Abstract
Pancreatic ductal adenocarcinoma (PDA) remains a lethal malignancy despite tremendous progress in its molecular characterization. Indeed, PDA tumors harbor four signature somatic mutations1–4, and a plethora of lower frequency genetic events of uncertain significance5. Here, we used Sleeping Beauty (SB) transposon-mediated insertional mutagenesis6,7 in a mouse model of pancreatic ductal preneoplasia8 to identify genes that cooperate with oncogenic KrasG12D to accelerate tumorigenesis and promote progression. Our screen revealed new candidates and confirmed the importance of many genes and pathways previously implicated in human PDA. Interestingly, the most commonly mutated gene was the X-linked deubiquitinase Usp9x, which was inactivated in over 50% of the tumors. Although prior work had attributed a pro-survival role to USP9X in human neoplasia9, we found instead that loss of Usp9x enhances transformation and protects pancreatic cancer cells from anoikis. Clinically, low USP9X protein and mRNA expression in PDA correlates with poor survival following surgery, and USP9X levels are inversely associated with metastatic burden in advanced disease. Furthermore, chromatin modulation with trichostatin A or 5-aza-2′-deoxycytidine elevates USP9X expression in human PDA cell lines to suggest a clinical approach for certain patients. The conditional deletion of Usp9x cooperated with KrasG12D to rapidly accelerate pancreatic tumorigenesis in mice, validating their genetic interaction. Therefore, we propose USP9X as a major new tumor suppressor gene with prognostic and therapeutic relevance in PDA.
The biological sequelae of PDA has been partially attributed to frequent and well characterized mutations in KRAS (>90%), CDKN2A (>90%), TP53 (70%) and SMAD4 (55%)1–4. Recent genome-wide analyses have uncovered numerous additional somatic genetic alterations, although the functional relevance of most remains uncertain5. To explore the molecular genesis of PDA we previously generated a mouse model of Pancreatic Intraepithelial Neoplasia (mPanIN) by conditionally expressing an endogenous KrasG12D allele in the developing pancreas8. Mice with mPanIN spontaneously progress to mouse PDA (mPDA) after a long and variable latency, providing an opportunity to characterize genes that cooperate with KrasG12D to promote early mPDA. We hypothesized that such genes could be directly identified by applying insertional mutagenesis strategies6,7,10,11 in our mPanIN model, and that these candidates could represent “drivers” of PDA development.
Accordingly, we interbred our mPanIN model with two distinct Sleeping Beauty (SB) transposon systems and monitored mice for early disease progression. Our initial approach utilized the well characterized CAGGS-SB10 transgenic allele to promote transposition6. Although CAGGS-SB10 promoted PDA, a variety of non-pancreatic neoplasms and a paucity of identified Common Insertion Sites (CISs) in the recovered pancreatic neoplasms precluded a comprehensive analysis, potentially reflecting the variegated expression of CAGGS-SB1012 (Supplementary Figures 1a and 2; Supplementary Tables 1 and 3b).
To increase the specificity and potency of SB mutagenesis, we generated a conditional SB13 mutant mouse by targeting the Rosa26 locus in embryonic stem cells (Supplementary Fig. 3a, b). The pancreatic specific expression and function of the conditional SB13 allele was confirmed (Supplementary Fig. 3c), and we found that SB13-induced transposition by itself did not promote lethality or pancreatic tumorigenesis (Fig. 1a, Supplementary Fig. 4a). In contrast, KCTSB13 mice (KrasLSL-G12D; Pdx1-cre; T2/Onc; Rosa26-LSL-SB13) rapidly progressed and succumbed to invasive pancreatic neoplasms (Fig. 1a–c). A cohort of 117 KCTSB13 mice (Supplementary Fig. 1b) was monitored for tumor development, and 103 of these mice were available for full necropsy and tissue procurement. The majority of such mice harbored multi-focal pancreatic tumors, and 198 distinct primary tumors and metastases were subjected to histological and molecular analysis. Most mice had invasive carcinomas (66/103) that consisted of classical mPDA (78.8%) or invasive cystic neoplasms (21.2%); 34.8% of mice also contained metastases predominantly in their liver and lungs (Supplementary Fig. 4c). The remainder of the mice (37/103) had preinvasive pancreatic tumors consisting of high grade mPanIN and cyst-forming papillary neoplasms (Supplementary Fig. 4b).
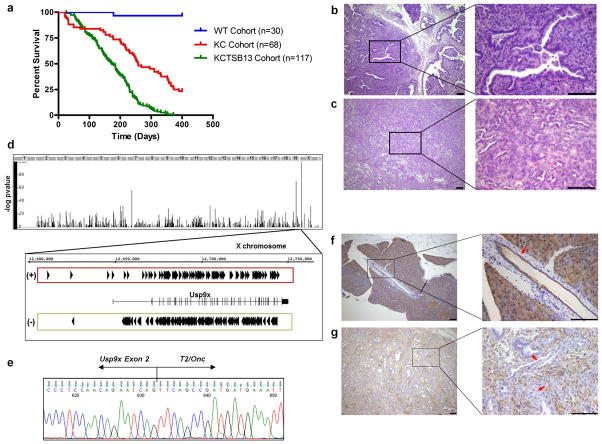
Figure 1. Transposon mutagenesis accelerates murine PDA and targets Usp9x.
a, Increased mortality of KCTSB13 mice compared to KC cohort (containing KCT, KCSB13 and KC mice) (172 vs. 257 days, p<0.001; long-rank test). Wild-type (WT) cohort is comprised of KTSB13 and CTSB13 mice. b–c, Invasive cystic neoplasm (b), and mPDA (c) in KCTSB13 mice. Scale bar: 100μm. d, Usp9x is the major CIS in KCTSB13 PDA tumors (X-axis denotes genome, Y-axis −log P-value), with bidirectional insertions. (+) parallel to Usp9x expression, (−) antiparallel. e, Usp9x Exon 2-T2/Onc chimeric mRNA in SB13 tumors. f–g, Usp9x protein expression in normal pancreatic ducts (arrow) (f), but not in neoplastic cells (g) (arrows) in SB13 PDA harboring Usp9x insertions. Scale bar: 100μm.
The candidate genes identified from the SB13 screen represented unanticipated candidates as well as many genes and pathways previously implicated in human PDA (Table 1, Supplementary Tables 2, 3a and 4). Indeed, various members of the TGFβ pathway, including Smad3, Smad4, Tgfbr1 and Tgfbr2, were collectively mutated in 32% of the tumors. Also, the Rb/p16Ink4a pathway was disrupted in 21% of the tumors. CISs representing the orthologues of additional human PDA genes included Fbxw7 (24.2%), Arid1a (19.1%), Acvr1b (19%), Stk11/Lkb1 (6.5 %), Mll3 (6%), Smarca4 (6%) and Pbrm1 (4.5%)5,13–15. Trp53 was the only commonly mutated PDA gene conspicuously absent, although the p53 regulatory deubiquitinase Usp7 was a CIS (6.5%)16. Several CISs previously noted in insertional mutagenesis screens for hepatocellular carcinoma or gastrointestinal tract adenomas, but not typically mutated in PDA, were also identified in this study, including Zbtb20, Nfib and Ube2h in liver tumors10; and Pten, Tcf12, Ppp1r12a, and Ankrd11 in gastrointestinal tract adenoma/adenocarcinoma11. This indicates that many tumor progression pathways may be common to pancreatic, liver and gastrointestinal/colorectal tumors.
Table 1. Top 20 candidate genes that cooperate with KrasG12D to promote mPDA in KCTSB13 mice.
CISs were scored by tumor frequency with the narrowest 15K kernel spatial distribution of insertion sites. Chr: chromosome; N: number of tumors from which the CIS was found; I, total number of insertions of the CIS in the indicated tumors.
| Gene | Chr | CIS Peak Location | CIS Height | N | I | Mutation in humans |
|---|---|---|---|---|---|---|
|
| ||||||
| Usp9x | X | 12691773 | 158.1266 | 101 | 341 | |
| Pten | 19 | 32872602 | 64.5204 | 61 | 96 | |
| Fndc3b | 3 | 27562591 | 13.7096 | 55 | 67 | |
| Setd5 | 6 | 113057997 | 35.6176 | 52 | 71 | |
| Arfip1|Fbxw7 | 3 | 84769635 | 21.6666 | 48 | 80 | Yes5 |
| Fam193a | 5 | 34705809 | 24.3555 | 45 | 78 | |
| Ctnna1 | 18 | 35342868 | 20.2017 | 45 | 50 | |
| Magi1 | 6 | 93859940 | 13.3715 | 43 | 57 | |
| Mkln1 | 6 | 31414109 | 16.5263 | 41 | 53 | |
| Pum1 | 4 | 130288478 | 12.7948 | 41 | 46 | Yes5 |
| Farp1 | 14 | 121587858 | 9.407 | 39 | 47 | |
| Foxp1 | 6 | 98921646 | 19.5831 | 38 | 60 | |
| Arid1a | 4 | 133268936 | 32.1628 | 38 | 47 | Yes5 |
| Acvr1b | 15 | 101024934 | 31.1752 | 38 | 47 | Yes15 |
| Map4k3 | 17 | 81109860 | 13.2385 | 38 | 45 | Yes5 |
| Stag2 | X | 39535994 | 16.8613 | 37 | 48 | |
| Mll5 | 5 | 22982314 | 16.0001 | 37 | 43 | Yes5 |
| Atxn2|Sh2b3 | 5 | 122267680 | 12.3174 | 37 | 41 | |
| Arhgap5 | 12 | 53644560 | 37.416 | 35 | 61 | |
| Gsk3b | 16 | 38106972 | 21.79 | 35 | 43 | |
Unexpectedly, the most frequent CIS observed was the X-linked deubiquitinase Usp9x, a gene that had not been previously associated with PDA or other types of carcinoma in humans or mouse models. Indeed, the COSMIC data base revealed only one USP9X mutation in a case of ovarian cancer, although the functional relevance of this mutation has not been characterized (COSMIC mutation ID: 73237). Usp9x was disrupted in over 50% of all tumors, with 341 insertions noted in the 101 tumors harboring this CIS (Fig. 1d, Table 1). Furthermore, Usp9x was also identified as a CIS in 4 samples from the initial SB10 screen (Supplementary Table 1), supporting its candidacy as a PDA genetic determinant. We confirmed that Usp9x was disrupted in tumors by isolating chimeric fusion mRNAs that spliced the Usp9x transcript to the T2/Onc transposon (Fig. 1e). In addition, the Usp9x protein was specifically absent in neoplastic cells in pancreatic tumors bearing intragenic insertions (Fig. 1f, g).
To characterize the cellular and molecular pathways affected by Usp9x in PDA, RNAi was used to deplete Usp9x in mPDA cell lines (Supplementary Fig. 5a). While Usp9x depletion did not affect the proliferation of monolayer cultures (Supplementary Fig. 5b), it significantly increased colony formation in soft agar (Fig. 2a, Supplementary Fig. 5c), compared to cells transfected with scrambled shRNAs. Additionally, Usp9x knock-down potently suppressed anoikis in mPDA cells (Fig. 2b). These properties of Usp9x were predominantly dependent upon its intrinsic deubiquitinase activity (Supplementary Fig. 6a, b).
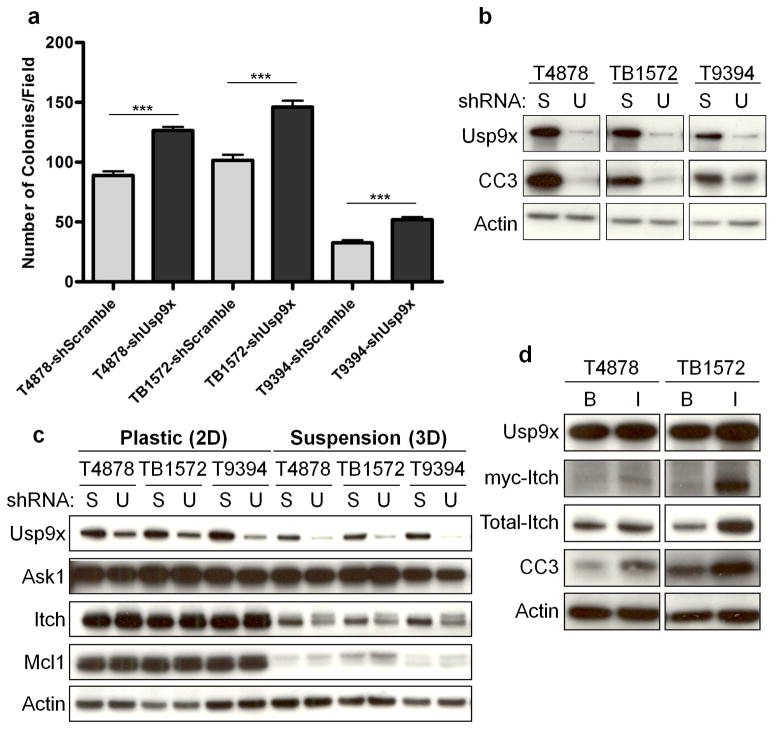
Figure 2. Usp9x regulates PDA cellular transformation and Itch.
a–b, Usp9x knock-down promotes anchorage-independent growth in three mPDA cell lines (a), and decreases anoikis denoted by cleaved caspase 3 (CC3) (b). The mean and s.e.m. of one representative experiment performed in triplicate are shown in (a) (***, p<0.001; Mann Whitney test). (S: Scramble; U: Usp9x). c, Usp9x knock-down decreases Itch but not Ask1 or Mcl1. Changes in Itch are more evident in suspension cultures, and the slower migrating band has the expected mobility of mono-ubiquitinated Itch. d, Ectopic Itch induces anoikis. (B: pBabe-neo; I: pBabe-neo-myc-Itch).
Since USP9X was previously reported to positively regulate SMAD4 transcriptional activity17 and SMAD4 is commonly mutated in PDA4, we hypothesized that Usp9x loss would attenuate Smad4 function or TGFβ responsiveness in PDA cell lines. However, irrespective of Usp9x expression level, mPDA cell lines expressed Smad4 and were equally sensitive to p21 induction, growth inhibition and morphological alterations following exposure to TGF-β1 (Supplementary Fig. 7a–d). Therefore we were unable to ascribe a specific role to Usp9x in the regulation of the Smad4/TGFβ pathway in mPDA cells or tumors.
We next investigated several additional proteins reported to be regulated by Usp9x and involved in pathways relevant to cellular transformation. Although USP9X was previously shown to bind to and regulate two proteins involved in cell survival, ASK118 and MCL19,19, we could not detect obvious changes in Ask1 or Mcl1 protein levels upon Usp9x loss (Fig. 2c). Usp9x has also been reported to deubiquitinate and thereby stabilize the E3 ligase Itch20, decreased protein levels of Itch were observed in mouse and human PDA cells upon the depletion of Usp9x (Fig. 2c, Supplementary Fig. 8a). Importantly, ectopic Itch expression was sufficient to promote anoikis in mPDA cells (Fig. 2d), and Itch was partially responsible for the ability of Usp9x to promote anoikis and suppress colony formation (Supplementary Fig. 6c, d). Since Itch is known to promote the degradation of several proteins relevant to cell proliferation and survival21, we evaluated the protein expression of likely candidates including c-Jun, p63 and c-FLIP but observed no alterations (Supplementary Fig. 8b). Furthermore, the Itch gene was identified as a CIS in 13% of cases (Supplementary Table 2). Therefore, Usp9x mutation may promote tumorigenesis in part by disabling Itch function, and the Usp9x/Itch pathway may work to constrain pancreatic tumorigenesis.
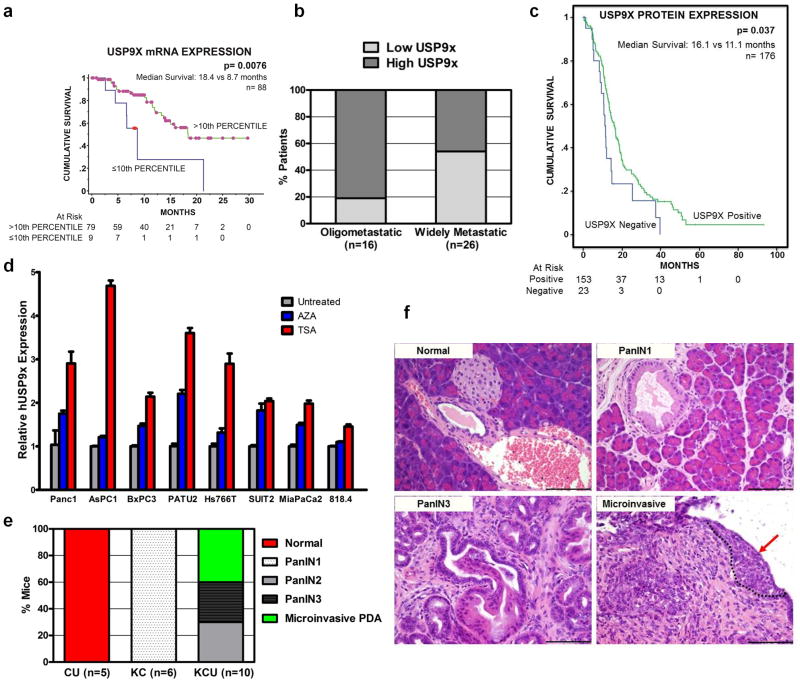
To determine whether USP9X expression is aberrant in human PDA, three distinct patient cohorts were assessed. First, we analyzed a cohort of 100 Australian patients who underwent surgery for localized PDA and had detailed information available concerning clinical-pathological characteristics and outcome (Supplementary Fig. 9, Supplementary Tables 5, 6). Tumor DNA from 88 patients in this cohort failed to yield somatic mutations in USP9X, consistent with prior reports5 (data not shown). Importantly, the low expression of USP9X mRNA correlated with poor survival following surgery (p=0.0076) (Fig. 3a), and multivariate analysis revealed that USP9X expression was an independent poor prognostic factor following surgery (Supplementary Table 7). We next analyzed autopsy specimens from a separate cohort of 42 American patients to determine that USP9X protein expression inversely correlated with a widespread metastatic pattern (p=0.0212) (Fig. 3b), and bore no relation to SMAD4 expression (Supplementary Table 8). A third collection of PDA specimens obtained from resected German patients (n=404) were used to determine that USP9X and ITCH protein levels were decreased (Supplementary Fig. 10a, b) and concordant (Spearman-Rho correlation: 0.47; p<0.01) (Supplementary Table 9a) in tumors compared to normal pancreatic tissue. Additionally, the proportion of tumors that had undetectable USP9X (13.6%) or ITCH (30.5%) protein correlated with a worse outcome (Supplementary Fig. 11, Supplementary Table 9b, c), particularly regarding USP9X in the subset of high grade tumors (Fig. 3c, Supplementary Tables 10 and 11). Collectively, these findings implicate the loss of USP9X expression as a relevant event in human pancreatic cancer progression.
Figure 3. USP9X loss promotes PDA.
a–c, Decreased USP9X expression correlates with shortened survival in Australian post-surgical cohort (a) (8.7 vs. 18.4 months, p=0.0076; log-rank test), increased metastatic burden in American autopsy series (b) (54% vs. 19%, p=0.0212; Fisher’s exact test), and diminished survival in German post-surgical cohort (c) (11.1 vs. 16.1 months, p=0.037; log-rank test). d, Trichostatin A (TSA, Red) and 5-Aza-2′-deoxycytidine (AZA, Blue) modestly increase USP9X mRNA expression. The mean and s.e.m. of one representative experiment performed in triplicate are shown. e, Usp9x deletion promotes mPanIN progression in KCU mice (p<0.0001; Fisher’s exact test). f, Representative normal pancreas (CU), mPanIN1 (KC), mPanIN3 (KCU) and microinvasive mPDA (KCU, arrow, circled). Scale bar: 100μm.
We found that USP9X was expressed throughout murine and human tumor development and lost focally in PDAs (Supplementary Figures 12, 13). Additionally, human PDA cell lines expressed lower levels of USP9X compared to non-PDA cancer cell lines (Supplementary Fig. 14). To investigate additional potential mechanisms of USP9X regulation in PDA, human cell lines were treated with the DNA methylase inhibitor 5-Aza-2′-deoxycytidine and the HDAC inhibitor trichostatin A. Both inhibitors modestly increased the USP9X mRNA and protein levels in most cell lines, suggesting that USP9X may be epigenetically silenced in vivo (Fig. 3d and Supplementary Fig. 15). Furthermore, although the promoter region of USP9X was not heavily methylated in tumor samples or PDA cells harboring low protein expression (data not shown), treatment with 5-Aza-2′-deoxycytidine did decrease colony formation of human PDA cells and this was partially reversed by concomitantly knocking-down USP9X (Supplementary Fig. 16).
To confirm that KrasG12D cooperated with Usp9x loss to promote pancreatic cancer, a conditional Usp9xflallele was generated (Supplementary Fig. 17a) and interbred with KrasLSL-G12D; Pdx1-cre mice to evaluate the impact on mPanIN progression. The mosaic expression of Usp9x in pancreas from Pdx1-cre; Usp9xfl/y mice was confirmed by immunohistochemistry (Supplementary Fig. 17b). We found that all hemizygous male mice and heterozygous female mice carriers of the Usp9xfl allele in the background of KrasLSL-G12D; Pdx1-cre rapidly developed advanced mPanIN and microinvasive neoplasms within 3 months of age (Fig. 3e–f, Supplementary Fig. 18). Immunohistochemical analysis of mPanINs from heterozygous female mice demonstrated absence of Usp9x expression in the preneoplastic and neoplastic cells (Supplementary Fig. 18), implicating additional events such as X inactivation of the other locus in female mice22,23. mPanINs in KrasLSL-G12D; Pdx1-cre; Usp9xfl mice expressed intranuclear Smad4, similar to KrasLSL-G12D; Pdx1-cre mice (Supplementary Fig. 19a). Additionally, early passage pancreatic cell cultures prepared from these mice confirmed the absence of the Usp9x protein and altered regulation of Itch (Supplementary Fig. 19b). Although some mice died of local or metastatic pancreatic cancer, aggressive oral papillomas often required the culling of young mice and demonstrated that KrasG12D and Usp9x loss also cooperated to transform keratinocytes (Supplementary Fig. 19c).
Although a recent report implicated USP9X as a pro-survival gene by stabilizing MCL19, potential inhibitors of USP9X should be developed with caution since we find that Usp9x has tissue specific effects including a tumor suppressor role in oncogenic Kras-initiated pancreatic carcinoma. USP9X is likely epigenetically silenced in a subset of PDA, thus explaining why prior DNA sequencing efforts have failed to identify this as participant in carcinogenesis, and offering the possibility that clinically available epigenome modulators may be useful agents to investigate in such patients. ITCH is a likely mediator of pancreatic tumor suppression by USP9X, and continued investigation of the USP9X/ITCH pathway is warranted. More generally, the identification of Usp9x through the use of transposon mutagenesis reaffirms the utility of in vivo mouse cancer screens to complement the direct investigation of human cancer.
Methods Summary
KrasLSL-G12D(K)24, Pdx1-Cre8 (C), T2/Onc6 (T), CAGGS-SB106 (SB10) and Rosa26-LSL-SB13 (SB13) strains were interbred to generate KrasLSL-G12D; Pdx1-Cre (KC), KrasLSL-G12D; Pdx1-Cre; T2/Onc; CAGGS-SB10 (KCTSB10) and KrasLSL-G12D; Pdx-1-Cre; T2/Onc; Rosa26-LSL-SB13 (KCTSB13) compound mutant mice. Non-quadruple mutant mice represented the comparison cohorts. KrasLSL-G12D and Pdx1-Cre mice were interbred with Usp9xfl mice to generate the KrasLSL-G12D; Pdx1-Cre; Usp9xfl/+ and KrasLSL-G12D; Pdx1-Cre; Usp9xfl/y (KCU) compound mutant mice, as well as the two control cohorts Pdx1-cre; Usp9xfl/y (CU) and KrasLSL-G12D; Pdx1-cre (KC). Usp9xfl mice were generated by Ozgene Pty. Ltd (Bentley, Australia). Mice were maintained in compliance with the UK home office regulations. Splinkerette PCRs were performed as described previously25,26. Reads from sequenced tumors were mapped to the mouse genome assembly NCBI m37 and merged together to identify SB insertion sites, as previously described25. Redundant sequences, as well as insertions in the En2 gene and in the T2/Onc donor concatemer resident chromosome (chromosome 1), were removed. Mouse survival curves and cell culture experiments were analyzed with the GraphPad prism program. The IHC histoscoring from the TMA samples and Kaplan-Meier survival curves were analyzed with SPSS18, and the Spearman-Rho correlation coefficient (2-sided) between USP9X and ITCH was calculated. The IHC USP9X histoscore and analysis was conducted using the Fishers Exact Test on post-mortem samples. The GEO accession number for the ICGC-APGI gene expression data is GSE36924.
Methods
Generation of Rosa26-LSL-SB13 knockin mice
TL1 ES cells27 were electroporated with linearized pRosa26-LSL-SA-SB13-BGHpolyA targeting construct and correctly targeted puromycin-resistant clones were identified by Southern blot. Two positives clones exhibiting a normal karyotype were used to generate chimeric mice by microinjection into C57BL/6 blastocysts. Germline transmission of the targeted allele was confirmed by Southern blot analysis of tail DNA from the agouti offspring.
T2/Onc excision PCR
Genomic DNAs were obtained from Pdx1-cre; T2/Onc; Rosa26-LSL-SB13 (CTSB13) and T2/Onc; Rosa26-LSL-SB13 (TSB13) mice and primers used to assess the excision of the T2/Onc concatemer in the CTSB13 mice were: 5′-TGTGCTGCAAGGCGATTA-3′ and 5′-ACCATGATTACGCCAAGC-3′.
CIS analysis
For the statistical analysis, 90,007 non-redundant insertion sites (Supplementary Table 3) were used to identify CISs using a Gaussian Kernel Convolution framework (GKC)28. An enhanced version of the framework was developed for SB screens to account for the local density of TA sites within the genome25. For example, a genomic region containing a large number of insertion sites but a low density of TA sites is considered to be significant and thereby identified as a candidate CIS. Conversely, a region with a large number of insertion sites but also containing a high density of TA sites is determined to be less significant, since the transposons have more “target” sites into which they can integrate. Multiple kernel scales were employed in the GKC framework (widths of 15K, 30K, 50K, 75K, 120K and 240K nucleotides). CISs predicted across multiple scales and overlapping in their genomic locations were clustered together, such that the CIS with the smallest genomic “footprint” was reported as the representative CIS. For highly significant CISs with narrow spatial distributions of insertion sites, the 15K kernel is typically the scale on which CISs are identified. Additional statistical analysis of insertion sites was performed using a Monte Carlo framework10. CISs were compared to previously published datasets of human pancreatic cancer genetics5,29,30.
Detection of Usp9x-T2/Onc fusion mRNA by RT-PCR in SB tumors
Total RNA was extracted from snap-frozen SB tumors using the RNeasy Mini Kit (Qiagen), and total RNA (1 μg) was reverse transcribed into cDNA using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). RT–PCR was carried out with a nested PCR approach using primers of mouse Usp9x exon 1 and the Carp-β-Actin splice acceptor sequence of the T2/Onc transposon cassette. cDNA was used as a template in a first round of PCR using specific primers corresponding to exon 1 of Usp9x (5-gagtctgcgctgccgctgctg-3′) and Carp-β-Actin splice acceptor sequence (5′-cataccggctacgttgctaa-3′). The product of this reaction was used as a template in a second round of nested PCR using an internal primer in the Usp9x exon 1 (5′-gctgccgctgctgttgctgc-3′) and a second primer in the Carp-β-Actin splice acceptor sequence (5′-acgttgctaacaaccagtgc-3′). PCR products were cloned into pCR 2.1-TOPO vector (Invitrogen) and positives clones sequenced.
Plasmids, shRNAs and transfections
pSuperRetro-PURO retroviral vector (Oligoengine) expressed a short hairpin against mouse and human USP9x (5′-gatgaggaacctgcatttc-3′), mouse Itch (5′-gacctgagaagacgtttgt-3′)31, and a scramble sequence (5′-gcgcgctttgtaggattcg-3′). pBabe-zeo-Ecotropic Receptor (ecoR) was obtained from Addgene (plasmid#10687). Myc-mItch cDNA was released from pCINeo-myc-Itch (Addgene plasmid#11427), and was subcloned in the retroviral vector pBabe-neo (Addgene plasmid#1767). KCU1 and KCU2 cell lines were transfected with pEF-DEST51-mUsp9x(WT)-V5 and pEF-DEST51-mUsp9x(C1566S)-V5 plasmids32,33. The plasmid pEF/GW-51/LacZ (Invitrogen) was used as control. Transfections were done using Lipofectamine 2000 (Invitrogen). 24 hours later, cells were selected with 5 μg/ml blasticidin (Invitrogen).
Cell culture
Tumor pancreatic cancer cell lines were established from KrasLSL-G12D; Pdx1-cre (T4878 and T9394), KrasLSL-G12D; P48-cre (TB1572) and KrasLSL-G12D; Pdx-1-Cre; Usp9xfl (KCU1 and KCU2) mice as described previously34. Cells were subsequently cultured in DMEM (Invitrogen), supplemented with 10% FCS (Hyclone). The normal human pancreatic ductal cell line HPDE was generously provided by Dr. Tsao and cultured as described previously35,36. The human pancreatic cancer cell lines AsPC1 (CRL-1682) and BxPC3 (CRL-1687) were acquired from ATCC and cultured according to instructions. The other cell lines were obtained from Clare Hall Laboratories (CRUK). The human cell lines Panc1, MiaPaCa2, 818.4, Hs766T, PATU2, SUIT2, FA6 and MDA-Panc3 (PDA); CaCO2 and SW1116 (colorectal cancer); SKBR3 (breast cancer) and A549 (lung cancer) were cultured in DMEM supplemented with 10% FCS. The human cell lines U937 (histiocytic lymphoma); RAMOS (Burkitt’s lymphoma); NCI-H2179 (lung cancer) and ZR75-1 (breast cancer) were cultured in RPMI (Invitrogen) supplemented with 10% FCS. Cells were treated with 1 μM trichostatin A (Sigma) for 24 hours or with 5 μM 5-Aza-2′-deoxycytidine (Sigma) for 96 hours where indicated to obtain RNA and protein lysates to assess USP9x expression. For anchorage-independent growth assay, cells were treated with 5 μM 5-Aza-2′-deoxycytidine (Sigma).
Retroviral infections
Phoenix cells were plated 24 hr before transfection using the ProFection Mammalian Transfection System Calcium Phosphate (Promega). Target cells were infected with retroviruses produced in the Phoenix packaging cells (24 and 48 hr after transfection) in the presence of 8 μg/ml polybrene (Sigma) and were selected with 2 μg/ml puromycin (Sigma) or 1 mg/ml G418 (Clontech). Experiments were performed using at least 2 independent cell line infected pools. Human PDA cells lines Panc1, SUIT2 and PATU2 infected with retroviral vectors expressed the Ecotropic Receptor (ecoR).
Transformation, Anoikis and EMT assays
Cell lines (1.5×104 cells) were plated in triplicate in 12-well plates and counted as indicated using a Z2 Coulter (Beckman). Cells were fed every other day. Anchorage-independent growth assay was assessed by colony formation in soft agar. Briefly, 15,000 cells were plated in duplicates in DMEM with 15% serum and 0.34% low–melting point agarose (LMP, BioGene) onto 6-cm dishes coated with 0.5% LMP. Cells were fed twice a week and grown for 2 weeks. For the anoikis assay, 105 cells/0.5 ml were plated in 24 well ultra low cluster plates (Costar) to allow them to grow in suspension for 4 days. Cells were harvested, washed with cold PBS and protein lysates were obtained. Cell line T4878 was cultured in matrigel as previously described37, plating 1000 cells/well. Cells were fed every 2 days and grown for 4 days. Epithelial-to-Mesenchymal Transition (EMT) was determined by plating 105 cells/6 well plates for 24 hr to allow attachment, followed by treatment with human TGF-β1 (5 ng/ml) (RD Systems) for 24 hr. p21 induction was assessed after treatment with human TGF-β1 (5 ng/ml) (RD Systems) for 2 hr.
Real-time PCR
Total RNA from human PDA cell lines was extracted using the Rneasy Mini Kit (Qiagen), and total RNA (1 μg) was reverse transcribed into cDNA using the High Capacity RNA-to-cDNA kit (Applied Biosystems). Human USP9x expression was analyzed by quantitative PCR (q-PCR) using TaqMan gene expressiom assays Hs00245009_m1 (Applied Biosystems) on a 7900HT Real-Time PCR system (Applied Biosystems). Gene expression was normalized to human β-ACTIN expression, assessed with the gene expression assays Hs99999903_m1 (Applied Biosystems), and shown relative to control samples.
Western blot analysis
Cells were washed three times in cold PBS and lysed with boiling lysis buffer (1% SDS; 10mM, pH 7.5 Tris; 50mM NaF; 1mM Na3VO4). Lysates were boiled 5 minutes, passed through a 26 gauge needle to shear genomic DNA and centrifuged for 10 minutes at 14,000 rpm. Equivalent amounts of protein were resolved in 4–12% gradient SDS-PAGE gels (Invitrogen), transferred to Immobilon–P Transfer Membranes (Millipore), and incubated with the corresponding antibodies including anti-Ask1 (NB110-55482, Novus Biologicals); anti-Mcl1 (5453, Cell Signaling); anti-Usp9x (A301-351A, Bethyl); anti-CC3 (9664, Cell Signaling); anti-Itch (611198, BD); anti-p21 (sc-6246, Santa Cruz); anti-Smad4 (sc-7966, Santa Cruz); anti-myc tag (2276, Cell Signaling); anti V5 tag (R960-25, Invitrogen); anti-c-FLIP (ALX-804-127, Enzo Life Sciences); anti-c-Jun (9165, Cell Signalling); anti-p63 (Ab110038, Abcam); anti a-Tubulin (T6074, Sigma) and anti-Actin (sc-1616, Santa Cruz Biotechnology). Reactive bands were visualized with ECL plus reagent (Amersham). Relative expression was quantified with Image Quant TL software (GE Healthcare)
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) mouse tissues were cut into 3-μm tissue sections, and antigen retrieval was performed in 10mM, pH 6.0 citric acid (for Usp9x and E-cadherin) or 10mM, pH 8.0 EDTA (for Smad4). Endogenous peroxidases were quenched in 3% H2O2/PBS for 20 minutes. Signal detection for immunohistochemistry was accomplished with biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA) using the Elite Vectastain ABC kit and peroxidase substrate DAB kit (Vector Laboratories, Burlingame, CA). Primary antibodies used were anti-Usp9x, 1:200 (A301-351A, Bethyl); E-cadherin, 1:200 (610182, BD) and anti-Smad4, 1:100 (sc-7966, Santa Cruz). Slides were counterstained with hematoxylin.
Clinical patient samples immunohistochemistry and analysis
Tissue microarrays (n=404) were prepared from patient samples obtained after appropriate informed consent in Dresden (Institute of Pathology, University Hospital Dresden), Regensburg (Institute of Pathology, University Hospital Regensburg) and Jena (Institute of Pathology, University Hospital Jena). Informed consent was obtained for each patient, following review by the human ethics committee Ethikkommission an der Technischen Universität Dresden The PDA tumor samples were collected from 1993–2009, and the majority of the patients (65%) did not undergo adjuvant chemotherapy. Those that did undergo adjuvant therapy (35%) were chiefly treated with 5FU or gemcitabine-based regimens, but in this subgroup there was no significant increase in patient survival. The median survival times of patients following surgery from each center were indistinguishable. Immunohistochemistry was performed on 5 μm sections that were prepared using silanized slides (Menzel Gläser, Braunschweig, Germany). Staining was performed with the Benchmark System (Ventana, Illkirch, France), using rabbit anti-USP9X antibody, 1:200 (A301-351A, Bethyl) and anti-ITCH, 1:200 (611198, BD); and the protocol UltraView HRP, with the CC1 modified protocol as pretreatment. Slides were counterstained with hematoxylin. Staining intensities were scored as absent (0), weak (1), medium (2) and strong (3). For further analysis the staining intensities were grouped as negative (0) and positive (1–3). The Cox regression model assumption of proportional hazard was tested using a plot of the cumulative hazards function.
A second cohort of patient samples was obtained from the Gastrointestinal Cancer Rapid Medical Donation Program in the Department of Pathology at Johns Hopkins Hospital, USA. Use of all human tissue samples from resection specimens and autopsy participants was approved by Johns Hopkins Institutional Review Board, and obtained after informed consent. All samples were collected within 12 hours postmortem and formalin fixed before paraffin embedding. 5 μm sections were cut from matched primary and metastasis samples onto glass slides. Slides were first incubated in Dako Target Retrieval Solution for antigen retrieval. Slides were then incubated with rabbit anti-USP9X antibody, 1:1000 (ab26334, Abcam) or 1:200 (NBP1-48321, Novus Bilogicals), and anti-SMAD4 as previously described38. Signal detection for immunohistochemistry was accomplished with Dako LSAB+System-HRP. Slides were counterstained with hematoxylin.
An additional cohort of pancreatic cancer resection samples was prospectively acquired through the Australian Pancreatic Cancer Network and the Australian Pancreatic Cancer Genome Initiative (http://www.pancreaticcancer.net.au/apgi). Consent was obtained for genomic sequencing through the Australian Pancreatic Cancer Genome Initiative (APGI) for each individual patient following approval from Human Research Ethics Committees (HREC) at participating sites (Sydney South West Area Health Service HREC Western Zone, 2006/054; Sydney Local Health Network HREC RPA Zone, X11-0220 and North Sydney Central Coast Health, Harbour HREC, 0612-251M). We extracted RNA from tumour samples using the Qiagen Allprep® Kit (Qiagen, Valencia, CA) in accordance with the manufacturer’s instructions, assayed for quality on an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA), and subsequently hybridized to Illumina Human HT-12 V4 microarrays. Raw idat files were processed using IlluminaGeneExpressionIdatReader (Cowley et al. manuscript in preparation). Following array quality control, these data were vs.t transformed, and then robust spline normalized, using the lumi R/Bioconductor package. For the ICGC-APGI cohort, we assumed a proportional hazard: that the probability of death is the same for those censored as for those remaining on study.
For the TMA and expression array cohorts, median survival was estimated using the Kaplan-Meier method and the difference was tested using the log-rank test. P-values of less than 0.05 were considered statistically significant. For the TMA cohort, as few parameters were significant in univariate analysis, all were initially considered for Cox Proportional Hazard multivariate analysis in a backward elimination model, and assessed with the SPSS18 Software (IBM, Ehningen, Germany) with overall survival used as the primary endpoint. For the ICGC-APGI cohort, clinico-pathologic variables analyzed with a P-value of less than 0.25 on log-rank test were entered into Cox Proportional Hazard multivariate analysis and the model was resolved using backward elimination. Statistical analysis was performed using StatView 5.0 Software (Abacus Systems, Berkeley, CA, USA). Disease-specific survival was used as the primary endpoint.
Supplementary Material
Acknowledgments
We thank Patricia Labosky for assistance in generating the Rosa26-LSL-SB13 mouse, B. Bhagavan for pathology consultation, Ming Tsao for kindly providing the HPDE cell line, and Neal Copeland and Karen Mann for sharing pre-published information. We thank Aarthi Gopinathan, Herve Tiriac, Danielle Engle, Derek Chan, Frances Connor, Sahra Derkits and other members of the Tuveson lab for assistance and advice, and the animal care staff and histology core at CRI, and The University of Minnesota’s Mouse Genetics Laboratory. This research was supported by the University of Cambridge and Cancer Research UK, The Li Ka Shing Foundation and Hutchison Whampoa Limited, the NIHR Cambridge Biomedical Research Centre, and the NIH (2P50CA101955 SPORE grant to DAT, DAL, CAI-D; grants CA62924, CA128920 and CA106610 to CAI-D; P50CA62924 SPORE grant to RHH and CAI-D; and CA122183 to LSC). DJA is supported by Cancer Research UK and the Wellcome Trust. LvdW is supported by the Kay Kendall Leukemia Fund. CP is supported by Wilhelm Sander Stiftung (2009.039.1) and DFG (PI 341/5-1). AVB, DKC, SMG and the APGI investigators are funded by the National Health and Medical research Council of Australia (NHMRC); Queensland Government; Cancer Council NSW; Australian Cancer Research Foundation; Cancer Institute NSW; The Avner Nahmani Pancreatic Cancer Research Foundation; and the R.T. Hall Trust. Additional support was obtained from Fundación Ibercaja (PAP-M). We regret that many primary references have been omitted due to space limitations.
Footnotes
Contributions: PAP-M performed the majority of all experiments, designed experiments, analyzed data, and wrote the manuscript. LvdW and JAB performed in vitro experiments. SS and SAW generated the conditional Usp9x mouse. LSC provided the CAGGS-SB10 and T2/Onc mice. AGR, ALS, KATS, JJtH, JdR and LFAW conducted the CIS data analysis. GK, RG, DA, PR, TK and CP generated data from resected pancreatic tumors. AL, RHH, RM, SK, JY, AL, AL, MG and CAI-D analyzed human samples from autopsy series, and analyzed mouse pathology and methylation studies. CH, DLS and RK sequenced PDA human samples from autopsy series. APK provided statistical analyses for the human PDA data sets. APGI, DKC, SMG and AVB generated and analysed data from ICGC (for APGI full list of contributors see http://www.pancreaticcancer.net.au/apgi/collaborators). DAL provided the CAGGS-SB10 and T2/Onc mice, and analyzed data. DJA and DAT designed the study, analyzed the data, and wrote the manuscript. All authors commented upon and edited the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Almoguera C, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 2.Caldas C, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 3.Redston MS, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- 4.Hahn SA, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 7.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 8.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 9.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 10.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier LS, et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 2009;69:8429–8437. doi: 10.1158/0008-5472.CAN-09-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avizienyte E, et al. LKB1 somatic mutations in sporadic tumors. Am J Pathol. 1999;154:677–681. doi: 10.1016/S0002-9440(10)65314-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su GH, et al. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci U S A. 2001;98:3254–3257. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 17.Dupont S, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 18.Nagai H, et al. Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol Cell. 2009;36:805–818. doi: 10.1016/j.molcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, et al. Bcr-Abl ubiquitination and Usp9x inhibition block kinase signaling and promote CML cell apoptosis. Blood. 2011;117:3151–3162. doi: 10.1182/blood-2010-03-276477. [DOI] [PubMed] [Google Scholar]
- 20.Mouchantaf R, et al. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- 21.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Soloway PD, Clark AG. Paternally biased X inactivation in mouse neonatal brain. Genome Biol. 2010;11:R79. doi: 10.1186/gb-2010-11-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nature genetics. 2011;43:1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uren AG, et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nature protocols. 2009;4:789–798. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tompers DM, Labosky PA. Electroporation of murine embryonic stem cells: a step-by-step guide. Stem Cells. 2004;22:243–249. doi: 10.1634/stemcells.22-3-243. [DOI] [PubMed] [Google Scholar]
- 28.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grutzmann R, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilarsky C, et al. Activation of Wnt signalling in stroma from pancreatic cancer identified by gene expression profiling. J Cell Mol Med. 2008;12:2823–2835. doi: 10.1111/j.1582-4934.2008.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberdoerffer P, et al. Efficiency of RNA interference in the mouse hematopoietic system varies between cell types and developmental stages. Mol Cell Biol. 2005;25:3896–3905. doi: 10.1128/MCB.25.10.3896-3905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nathan JA, et al. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray RZ, Jolly LA, Wood SA. The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and beta-catenin. Mol Biol Cell. 2004;15:1591–1599. doi: 10.1091/mbc.E03-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang H, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157:1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furukawa T, et al. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 37.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 38.Iacobuzio-Donahue CA, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.