Abstract
BACKGROUND
People with the human neutrophil antigen (HNA)-3b/3b type can make HNA-3a antibodies, which have been reported to cause immune neutropenia disorders, and are especially prone to cause severe cases of transfusion-related acute lung injury (TRALI). However, knowledge of HNA-3 allele frequencies outside Caucasian populations is limited. We developed a high-throughput genotyping assay and determined the HNA-3a/3b genotype frequencies in 6 different racial and ethnic groups.
STUDY DESIGN AND METHODS
Genotyping utilized Taqman 5’ exonuclease chemistry and real-time PCR. A total of 742 DNA samples from 6 different racial and ethnic groups were genotyped for HNA-3a and HNA-3b.
RESULTS
The genotyping assay showed 100% sensitivity and specificity compared to sequencing and phenotyping and had high throughput. A significant percentage of Caucasians (6.5%), Han Chinese (16%), and Asian Indians (6%) typed HNA-3b/3b, but only a small percentage of Hispanics (1%) and no African or Native Americans.
CONCLUSIONS
The HNA-3 genotyping assay had high sensitivity, specificity, and sample throughput. HNA-3b/b genotype results determined for 742 individuals representing 6 different racial and ethnic groups showed that there could be a significant risk of producing anti-HNA-3a in Chinese, as well as in Caucasian and Asian Indian blood donor populations, but a very low risk in Hispanic, African or Native American populations.
INTRODUCTION
To date, nine different human neutrophil alloantigens (HNA) have been identified, HNA-1a,-1b, -1c, HNA-3a, -3b, HNA-4a, -4b, and HNA-5a, -5b. The antigen HNA-2a is a misnomer since HNA-2a antibodies are technically isoantibodies that recognize the CD177 protein, which is missing from neutrophils of immunized individuals. HNA-1, -4, and -5 are expressed on glycoproteins CD16 (FcγRIIIb), CD11b/18 (Mac-1), and CD11a/18 (LFA-1), respectively, and single nucleotide polymorphisms (SNPS) in the encoding genes determine the different allelic forms. This has enabled the development of DNA genotyping assays for the SNPs. Only recently, it was shown that a G461A SNP in SLC44A2 encodes R154 and Q154 in choline transporter-like protein 2 (CTL2) and the HNA-3a, HNA-3b antigens, respectively.1,2 This finding now makes it possible to perform genotyping for HNA-3. Individuals homozygous for HNA-3a or HNA-3b can be immunized and produce antibodies when exposed to the cognate antigen through blood transfusion, pregnancy, or possibly even naturally occurring antigens in the environment.3
HNA-3a antibodies have been reported to cause, Neonatal Alloimmune Neutropenia (NAN) and Transfusion-Related Acute Lung Injury (TRALI). NAN occurs when a mother is immunized against an incompatible paternal HNA inherited by the fetus and antibodies produced cross the placenta and destroy fetal neutrophils. TRALI is currently the number one cause of transfusion-related death.4 TRALI reactions are believed to occur when leukocyte antibodies in a transfused blood react with antigens on the recipient’s neutrophils or monocytes. Both HLA and HNA antibodies have been implicated in causing TRALI, and HNA-3a antibodies are especially prone to cause severe and often fatal TRALI reactions.5-7 Genotyping for HNA-3 a and HNA-3b could, therefore, be important in prenatal determination of a fetus at risk of developing NAN, to confirm the HNA-3a or HNA-3b specificity of antibodies in maternal plasma, and in identifying blood donors at risk for producing antibodies against HNA-3a and -3b that could cause TRALI.
Studies of Caucasian populations indicate that approximately 5 % of individuals are negative for HNA-3a and are therefore at risk to be immunized against this antigen. However, the frequency of HNA-3 alleles is not well defined for non-Caucasian populations.8,9 The fact that HNA-3 antibodies are likely to cause severe forms of TRALI5-7 highlights the need to determine the HNA-3 allele frequencies in other populations. We, therefore, developed a relatively high-throughput 5’ exonuclease SNP genotyping assay (SGA) and determined the HNA-3a and -3b genotype frequencies in six different racial and ethnic groups.
MATERIALS AND METHODS
Donor DNA samples
DNA from a total of 742 individuals representing 6 different racial groups was isolated from EDTA whole blood using the QIAamp 96 DNA Blood Kit (Qiagen, Valencia, CA). Caucasian and African American samples were collected from normal volunteer blood donors at BloodCenter of Wisconsin. Han Chinese blood samples were kindly provided by Dr. Guo Guang Wu, NanNing, China. Asian Indian, Hispanic/Latino Americans, and Native American samples were from those used in previous reports.10,11 Race was assigned on the basis of each donor’s self-identification. DNA samples with T457 in SLC44A2 were kindly provided by Dr. Brigitte Flesch, Bad Kreuznach, Germany and Dr. Sentot Santoso, Giessen, Germany. Studies were approved by the Internal Review Board, BloodCenter of Wisconsin.
Reagents
Commercial gene-specific forward and reverse primers and fluorescent labeled allele-specific hydrolysis probes (HNA-3a/3b PCR primer-probe mix) were purchased from Life Technologies/Applied Biosystems™ (Carlsbad, CA). One probe is labeled with the fluorochrome FAM (6-carboxyfluorescein) and specific for the HNA-3a allele (G461 of SLC44A2). The other probe is labeled with the fluorochrome VIC and specific for the HNA-3b allele (A461 of SLC44A2). Primer and probe sequences are proprietary and not disclosed by the manufacturer. The HNA-3 genotyping PCR pre- mix was made in-house and a single reaction consisted of: 18 μL PCR grade water, 2.5 μL of 10x PCR buffer without MgCl2, 2 μL of 25 mM MgCl2, and 0.13 μL of 25 mM dNTPs.
HNA-3 SNP Genotyping Assay (SGA)
Genotyping of SLC44A2 for the HNA-3a (G461, SLC44A2*1) and HNA-3b (A461, SLC44A2*2) alleles was performed using a SNP genotyping assay (SGA). The assay utilized TaqMan® 5’ exonuclease chemistry, and consisted of PCR amplification of 1 μL of genomic DNA, 1.25 μL of HNA-3a/3b PCR primer-probe mix, 22.63 μL of PCR premix, and 0.12 μL Taq enzyme (Roche, Madison, WI) + Taq antibody (Clonetech Labs, Mountainview, CA). PCR reactions were performed in a single well of a 96-well plate on a LightCycler™ (LC) 480 real-time PCR instrument (Roche). Four control DNA samples were tested in each plate and consisted of HNA-3a/3a, HNA-3a/3b, and HNA-3b/3b positive controls and a water (no-template) negative control. FAM vs. VIC fluorescence (FL) was plotted using instrument software to make HNA-3a/3b genotype calls.
DNA Sequence Analysis
Genomic DNA was amplified by PCR using primers designed to amplify exon 7 of SLC44A2 (reference sequence National Center for Biotechnology database, www.ncbi.nlm.nih.gov/), as described previously.1 Primer sequences are shown below:
Forward Primer: 5’-GTA AAA CGA CGG CCA GTT CCT GAG AGC ACA GGT ATG G-3’
Reverse Primer: 5’-CAG GAA ACA GCT ATG ACC ACT GCA TGG AGC AGA GGA TG-3’
Automated sequence analysis of PCR products was performed in both directions with the Big Dye Terminator v.3.1 Cycle Sequencing kit on an ABI 3130XL genetic analyzer (Applied Biosystems, Foster City, CA).
Statistics
Statistical analysis was performed using R 2.12 software (R Foundation for Statistical Computing, Vienna, Austria) with libraries ‘VGAM’ (vector generalized additive models) for testing Hardy Weinberg equilibrium (HWE) and fitting the model, and ‘multcomp’ for multiple comparison adjustment. A likelihood ratio test was first performed on genotype frequency data to examine deviation from HWE simultaneously over all groups. A multinomial regression model assuming HWE was then fitted, followed by pair-wise comparisons of the estimated log-odds from each of the 6 groups adjusted for multiple testing using the single-step method to examine differences in HNA-3b allele frequencies between groups.
RESULTS
HNA-3 genotype assay validation and performance
HNA-3 genotype calls by SGA could be easily determined from FAM/VIC fluorescence plots of the data (Figure 1). However, genotype calls were standardized by establishing FAM/VIC FL ratio (FLR) ranges for each genotype: ≥ 3.68, HNA-3a/3a; 0.67 – 1.44, HNA-3a/3b; ≤ 0.04, HNA-3b/3b. The SGA was validated by comparing HNA-3a/3b genotype results for 16 subjects to phenotype and genotype results previously determined by flow cytometry and sequencing, respectively.1 The 16 samples gave identical HNA-3a/3a, HNA-3a/3b, and HNA-3b/3b types by all 3 methods (Table 1) corresponding to 100% analytic sensitivity and specificity for the SGA. Testing of serially diluted DNA samples of each HNA-3 genotype showed an SGA detection limit of 1ng/μl (data not shown). Assay throughput is 388 isolated DNA samples typed by a single technologist in 8 hours with only 1.5 hours of hands-on time.
Figure 1. FAM/VIC fluorescence (FL) plot of HNA-3a/HNA-3b genotyping results using the SNP genotyping assay (SGA).
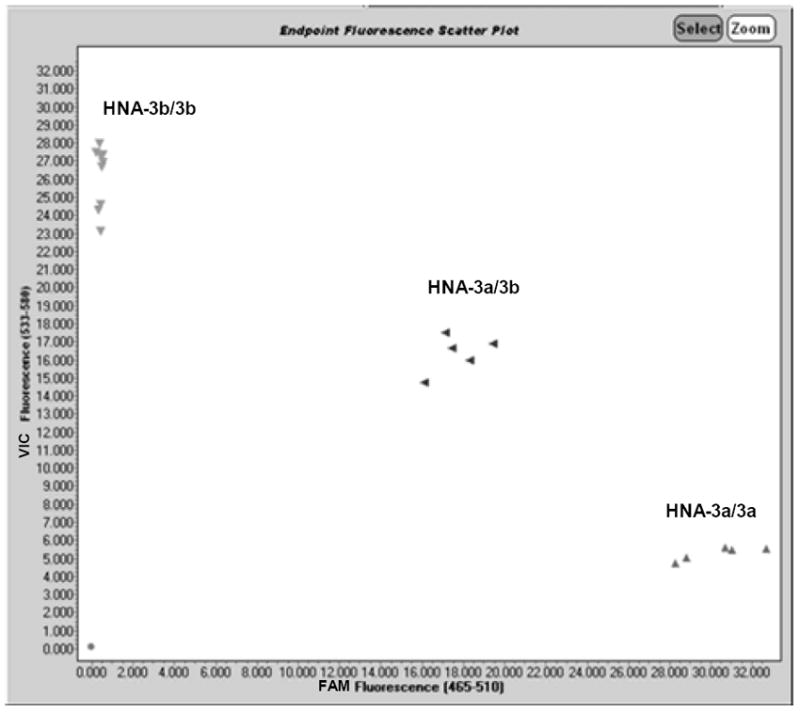
The assay employed TaqMan® 5’ exonuclease chemistry and real-time PCR amplification of genomic DNA with specific primers and detection of amplicons with allele-specific fluorescence (FL) hydrolysis probes, and FAM/VIC fluorescence scatter plots generated by instrument software as described in materials and methods. HNA-3 genotype results are shown for 16 normal blood donors and 3 control samples consisting of HNA-3a/3a (triangles, lower right plot area), HNA-3a/3b (triangles, middle plot area), and HNA-3b/3b genotypes (triangles, upper left plot area). Some FL values were identical resulting in overlap of data points in the HNA-3b/3b genotype area of the plot.
Table 1.
HNA 3a/3b genotype results comparison.
| Subject | HNA-3 Phenotype | HNA-3 Sequencing | HNA-3 SGA |
|---|---|---|---|
| 1 | 3a/a | 3a/a | 3a/a |
| 2 | 3a/a | 3a/a | 3a/a |
| 3 | 3a/a | 3a/a | 3a/a |
| 4 | 3a/a | 3a/a | 3a/a |
| 5 | 3a/b | 3a/b | 3a/b |
| 6 | 3a/b | 3a/b | 3a/b |
| 7 | 3a/b | 3a/b | 3a/b |
| 8 | 3a/b | 3a/b | 3a/b |
| 9 | 3b/b | 3b/b | 3b/b |
| 10 | 3b/b | 3b/b | 3b/b |
| 11 | 3b/b | 3b/b | 3b/b |
| 12 | 3b/b | 3b/b | 3b/b |
| 13 | 3b/b | 3b/b | 3b/b |
| 14 | 3b/b | 3b/b | 3b/b |
| 15 | 3b/b | 3b/b | 3b/b |
HNA-3 phenotype determined serologically by flow cytometry
SGA = SNP genotyping assay
HNA-3 genotype frequencies for 6 different racial groups
SGA genotype results for six different racial and ethnic groups are shown in Table 2. A likelihood ratio test showed no deviation from Hardy-Weinberg equilibrium (p = 0.52). HNA-3 genotype frequencies were similar for Asian Indians, Caucasian Americans and Hispanic/Latino Americans and for African Americans and Native American Indians, but the Han Chinese cohort was significnatly different from the rest. Analysis of variance for all groups showed significant differences (p < 0.05) between specific groups for HNA-3b frequency (Table 2). HNA-3b genotype frequency was similar for Asian Indian (24.2%), Caucasian (23.7%), and Hispanic (17.1%) populations, but significantly higher in Han Chinese (38.2%), and no HNA-3b/3b individuals were found in African American or Native American Inidans.
Table 2.
HNA-3 genotype frequencies by SGA for 6 different racial groups.
| HNA-3 genotype frequency (%) | ||||||
|---|---|---|---|---|---|---|
| Population | N | 3a/3a | 3a/3b | 3b/3b | HNA-3b Allele Frequency | *Significance |
| African American | 161 | 138 (86) | 23 (14) | 0(0) | 7.1% | a |
| Asian Indian | 91 | 52(57) | 34 (37) | 5(6) | 24.2% | b |
| Caucasian | 186 | 110 (59.1) | 64 (34.4) | 12 (6.5) | 23.7% | b |
| Han Chinese | 119 | 47 (39) | 53 (45) | 19 (16) | 38.2% | c |
| Hispanic/Latino American | 92 | 61 (66) | 30 (33) | 1(1) | 17.4% | b |
| Native American Indian | 93 | 83 (89) | 10 (11) | 0(0) | 5.4% | a |
groups with different letters are significantly different from each other
SGA = SNP genotyping assay
No subjects detected with T457 in SLC44A2
Recently, the SNP C457T in SLC44A2 that encodes L153F in CTL2 was reported to cause false negative HNA-3a genotype results when sequence-specific primers (SSP) were employed that encompass the SNP.12 T457 has a frequency in German blood donors of ~1-2%.13,14 Previous studies showed that errors in HNA-3 genotyping occurred with DNA samples that were heterozygous A461/G461 (HNA-3a/3b) and heterozygous C457/T457 in SLC44A2. These samples mistyped as HNA-3b/3b.8,12 We therefore specifically sequenced exon 7 of SLC44A2 for 140 donors with HNA-3b/b and HNA-3a/b genotypes. Sequencing results matched HNA-3 genotypes determined by SGA for all 140 samples and none had T457 (Table 3). To further determine whether our SGA assay could be adversely affected by the presence of T457, we genotyped two HNA-3a/3b DNA samples with T457, and that had been previously reported to cause mistyping by PCR-SSP assays (Table 3).8,12 An HNA-3a/3b genotype call could not be made for either sample by SGA, because the FAM/VIC FLR (0.21) fell into a “gray zone” between HNA-3a/3b and HNA-3b/3b (Figure 2). However, the samples would not have been mistyped on a routine run but instead flagged for genotype determination by sequencing. DNA sequencing of both samples confirmed their genotypes as A461/G461 (HNA-3a/3b), C457/T457 in SLC44A2 (results not shown).
Table 3.
HNA-3a/3b and C457T sequencing results for 140 samples
| HNA-3a/3b Genotype | C45T Genotype | |||
|---|---|---|---|---|
| Population | N | SGA | Sequencing | Sequencing |
| African American | 23 | a/b | a/b | C/C |
| Asian Indian | 5 | b/b | b/b | C/C |
| Caucasian American | 12 | b/b | b/b | C/C |
| Caucasian American | 66 | a/b | a/b | C/C |
| Caucasian (German) | 2 | * | a/b | C/T |
| Han Chinese | 19 | b/b | b/b | C/C |
| Hispanic | 1 | b/b | b/b | C/C |
| Native American | 10 | a/b | a/b | C/C |
HNA-3 genotype could not be called since fluorescence ratio was 0.21
Figure 2. FAM/VIC fluorescence (FL) plot of HNA-3a/HNA-3b genotyping results using the SNP genotyping assay (SGA) with samples from individuals with T457 in SLC44A2.
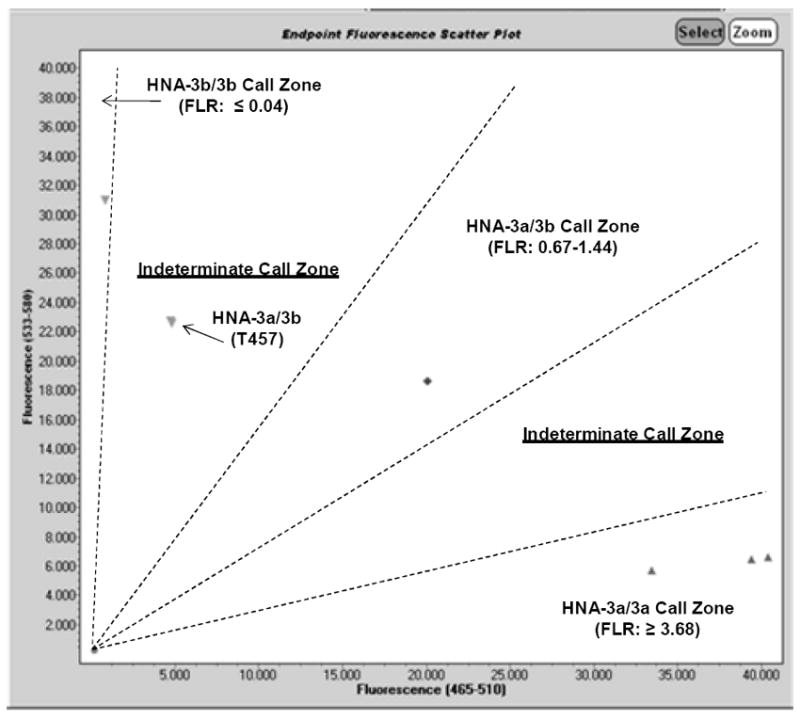
The SGA was performed with 3 HNA-3a/3a (symbols, lower right plot area), 1 HNA-3a/3b (symbol, middle plot area), and 1HNA-3b/3b (symbol, upper left lot area) control DNA samples, and 2 HNA-3a/3b DNA samples from subjects previously shown to have the T457 SNP (labeled on plot). HNA-3 genotype “call zones” and associated ranges of fluorescence ratios (FLR) are labeled on the plot. HNA-3 genotype results by SGA for the two samples with T457 fell into an “indeterminate call zone” (FLR = 0.21 for both samples) between the HNA-3a/3b and HNA-3b/3b call zones that triggered DNA sequencing to determine the true HNA-3a/3b genotypes (results not shown) for these samples.
DISCUSSION
We developed and validated a high-throughput SNP genotyping assay (SGA) for the HNA-3 alleles. Assay sensitivity and specificity was 100% compared to HNA-3 typing by both serology and sequencing. However, it was difficult to make a genotype call by SGA for HNA-3a/b samples with the T457 SNP in SLC44A2. Samples with this SNP mistype as HNA-3b/3b using PCR-SSP12 and real-time PCR8 assays. We typed two such samples by SGA, and obtained “indeterminate” results, but both samples were appropriately reflexed to DNA sequencing, ensuring an accurate HNA-3a/3b genotype.
The SGA was then used to type 742 individuals from 6 different racial and ethnic groups: African Americans (N = 161), Caucasian Americans (N = 186), Asian Indians (N = 91), Han Chinese (N = 119), Hispanic/Latino Americans (N = 92), and Native American Indians (N = 93). Results showed HNA-3 genotype frequencies for Caucasian and African Americans comparable to those reported in previous studies2,8,12,15 and to those reported for Caucasians (CEU), African Americans from the southwestern United States (ASW), and sub-Saharan Africans (YRI) in db SNP (cluster rs2288904). HNA-3 genotype frequencies for Han Chinese were also quite similar to those reported in db SNP (cluster rs2288904, HCB and HCD), but differed from a recent report by Xia et al,9 in which they found a HNA-3b/b genotype frequency (5.64%) in Han Chinese that was almost one third that found in our study (16%). One possible reason for this discrepancy could have been that a significant number of the subjects we genotyped were actually HNA-3a/3b and had the T457 SNP, which as previously described can cause mistyping as HNA-3b/3b. To exclude this possibility, we sequenced DNA from all 19 HNA-3b/b Chinese subjects, but none were found to have T457 and all confirmed HNA-3b/3b (Table 3).
We report for the first time the HNA-3 genotype frequencies in Asian Indians, Hispanic/Latino Americans, and Native American Indians and found Asian Indian and Hispanic/Latino American HNA-3 genotype frequencies similar to those for Caucasians and those for Native American Indians similar to African Americans. Individuals with the HNA-3b/3b genotype can be immunized and make HNA-3a antibodies, which have been reported to cause NAN16 and severe, often fatal TRALI.5-7 Our results show a significant percentage of Caucasians (6.5%), Han Chinese (16%), and Asian Indians (6%) have the genotype HNA-3b/3b, but only a small percentage of Hispanic/Latino Americans (1%) and no African Americans or Native American Indians. These results indicate that alloimmunization against HNA-3a could be as or more frequent in Chinese and Asian Indians, as in Caucasian populations. With respect to TRALI, it may therefore be prudent to consider HNA-3a antibody screening of blood donors in these groups, as is currently done for class I and II HLA antibodies17 to further reduce the risk of TRALI. Unfortunately, development of a practical high-throughput screening assay for HNA-3 antibodies has proven difficult due to the complex nature of the CTL2 molecule.18,19 The CTL2 protein passes through the cell membrane 10 times resulting in five extracellular loops with the HNA-3a/3b epitopes predicted to be in the first loop.1,20 An assay utilizing isolated CTL2 to exclude detection of HLA antibodies would be ideal, but the structural intricacies of CTL2 make it difficult to isolate while still preserving the HNA-3a/3b epitopes necessary for antibody detection.3 For these reasons, efforts to use chemically synthesized small CTL2 peptides21 and GST-fusion proteins22 containing both R154 (HNA-3a) and Q154 (HNA-3b) have proven ineffective to detect and distinguish HNA-3 antibodies. Recently, two groups have successfully expressed recombinant R154 and Q154 forms of full length CLT2 in a human cell line that appears to maintain the confirmation required to detect all HNA-3a and HNA-3b antibodies tested,14,23 but further work is required to apply these findings to development of an effective high-throughput antibody screening assay. Until these issues are resolved, practical screening of blood donors for HNA-3 antibodies must be performed using conventional methods. This is made possible by first genotyping to identify HNA-3b/3b blood donors who are at risk of producing anti-HNA-3a to narrow down the number of donors requiring antibody screen using conventional methods, e.g. flow cytometry or granulocyte agglutination. For this to be practical, HNA-3 genotyping assays must be truly high-throughput to enable typing the large numbers of blood donors required. However, HNA-3 genotyping assays published to date8,9,14,15 give no specifics of their throughput, and those using PCR-SSP with highly manual gel endpoints and subjective results interpretation are not practical. The SGA described here is not only highly sensitive and specific, but also relatively high-throughput enabling HNA-3 genotyping of a minimum of 388 isolated DNA samples typed by a single technologist in 8 hours with only 1.5 hours of hands-on time. Therefore, the SGA could be immediately employed to screen blood donors for HNA-3a and -3b to identify those at risk to produce anti-HNA-3a, and allow for manageable testing of their plasma for antibodies using conventional serologic methods.
In summary, we developed a highly sensitive, specific, high-throughput HNA-3 genotyping assay and determined the HNA-3a and HNA-3b genotype frequencies for 6 different racial and ethnic groups that will be useful in determining the risk for HNA-3 alloimmunization in these populations.
Acknowledgments
The authors wish to thank Dr. Guo Guang Wu for his generosity in providing the Han Chinese blood samples. We also wish to thank the Department of Biostatistics, Medical College of Wisconsin, Wauwatosa, WI for performing statistical analysis of the data.
Supported, in part, by grant 1UL1RR031973 from the Clinical and Translational Science Award (CTSI) program of the National Center for Research Resources, National Institutes of Health.
Footnotes
Authors Contributions: KLB obtained reagents, designed and performed experiments, collected data, and reviewed manuscript. MJS designed and performed experiments and collected data. BRC obtained reagents and samples, designed experiments, analyzed data, wrote and reviewed the manuscript.
Conflict of interest: Authors declare no conflicts of interest relevant to the content of this manuscript.
References
- 1.Curtis BR, Cox NJ, Sullivan MJ, et al. The neutrophil alloantigen HNA-3a (5b) is located on choline transporter-like protein 2 and appears to be encoded by an R>Q154 amino acid substitution. Blood. 2010;115:2073–6. doi: 10.1182/blood-2009-11-248336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greinacher A, Wesche J, Hammer E, et al. Characterization of the human neutrophil alloantigen-3a. Nat Med. 2010;16:45–8. doi: 10.1038/nm.2070. [DOI] [PubMed] [Google Scholar]
- 3.Curtis BR. Another blow to leukocyte antibody-mediated TRALI. Expert Rev Hematol. 2010;3:373–5. doi: 10.1586/ehm.10.38. [DOI] [PubMed] [Google Scholar]
- 4.FDA/CBER. Fatalities reported to FDA following blood collection and transfusion. Annual summary for fiscal year 2010. 2010 http://www.fda.gov/BiologicsBloodVaccines/SafetyAvailability/ReportaProblem/TransfusionDonationFatalities/ucm254802.htm [monograph on the internet]
- 5.Kopko PM, Marshall CS, MacKenzie MR, et al. Transfusion-related acute lung injury: report of a clinical look-back investigation. JAMA. 2002;287:1968–71. doi: 10.1001/jama.287.15.1968. [DOI] [PubMed] [Google Scholar]
- 6.Davoren A, Curtis BR, Shulman IA, et al. TRALI due to granulocyte-agglutinating human neutrophil antigen-3a (5b) alloantibodies in donor plasma: a report of 2 fatalities. Transfusion. 2003;43:641–5. doi: 10.1046/j.1537-2995.2003.00374.x. [DOI] [PubMed] [Google Scholar]
- 7.Reil A, Keller-Stanislawski B, Gunay S, Bux J. Specificities of leucocyte alloantibodies in transfusion-related acute lung injury and results of leucocyte antibody screening of blood donors. Vox Sang. 2008;95:313–7. doi: 10.1111/j.1423-0410.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 8.Huvard MJ, Schmid P, Stroncek DF, Flegel WA. Frequencies of SLC44A2 alleles encoding human neutrophil antigen-3 variants in the African American population. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03396.x. article first published online: 1 NOV 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia W, Bayat B, Sachs U, et al. The frequencies of human neutrophil alloantigens in the Chinese Han population of Guangzhou. Transfusion. 2010;51:1271–7. doi: 10.1111/j.1537-2995.2010.02979.x. [DOI] [PubMed] [Google Scholar]
- 10.Hessner MJ, Curtis BR, Endean DJ, Aster RH. Determination of neutrophil antigen gene frequencies in five ethnic groups by polymerase chain reaction with sequence-specific primers. Transfusion. 1996;36:895–9. doi: 10.1046/j.1537-2995.1996.361097017176.x. [DOI] [PubMed] [Google Scholar]
- 11.Hessner MJ, Shivaram SM, Dinauer DM, et al. Neutrophil antigen (FcgammaRIIIB) SH gene frequencies in six racial groups. Blood. 1999;93:1115–6. [PubMed] [Google Scholar]
- 12.Flesch BK, Reil A, Bux J. Genetic variation of the HNA-3a encoding gene. Transfusion. 2011;51:2391–7. doi: 10.1111/j.1537-2995.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- 13.Bierling P, Bux J, Curtis B, et al. Recommendations of the ISBT Working Party on Granulocyte Immunobiology for leucocyte antibody screening in the investigation and prevention of antibody-mediated transfusion-related acute lung injury. Vox Sang. 2008 doi: 10.1111/j.1423-0410.2008.01144.x. [DOI] [PubMed] [Google Scholar]
- 14.Bayat B, Tjahjono Y, Werth S, et al. Implication of transfected cell lines for the detection of alloantibodies against human neutrophil antigen-3. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03303.x. Article first published online: 24 AUG 2011. [DOI] [PubMed] [Google Scholar]
- 15.Reil A, Wesche J, Greinacher A, Bux J. Geno- and phenotyping and immunogenicity of HNA-3. Transfusion. 2011;51:18–24. doi: 10.1111/j.1537-2995.2010.02751.x. [DOI] [PubMed] [Google Scholar]
- 16.de Haas M, Muniz-Diaz E, Alonso LG, et al. Neutrophil antigen 5b is carried by a protein, migrating from 70 to 95 kDa, and may be involved in neonatal alloimmune neutropenia. Transfusion. 2000;40:222–7. doi: 10.1046/j.1537-2995.2000.40020222.x. [DOI] [PubMed] [Google Scholar]
- 17.Kleinman S, Grossman B, Kopko P. A national survey of transfusion-related acute lung injury risk reduction policies for platelets and plasma in the United States. Transfusion. 2010;50:1312–21. doi: 10.1111/j.1537-2995.2010.02659.x. [DOI] [PubMed] [Google Scholar]
- 18.Kommareddi PK, Nair TS, Thang LV, et al. Isoforms, expression, glycosylation, and tissue distribution of CTL2/SLC44A2. Protein J. 2010;29:417–26. doi: 10.1007/s10930-010-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair TS, Kozma KE, Hoefling NL, et al. Identification and characterization of choline transporter-like protein 2, an inner ear glycoprotein of 68 and 72 kDa that is the target of antibody-induced hearing loss. J Neurosci. 2004;24:1772–9. doi: 10.1523/JNEUROSCI.5063-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Regan S, Traiffort E, Ruat M, et al. An electric lobe suppressor for a yeast choline transport mutation belongs to a new family of transporter-like proteins. Proc Natl Acad Sci U S A. 2000;97:1835–40. doi: 10.1073/pnas.030339697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis BR, Sullivan MJ, Holyst MT, et al. HNA-3a-specific antibodies recognize choline transporter-like protein-2 peptides containing arginine, but not glutamine at Position 154. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03145.x. Article first published online: 22 APR 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berthold T, Wesche J, Kuhnert K, et al. Epitope mapping of antibodies directed against the human neutrophil alloantigen 3a. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03115.x. Article first published online: 24 MAR 2011. [DOI] [PubMed] [Google Scholar]
- 23.Kanack AJ, Peterson JA, Sullivan MJ, et al. Full-length recombinant choline transporter like protein 2 containing arginine 154 reconstitutes the epitope recognized by HNA-3a antibodies. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03411.x. Article first published online : 27 OCT 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]


