Abstract
Bacteria synthesize and sense low molecular weight signaling molecules, termed autoinducers, to measure their population density and community complexity. One class of autoinducers, the α-hydroxyketones (AHKs), is produced and detected by the water-borne opportunistic pathogens Legionella pneumophila and Vibrio cholerae, which cause Legionnaires’ disease and cholera, respectively. The “Legionella quorum sensing” (lqs) or “cholera quorum sensing” (cqs) genes encode enzymes that produce and sense the AHK molecules “Legionella autoinducer-1” (LAI-1; 3-hydroxypentadecane-4-one) or cholera autoinducer-1 (CAI-1; 3-hydroxytridecane-4-one). AHK signaling regulates the virulence of L. pneumophila and V. cholerae, pathogen-host cell interactions, formation of biofilms or extracellular filaments, expression of a genomic “fitness island” and competence. Here, we outline the processes, wherein AHK signaling plays a role, and review recent insights into the function of proteins encoded by the lqs and cqs gene clusters. To this end, we will focus on the autoinducer synthases catalysing the biosynthesis of AHKs, on the cognate trans-membrane sensor kinases detecting the signals, and on components of the down-stream phosphorelay cascade that promote the transmission and integration of signaling events regulating gene expression.
Keywords: autoinducer synthase, cell-cell signaling, horizontal gene transfer, pathogen-host interactions, response regulator, quorum sensing, sensor kinase, two-component system, virulence
1. Introduction
Quorum sensing (QS) is a mode of cell-cell communication that allows microbial communities in a given niche to gauge their population density and complexity [1]. QS-dependent processes are based on molecular signaling circuits that synthesize, detect and integrate small diffusible molecules termed autoinducers (AIs) [2,3]. AIs are effective only above a certain threshold concentration, which is reached at a specific cell density of a bacterial population, termed the “quorum”. Accordingly, QS allows bacteria to co-ordinately adjust their behaviour to environmental conditions. AI signaling regulates gene expression of various complex bacterial processes, including biofilm formation, virulence, sporulation, bioluminescence and competence [3]. AI molecules can be grouped into distinct classes according to their basic chemical design. Carbohydrates, lipids (mainly fatty acids), quinolones and small peptides are used as chemical signals in microbial intra- and interspecies communication [4,5].
Species-specific modifications and derivatives of these basic compounds are found in virtually every bacterial genus, thus yielding a rich microcosm of microbial signal molecules. Many Gram-negative bacteria generate AIs that belong to the first discovered class of N-acyl-L-homoserine lactones (AHLs) [2], and/or they produce the almost universally used furanosyl borate diester termed autoinducer-2 (AI-2) [6,7]. Some bacteria utilize additional AIs to regulate gene expression. A class of α,β-unsaturated fatty acids termed diffusible signal factors (DSFs) plays a prominent role in soil- and plant-associated bacteria such as Xanthomonas, Burkholderia or Pseudomonas spp. [8]. Finally, the recently discovered α-hydroxyketones (AHKs) are predominantly produced by aquatic γ-proteobacteria, including Legionella and Vibrio spp. (Figure 1(a)) [9].
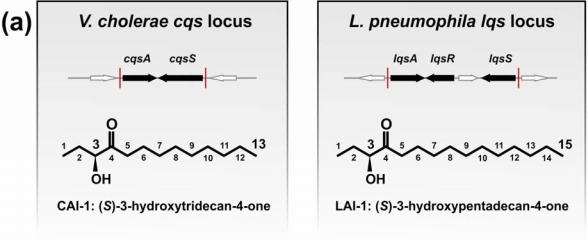
Figure 1.
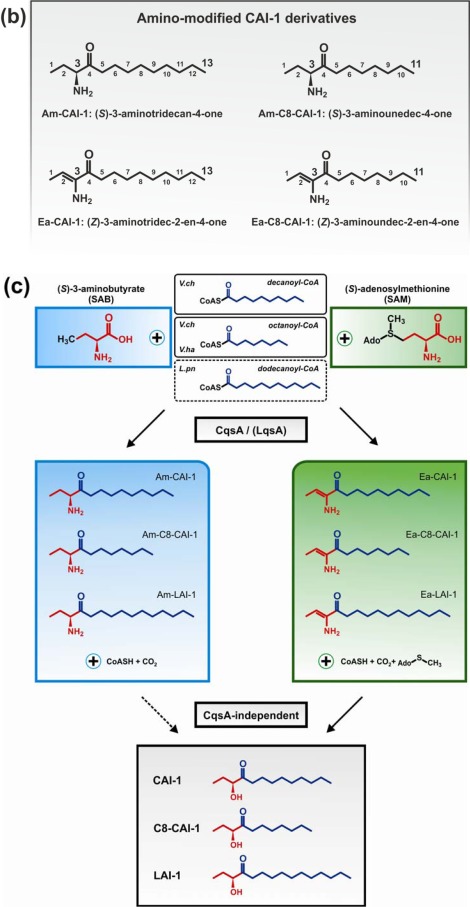
Genetic organization of the V. cholerae (cqs) and L. pneumophila (lqs) QS locus, and biosynthesis of AIs produced by CqsA and LqsA. (a) The cqs and lqs loci harbour AI synthases (cqsA, lqsA), cognate sensor kinases (cqsS, lqsS) and a response regulator (lqsR). AI molecules synthesized by V.chCqsA and L.pnLqsA: (a) CAI-1 and LAI-1 AHKs, and (b) C3-amino-derivatives of CAI-1 (Am-CAI-1 and Ea-CAI-1). (c) Biosynthesis of CAI-1 and C3-amino-derivatives by PLP-dependent V. cholerae and V. harveyi CqsA and presumably L. pneumophila LqsA using (S)-3-aminobutyrate (SAB) or (S)-adenosylmethionine (SAM) and acyl-CoAs. V.chCqsA utilizes decanoyl-CoA (C10) or octanoyl-CoA (C8) to produce Am-CAI-1/Ea-CAI-1 (C13) or Am-C8-CAI-1/Ea-C8-CAI-1 (C11). Ea-CAI-1 is converted into CAI-1 by spontaneous hydrolysis and a dehydrogenase. The intermediate Am-CAI-1 is converted into CAI-1 by an unknown mechanism. V.haCqsA utilizes only octanoyl-CoA to yield Am-C8-CAI-1/Ea-C8-CAI-1 and C8-CAI-1 (C11). Synthesis of LAI-1 (C15) by L.pnLqsA is not elucidated, but might use SAM (or SAB) and dodecanoyl-CoA (C12).
The Gram-negative bacteria Legionella pneumophila and Vibrio cholerae colonize several environmental niches and are important opportunistic human pathogens that cause the life-threatening diseases “Legionnaires’ pneumonia” or “cholera”, respectively [10,11]. In this review we summarize recent insights into the molecular structure of the Legionella and Vibrio AHK signaling circuits, discuss the mechanisms of AHK production and sensing and outline their role for pathogen-host interactions, biofilm formation and competence.
2. Virulence of Legionella pneumophila and Vibrio cholerae
Legionella and Vibrio spp. are water-borne bacteria that persist in aquatic freshwater or brackwater habitats, either as individual planktonic cells, or attached to biotic/abiotic surfaces, where they are part of multispecies biofilm communities [12–14]. Remarkably, Legionella and Vibrio spp. also evolved specific and distinct mechanisms, which support their survival and growth in eukaryotic hosts ranging from protozoa and nematodes to animals and humans [15–17]. Pathogenic species of both bacterial genera evolved from environmental non-pathogenic progenitor strains by acquiring virulence determinants through horizontal gene transfer [18,19].
2.1. Virulence of Legionella pneumophila
Legionella spp. cause a severe pneumonia called Legionnaires’ disease or a milder, flu-like ailment termed Pontiac fever [20]. The sources for Legionella infections can be natural aquatic biotopes, but primarily are anthropogenic water supply systems. Legionellosis occurs through the inhalation of contaminated aerosols, and its epidemiology is strongly linked to the use of technical appliances in developed countries. There are no reports of human to human transmission, and therefore, humans are considered dead-end hosts for Legionella species. Approximately half of the more than 50 Legionella spp. described so far have been reported to infect humans. Yet, L. pneumophila (serogroup 1) and L. longbeachae account for the majority of clinical cases [11]. Elderly and immuno-compromised individuals are predominantly prone to Legionnaires’ disease, since their innate immune system is not able to efficiently clear the bacterial infection [21]. In addition to sporadic epidemic outbreaks, nosocomial pneumonia due to L. pneumophila is a major issue for public health services around the world [22].
L. pneumophila is a facultative intracellular bacterium that thrives within a broad spectrum of environmental phagocytes, including at least 15 protozoan genera, e.g., Acanthamoeba or Hartmanella spp. and Dictyostelium discoideum [16]. The co-evolution of L. pneumophila with phagocytic protozoa most likely selected for virulence traits that also allow infection and growth in functionally related professional phagocytes of the human innate immune system, in particular alveolar macrophages [23,24]. The infectious cycle of L. pneumophila is a multistage process, where virulent, transmissive bacteria induce their uptake, avoid lysosomal degradation, establish an intracellular replicative niche and finally exit the spent host cell again [25].
A key virulence factor of L. pneumophila is a type IV secretion system (T4SS) encoded by 25 different icm/dot genes. The Icm/Dot T4SS is essential to establish an intracellular replication-permissive niche, termed the Legionella-containing vacuole (LCV) [26,27]. A set of more than 250 distinct “effector proteins” is translocated by the T4SS into host cells, where they manipulate conserved cellular processes, such as signal transduction and vesicle trafficking pathways [28–33]. To this end, Icm/Dot-translocated effector proteins target host small GTPases and phosphoinositide lipids, as well as ubiquitinylation and apoptosis factors. Strikingly, many of these effector proteins share sequence homologies with eukaryotic proteins or harbour functional eukaryotic domains, which likely were acquired by trans-kingdom horizontal gene transfer [14,24,34].
2.2. Virulence of Vibrio cholerae
In contrast to the facultative intracellular bacterium L. pneumophila, V. cholerae is an extracellular bacterium. The toxigenic V. cholerae serogroups O1 and O139 cause the vast majority of cholera cases [35]. Cholera is a dehydrating severe diarrhea, frequently fatal when left untreated. Infection with V. cholerae occurs via the consumption of contaminated water or food and subsequent bacterial colonization of the epithelium in the upper small intestine [10]. Poor sanitary standards and uncontrolled dissemination of bacteria by diarrhea into ground and surface water can lead to epidemic outbreaks. V. cholerae virulence is a multifactorial process that relies on the interplay of environmentally selected traits such as motility, surface interaction or biofilm formation, combined with the acquisition of phage-derived virulence genes. V. cholerae utilizes flagellar motility and secreted proteinases to reach and penetrate the glycokalyx of the intestinal mucosa and binds via N-acetylglucosamine-binding protein A (GbpA) to the epithelium of the small intestine [10]. Efficient colonization of the intestine requires the subsequent expression of the ToxR regulon, which includes the genes coding for two critical virulence factors, the phage-acquired cholera enterotoxin (CTX) and the toxin co-regulated pilus (TCP) [36].
CTX is a secreted AB5 subunit protein, which enters intestinal epithelial cells via receptor-mediated endocytosis and disturbs intracellular cAMP signaling. Constitutive cAMP production results in a drastic ion efflux that leads to solute imbalance and massive secretion of water into the intestinal lumen. CTX function is responsible for a profuse secretory diarrhea and is assumed to facilitate transmission of hyper-virulent bacteria [10]. TCP is an auto-interacting type IV pilus that connects V. cholerae cells, a process resulting in aggregation and microcolony forming in the mucosa [37]. Importantly, TCP also acts as a receptor for the bacteriophage CTXphi that carries and transduces the CTX genes [38].
Once established, V. cholerae proliferates in the nutrition-rich small intestine. At late stages of the infection, parameters such as host-specific stimuli, availability of nutrients and increasing bacterial cell density coordinate an “exit response” that promotes detachment of the bacteria, survival in the aquatic environment and spread to new hosts [10]. This stage involves QS-dependent repression of ToxR-controlled genes, activation of the haemagglutinin/protease HapA and the motility apparatus [39,40].
3. QS in Vibrio and Legionella spp
The facultative pathogenic potential of V. cholerae and L. pneumophila implies a biphasic life style and a periodic transition between environmental and host-associated niches [10,14]. The transition between different habitats is reflected by a prominent switch in gene expression that leads to niche-specific expression patterns [24,41]. Accordingly, V. cholerae and L. pneumophila reciprocally express genes, which either support bacterial growth under distinct environmental conditions, or promote host colonization and virulence. Under laboratory conditions, these transcriptional programs can be (at least partially) mimicked in broth cultures. In V. cholerae the reciprocal gene expression pattern is responsive to the cell density and, via cAMP and cAMP receptor protein (CRP), also dependent on the metabolic state [3,42,43]. In L. pneumophila the biphasic gene expression is regulated by the growth phase [24,25]. Noteworthy, in both species the transition between niche-specific gene expression patterns is dependent on two-component (TC) systems (VarA/VarS in V. cholerae, LetA/LetS in L. pneumophila) and the global regulator CsrA [44–48]. The VarAS-CsrA or LetAS-CsrA systems integrate metabolic signals such as nutrient depletion and carbon availability, which are strongly linked to the bacterial cell density and growth phase.
Colonizing a new niche provides the opportunity to exploit different nutrition-rich sites, yet the bacteria also have to continuously adapt to changing environmental conditions. Accordingly, V. cholerae and L. pneumophila need to integrate complex patterns of environmental and host-derived stimuli to ensure efficient replication within a niche, progression to subsequent infection stages within a host and transition between ecological niches. To this end, a fine-tuned sensor machinery is essential to constantly monitor endogenous and environmental cues, transmit and integrate the signals and respond by regulating gene expression. V. cholerae and L. pneumophila employ QS circuits in concert with additional sensor systems to coordinate various processes during their life cycle, including pathogen-host interaction and production of virulence factors [39,49–51], biofilm and filament formation [40,52–54], the regulation of a genomic “fitness island” [54] and natural competence [55–57].
The molecular structure of QS signaling units basically comprises an AI synthase and a cognate sensor protein, which is coupled to a signal transduction phosphorelay that links the detection of AIs to gene regulation [3]. QS circuit components differ significantly between distinct systems and bacterial species. LuxI/LuxR-like systems, first described in Vibrio fischeri, synthesize and detect freely diffusible AHLs [58,59]. Here, the AHL ligand binds the LuxR regulator directly in the cytosol and, depending on the system, either destabilizes or stabilizes LuxR dimerization and DNA binding [2]. Other QS systems adopt membrane-bound sensor kinases derived from TC systems to detect and transmit an AI signal, e.g., the Vibrio-specific LuxM/LuxN AI synthase/sensor kinase system that produces and responds to an AHL compound termed harveyi autoinducer-1 (HAI-1), or the broadly-used LuxS/LuxPQ system that signals through AI-2 [3].
Bacterial TC systems are sensor units composed of an inner membrane-associated sensor histidine kinase and a downstream response regulator [60]. TC sensor proteins recognize a wide range of chemical and physical stimuli in the periplasmic space, including osmolarity, pH, envelope damage, as well as chemical signaling molecules. The common feature of these bacterial sensors is the conversion of ligand binding into phosphorylation events, which are integrated and transduced via phosphorelay pathways to downstream regulatory proteins [61,62]. The genomic organization of TC genes, encoding a sensor kinase and a response regulator, can either constitute tandem pairs controlled as an operon or represent distantly located “orphans” used by different transcriptional programs [63,64]. QS genes, encoding a AI synthase and a cognate sensor protein, are usually located adjacently or in close vicinity of each other in the bacterial genome, and therefore, are believed to retain their pairwise functional relationships through co-evolution as a single cassette [65,66]. Multiple sensors of TC and QS systems can be integrated and may converge on shared response regulators.
4. AHK Signaling in V. cholerae and L. pneumophila
4.1. Components and Integration of AHK Signaling
While AHL and AI-2 signaling circuits are broadly used by many bacterial species [67,68], the usage of AHKs as small signaling molecules is apparently restricted to a group of aquatic γ-proteobacteria, including Vibrio and Legionella spp. [9]. The V. cholerae and L. pneumophila AHK signaling circuits are constituted by the cqs or lqs (cholera/Legionella quorum sensing) gene clusters, respectively [9]. These clusters encode cognate pairs of an AI synthase and a sensor histidine kinase (CqsA/CqsS, LqsA/LqsS), which produce and detect the corresponding AHKs CAI-1 (cholera autoinducer-1) [69] or LAI-1 (Legionella autoinducer-1) [70] (Figure 1(a)). In addition, the lqs gene cluster encodes the putative response regulator LqsR [50].
Vibrio spp. utilize the Cqs system together with additional QS systems. V. cholerae and V. harveyi use two or even three QS systems operating in parallel to integrate AI signals in a joint phosphorylation cascade. Thus, V. cholerae employs CqsA/CqsS, as well as LuxS/LuxPQ to signal through CAI-1 and AI-2 [6,49,68,69]. In addition to these two systems, V. harveyi uses a third, LuxM/LuxN-type system, which signals through the AHL molecule HAI-1 [71,72]. QS signals received by all sensor kinases (CqsS, LuxQ and LuxN) are channelled onto the phosphorelay protein LuxU that transfers the phosphate to the response regulator LuxO [73,74].
A common feature of the sensor histidine kinases CqsS, LuxQ and LuxN is their ability to switch between kinase and phosphatase function. The LuxQ [75] and LuxN [76] sensor kinases have been instrumental to study mechanistic aspects of coupling the binding of an AI ligand to the switch from kinase to phosphatase activity. High-resolution crystal structures of the periplasmic binding protein LuxP and a fragment of the sensor kinase LuxQ in complex with or without its ligand AI-2 revealed that binding of AI-2 to LuxP disrupts tetrameric complexes of LuxPQ dimers, thus causing a switch from kinase to phosphatase activity. Similarly, a switch from kinase to phosphatase activity is also proposed for CqsS upon interaction with its ligand CAI-1 (Figure 2). However, the exact mechanism of how CAI-1 binding alters the oligomerization state and the activity of a putative CqsS dimer has to be elucidated. At low AI concentration the sensor kinase activity will lead to phosphorylation and activation of the downstream targets LuxU and LuxO. Conversely, at high AI concentration the sensor phosphorylation activity will result in the dephosphorylation and inactivation of LuxU and LuxO.
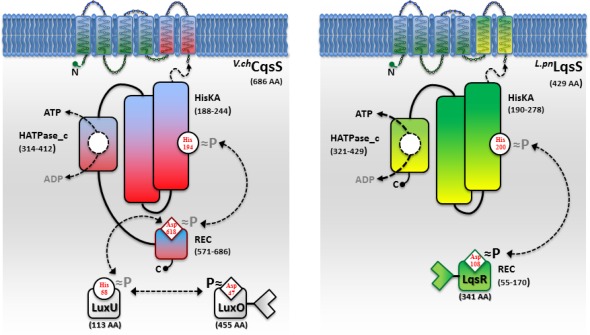
Figure 2.
Signal transduction by the V. cholerae CqsS and L. pneumophila LqsS sensor kinases. V.chCqsS and L.pnLqsS are six trans-membrane helix sensor kinases with C-terminal cytoplasmic signal transduction domains. Predicted sub-domains (amino acids in parenthesis): HisKA (histidine kinase A domain with conserved histidine phospho-acceptor site), HATPase_c (catalytic ATP binding and transferase domain, C-terminal); REC (receiver domain with conserved aspartate). CqsS is a hybrid histidine kinase coupled to a phosphorelay system. The HisKA/HATPase_C domain catalyzes the autophosphorylation by ATP at H194 and the phosphotransfer to D618 in the REC domain. The phosphoryl group is then shuttled via the orphan phosphorelay protein LuxU (H58) to the REC domain of the response regulator LuxO (D47). In LqsS, after phosphorylation of H200, the phosphate is presumably transferred to D108 in the REC domain of the response regulator LqsR. CqsS and LqsS are likely bifunctional kinases/phosphatases.
Below the AI threshold concentration, phosphorylated LuxO represses the activity of the QS master regulator HapR in V. cholerae [49] or LuxR in V. harveyi [77], respectively. The repression of HapR is achieved via coordinated LuxO-RpoN-Fis-dependent expression of four functionally redundant small regulatory RNAs termed Qrr1-4. The Qrr sRNAs act in concert with the RNA chaperonine Hfq to destabilize hapR mRNA and prevent its translation [45,78–80]. In contrast, high AI concentrations deactivate LuxO and release the repression of HapR and LuxR. At high AI concentration, virulence and biofilm formation in Vibrio spp. is repressed by HapR and LuxR, while the haemagglutinin/protease HapA, motility and competence in V. cholerae or bioluminescence in V. harveyi are induced [3,10,55].
The genes for LuxU, LuxO and HapR are not associated with the cqs genes or the other QS genes in the V. cholerae genome. This scattered genomic organization reflects the shared usage of the genes by parallel QS systems in Vibrio spp. Thus, LuxU, LuxO and HapR are also targets for other sensor or regulatory system systems, including the VarAS-CsrA system, VpsRS and cyclic-di-GMP, which converge on LuxO and control HapR-regulated genes [45,81,82].
In contrast to Vibrio spp, L. pneumophila apparently employs only the AHK-based Lqs QS system. The lqs genes are not present in the L. longbeachae genome [83], and lqsA was not detected by PCR using genomic DNA of several non-pneumophila strains [70]. In L. pneumophila AHK signaling is assumed to be mediated, at least partially, by the putative response regulator LqsR [50] (Figure 2). This prototypic member of a novel family of response regulators harbours a canonical, N-terminal receiver domain (amino acids 80–160) including the conserved aspartate residue (D108). The C-terminal fragment of LqsR does not show homology to any known signal output domains, and its function is unknown. The clustering of the lqsR gene with lqsA and lqsS is conserved in bacterial species that harbour an LqsR homologue, suggesting an evolutionarily conserved functional correlation between LqsA/LqsS and LqsR [9,50]. Similar to the modification of Vibrio LuxO function by a range of regulatory factors, LqsR is also intimately connected to other regulatory networks. The production of LqsR is dependent on the stationary sigma factor RpoS, which in the L. pneumophila life cycle controls the reversible transition from the replicative to the virulent phase, as well as pathogen-host interactions [9,50].
L. pneumophila strains lacking lqsS or lqsR show severe defects for uptake by phagocytes, intracellular growth and the production of extracellular filaments [50,54]. In contrast, L. pneumophila lacking lqsA is only mildly impaired for host cell infection, similar to V. cholerae lacking cqsA. Yet, the overexpression of lqsA in L. pneumophila lqsS or lqsR mutant strains restores the virulence defects of these mutant strains to wild-type levels. These results suggest that the LqsA product LAI-1 indeed regulates virulence and that L. pneumophila employs multiple and redundant LAI-1-responsive signaling pathways [54]. It is tempting to speculate that the LCV represents a LAI-1-impermeable compartment and that the cell density-dependent concentration of AHK signaling molecules within this pathogen vacuole regulates bacterial virulence and motility, as well as cycling from the transmissive to the replicative growth phase.
4.2. CAI-1/LAI-1 AHK Signal Production by CqsA/LqsA AI Synthases
The CqsA/CqsS circuit signals through the major V. cholerae AI, the AHK molecule CAI-1 (3-hydroxytridecan-4-one) [69], whereas the L. pneumophila LqsA/LqsS system mainly produces and presumably responds to LAI-1 (3-hydroxypentadecan-4-one) (Figure 1(a)) [70]. The AI synthases CqsA and LqsA are 45% identical, and both enzymes are predicted to be related to pyridoxal-5′-phosphate (PLP)-dependent aminotransferases [49,50,69]. Accordingly, CqsA and LqsA contain conserved lysine residues (CqsA_K236, LqsA_K258), which covalently bind PLP [70,84,85]. Moreover, LqsA and CqsA are also functionally similar, as lqsA partially complements the deletion of cqsA in V. cholerae [70].
Crystallographic and biochemical characterization of CqsA confirmed its function as a PLP-dependent aminotransferase-like enzyme [84,85]. Studies with purified CqsA revealed that in vitro the enzyme catalyses the formation of a carbon-carbon bond between (S)-2-aminobutyrate (SAB) and decanoyl-Coenzyme A (CoA) to produce (S)-3-aminotridecan-4-one (Am-CAI-1) (Figure 1(b,c)). Synthetic Am-CAI-1 and CAI-1 both bind to CqsS, as detected by a bioluminescence assay. However, CAI-1 is the prominent AHK compound in V. cholerae culture supernatants, suggesting that Am-CAI-1 is an intermediate, which is converted to CAI-1 by the bacteria [84].
In recent studies dissecting the biosynthetic route of CAI-1, (S)-adenosylmethionine (SAM) was identified as a much more efficient biosynthetic co-substrate than SAB for CqsA to produce the novel compound 3-aminotridec-2-en-4-one (Ea-CAI-1) (Figure 1(b,c)) [86,87]. Isotope labelling experiments confirmed that Ea-CAI-1 is synthesized by CqsA in vitro, as well as in vivo by coupling SAM and decanoyl-CoA. The unstable enamine Ea-CAI-1 is converted to CAI-1 via the spontaneous conversion into tridecane-3,4-dione (DK-CAI-1) followed by an NADPH-dependent reduction catalysed by a dehydrogenase [86,87]. The use of SAM as a co-substrate in the synthesis of AHKs is interesting, as it indicates that bacteria employ this abundant metabolite for the synthesis of at least three classes of QS signaling molecules: AHKs, AHLs and AI-2. The abundance and physiological importance of SAM poses several advantages for its usage as an AI substrate, since (i) this metabolite represents a constant source of QS signals and thus is a reliable parameter for cell density, and (ii) it reflects the physiological status of a bacterial population due to its prominent role in housekeeping reactions [86]. Thus, SAM-based QS systems link an important parameter of bacterial physiology to population density.
In addition to CAI-1 derivatives harbouring a C10 acyl tail (Am-CAI-1, Ea-CAI-1, DK-CAI-1, CAI-1), V. cholerae produces and responds to AHKs with other acyl tail lengths [69,70,84]. V. cholerae CqsA shows promiscuous substrate specificity and also forms Ea-C8-CAI-1 from SAM and octanoyl-CoA. Still, V. cholerae CqsA prefers the acyl CoA substrate C10-CoA over C8-CoA, and this substrate preference is also reflected by the in vivo production profiles of CAI-1 molecules detected in native cultures supernatants (concentration ratios of C8-CAI-1:Ea-CAI-1:CAI-1 = 1:7.4:11.8).
Relaxed specificity is also observed for L. pneumophila LqsA, which produces either LAI-1 (C12 acyl tail), or to a smaller extent CAI-1 (C10 acyl tail) and derivatives with C11 or C13 acyl tails [70]. However, this relaxed product specificity was observed upon heterologous production of LqsA in E. coli. The physiological specificity of LqsA might be more stringent, and presumably, dodecanoyl-CoA is used preferentially by L. pneumophila as a co-substrate to synthesize LAI-1(Figure 1(c)). The in vivo pathway for the biosynthesis of LAI-1 in L. pneumophila has yet to be elucidated, and it is not known, whether LqsA also produces Ea-LAI-1 or Am-LAI-1.
In contrast to the relaxed specificities of V. cholerae CqsA and L. pneumophila LqsA for different CoA co-substrates, the V. harveyi CqsA homologue exhibits a rather stringent substrate specificity, using only octanoyl-CoA together with SAM or SAB to produce Ea-C8-CAI-1 and C8-CAI-1, respectively [87]. In line with the stringent AI production, V. harveyi CqsS preferentially detects C8-CAI-1 derivatives.
4.3. AHK Signal Perception by CqsS/LqsS Sensor Kinases
The CAI-1 receptor CqsS and the potential LAI-1 receptor LqsS belong to the class of six trans-membrane helix TC sensor histidine kinases (Figure 3(a)). CqsS and LqsS couple the detection of the AI molecules via a receptor domain at the N-terminus to signal transduction modules at the C-terminal part of the protein [9]. TC sensor proteins are mainly located in the inner bacterial membrane, where they sense specific environmental signals present in the periplasmic space [62]. However, the molecular nature and the exact mechanisms of how these signals interact with the corresponding TC sensor kinases are poorly understood. Only a small fraction of TC system ligands has been identified so far, and therefore, AI molecules such as AHKs are valuable research tools to study membrane-bound sensors [3].
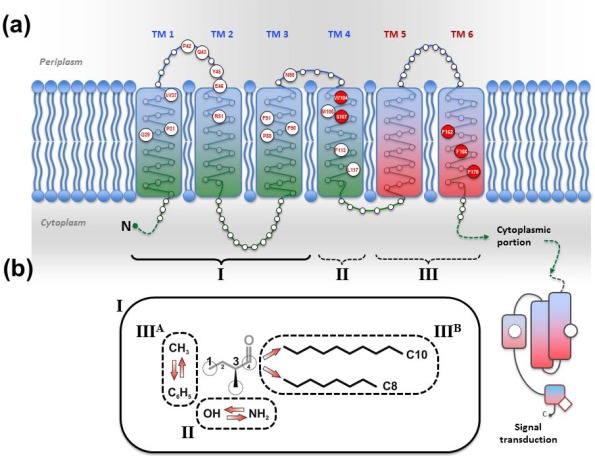
Figure 3.
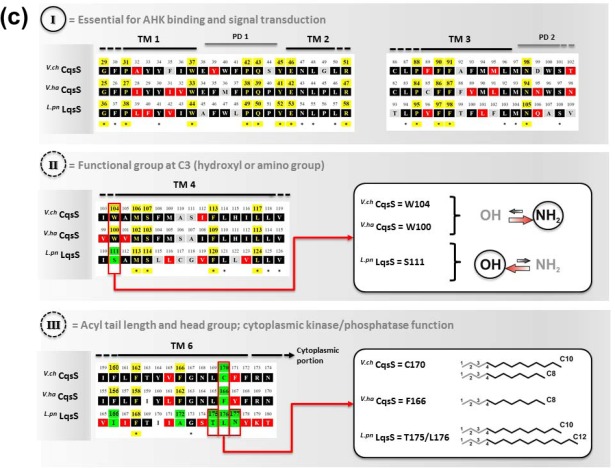
Signal recognition by the V. cholerae CqsS and L. pneumophila LqsS sensor kinases. (a) Topology model of the V. cholerae CqsS trans-membrane sensor domain based on the secondary structure prediction algorithms TMPRED and TMHMM. The N-terminal sensor domain encompasses six trans-membrane α-helices and defines the specificity and sensitivity for distinct AHK molecules (enlarged circles, amino acid position and one-letter code). (b) Sensor domain regions (I, II, III) define the interactions with functional groups of AHK ligands. (I) TM 1–3 and periplasmic domain PD 1 contain conserved amino acid clusters essential for ligand binding and signal transduction (cytoplasmic domain CD 1). (II) TM 4 (W104, S107) discriminates between hydroxyl and amino groups at C3 of AHK ligands. (III) TM 6 (F162, C170) determines binding to the head group (methyl vs. phenyl) and the acyl tail of AHK ligands. (c) Amino acid sequence alignment (Clustal Omega algortithm) of the sensor domains of V. cholerae (V.ch) CqsS, V. harveyi (V.ha) CqsS and L. pneumophila (L.pn) LqsS. The positions of species-specific amino acids in the sensor domain regions (I, II, III) are indicated, and stars denote conserved amino acids. Color scheme: black (2/3 conserved); red (similar amino acids); grey (different amino acids); yellow (amino acids defining ligand sensitivity, specificity and signal transduction motifs); green (amino acid exchange results in altered ligand specificity). Boxes illustrate altered ligand preferences due to site-specific amino acid polymorphisms in sensor domain region II and III.
Structure-function and ligand interaction analysis of intrinsic polytopic membrane proteins is challenging, yet recent studies used an elegant biochemical/genetic strategy to investigate CAI-1 recognition by CqsS sensor kinases [87–89]. The pairwise approach is based on defined CqsS sensor mutants harboring amino acid substitutions at distinct positions within the six trans-membrane helices, combined with probing these mutant receptors with natural and modified CAI-1 molecules. This approach allowed the identification of sensitivity and specificity determinants of CqsS that govern the discrimination between distinct CAI-1 molecules. The first three trans-membrane helices were found to harbor motifs essential for ligand binding and signal transduction. These motifs are conserved in all Vibrio CqsS homologues and in L. pneumophila LqsS, suggesting that they constitute a common ligand-binding domain (Figure 3(a)).
Some motifs in the first half of the N-terminal receptor region are strictly conserved, indicating an interaction with chemical characteristics present in all AHK molecules, such as the C3/C4 modifications in the hydrophilic head group and the hydrophobic acyl tail. The second half of the CqsS receptor domain is more variable among different Vibrio spp. and thus defines the binding specificity of the receptor for distinct natural and synthetic CAI-1 analogues. The amino acids W104 and S107 in V. cholerae CqsS determine the specificity for the CAI-1 head group, such that the amino modification at position C3 is (slightly) favored over the hydroxyl modification (Figure 3(b)). By contrast, any alteration of W104 results in an increased preference for a hydroxyl over an amino modification at the C3 position [88]. Moreover, the residues F160/F162/F166 and C170 in V. cholerae CqsS determine the size of the polar head group and the length of the acyl tail of the AHK ligand to be bound. This proposed “gatekeeper” function likely controls the access to the binding site, thereby preventing cross-talk between related signals and reducing signal to noise from similar molecules in the environment. A gatekeeper is an essential regulatory mechanism for sensing systems, because it prevents inappropriate activation or inhibition of signaling, and thus, maintains the stringency of QS input and responses [3]. The motif F160/F162/F166 is conserved in Vibrio spp., and F162 is essential for proper interaction with the head group of CAI-1 molecules. Pairwise screening of CqsS sensor mutants with a synthetic library of modified CAI-1 analogues showed that amino acid exchanges at residues F160/F162/F166 can facilitate the binding of inhibitory CAI-1-like molecules carrying a phenyl moiety at the head group. These mutants are impaired for downstream kinase activity, and thus, residues F160/F162/F166 are suggested to act in concert to prevent binding of inhibitory non-natural CAI-1 molecules [88].
The position corresponding to V. cholerae C170 is highly variant in different Vibrio spp. V. cholerae C170 determines the preference for a C10 acyl tail, but with lower affinity also accepts a C8 acyl tail (Figure 3(b)). Thus, in line with the promiscuous substrate selection of V. cholerae CqsA utilizing C10-CoA as well as C8-CoA [86], V. cholerae CqsS displays a relaxed ligand specificity. Yet, V. cholerae CqsS does not recognize LAI-1 with a C12 acyl tail [70]. Conversely, the strict substrate specificity of V. harveyi CqsA is reflected in the presence of a bulky phenylalanine at the corresponding position (F166), which only allows binding of CAI-1 derivatives with a C8 but not with longer acyl tails [87]. Furthermore, V. harveyi CqsS preferentially binds Ea-C8-CAI-1, and the presence of the enamine group at the C3 moiety is critical for downstream signaling.
Direct binding and stimulation of the L. pneumophila sensor kinase LqsS by LAI-1 has not been shown experimentally. However, in analogy to V. cholerae CqsS, the motifs located in the first three trans-membrane domains that are essential for the interaction with CAI-1, are conserved at the corresponding sites in LqsS (Figure 3(a)). Moreover, the motifs in the second part of the CqsS sensor domain, which define the chemical characteristics of bound AHKs, are also similar to the corresponding motifs in LqsS. Accordingly, the serine at position 107 is conserved in the sensor kinases in Vibrio spp., as well as in L. pneumophila (S114). In contrast, the tryptophan at position 104 in CqsS is replaced by a serine in LqsS (S111), which possibly indicates a preference for LAI-1 molecules with hydroxyl modifications. Furthermore, at the position that discriminates acyl tail length in Vibrio spp. (C170 in V. cholerae CqsS, F175 in V. harveyi CqsS), LqsS harbors small amino acids (T175/L176), suggesting that the LqsA/LqsS system might show relaxed specificity similar to V. cholerae CqsA/CqsS. Finally, LqsS and CqsS also differ in the gatekeeper motif, which defines binding of the polar head group (F160/F162/F166 vs. I166/F168/A172).
In summary, the V. cholerae CqsA/CqsS system produces and detects three different AHK molecules: (1) CAI-1, (2) amino-modified CAI-1 analogues (Am-CAI-1, Ea-CAI-1), and (3) Ea-C8-CAI-1 (but not C8-CAI-1). V. cholerae CqsA prefers the acyl CoA substrate C10-CoA over C8-CoA, and this substrate preference also correlates with the ligand selectivity of CqsS, which binds CAI-1 with C10 acyl tails more efficiently than molecules with C8 acyl tails. The chain length discrimination is defined by the amino acid residue at position 170 located in the sixth trans-membrane helix of the CqsS receptor domain. V. cholerae C170 allows the detection of ligands with C10 or C8 but not shorter (C6, C4) or longer (C12) acyl tails. The motifs F160/F162/F166 (polar head group) and W104/S107 (functional group at C3 moiety) ensure the exclusive binding of CAI-1 analogues with a relaxed specificity for amino and hydroxyl modification.
5. Ecological and Evolutionary Implications of AHK Signaling
The V. cholerae CqsA/CqsS system shows a rather relaxed signal discrimination, and multiple CAI analogues can interact with the sensor kinase CqsS, including molecules produced by other Vibrio spp. Since V. cholerae and V. harveyi produce and detect Ea-C8-CAI-1, this CAI-1-derivative might be used for communication among different Vibrio spp. Potential ecological interactions among Vibrio spp. might indeed take place in river deltas and other aquatic ecosystems [10]. Still, the CqsS sensors of different Vibrio spp. are most sensitive for AHK molecules produced by their cognate CqsA AI synthases. The coordination and fine-tuning of cognate AI synthases and AI receptors ensures efficient ligand-receptor interactions, avoids inappropriate communication and maintains high fidelity QS signaling.
V. harveyi produces higher concentrations of the hydroxyl-modified C8-CAI-1 compared to Ea-C8-CAI-1, even though the hydroxyl molecule is only a weak agonist. Similar CAI-1 production profiles were found in supernatants of several other marine Vibrio spp., e.g., V. parahaemolyticus, V. alginolyticus or V. anguillarum [72,87]. Furthermore, all these Vibrio spp. employ three parallel QS systems (i.e., CqsA/CqsS, LuxS/LuxPQ and LuxM/LuxN). As LuxS/LuxPQ-based AI-2 signaling is broadly used, the stringency imposed by the AHK (and AHL) systems might be essential for an accurate integration of multiple and similar AI signals.
In contrast to Vibrio spp., which employ several QS systems, L. pneumophila apparently uses only the Lqs system. Perhaps, this reflects the distinct lifestyles of the bacteria. While Legionella spp. can colonize extracellular niches, such as multi-species biofilms, their preferential habitat is likely the resident amoebal fauna in biofilms [14,90]. Thus, intracellular L. pneumophila might avoid competitors, which are normally encountered in nutrition-rich extracellular ecological niches, and consequently, the bacteria might have lost (or never acquired) QS systems that support broad inter-species communication. In agreement with the notion that (inter-species) communication systems are dispensable for intracellular bacteria, neither L. longbeachae [83], nor Legionella spp. other than L. pneumophila apparently possess the lqs system [9,70,91].
The lqs genes are located in a genomic region of L. pneumophila that seems to represent a hot spot for recombination. The corresponding site in L. longbeachae harbours a homologue of the putative effector gene legG2 (llo0327, lpg0267), which is flanked by transposase elements and localizes upstream of a homologue of E. coli hdeD. The function of L. pneumophila hdeD is unknown [54], but interestingly, the gene interrupts the convergently transcribed lqsS and lqsR genes (Figure 1(a)) [50]. Thus, the discrete genes for lqsS and lqsR might originate from a larger cqsS-like ancestor gene, which was split by the resident hdeD locus by recombinatory events that integrated a cqs-like cassette into the L. pneumophila genome.
Gene clusters including homologues of lqsA and lqsS are present not only in L. pneumophila and Vibrio spp., but also in several other environmental bacteria, including Ralstonia eutropha, Nitrococcus mobilis, Burkholderia xenovorans and Polaromonas spp. [9,50]. While most of these gene clusters harbor only an lqsA and lqsS homologue, some also contain an lqsR homologue, but the hdeD gene is present only in the L. pneumophila lqs cluster. Likely, L. pneumophila acquired the lqsA-lqsR-lqsS cluster by horizontal gene transfer from other environmental bacteria, such as Nitrococcus, Burkholderia or Polaromonas spp. The prevalence of putative AHK AI synthases and sensors suggests that intra- and inter-species AHK-based cell-cell communication is wide-spread among bacteria.
6. Conclusions/Outlook
The water-borne opportunistic pathogens L. pneumophila and V. cholerae harbor the lqs or cqs gene clusters and engage in cell-cell communication through the production and sensing of the AHKs LAI-1 or CAI-1, respectively. These small signaling molecules regulate virulence and pathogen-host interactions, formation of biofilms or extracellular filaments, competence, and the expression of a genomic “fitness island”. Recent studies elucidated the PLP-dependent biosynthesis of CAI-1 and amino-derivatives by the AI synthase CqsA using SAM and decanoyl-CoA as substrates. CAI-1-related AIs and probably also the corresponding LAI-1 molecules signal through CqsS/LqsS sensor kinases via a phosphorelay cascade, which converges on the V. cholerae master regulator HapR or the L. pneumophila response regulator LqsR. The continued analysis of the Lqs and Cqs QS systems (i) yields insights into a QS strategy likely used by a number of environmental bacteria, (ii) provides tools to analyze the mechanism of signal transduction by AHK ligands and TC system sensor kinases, and (iii) might lead to the development of compounds useful in clinical or environmental settings to interfere with virulence and persistence of important human pathogens.
Acknowledgments
Work in the group of H.H. was supported by the Max von Pettenkofer Institute, Ludwig-Maximilians University Munich, the German Research Foundation (DFG, HI 1511/2-1) and the Swiss National Science Foundation (SNF, CRSI33_130016).
References
- 1.Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Fuqua C., Greenberg E.P. Listening in on bacteria: Acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 3.Ng W.L., Bassler B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler B.L., Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Shank E.A., Kolter R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 7.Galloway W.R., Hodgkinson J.T., Bowden S.D., Welch M., Spring D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 8.Deng Y., Wu J., Tao F., Zhang L.H. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem. Rev. 2011;111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- 9.Tiaden A., Spirig T., Hilbi H. Bacterial gene regulation by α-hydroxyketone signaling. Trends Microbiol. 2010;18:288–297. doi: 10.1016/j.tim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Nelson E.J., Harris J.B., Morris J.G., Jr., Calderwood S.B., Camilli A. Cholera transmission: The host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 2009;7:693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton H.J., Ang D.K., van Driel I.R., Hartland E.L. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 2010;23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yildiz F.H., Visick K.L. Vibrio biofilms: So much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Declerck P. Biofilms: The environmental playground of Legionella pneumophila. Environ. Microbiol. 2010;12:557–566. doi: 10.1111/j.1462-2920.2009.02025.x. [DOI] [PubMed] [Google Scholar]
- 14.Hilbi H., Hoffmann C., Harrison C.F. Legionella spp. outdoors: Colonization, communication and persistence. Environ. Microbiol. Rep. 2011;3:286–296. doi: 10.1111/j.1758-2229.2011.00247.x. [DOI] [PubMed] [Google Scholar]
- 15.Greub G., Raoult D. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilbi H., Weber S.S., Ragaz C., Nyfeler Y., Urwyler S. Environmental predators as models for bacterial pathogenesis. Environ. Microbiol. 2007;9:563–575. doi: 10.1111/j.1462-2920.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- 17.Steinert M. Pathogen-host interactions in Dictyostelium, Legionella, Mycobacterium and other pathogens. Semin. Cell Dev. Biol. 2011;22:70–76. doi: 10.1016/j.semcdb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Raskin D.M., Seshadri R., Pukatzki S.U., Mekalanos J.J. Bacterial genomics and pathogen evolution. Cell. 2006;124:703–714. doi: 10.1016/j.cell.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Valero L., Rusniok C., Jarraud S., Vacherie B., Rouy Z., Barbe V., Medigue C., Etienne J., Buchrieser C. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields B.S., Benson R.F., Besser R.E. Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massis L.M., Zamboni D.S. Innate immunity to Legionella pneumophila. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carratala J., Garcia-Vidal C. An update on Legionella. Curr. Opin. Infect. Dis. 2010;23:152–157. doi: 10.1097/QCO.0b013e328336835b. [DOI] [PubMed] [Google Scholar]
- 23.Cazalet C., Rusniok C., Brüggemann H., Zidane N., Magnier A., Ma L., Tichit M., Jarraud S., Bouchier C., Vandenesch F., et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 24.Brüggemann H., Cazalet C., Buchrieser C. Adaptation of Legionella pneumophila to the host environment: Role of protein secretion, effectors and eukaryotic-like proteins. Curr. Opin. Microbiol. 2006;9:86–94. doi: 10.1016/j.mib.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Molofsky A.B., Swanson M.S. Differentiate to thrive: Lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- 26.Isberg R.R., O’Connor T.J., Heidtman M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat. Rev. Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urwyler S., Nyfeler Y., Ragaz C., Lee H., Mueller L.N., Aebersold R., Hilbi H. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic. 2009;10:76–87. doi: 10.1111/j.1600-0854.2008.00851.x. [DOI] [PubMed] [Google Scholar]
- 28.Hubber A., Roy C.R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 29.Hilbi H., Weber S., Finsel I. Anchors for effectors: Subversion of phosphoinositide lipids by Legionella. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber S.S., Ragaz C., Hilbi H. Pathogen trafficking pathways and host phosphoinositide metabolism. Mol. Microbiol. 2009;71:1341–1352. doi: 10.1111/j.1365-2958.2009.06608.x. [DOI] [PubMed] [Google Scholar]
- 31.Urwyler S., Brombacher E., Hilbi H. Endosomal and secretory markers of the Legionella-containing vacuole. Commun. Integr. Biol. 2009;2:107–109. doi: 10.4161/cib.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Valero L., Rusniok C., Cazalet C., Buchrieser C. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., Luo Z.Q. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franco I.S., Shuman H.A., Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 35.Sack D.A., Sack R.B., Nair G.B., Siddique A.K. Cholera. Lancet. 2004;363:223–233. doi: 10.1016/s0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 36.Matson J.S., Withey J.H., DiRita V.J. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 2007;75:5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirn T.J., Lafferty M.J., Sandoe C.M., Taylor R.K. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- 38.McLeod S.M., Kimsey H.H., Davis B.M., Waldor M.K. CTXphi and Vibrio cholerae: Exploring a newly recognized type of phage-host cell relationship. Mol. Microbiol. 2005;57:347–356. doi: 10.1111/j.1365-2958.2005.04676.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J., Miller M.B., Vance R.E., Dziejman M., Bassler B.L., Mekalanos J.J. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J., Mekalanos J.J. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 41.Schild S., Tamayo R., Nelson E.J., Qadri F., Calderwood S.B., Camilli A. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang W., Pascual-Montano A., Silva A.J., Benitez J.A. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 43.Liang W., Sultan S.Z., Silva A.J., Benitez J.A. Cyclic AMP post-transcriptionally regulates the biosynthesis of a major bacterial autoinducer to modulate the cell density required to activate quorum sensing. FEBS Lett. 2008;582:3744–3750. doi: 10.1016/j.febslet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammer B.K., Tateda E.S., Swanson M.S. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 2002;44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 45.Lenz D.H., Miller M.B., Zhu J., Kulkarni R.V., Bassler B.L. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 46.Tsou A.M., Liu Z., Cai T., Zhu J. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology. 2011;157:1620–1628. doi: 10.1099/mic.0.046235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molofsky A.B., Swanson M.S. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 2003;50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 48.Sahr T., Brüggemann H., Jules M., Lomma M., Albert-Weissenberger C., Cazalet C., Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 2009;72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller M.B., Skorupski K., Lenz D.H., Taylor R.K., Bassler B.L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 50.Tiaden A., Spirig T., Weber S.S., Brüggemann H., Bosshard R., Buchrieser C., Hilbi H. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol. 2007;9:2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- 51.Tiaden A., Spirig T., Carranza P., Brüggemann H., Riedel K., Eberl L., Buchrieser C., Hilbi H. Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions. J. Bacteriol. 2008;190:7532–7547. doi: 10.1128/JB.01002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammer B.K., Bassler B.L. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 53.Waters C.M., Lu W., Rabinowitz J.D., Bassler B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiaden A., Spirig T., Sahr T., Walti M.A., Boucke K., Buchrieser C., Hilbi H. The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ. Microbiol. 2010;12:1243–1259. doi: 10.1111/j.1462-2920.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- 55.Meibom K.L., Blokesch M., Dolganov N.A., Wu C.Y., Schoolnik G.K. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 56.Antonova E.S., Hammer B.K. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol. Lett. 2011;322:68–76. doi: 10.1111/j.1574-6968.2011.02328.x. [DOI] [PubMed] [Google Scholar]
- 57.Suckow G., Seitz P., Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J. Bacteriol. 2011;193:4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 59.Engebrecht J., Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beier D., Gross R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Mascher T., Helmann J.D., Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitrophanov A.Y., Groisman E.A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitworth D.E., Cock P.J. Evolution of prokaryotic two-component systems: Insights from comparative genomics. Amino Acids. 2009;37:459–466. doi: 10.1007/s00726-009-0259-2. [DOI] [PubMed] [Google Scholar]
- 64.Barakat M., Ortet P., Whitworth D.E. P2CS: A database of prokaryotic two-component systems. Nucleic Acids Res. 2011;39:D771–D776. doi: 10.1093/nar/gkq1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray K.M., Garey J.R. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology. 2001;147:2379–2387. doi: 10.1099/00221287-147-8-2379. [DOI] [PubMed] [Google Scholar]
- 66.Lerat E., Moran N.A. The evolutionary history of quorum-sensing systems in bacteria. Mol. Biol. Evol. 2004;21:903–913. doi: 10.1093/molbev/msh097. [DOI] [PubMed] [Google Scholar]
- 67.Case R.J., Labbate M., Kjelleberg S. AHL-driven quorum-sensing circuits: Their frequency and function among the proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- 68.Xavier K.B., Bassler B.L. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Higgins D.A., Pomianek M.E., Kraml C.M., Taylor R.K., Semmelhack M.F., Bassler B.L. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 70.Spirig T., Tiaden A., Kiefer P., Buchrieser C., Vorholt J.A., Hilbi H. The Legionella autoinducer synthase LqsA produces an α-hydroxyketone signaling molecule. J. Biol. Chem. 2008;283:18113–18123. doi: 10.1074/jbc.M801929200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao J.G., Meighen E.A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 72.Henke J.M., Bassler B.L. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freeman J.A., Bassler B.L. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 74.Freeman J.A., Bassler B.L. Sequence and function of LuxU: A two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 1999;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neiditch M.B., Federle M.J., Pompeani A.J., Kelly R.C., Swem D.L., Jeffrey P.D., Bassler B.L., Hughson F.M. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swem L.R., Swem D.L., Wingreen N.S., Bassler B.L. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–473. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pompeani A.J., Irgon J.J., Berger M.F., Bulyk M.L., Wingreen N.S., Bassler B.L. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol. Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bardill J.P., Zhao X., Hammer B.K. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol. 2011;80:1381–1394. doi: 10.1111/j.1365-2958.2011.07655.x. [DOI] [PubMed] [Google Scholar]
- 79.Lenz D.H., Bassler B.L. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 2007;63:859–871. doi: 10.1111/j.1365-2958.2006.05545.x. [DOI] [PubMed] [Google Scholar]
- 80.Lenz D.H., Mok K.C., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 81.Shikuma N.J., Fong J.C., Odell L.S., Perchuk B.S., Laub M.T., Yildiz F.H. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bacteriol. 2009;191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srivastava D., Harris R.C., Waters C.M. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 2011;193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cazalet C., Gomez-Valero L., Rusniok C., Lomma M., Dervins-Ravault D., Newton H.J., Sansom F.M., Jarraud S., Zidane N., Ma L., et al. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelly R.C., Bolitho M.E., Higgins D.A., Lu W., Ng W.L., Jeffrey P.D., Rabinowitz J.D., Semmelhack M.F., Hughson F.M., Bassler B.L. The Vibrio cholerae quorum-sensing autoinducer CAI-1: Analysis of the biosynthetic enzyme CqsA. Nat. Chem. Biol. 2009;5:891–895. doi: 10.1038/nchembio.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jahan N., Potter J.A., Sheikh M.A., Botting C.H., Shirran S.L., Westwood N.J., Taylor G.L. Insights into the biosynthesis of the Vibrio cholerae major autoinducer CAI-1 from the crystal structure of the PLP-dependent enzyme CqsA. J. Mol. Biol. 2009;392:763–773. doi: 10.1016/j.jmb.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 86.Wei Y., Perez L.J., Ng W.L., Semmelhack M., Bassler B.L. Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem. Biol. 2011;6:356–365. doi: 10.1021/cb1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ng W.L., Perez L.J., Wei Y., Kraml C., Semmelhack M.F., Bassler B.L. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 2011;79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng W.L., Wei Y., Perez L.J., Cong J., Long T., Koch M., Semmelhack M.F., Wingreen N.S., Bassler B.L. Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. Proc. Natl. Acad. Sci. USA. 2010;107:5575–5580. doi: 10.1073/pnas.1001392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bolitho M.E., Perez L.J., Koch M.J., Ng W.L., Bassler B.L., Semmelhack M.F. Small molecule probes of the receptor binding site in the Vibrio cholerae CAI-1 quorum sensing circuit. Bioorg. Med. Chem. 2011;19:6906–6918. doi: 10.1016/j.bmc.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taylor M., Ross K., Bentham R. Legionella, protozoa, and biofilms: Interactions within complex microbial systems. Microb. Ecol. 2009;58:538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- 91.Chien M., Morozova I., Shi S., Sheng H., Chen J., Gomez S.M., Asamani G., Hill K., Nuara J., Feder M., et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]