioglitazone plus fenofibrate group. Our study indicates that this combination therapy was more effective in normalizing the impaired gene expression pattern in insulin-resistant subjects, and may prove more effective in prevention or treatment of T2D in larger clinical trials.
PPARγ is expressed predominantly in adipose tissue, and a recent study in transgenic mice showed that selective activation of PPARγ in adipocytes is sufficient for whole-body insulin sensitization equivalent to systemic TZD treatment [32]. Thus, we have not studied the effect of pioglitazone monotherapy on skeletal muscle transcripts. Since PPARα is highly expressed in muscle, in our current study we analyzed the effect of fenofibrate alone or in combination with pioglitazone on the muscle transcriptome. Fenofibrate monotherapy did not improve insulin sensitivity, unlike the combination of fenofibrate and pioglitazone.
It may appear surprising that we found no statistically significantly enriched pathway in the skeletal muscle transcriptome with PPAR agonist treatment, even though there was a significant increase in insulin sensitivity (SI), which reflects peripheral glucose disposal. Our muscle sample set had 85% power (at per gene α = 0.05 and SD[log2] of gene intensity = 0.7) to detect a 2-fold change in gene expression; the power of our sample set was low to detect changes smaller than that. This relative lack of power may explain some of our inability to detect differentially expressed transcripts in muscle. As noted in the Results section, several genes involved in lipid metabolism, including FABP4 and DGAT2, showed differential expression after PPAR agonist treatment in muscle, but failed to meet our FDR criteria. However, the statistical power of our adipose data set was only marginally higher than our muscle data set. Thus, we speculate that the PPAR agonist-mediated improvement in peripheral insulin resistance is independent of its role in modulating transcription in muscle and could be posttranscriptional. Several recent studies pointed out PPAR-dependent and independent non-genomic mechanisms that may explain PPAR agonist-mediated improvement in insulin sensitivity [33]. Those studies indicated that insulin-sensitizing effects of PPAR gamma agonists in human skeletal muscle are partly dependent on activation of the fuel-sensing enzyme AMPK (α-Thr172 phosphorylation of AMP-activated protein kinase directly, or indirectly via increase in adiponectin from adipocytes), phosphorylation (inactivation) of acetyl-coA carboxylase (ACC), and inhibition of pyruvate dehydrogenase activity in skeletal muscle[34, 35].
Alternatively, the changes in adipose tissue alone probably can drive peripheral glucose disposal. Our earlier study showed that pioglitazone improved insulin sensitivity significantly along with a 34% decrease in intramyocellular lipids [4]; however this study found no change in the expression of genes involved in lipid oxidation in muscle. On the other hand, as discussed above, treatment with PPARγ agonists induced significant upregulation of genes in lipid metabolism in subcutaneous adipose tissue, supporting the hypothesis that the primary mechanism of TZD–induced skeletal muscle insulin sensitivity is enhanced partitioning of lipids into adipose tissue, relieving lipotoxicity in muscle. However, a recent study by Sears et al [36] in a cohort of insulin-sensitive and -resistant subjects (including diabetics) concluded that TZD (pioglitazone, rosiglitazone or troglitazone)-mediated insulin sensitization is partly mediated by upregulation of transcripts involved in adipogenesis in muscle or intramuscular adipocyte differentiation. Higher expression of genes in lipid uptake/storage and adipogenesis (including FABP4, PLIN and CEBPA) was observed in insulin-resistant versus insulin-sensitive subjects, and the difference was increased upon TZD treatment. That study did not indicate enrichment of any biological pathway among genes differentially expressed by TZD in muscle, but showed amelioration of defective insulin-stimulated HK2 and PDK4 expression (genes in glycolytic flux) in insulin-resistant subjects treated with TZD, in a post-hyperinsulinemic clamp condition. A different study in Wistar rats indicated that TZD increased fatty acid uptake (via upregulation of CD36) and triglyceride accumulation in skeletal muscle when stimulated by insulin [37]. We analyzed our transcript profiles in fasting conditions, and in non-diabetic subjects, and thus may have failed to detect changes observed in other studies that included diabetic patients and which examined insulin-stimulated conditions.
We observed an overlap between biological pathways dysregulated in insulin-resistant subjects that were modulated upon PPAR ligand treatment in adipose tissue. This analysis helped us refine the biochemical and physiological mechanisms involved in PPAR agonist-mediated improvement of insulin sensitivity in IGT subjects. Several cell biological, metabolomic and animal studies also support the role of these pathways (including BCAA metabolism, PPAR signaling, AMPK- and cAMP-mediated signaling, and fatty acid and propanoate metabolism) in insulin resistance [38–41].
Our study also provides evidence for several novel mechanisms involved in PPAR agonist-mediated improvement of insulin sensitivity in human subjects. Two novel regulatory networks involved in lipid metabolism were modulated in subcutaneous adipose tissue of subjects treated with PPARγ agonists in our study. Network 4 indicates a link between PPAR agonist-mediated upregulation of adiponectin and upregulation of lipid mobilization factor zincα2-glycoprotein. This network was predicated to alleviate inflammation by downregulating macrophage scavenger receptor (MSR1). Network 14 supports a regulatory interaction between PPARγ agonist-mediated upregulation of lipid-metabolizing enzymes (ACACA, CETP, DGAT2, FASN and SCD) and nuclear receptors (NCOA4, NCOR1, NR1D2 and NR1H3). Additionally, we observed enrichment of genes in several pathways, including nuclear factor of activated T cells (NFAT) signaling, relaxin signaling, renin-angiotensin signaling, nitric oxide signaling, and molecular mechanisms of cancer that are modulated by PPAR agonists in adipose but were not differentially expressed in insulin-resistant subjects. We speculate that these pathways may be involved in PPARγ agonist-mediated adverse effects. Future efforts of designing new oral antidiabetic agents should devise strategies to avoid activating these pathways.
Our integrative genomic analysis indicated that SNPs in PPAR agonist-modulated genes are associated with glucose homeostasis traits and may modulate insulin sensitivity. Transcript-level expression of several genes, including PRKAR1A and CD36, in these important pathways are also regulated by cis regulatory SNPs. CD36 is a high-affinity receptor for lipoproteins (including oxidized LDL), and a recent study confirmed the role of polymorphisms in regulating its transcription and protein expression [42, 43]. Further studies are required to determine if these regulatory polymorphisms can explain the observed inter-individual variability in PPAR agonist-mediated transcriptional response.
There is precedent for the successful application of pharmacogenetic concepts to monogenic forms of diabetes, such as maturity-onset diabetes of the young. A similar strategy for the common form of T2D is currently lacking [44]. Applying integrative genomics approaches [45] that leverage the utilization of data from GWAS studies of disease or drug response traits; eQTL data for drug target tissues; information on drug-mediated perturbation of pathways and biological networks; and effects of eQTLs on drug-mediated modulation those networks may improve our understanding of individual-level variability in drug response. Although preliminary so far, our study shows promise that such integrative strategy may be useful in: i) more effective use of existing antidiabetic agents, ii) designing new drug molecules that bypass shortcomings of currently available ones; iii)developing new therapeutic modalities including combination therapy; and iv) better understanding of the underlying disease mechanisms.
Conclusions
Using an integrative biology approach, we present a comprehensive characterization of PPAR agonist-mediated transcriptional response in adipose and muscle tissues of individuals with impaired glucose tolerance. Our study defined biological pathways and gene networks in adipose tissue that are changed following improvement of insulin sensitivity with PPAR treatment, and these adipose tissue changes correspond well with biological pathway differences that distinguished insulin-resistant from insulin-sensitive subjects. However, PPAR treatment resulted in remarkably few changes in muscle gene expression. We identified cis-regulatory SNPs associated with the expression of PPAR agonist-modulated transcripts. SNPs in a subset of PPAR agonist-modulated genes were associated with glucose homeostasis traits and may modulate insulin sensitivity. These SNPs are important candidates for future pharmacogenomic studies that may explain the inter-individual variability in TZD and fenofibrate response to treat or prevent diabetes.
Supplementary Material
Acknowledgments
This study was supported by a VA merit grant (NR), a grant from the Sturgis Foundation to the University of Arkansas for Medical Sciences, NIH grants DK039311 (SKD and SCE), DK071349 (PAK), M01RR14288, and UL1RR033173, and Wake Forest School of Medicine research development funds (SCE and SKD). We thank the dedicated staff of the Clinical Research Center at UAMS for support of the clinical studies and we thank Karen Klein (Research Support Core, Office of Research, Wake Forest School of Medicine) for critical reading and editing of our manuscript. The medications used in this study were generously provided by Takeda and Abbott Pharmaceuticals.
Footnotes
Conflicts of interest: NR has received investigator-initiated grants from Takeda and Abbott Pharmaceuticals. PAK has an investigator-initiated grant from Glaxo Smith Kline. SKD, SCE, and NKS declare no conflicts of interest.
References
- 1.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 2.Yki-Jarvinen H. Thiazolidinediones. N Engl J Med. 2004;351:1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 3.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 4.Rasouli N, Raue U, Miles LM, Lu T, Di Gregorio GB, Elbein SC, et al. Pioglitazone improves insulin sensitivity through reduction in muscle lipid and redistribution of lipid into adipose tissue. Am J Physiol Endocrinol Metab. 2005;288:E930–E934. doi: 10.1152/ajpendo.00522.2004. [DOI] [PubMed] [Google Scholar]
- 5.Di Gregorio GB, Yao-Borengasser A, Rasouli N, Varma V, Lu T, Miles LM, et al. Expression of CD68 and macrophage chemoattractant protein-1 genes in human adipose and muscle tissues: association with cytokine expression, insulin resistance, and reduction by pioglitazone. Diabetes. 2005;54:2305–2313. doi: 10.2337/diabetes.54.8.2305. [DOI] [PubMed] [Google Scholar]
- 6.Nathan DM, Davidson MB, DeFronzo RA, Heine RJ, Henry RR, Pratley R, et al. Impaired fasting glucose and impaired glucose tolerance: implications for care. Diabetes Care. 2007;30:753–759. doi: 10.2337/dc07-9920. [DOI] [PubMed] [Google Scholar]
- 7.DeFronzo RA, Abdul-Ghani M. Type 2 diabetes can be prevented with early pharmacological intervention. Diabetes Care. 2011;34(Suppl 2):S202–S209. doi: 10.2337/dc11-s221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFronzo RA, Tripathy D, Schwenke DC, Banerji M, Bray GA, Buchanan TA, et al. Pioglitazone for diabetes prevention in impaired glucose tolerance. N Engl J Med. 2011;364:1104–1115. doi: 10.1056/NEJMoa1010949. [DOI] [PubMed] [Google Scholar]
- 9.Igarashi M, Jimbu Y, Kimura M, Hirata A, Yamaguchi H, Tominaga M. Effect of pioglitazone on atherogenic outcomes in type 2 diabetic patients: a comparison of responders and non-responders. Diabetes Res Clin Pract. 2007;77:389–398. doi: 10.1016/j.diabres.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Choi WS, Lee JJ, Kim Y, Kim IS, Zhang WY, Myung CS. Synergistic improvement in insulin resistance with a combination of fenofibrate and rosiglitazone in obese type 2 diabetic mice. Arch Pharm Res. 2011;34:615–624. doi: 10.1007/s12272-011-0412-9. [DOI] [PubMed] [Google Scholar]
- 11.Han KL, Choi JS, Lee JY, Song J, Joe MK, Jung MH, et al. Therapeutic potential of peroxisome proliferators--activated receptor-alpha/gamma dual agonist with alleviation of endoplasmic reticulum stress for the treatment of diabetes. Diabetes. 2008;57:737–745. doi: 10.2337/db07-0972. [DOI] [PubMed] [Google Scholar]
- 12.Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, et al. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes. 2011;60:1019–1029. doi: 10.2337/db10-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, et al. GeneTrail--advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 21.Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bing C, Mracek T, Gao D, Trayhurn P. Zinc-alpha2-glycoprotein: an adipokine modulator of body fat mass? Int J Obes (Lond) 2010;34:1559–1565. doi: 10.1038/ijo.2010.105. [DOI] [PubMed] [Google Scholar]
- 24.Ouchi N, Kihara S, Arita Y, Nishida M, Matsuyama A, Okamoto Y, et al. Adipocyte-derived plasma protein, adiponectin, suppresses lipid accumulation and class A scavenger receptor expression in human monocyte-derived macrophages. Circulation. 2001;103:1057–1063. doi: 10.1161/01.cir.103.8.1057. [DOI] [PubMed] [Google Scholar]
- 25.Gauthier MS, O’Brien EL, Bigornia S, Mott M, Cacicedo JM, Xu XJ, et al. Decreased AMP-activated protein kinase activity is associated with increased inflammation in visceral adipose tissue and with whole-body insulin resistance in morbidly obese humans. Biochem Biophys Res Commun. 2011;404:382–387. doi: 10.1016/j.bbrc.2010.11.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeBrasseur NK, Kelly M, Tsao TS, Farmer SR, Saha AK, Ruderman NB, et al. Thiazolidinediones can rapidly activate AMP-activated protein kinase in mammalian tissues. Am J Physiol Endocrinol Metab. 2006;291:E175–E181. doi: 10.1152/ajpendo.00453.2005. [DOI] [PubMed] [Google Scholar]
- 27.Saha AK, Avilucea PR, Ye JM, Assifi MM, Kraegen EW, Ruderman NB. Pioglitazone treatment activates AMP-activated protein kinase in rat liver and adipose tissue in vivo. Biochem Biophys Res Commun. 2004;314:580–585. doi: 10.1016/j.bbrc.2003.12.120. [DOI] [PubMed] [Google Scholar]
- 28.Daval M, Foufelle F, Ferre P. Functions of AMP-activated protein kinase in adipose tissue. J Physiol. 2006;574:55–62. doi: 10.1113/jphysiol.2006.111484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das SK, Sharma NK, Hasstedt SJ, Mondal AK, Ma L, Langberg KA, et al. An integrative genomics approach identifies activation of thioredoxin/thioredoxin reductase-1-mediated oxidative stress defense pathway and inhibition of angiogenesis in obese nondiabetic human subjects. J Clin Endocrinol Metab. 2011;96:E1308–E1313. doi: 10.1210/jc.2011-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 32.Sugii S, Olson P, Sears DD, Saberi M, Atkins AR, Barish GD, et al. PPARgamma activation in adipocytes is sufficient for systemic insulin sensitization. Proc Natl Acad Sci U S A. 2009;106:22504–22509. doi: 10.1073/pnas.0912487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luconi M, Cantini G, Serio M. Peroxisome proliferator-activated receptor gamma (PPARgamma): Is the genomic activity the only answer? Steroids. 2010;75:585–594. doi: 10.1016/j.steroids.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Coletta DK, Sriwijitkamol A, Wajcberg E, Tantiwong P, Li M, Prentki M, et al. Pioglitazone stimulates AMP-activated protein kinase signalling and increases the expression of genes involved in adiponectin signalling, mitochondrial function and fat oxidation in human skeletal muscle in vivo: a randomised trial. Diabetologia. 2009;52:723–732. doi: 10.1007/s00125-008-1256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fediuc S, Pimenta AS, Gaidhu MP, Ceddia RB. Activation of AMP-activated protein kinase, inhibition of pyruvate dehydrogenase activity, and redistribution of substrate partitioning mediate the acute insulin-sensitizing effects of troglitazone in skeletal muscle cells. J Cell Physiol. 2008;215:392–400. doi: 10.1002/jcp.21321. [DOI] [PubMed] [Google Scholar]
- 36.Sears DD, Hsiao G, Hsiao A, Yu JG, Courtney CH, Ofrecio JM, et al. Mechanisms of human insulin resistance and thiazolidinedione-mediated insulin sensitization. Proc Natl Acad Sci U S A. 2009;106:18745–18750. doi: 10.1073/pnas.0903032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd MK, Watt MJ, Le J, Hevener AL, Turcotte LP. Thiazolidinediones enhance skeletal muscle triacylglycerol synthesis while protecting against fatty acid-induced inflammation and insulin resistance. Am J Physiol Endocrinol Metab. 2007;292:E485–E493. doi: 10.1152/ajpendo.00080.2006. [DOI] [PubMed] [Google Scholar]
- 38.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah SH, Svetkey LP, Newgard CB. Branching out for detection of type 2 diabetes. Cell Metab. 2011;13:491–492. doi: 10.1016/j.cmet.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh A, Murugesan G, Chen K, Zhang L, Wang Q, Febbraio M, et al. Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood. 2011;117:6355–6366. doi: 10.1182/blood-2011-02-338582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gautam S, Banerjee M. The macrophage Ox-LDL receptor, CD36 and its association with type II diabetes mellitus. Mol Genet Metab. 2011;102:389–398. doi: 10.1016/j.ymgme.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Huang C, Florez JC. Pharmacogenetics in type 2 diabetes: potential implications for clinical practice. Genome Med. 2011;3:76. doi: 10.1186/gm292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasarskis A, Yang X, Schadt E. Integrative genomics strategies to elucidate the complexity of drug response. Pharmacogenomics. 2011;12:1695–1715. doi: 10.2217/pgs.11.115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.