Abstract
Pregnancy alters the rate and extent of drug metabolism, but little is known about the underlying molecular mechanism. We have found that 17β-estradiol (E2) upregulates expression of the major drug-metabolizing enzyme CYP2B6 in primary human hepatocytes. Results from promoter reporter assays in HepG2 cells revealed that E2 activates constitutive androstane receptor (CAR) and enhances promoter activity of CYP2B6, for which high concentrations of E2 reached during pregnancy were required. E2 triggered nuclear translocation of CAR in primary rat hepatocytes that were transiently transfected with human CAR as well as in primary human hepatocytes, further confirming transactivation of CAR by E2. E2-activated estrogen receptor (ER) also enhanced CYP2B6 promoter activity. The DNA-binding domain of ER was not required for the induction of CYP2B6 promoter activity by E2, suggesting involvement of a non-classical mechanism of ER action. Results from deletion and mutation assays as well as electrophorectic mobility shift and supershift assays revealed that two AP-1 binding sites (−1782/−1776 and −1664/−1658 of CYP2B6) are critical for ER-mediated activation of the CYP2B6 promoter by E2. Concurrent activation of both ER and CAR by E2 enhanced CYP2B6 expression in a synergistic manner. Our data demonstrate that at high concentrations reached during pregnancy, E2 activates both CAR and ER that synergistically induce CYP2B6 expression. These results illustrate pharmacological activity of E2 that would likely become prominent during pregnancy.
Keywords: Cytochrome P450 2B6, estradiol, nuclear receptors, pregnancy
1. Introduction
Over 50% of pregnant women take at least one medication, excluding perinatal vitamins and iron [1]. Dosage regimens used during pregnancy are typically determined based on pharmacokinetic and pharmacodynamic studies obtained in men and/or nonpregnant women. Data from limited clinical studies, however, suggest that pregnancy alters pharmacokinetic profiles of drugs, especially the rate of hepatic drug metabolism. For example, pregnancy is known to increase elimination of drugs metabolized by CYP2A6 and CYP3A4, while decreasing elimination of CYP1A2 substrates [2]. However, little is known about molecular mechanisms responsible for the altered drug metabolism during pregnancy.
One notable change during pregnancy is a dramatic increase (up to 100-fold) in plasma concentration of female hormones such as estrogen [3]. Multiple studies have examined the effects of estrogen on hepatic drug metabolism in rodent models (reviewed in [4]); however, due to significant divergence in hepatic drug-metabolizing enzymes between humans and rodents [5, 6] as well as interspecies differences in pregnancy-related physiology (e.g., <10-fold increases in plasma concentrations of estrogen in rodents [7] vs. ~100-fold increase in pregnant women), the results are difficult to be extrapolated to humans. In human cells, 17β-Estradiol (E2), a major estrogen elevated during pregnancy [3], is shown to induce the transcription of CYP2A6 and UGT1A4 (a conjugating drug-metabolizing enzyme) by activating estrogen receptor (ER)α [8, 9]. To date, the effects of E2 on other major drug-metabolizing enzymes and whether estrogen plays a role in altered drug metabolism during pregnancy remain to be determined.
The biological effects of E2 are typically mediated through two cognate nuclear receptors, ERα and ERβ, among which ERα is the major subtype expressed in the liver [10]. ER, being activated upon association with estrogen, may regulate expression of its target genes in different ways: it may bind to a cis-element, called the estrogen response element (ERE), of target genes, or it may associate with other transcriptional regulators and indirectly modulate expression of target genes (reviewed in [11]). In the latter case, the activation of estrogen target genes occurs in an ERE-independent manner, and transcriptional regulators belonging to the activator protein-1 (AP-1) family can play an important role [12]. The transcriptional regulators in the AP-1 family include c-Jun and c-Fos, which bind to an AP-1 motif in the upstream region of target genes and regulate gene expression.
Major transcription factors regulating expression of drug-metabolizing enzymes include pregnane X receptor (PXR) and constitutive androstane receptor (CAR). PXR and CAR are activated upon binding of the ligands rifampin and CITCO, respectively, and subsequently translocate into the nucleus where the activated PXR and CAR control expression of its target genes, such as CYP3A4 and CYP2B6 [13, 14]. CYP3A4 and CYP2B6 account for ~50% and ~15% of overall CYP-mediated metabolism of marketed drugs, respectively, playing a critical role in the metabolism of an increasing number of clinically important drugs [15]. While pregnancy is known to increase CYP3A4-mediated drug metabolism, whether pregnancy influences elimination of CYP2B6 substrates is unknown.
In this study, using primary human hepatocytes as a liver model, we have examined the effects of E2 on expression of hepatic genes, including genes for drug-metabolizing enzymes. Our results revealed that E2 significantly induces expression of CYP2B6. We further investigated the mechanism underlying the CYP2B6 induction by E2.
2. Materials and methods
2.1. Chemicals and reagents
S-Mephenytoin, S-nirvanol, phenytoin, 17β-estradiol, PK11195, phenobarbital, bupropion and rifampin were obtained from Sigma (St. Louis, MO). CITCO, i.e., 6-(4-Chlorophenyl) imidazo[2,1-b] [1,3]thiazole-5-carbaldehyde O-3,4-dichlorobenzyl) oxime, was purchased from Biomol (Plymouth Meeting, PA). ICI182,780 and hydroxybupropion were purchased from Tocris Bioscience (Ellisville, MO).
2.2. Cell culture
Freshly isolated human hepatocytes (HHs) from females (age ranging from 29 to 50; no known history of liver disease, alcohol abuse or hepatitis B or C infection) were obtained from CellzDirect (Pittsboro, NC) or Liver Tissue Cell Distribution System (Pittsburgh, PA). Upon receipt, media were replaced with serum-free Williams’ E media as previously described [16]. HepG2 cells from ATCC (Manassas, VA) were cultured as previously described [16]. HepG2 cells stably expressing ERα (HepG2-ER) were kindly provided by Dr. David Shapiro (University of Illinois at Urbana Champaign) and cultured in complete DMEM supplemented with 10% charcoal/dextran-stripped fetal bovine serum (Gemini, Woodland, CA), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1% MEM nonessential amino acids.
2.3. Plasmids
To construct the pGL3-CYP2B6 plasmid, the upstream region of CYP2B6 (−3290 to +8) was PCR-amplified using human genomic DNA as template and a pair of primers listed in supplemental Table 1. The PCR product was digested by KpnI and HindIII restriction enzymes and cloned into promoterless pGL3-basic vector (Promega, Madison, WI) digested by the same enzymes, yielding pGL3-CYP2B6. CYP2B6 deletion constructs with different lengths of the 5′-flanking region of the human CYP2B6 gene (−1724 to +8, −1461 to +8, and −845 to +8) for deletion assay were prepared by PCR-amplification of relevant regions (using pGL3-CYP2B6 as template) followed by restriction digestion using KpnI and HindIII and cloning into pGL3-basic plasmid. The PCR primers of the pGL3-CYP2B6 deletion constructs are listed in supplemental Table 1.
pGL3-ERE3 and expression vectors for ERα mutants [9], wild-type ER expression vector [17], PXR expression vector (pcDNA3-PXR) [17], and CAR expression vector (pcDNA3-CAR) [16] were previously described. pGL3-CYP2B6 [−1839/−12] and pEGFP-CAR (expressing enhanced green fluorescent protein-tagged human CAR) were generously provided by Dr. Masahiko Negishi (NIEHS). pTracerCMV2-CAR3 (expressing CAR3) was provided by Dr. Curtis J. Omiecinski (Pennsylvania State University).
Mutation constructs of pGL3-CYP2B6 (i.e., mutations at A1 and/or A2 sites or a mutation at ERE (see Fig. 4C for their locations)) were prepared using a QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer’s protocol. All sequences were confirmed by sequencing.
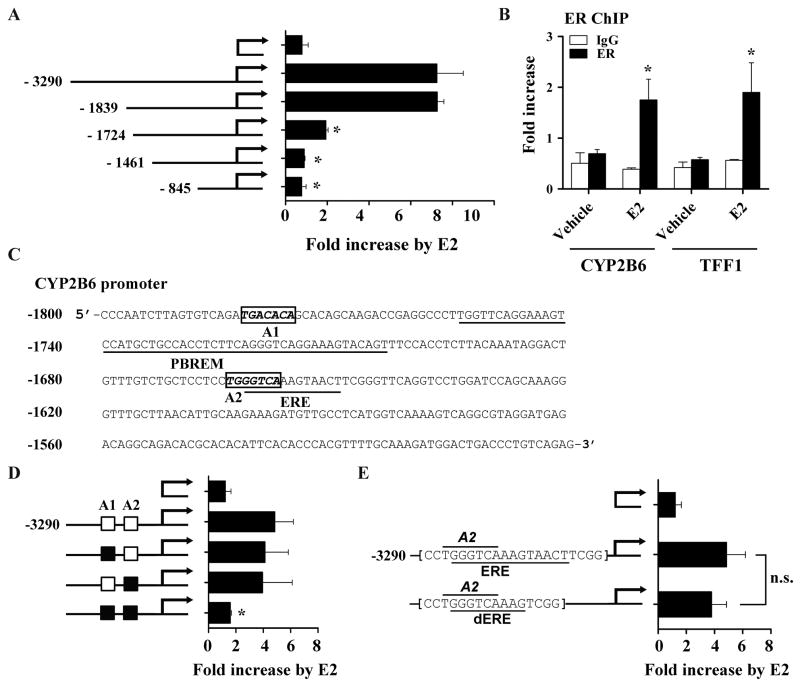
Fig. 4.
Putative AP-1 binding sites are potentially responsible for CYP2B6 induction by E2 in HepG2 cells. (A) HepG2 cells were co-transfected with a 5′-deletion construct of pGL3-CYP2B6 (or pGL3-basic), ER expression plasmid, and β-galactosidase expression vector. The transfected HepG2 cells were treated with ethanol or E2 (1 μM) for 24 hr, and luciferase assay was performed. (B) HepG2-ER cells were treated with E2 for 45 min. ChIP assays were carried out using rabbit IgG or an antibody against ER, and the amount of ER-bound DNA in CYP2B6 or TFF1 upstream regions was examined by qRT-PCR. For CYP2B6, a primer set that amplifies −1803/−1729 of the gene was used (supplemental Table 1). (C) Putative AP-1 binding sites (A1 and A2), PBREM [13], and ERE [32] within −1800/−1500 upstream region of CYP2B6 are shown. NCBI reference sequence NG_007929.1 was used. (D) HepG2 cells were co-transfected with pGL3-CYP2B6 harboring mutations in A1 and/or A2 (represented by a black box), ER expression plasmid, and β-galactosidase expression plasmid. The transfected cells were treated with vehicle (ethanol) or E2 (1 μM), and luciferase assay was performed. (E) HepG2 cells were transfected with pGL3-CYP2B6 harboring a deletion of part of a putative ERE (i.e., dERE) or wild type sequences. The transfected cells were treated with ethanol or E2 (1 μM), and luciferase assay was performed. Results shown are fold increases in CYP2B6 promoter activity by E2-treatment. n.s., statistically not significant.
2.4. DNA microarray experiment and analysis
After treatment of primary human hepatocytes with either ethanol or E2 (1 μM), RNAs were isolated using Trizol and then DNase I-treated. cDNA synthesis, modification, hybridization, and labeling on Affymetrix Human Genome U133 Plus 2.0 oligonucleotide arrays were performed using kits from Affymetrix (Santa Clara, CA) as described in the manufacturer’s instructions. Six microarray experiments were conducted on three hepatocytes isolated from different donors. The image data were analyzed using Affymetrix Gene Chip Operating System Software, and Partek Genomic Solution Software 6.4 was used for statistical analysis. The relative change in expression (ratio > 1.5) was used to obtain a list of differentially expressed genes between ethanol- and E2-treated hepatocytes.
2.5. RNA isolation and quantitative real time-PCR (qRT-PCR)
Total RNAs were isolated from cell lysates using Trizol (Invitrogen, Carlsbad, CA). qRT-PCR was performed using StepOnePlus Real-Time PCR System and TaqMan® Gene expression assays (Applied Biosystems), and fold changes in mRNA levels of genes were determined after normalizing the gene expression levels by those of GAPDH or PPIA (2−Δ ΔCt method) as previously done [18].
2.6. Determination of CYP2B6 activity
S-Mephenytoin (100 μM) or bupropion (10 μM) was individually added to the primary human hepatocytes culture after 72 hours of hormone treatment, and media were sampled at various time points for up to 6 hours. Concentrations of metabolites of each drug were analyzed by LC/MS/MS as described previously [18].
2.7. Luciferase reporter assay
Luciferase reporter assays were performed in HepG2 cells as previously described [9, 16]. At least two independent experiments were performed in triplicate.
2.8. CAR nuclear translocation assay in rat hepatocytes
Primary rat hepatocytes were isolated and cultured in 6-well plates as previously described [19]. Rat hepatocytes were transfected with 2 μg of pEGPF-CAR expression plasmid using Lipofectamine 2000 (Invitrogen). After 24 hr incubation, cells were treated with E2 (1 μM), CITCO (100 nM), or DMSO (vehicle). After 4 hr incubation with drugs, cells were fixed with 4% paraformaldehye and nuclei were stained with Hoechst 33258 dye. The intracellular localization of EGFP-CAR fusion protein was examined using confocal microscopy.
2.9. Quantitation of nuclear CAR by LC/MS/MS
The nuclear proteins from drug-treated primary human hepatocytes were resolved on a SDS-10% polyacrylamide gel, and the gel segments for ~46 kD (the expected size for CAR) was sliced. The gel slices were digested using 12.5 ng/μl of trypsin (Promega, WI). Each sample was spiked with synthetic AngII peptide (DRVYIHPF; Sigma-Aldrich) as an internal standard, and separated by HPLC (Eksigent-Nano-HPLC system; Eksigent Technologies, CA) using a silica microcapillary column (12 cm x 75 μm) packed with C18 resin (5 mm, 300 A, Alltech, KY). The following mobile phase was used at a flow rate of 300 nl/min: a linear gradient of buffer B (3% to 40% over 35 min) where buffer A is 0.1% formic acid in H2O and buffer B 0.1% formic acid in acetonitrile. The samples were analyzed by LTQ XL ion-trap mass spectrometer (Thermo Finnigan, West Palm Beach, FL). The MS signal intensities from CAR peptides were normalized by peak area corresponding to from AngII peptide for comparison of CAR quantity among hepatocytes treated with different drugs.
Peptides corresponding to CAR protein (NP_001070948) were identified using TurboSEQUEST (Thermo Finnigan) and SEQUEST algorithm [20, 21], which was further validated using Scaffold software (Proteome Software, Portland, OR; version 3_00_02) [22]. Probabilities that the identified peptide and protein are correctly from CAR were >95% and >99%, respectively.
2.10. Electrophoretic mobility shift assay (EMSA)
Nuclear proteins were prepared from HepG2 or HepG2-ER cells using the CelLytic Nuclear Extraction Kit (Sigma-Aldrich) following manufacturer’s protocol. EMSA was performed using Gel Shift Assay Systems (Promega) following manufacturer’s protocol. Briefly, nuclear proteins (50 μg) were preincubated with the provided reaction buffer at room temperature in the presence or absence of nonradioactive DNA probe (for competition) or antibody (for supershift experiment) as indicated. DNA probe sequences are listed in supplemental Table 1. For supershift experiment, the following antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were used: ER (sc-543x), c-Jun (sc-44x), and c-Fos (sc-52x). After 10 min, the binding reaction was initiated by adding 1.75 pmol of 5′-end 32P-labeled CYP2B6 probes harboring putative AP-1 binding sequences. The reaction mixture was incubated at room temperature for 20 min. Protein-bound probes were separated from free probes on 4% (w/v) nondenaturing polyacrylamide gel. The gel was dried, and radioactivity visualized by using PhosphorImager.
2.11. Chromatin immunoprecipitation (ChIP) assay
HepG2 or HepG2-ER cells were seeded in 10-cm dishes and grown in phenol-red free media containing 5% charcoal-dextran stripped FBS for 72 hr. On reaching 90% confluency, cells were treated with vehicle or E2 (1 μM) for 45 min. Crosslinking was carried out with 1.5% formaldehyde for 15 min at room temperature. Cells were lysed with SDS Lysis Buffer (1% SDS, 10 mM EDTA pH 8.0, and 50 mM Tris-HCl pH 8.1) and chromatin was sonicated for 4 min twice using S-4000 sonicator (Misonix, Qsonica, CT). Protein A-coated magnetic beads washed three times with 5% BSA in PBS were incubated overnight at 4°C with 1 μg of ER antibody per pulldown (sc-543x; Santa Cruz Biotechnology) or normal rabbit IgG. Antibody-bound beads were washed two times with 5% BSA in PBS and incubated overnight at 4°C with sonicated chromatin diluted in Dilution Buffer (1% TritonX-100, 2 mM EDTA, 20 mM Tris-HCl, and 150 mM NaCl). Beads were washed three times using RIPA-LiCl Wash Buffer (50 mM Hepes, 0.5 M LiCl, 1% NP40, 0.7% deoxycholic acid, and 1 mM EDTA), and two times with Tris-EDTA (10 mM Tris-HCl and 1 mM EDTA). DNA was eluted from beads in elution buffer containing 0.1 M sodium bicarbonate and 1% SDS. Decrosslinking was carried out for 16 hr at 65°C. DNA was purified using Qiaquick columns (Qiagen, Valencia, CA) and eluted in pre-warmed water. To generate a standard curve for input DNA amount, ~5% of cell lysate was saved prior to immunoprecipitation. The cell lysate was diluted (1:20, 1:200, and 1:2000) and used to calculate the percent of input in each pull-down using qRT-PCR. The % maximum occupancy for each assay was then calculated relative to the treatment with the highest fold change, which was set to 100%. Primers for ChIP qRT-PCR are listed in supplemental Table 1.
2.12. Statistical analyses
Data are expressed as means ± standard deviation from triplicate experiments. Statistical analyses were performed by using Student’s t-test using the Prism analysis program (Graphpad Inc., San Diego, CA, USA). The level of significance used for hypothesis testing was 0.05.
3. Results
3.1. E2 upregulates expression of CYP2B6 in primary human hepatocytes
To determine the effects of E2 on expression of hepatic genes, we treated 3 different batches of primary human hepatocytes with E2 (or vehicle) and compiled a list of differentially regulated genes by using microarray. E2 added to the culture media was rapidly metabolized by the primary hepatocytes (first order elimination half-life 37 min; data not shown). Accordingly, culture media containing 1 μM E2 were replenished regularly (every 6 hr during daytime and 12 hr for overnight incubation) to maintain average E2 concentrations during the experimental period of 72 hr to ~100 nM. This average concentration, although ~10-fold lower than the initial E2 concentration at the time of media change (1 μM), approximates the plasma concentration at term pregnancy [3]. Throughout the study, the E2 concentration of 1 μM was used to reflect the reported plasma concentration at term, i.e., 100 nM.
Through the microarray experiment, 34 genes (Table 1) were identified as differentially regulated by E2. This list included drug-metabolizing enzymes, such as CYP2A6 and CYP2B6, in part confirming the previous finding that CYP2A6 expression is induced by E2 [8]. Follow-up studies were focused on CYP2B6 considering its significant contribution to overall drug metabolism.
Table 1.
List of genes differentially regulated by E2 in human hepatocytes
| No. | Probe set | Gene Title | Gene Symbol | Fold Change (E2/Control) |
|---|---|---|---|---|
| 1 | 205862_at | GREB1 protein | GREB1 | 20.10 |
| 2 | 209987_s_at, 209988_s_at, 213768_s_at | achaete-scute complex homolog 1 (Drosophila) | ASCL1 | 9.54 |
| 3 | 211295_x_at, 1494_f_at, 214320_x_at, 207244_x_at | cytochrome P450, family 2, subfamily A, polypeptide 6 | CYP2A6 | 7.20 |
| 4 | 207718_x_at | cytochrome P450, family 2, subfamily A, polypeptide 7 | CYP2A7 | 6.85 |
| 5 | 210272_at | cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1 | CYP2B7P1 | 6.33 |
| 6 | 206755_at | cytochrome P450, family 2, subfamily B, polypeptide 6 | CYP2B6 | 4.98 |
| 7 | 206325_at | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 | SERPINA6 | 4.78 |
| 8 | 225987_at | STEAP family member 4 | STEAP4 | 2.50 |
| 9 | 210521_s_at | fetuin B | FETUB | 2.46 |
| 10 | 206292_s_at, 206293_at | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 | SULT2A1 | 2.42 |
| 11 | 218245_at | tsukushin | TSKU | 2.16 |
| 12 | 208383_s_at | phosphoenolpyruvate carboxykinase 1 (soluble) | PCK1 | 2.15 |
| 13 | 229331_at | spermatogenesis associated 18 homolog (rat) | SPATA18 | 2.04 |
| 14 | 239594_at | hypothetical LOC145837 | LOC145837 | 1.96 |
| 15 | 222102_at | glutathione S-transferase alpha 3 | GSTA3 | 1.92 |
| 16 | 215506_s_at | DIRAS family, GTP-binding RAS-like 3 | DIRAS3 | 1.77 |
| 17 | 239650_at | Nck-associated protein 5 | NAP5 | 1.70 |
| 18 | 228489_at | transmembrane 4 L six family member 18 | TM4SF18 | 1.69 |
| 19 | 203362_s_at | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | 1.67 |
| 20 | 208327_at | cytochrome P450, family 2, subfamily A, polypeptide 13 | CYP2A13 | 1.66 |
| 21 | 227198_at | AF4/FMR2 family, member 3 | AFF3 | 1.65 |
| 22 | 225511_at | G protein-coupled receptor, family C, group 5, member B | GPRC5B | 1.64 |
| 23 | 230009_at | family with sequence similarity 118, member B | FAM118B | 1.63 |
| 24 | 213293_s_at | tripartite motif-containing 22 | TRIM22 | 1.62 |
| 25 | 203196_at | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | ABCC4 | 1.62 |
| 26 | 226028_at | roundabout homolog 4, magic roundabout (Drosophila) | ROBO4 | 1.58 |
| 27 | 202731_at, 202730_s_at, 212593_s_at | programmed cell death 4 (neoplastic transformation inhibitor) | PDCD4 | 1.56 |
| 28 | 223652_at | arsenic (+3 oxidation state) methyltransferase | AS3MT | 1.56 |
| 29 | 216687_x_at | UDP glucuronosyltransferase 2 family, polypeptide B15 | UGT2B15 | 1.56 |
| 30 | 202878_s_at | cytohesin 1 | CYTH1 | 1.56 |
| 31 | 218096_at | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) | AGPAT5 | 1.52 |
| 32 | 243998_at | schlafen family member 5 | SLFN5 | 1.51 |
| 33 | 204521_at | chromosome 12 open reading frame 24 | C12orf24 | 1.51 |
| 34 | 227425_at, 242571_at | RALBP1 associated Eps domain containing 2 | REPS2 | 1.50 |
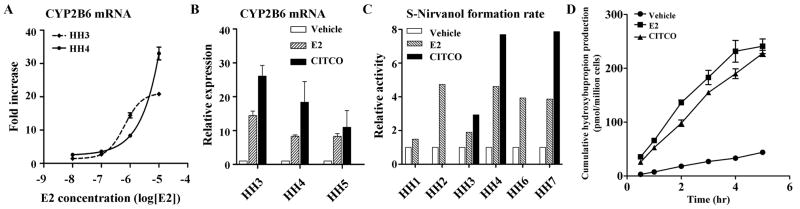
Further verification of the microarray result revealed that E2 promotes expression of CYP2B6 in a concentration dependent manner in primary human hepatocytes (Fig. 1A). The effects of E2 on mRNA levels of CYP2B6 were of similar magnitude as the induction by CITCO, a well-known CYP2B6 inducer (Fig. 1B). E2 treatment also increased CYP2B6 enzyme activities against a typical CYP2B6 probe drug, S-mephenytoin (Fig. 1C). Metabolism of bupropion, another CYP2B6 substrate drug commonly prescribed during pregnancy, was also enhanced in E2-treated hepatocytes (Fig. 1D). Taken together, our results showed that E2, at the high concentration reached during pregnancy, enhances CYP2B6 expression and leads to corresponding increases in enzyme activity.
Fig. 1.
E2 enhances CYP2B6 expression. (A) Primary human hepatocytes from 2 donors (HH3 and HH4) were treated with increasing concentrations of E2 for 72 hr. mRNA expression level of CYP2B6 was determined by qRT-PCR. (B) Primary human hepatocytes from 3 donors (HH3, HH4, and HH5) were treated with ethanol, E2 (1 μM), or CITCO (100 nM) for 72 hr and mRNA expression level of CYP2B6 was determined. (C and D) Primary human hepatocytes were treated with ethanol, E2 (1 μM), or CITCO (100 nM) for 72 hr. CYP activity was then determined by adding a CYP2B6-specific probe drug [100 μM S-mephenytoin (C) or 10 μM bupropion (D)] to the media and measuring concentrations of S-nirvanol (C) or hydroxybupropion (D) in the media. Data shown are relative S-nirvanol formation rate in drug-treated hepatocytes as compared to vehicle-treated cells (C) and cumulative hydroxybupropion production in HH8 (D). CYP2B6 activities were not determined in CITCO-treated hepatocytes #1, 2, and 6.
3.2. E2 enhances CYP2B6 promoter activity via CAR transactivation
To examine involvement of PXR and/or CAR in CYP2B6 induction by E2, promoter reporter assays were performed in HepG2 cells. To this end, pGL3-CYP2B6 [−1839/−12] was used, which is a luciferase reporter plasmid harboring the luciferase gene driven by the phenobarbital responsive element module (PBREM) to which CAR or PXR binds to activate the transcription of a downstream gene [23]. HepG2 cells were cotransfected with pGL3-CYP2B6 [−1839/−12] and PXR (or CAR expression plasmid). The transfected cells were treated with vehicle, E2, rifampin (a PXR activator) or CITCO (a CAR activator); then luciferase activities were measured. In PXR-transfected cells, E2 (at concentrations up to 1 μM) did not induce the promoter activity (data not shown). On the other hand, in CAR-transfected cells, E2 at 1 μM enhanced PBREM-driven promoter activity by 2.1-fold as compared to vehicle treatment, as CITCO did (Fig. 2A). E2 at lower concentrations (less than 1 μM), however, did not enhance the promoter activity (data not shown).
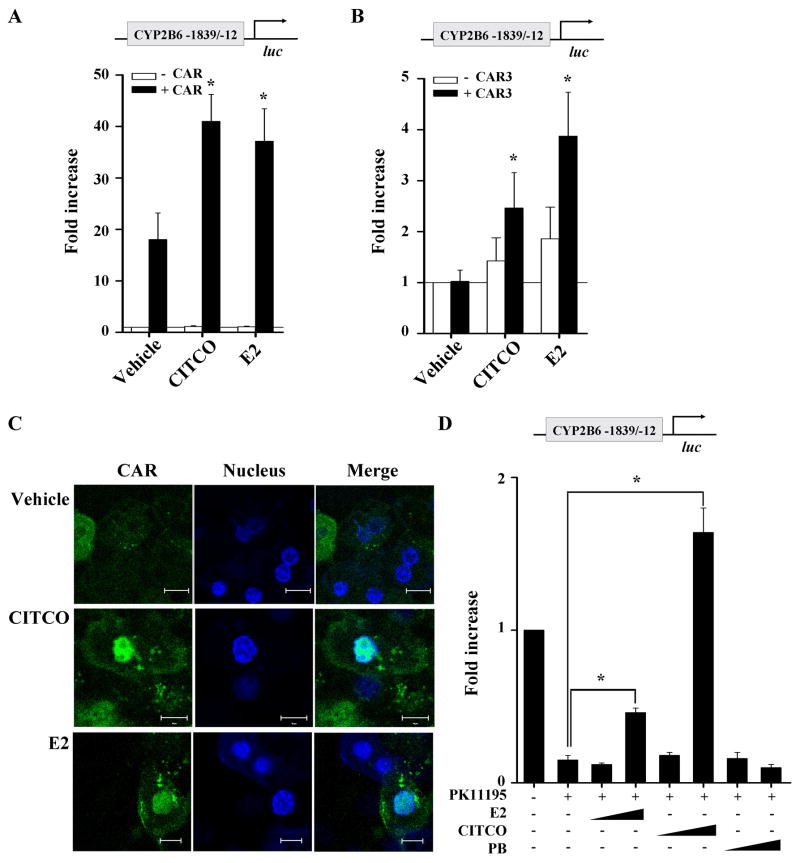
Fig. 2.
E2 activates CAR. (A and B) HepG2 cells were transfected with pGL3-CYP2B6 [−1839/−12], CAR expression plasmid (or pcDNA3) (A) or CAR3 expression plasmid (or pTracerCMV2) (B), and β-galactosidase expression vector. The transfected HepG2 cells were treated with ethanol, E2 (1 μM), or CITCO (100 nM) for 24 hr, and luciferase assay was performed. (C) Primary rat hepatocytes were transfected with pEGFP-CAR plasmid, and treated with DMSO, E2 (1 μM), or CITCO (100 nM). After 4 hr incubation with drugs, cells were fixed and nuclei were stained. The cells were visualized using confocal microscopy. Scale bar represents 10 μm. (D) HepG2 cells were transfected with pGL3-CYP2B6 [−1839/−12], CAR expression plasmid, and β-galactosidase expression plasmid. The transfected HepG2 cells were treated with vehicle (DMSO), various concentrations of E2 (1 or 10 μM), or CITCO (0.1 or 1 μM), or phenobarbital (0.1 or 1 mM) in the presence or absence of PK11195 (10 μM) for 24 hr, and luciferase assay was performed. Results represent fold changes in luciferase activity by drug treatment relative to vehicle treatment.
CAR transfection itself dramatically increased CYP2B6 promoter activity in HepG2 cells (Fig. 2A), consistent with results from a previous study where ectopically expressed CAR exhibits constitutive activation in transformed cells [24]. To rule out the effect of basal CAR activation on the E2 action, we repeated the promoter reporter assay with the CAR expression vector substituted by a plasmid expressing CAR3. CAR3 contains an extra 5 amino acid residues inserted within the ligand-binding domain, which abolishes the constitutive activation of CAR in HepG2 [24]. In CAR3-transfected cells, E2 significantly increased CYP2B6 promoter activity to a level similar to that by CITCO (Fig. 2B).
In primary rodent or human hepatocytes, CAR translocates into the nucleus from the cytosol only when activated by its cognate ligand [25, 26]. To determine whether E2 triggers the nuclear translocation of CAR, primary rat hepatocytes were transfected with EGFP-tagged human CAR or the control vector; then the transfected cells were treated with vehicle, E2, or CITCO. The nuclei of the drug-treated cells were stained with Hoechst dye (blue) and visualized using confocal microscopy. As expected, in vehicle-treated hepatocytes, CAR (green signal) remained in the cytosol; and in CITCO-treated hepatocytes, CAR translocated into the nucleus (i.e., co-localization of blue and green signals) (Fig. 2C). In the E2-treated hepatocytes, co-localization of CAR and nuclear signals was observed, indicating that E2 triggers the nuclear translocation of CAR. Considering potential interspecies differences in actions of gene regulatory factors between rat and human hepatocytes, the results were further verified in a batch of primary human hepatocytes. Primary human hepatocytes were treated with vehicle, E2, or CITCO, and the amount of CAR protein in the nuclear fractions was quantified using LC/MS/MS (due to a lack of antibodies that can specifically detect human CAR). Signals of nuclear CAR, normalized to the signal from spiked internal control, were 1.29 and 0.47 in the CITCO- or E2-treated hepatocytes, respectively (data not shown). In the vehicle-treated hepatocytes, CAR was not detected.
CAR can be activated either directly by binding to its ligand (e.g., CITCO) or indirectly by altering cell signaling (e.g., phenobarbital) [19, 27]. To differentiate these two possibilities, we tested whether E2 can reverse the PK11195-mediated deactivation of CAR. PK11195 is a CAR-specific antagonist that represses the constitutive activity of CAR in immortalized cells such as HepG2, and the antagonistic activity of PK11195 can be abrogated only by direct CAR activators [28]. In CAR-transfected HepG2 cells, PK11195 repressed PBREM-driven CYP2B6 promoter activity by 85% (Fig. 2D), and phenobarbital did not reverse PK11195-mediated deactivation. On the other hand, CITCO (1 μM) or E2 (10 μM) reversed the repression on promoter activity (Fig. 2D). The concentration of CITCO that effectively reverses the antagonistic activity of PK11195 on the PBREM-driven promoter was higher than the previously reported EC50 values for CAR activation (i.e., 25 nM) [19], indicating a right-shift in concentration-response curve for CITCO-mediated CAR activation. Taken together, these results suggest that E2 is likely a direct activator of CAR.
3.3. E2 enhances transcription of CYP2B6 via ER/AP-1 Pathway
While E2 exerts many of its biological effects via activation of ER, its effect can be also mediated in an ER-independent manner [29]. To determine whether ER plays an important role in CYP2B6 induction by E2, we examined whether ER expression is obligatory for the E2 action. To this end, HepG2 cells stably expressing ER (HepG2-ER) and primary human hepatocytes were treated with E2 (or vehicle) in the absence or presence of the ER-degrading antiestrogen ICI182,780 [30]; then CYP2B6 mRNA expression levels were determined. In HepG2-ER cells, E2 treatment enhanced CYP2B6 expression by 4-fold as compared to that of the control, and this induction was completely abrogated by ICI182,780 treatment (Fig. 3A). A similar pattern was observed for a known ER-responsive gene TFF1 (Fig. 3A). The E2 treatment in parental (ER-negative) HepG2 cells did not enhance expression of CYP2B6 nor TFF1 (data not shown). Also, in primary human hepatocytes, similar results as in HepG2-ER were obtained for CYP2B6 and TFF3 (an ER-responsive gene used in replacement of TFF1 due to minimal expression of TFF1 in primary human hepatocytes) (Fig. 3B). Taken together, these results demonstrate that the CYP2B6 induction by E2 requires ER.
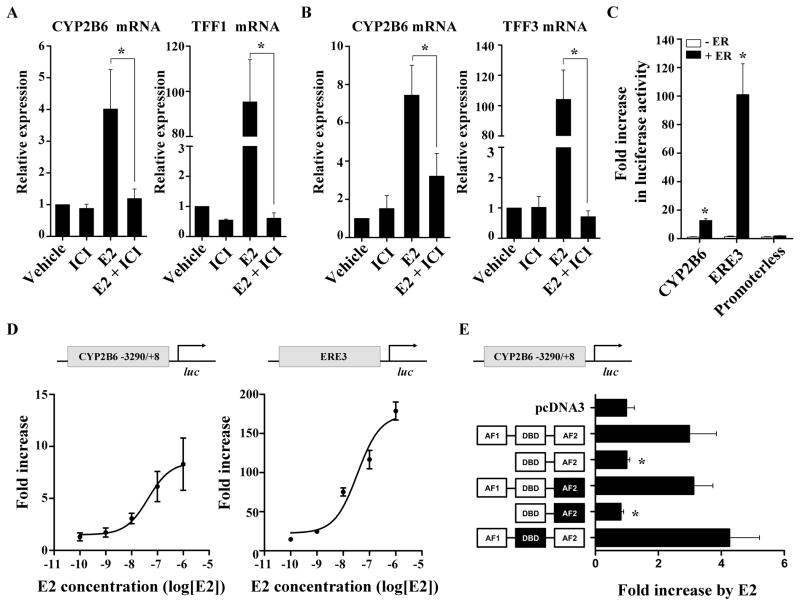
Fig. 3.
E2 enhances CYP2B6 expression via ER. (A and B) HepG2-ER cells (A) or primary human hepatocytes (B) were treated with ethanol or E2 (1 μM), in the presence or absence of ICI182,780 (10 μM), for 48 hr, and mRNA expression was determined. (C) HepG2 cells were co-transfected with a luciferase vector [pGL3-CYP2B6, pGL3-ERE3, or pGL3-basic], ER expression plasmid (or pcDNA3), and β-galactosidase expression vector. The transfected HepG2 cells were treated with ethanol or E2 (1 μM) for 24 hr, and luciferase assay was performed. (D) HepG2 cells were co-transfected with a luciferase vector [pGL3-CYP2B6 or pGL3-ERE3], ER expression plasmid, and β-galactosidase expression vector. The transfected cells were treated with E2 ranging from 100 pM to 1 μM for 24 hr. (E) HepG2 cells were co-transfected with pGL3-CYP2B6 and one expression vector for an ER mutant (see text for details) along with β-galactosidase expression plasmid. The ER domain whose function is inactivated by point mutations is shown in black. The transfected cells were treated with ethanol or E2 (1 μM), and luciferase assay was performed. Results shown are fold increases in CYP2B6 promoter activity in E2-treated cells relative to the vehicle-treated control.
To determine the effects of E2 on transcriptional activity of CYP2B6, luciferase reporter assays were performed. A reporter plasmid pGL3-CYP2B6 was constructed that carries the luciferase gene whose expression is driven by the upstream regulatory region of CYP2B6 (from −3290 to +8). HepG2 cells were cotransfected with pGL3-CYP2B6 and ER expression vector (or pcDNA3). The transfected cells were treated with E2 or vehicle, after which the luciferase activity was measured. In ER-transfected cells, E2 treatment enhanced CYP2B6 promoter activity by 12.5-fold as compared to vehicle treatment (Fig. 3C), whereas E2 had no effect in pcDNA3-transfected cells. A similar finding was observed for pGL3-ERE3, where luciferase expression is driven by 3 copies of vitellogenin ERE (Fig. 3C). Further, the induction of CYP2B6 promoter activity by E2 displayed a concentration-dependency (Fig. 3D); the estimated EC50 was 44 nM, which is similar to the EC50 value (35 nM) estimated for pGL3-ERE3.
Regulation of ER target genes by tethering of ER to other transcription factors (such as AP-1) does not require the DNA-binding domain of ER. To determine whether the direct or indirect binding of ER (to target gene promoter) is involved in CYP2B6 regulation, we ectopically expressed a mutant ER and examined induction of CYP2B6 promoter activity by E2. The mutant ERs carry a deletion of activation function (AF)1, point mutations in the DNA-binding domain (DBD), or point mutations in AF2. The point mutations result in loss of functionality in the relevant domains [31]. The CYP2B6 induction by E2 was not observed in HepG2 cells expressing the mutant ER with deletion of the AF1 domain (Fig. 3E), whereas induction was retained in HepG2 cells expressing the mutant ER with a nonfunctional DBD. This result suggests that E2-activated ER mediates the CYP2B6 induction in an indirect manner, not involving direct ER binding to DNA.
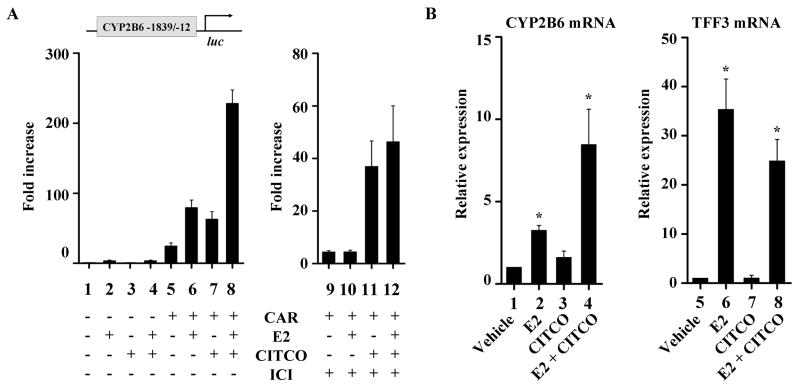
To map potential cis-elements responsible for CYP2B6 induction by E2, a series of luciferase vectors each containing a different segment of the ~3.3kb upstream region of CYP2B6 (−3290/+8, −1839/−12, −1724/+8, −1461/+8, and −845/+8) were constructed. Each luciferase vector was transfected into HepG2 cells along with an ER expression vector and treated with E2 (or vehicle), after which luciferase activities were measured. E2-responsiveness was maintained when the far upstream region of CYP2B6 (from −3290 to −1840) was deleted (Fig. 4A). Deletion of the region from −1839 to −1725 led to a significant decrease in the E2-responsiveness, and further deletion of the region from −1724 to −1462 completely abolished the E2 responsiveness. This result suggests that the upstream region from −1839 to −1462 is critical for CYP2B6 induction by E2 in HepG2 cells and contains potential cis-elements mediating E2 activity. Recruitment of ER on the mapped upstream region of CYP2B6 was verified by chromatin immunoprecipitation assay (Fig. 4B).
In silico analysis of the upstream region (−1839/−1462) of CYP2B6 using MatInspector (Genomatix, Munich, Germany) revealed two putative binding sites for AP-1 protein, which is known to associate with ER for regulation of target gene expression (Fig. 4C). To determine the role of the identified AP-1 motifs (called CYP2B6/A1 for distal site and CYP2B6/A2 for proximal site), we mutated A1 or A2 (individually or both) and examined their effect on CYP2B6 induction by E2 using the luciferase reporter assays. Mutations in individual AP-1 binding sites had no effects on the induction in CYP2B6 transcriptional activity by E2 (Fig. 4D); however, the mutation of both AP-1 binding sites resulted in complete loss of the E2-responsiveness of the CYP2B6 promoter. During our study, Lo et al. reported that E2-activated ER enhances CYP2B6 promoter activity via an ERE located at −1662/−1650 of CYP2B6 in Huh7 hepatoma cells [32]. However, deletion of part (−1654/−1650) of the previously reported ERE (i.e., rendering it a half-ERE and 50- to 100-fold decreased binding to ER [33]) did not affect the E2-responsiveness of CYP2B6 (Fig. 4E). These results suggest that two AP-1 binding sites identified are critical for the CYP2B6 induction by E2 in HepG2 cells.
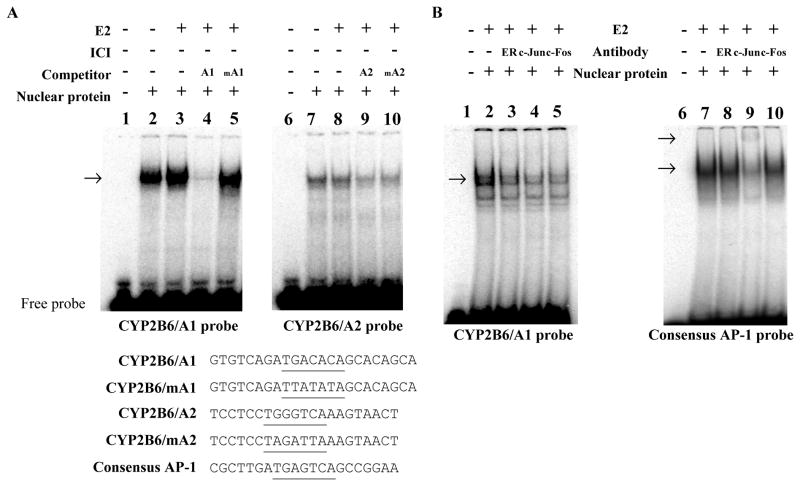
To further examine the role of the identified AP-1 binding sites, we performed electrophoretic mobility shift assays. Nuclear extracts, each prepared from HepG2-ER cells treated with vehicle or E2, were incubated with the radio-labeled probe encompassing the distal CYP2B6/A1 or proximal CYP2B6/A2, respectively, in the presence or absence of indicated unlabeled competitors. Then, the binding reaction samples were resolved on non-denaturing gels and visualized by using PhosphorImager. Binding complexes between the nuclear extract from E2-treated HepG2-ER and the CYP2B6 probes were observed (Fig. 5A, lanes 2 and 7). The binding signal intensity for CYP2B6/A1 was markedly reduced when unlabeled CYP2B6/A1 probe was added (Fig. 5A, lane 4), but not by CYP2B6/mA1 probe harboring a mutated A1 sequence (Fig. 5A, lane 5). On the other hand, the binding of nuclear proteins to CYP2B6/A2 probe was inhibited by unlabeled competitors of either CYP2B6/A2 or CYP2B6/mA2 (Fig. 5A, lanes 9 and 10), suggesting nonspecific or very weak binding of nuclear proteins to CYP2B6/A2 in vitro. These results suggest that CYP2B6/A1 is the primary cis-element mediating CYP2B6 induction by E2.
Fig. 5.
ER and AP-1 proteins form a complex at A1 site. (A) Nuclear proteins from HepG2-ER cells treated with ethanol or E2 (1 μM) were prepared and incubated with 32P-labeled CYP2B6 probe containing CYP2B6/A1 (left) or CYP2B6/A2 (right panel). For indicated samples, a 33-fold molar excess of unlabeled DNA probe was added to the mixture as competitors. The DNA and DNA/protein complex were resolved on non-denaturing gel. The arrow indicates location of shifted bands by apparent AP-1 binding to DNA. DNA sequences for probes are listed below the gel images, and the putative AP-1 binding sites are underlined. (B) Nuclear proteins from E2-treated HepG2-ER cells were incubated with a 32P-labeled CYP2B6/A1 or consensus AP-1 probe in the presence or absence of various antibodies, and resolved on non-denaturing gel. The arrow indicates location of shifted or super-shifted bands by apparent AP-1 binding to DNA.
The nuclear proteins from E2-treated HepG2-ER cells showed stronger binding to CYP2B6/A1 (Fig. 5A, lane 3) as compared to that from vehicle-treated cells (Fig. 5A, lane 2), suggesting that E2 enhanced the binding of nuclear proteins to CYP2B6/A1. Nuclear proteins from ER-negative parental HepG2 cells showed weak binding to CYP2B6/A1, and this was not enhanced by E2 treatment (data not shown). To further determine involvement of ER and AP-1 complex in binding to CYP2B6/A1, we performed an electrophoretic mobility supershift assay. Nuclear proteins prepared from HepG2-ER cells treated with E2 were incubated with radio-labeled CYP2B6/A1 probes in the presence or absence of antibodies against ER or components of AP-1 complex (i.e., c-Jun and c-Fos). Antibodies against ER, c-Jun, or c-Fos significantly decreased the binding signal between nuclear proteins and CYP2B6/A1 (Fig. 5B, lanes 3–5), suggesting inhibition of AP-1 binding to CYP2B6/A1 by the antibodies. Supershifted bands were not observed likely due to the big size of the ternary complex of ER and AP-1 proteins with the DNA probe, consistent with results previously reported [34]. Of note, antibody against ER did not decrease the binding for consensus AP-1 probe (Fig. 5B, lane 8), indicating that ER association in AP-1 complex is specific for the CYP2B6/A1 site. Antibody against c-Jun further shifted the retarded band for consensus AP-1 probe (Fig. 5B, lane 9) whereas antibody against c-Fos slightly decreased the signal of retarded band (Fig. 5B, lane 10). This indicates that proteins bound to the consensus AP-1 in HepG2-ER cells are composed of c-Jun that may form homodimers or heterodimers with AP-1 proteins other than c-Fos [35]. Together, these data suggest that AP-1 proteins bind to CYP2B6/A1 and that upregulation of CYP2B6 by E2 is mediated by activation of an ER/AP-1 pathway in HepG2 cells.
3.4. Transactivation of ER and CAR synergistically induces CYP2B6 expression
To examine potential crosstalk between ER and CAR, we compared the CYP2B6 promoter activity under conditions where ER and CAR are activated individually or concurrently. As an activator of ER, E2 was used at 100 nM, i.e., a concentration that activates ER but not CAR while CITCO was used as an activator of CAR. HepG2-ER cells were co-transfected with the pGL3-CYP2B6 [−1839/−12] and CAR expression vector (or the empty vector pcDNA3). The transfected cells were treated with E2 and/or CITCO. In the CAR-transfected cells treated with either E2 or CITCO, CYP2B6 promoter activity increased by 3.2- and 2.5-fold, respectively, as compared to that in the vehicle-treated CAR-transfected cells (Fig. 6A, lane 5 vs. 6 and lane 5 vs. 7, respectively). Interestingly, co-treatment of CAR-transfected cells with E2 and CITCO increased the promoter activity by 9.1-fold (Fig. 6A, lane 5 vs. 8), which was greater than the expected additive effects of individual drug treatments. The apparent synergy was abolished in the absence of ER expression by treatment with ICI182,780 (Fig. 6A, right).
Fig. 6.
ER and CAR transactivation synergistically enhances CYP2B6 expression. (A) HepG2-ER cells were co-transfected with pGL3-CYP2B6 [−1839/−12], CAR expression vector (or pcDNA3), and β-galactosidase expression vector. The cells were treated overnight with ethanol, E2 (100 nM), CITCO (100 nM), or both E2 and CITCO, and then luciferase assay was performed. Results shown are fold increases in CYP2B6 promoter activity in drug-treated cells relative to the vehicle-treated control. (B) Primary human hepatocytes were treated with ethanol, E2 (100 nM), CITCO (100 nM), or both E2 and CITCO for 72 hr, and mRNA levels were determined.
To further confirm the synergistic action of ER and CAR on CYP2B6 expression, we examined the effects of E2 and CITCO on CYP2B6 expression in primary hepatocytes. Similar to the findings in HepG2-ER cells, co-treatment of E2 and CITCO enhanced CYP2B6 expression (8.5-fold), the extent of which was greater than the expected additive induction by individual drug treatments (3.3- and 1.6-fold by E2 and CITCO, respectively) (Fig. 6B). On the other hand, co-treatment with E2 and CITCO slightly decreased the E2-mediated induction in expression TFF3 (an ER-responsive gene not regulated by CAR) (Fig. 6B), presumably due to sequestration of coactivators by CAR as previously reported [36]. Together, these results suggest that activated ER and CAR synergistically enhance CYP2B6 promoter activity.
4. Discussion
In this study, using primary human hepatocytes, we have examined the effects of the major estrogen E2 on hepatic gene expression and showed that E2 enhances CYP2B6 expression, subsequently leading to increased enzyme activity. CYP2B6 accounts for 15% of overall CYP-mediated metabolism of marketed drugs, and plays a critical role in metabolism of an increasing number of clinically important drugs including anti-depressants (e.g., bupropion) and anesthetics [13]. Our results in primary human hepatocytes show that E2 upregulates expression of CYP2B6 at concentration reached during pregnancy, suggesting potential increases in CYP2B6-mediated drug metabolism in pregnant women. Estrone, another endogenous estrogen whose concentration rises (~100-fold) during pregnancy [3], also induces CYP2B6 expression in primary hepatocytes to a similar extent to that by E2 (data not shown). Whether pregnancy indeed increases elimination of CYP2B6 substrates (such as bupropion) awaits clinical confirmation.
Our results from luciferase assays and nuclear translocation assays suggest that E2 is an activator of human CAR. Of note, human and mouse CARs exhibit a significant interspecies difference with respect to ligand specificity; for instance, a mouse CAR ligand, TCPOBOP, does not serve as a human CAR ligand [27]; and a human CAR ligand, CITCO, does not activate mouse CAR [19]. Despite prominent interspecies differences in CAR ligand specificity, interestingly E2 appears to serve as a CAR activator in both mice and humans; a previous study has shown that E2 upregulates Cyp2b10 expression by activating the constitutive androstane receptor (CAR) in mice [37]. CAR is known to activate expression of genes involved in drug elimination, as well as genes modulating lipid and glucose homeostasis (reviewed in [38]). It appears plausible that the role of E2 as a CAR activator is conserved in mice and humans (unlike other CAR activators), potentially because certain functions of CAR-regulated genes are essential for maintenance of normal pregnancy in both humans and mice.
Our results in HepG2 cells indicate that ER activation by E2 enhances transcriptional activity of CYP2B6 through a nonclassical mechanism involving AP-1 and CYP2B6/A1. Although involvement of CYP2B6/A2 is questionable based on our results from EMSA, it may be possible that certain proteins bind to CYP2B6/A2 only through interactions with transcription regulators bound in the upstream regulatory region of CYP2B6; the results from the luciferase reporter assays (Fig. 4D) show that the CYP2B6/A2 site is involved in CYP2B6 induction by E2. Our results indicating CYP2B6 induction by E2 via AP-1 pathway is somewhat different from the report of a recent study in Huh7 cells, where Lo et al. showed that E2 induces CYP2B6 expression through the classical mechanism of ER action, i.e., in an ERE-dependent manner [32]. However, partial deletion of the ERE located upstream of CYP2B6 did not affect E2 induction of CYP2B6 in HepG2 cells (Fig. 4E). This apparent discrepancy with respect to the role of the ERE may be due in part to the differential activities of AP-1 proteins or other mediators of ER action in different cell lines. Cell-line dependent differential activity of c-Jun (a principal component of AP-1 complex) has been previously reported [39] although in HepG2 and Huh7 cells, we did not find significant differences in AP-1 activity in their nuclear extracts (data not shown). On the other hand, studies have suggested cell line-dependent activities of other mediators of ER action such as coactivators ([40] and references therein). Therefore, it appears possible that ER-mediated regulatory mechanism for CYP2B6 expression may vary in different cell lines and multiple pathways play a role in CYP2B6 induction in an in vivo system.
Ligand-activated CAR is known to antagonize ER activity in HepG2 cells by squelching limited amounts of a shared coactivator [36], suggesting potential antagonism between ER and CAR upon concurrent activation by E2 (at high concentrations ≥ 1 μM). If this occurs for CYP2B6 expression, the overall contribution of ER and CAR to CYP2B6 induction by E2 could be minor. However, our results demonstrate that their actions on CYP2B6 expression are in fact synergistic. Although a detailed molecular mechanism underlying the cooperative actions of ER and CAR on the CYP2B6 promoter requires further investigation, it can be speculated as follows. Conventional notion indicates that multiple transcriptional factors bind to DNA independently, and this leads to an exponential increase in affinity between the transcription factors and basal transcriptional machinery by thermodynamically stabilizing the complex [41]. Alternatively, CAR and ER may physically interact through intermediary factors, based on proximity of the AP-1 motif and PBREM, stabilizing each other’s binding to DNA and potentiating transcriptional activation of CYP2B6.
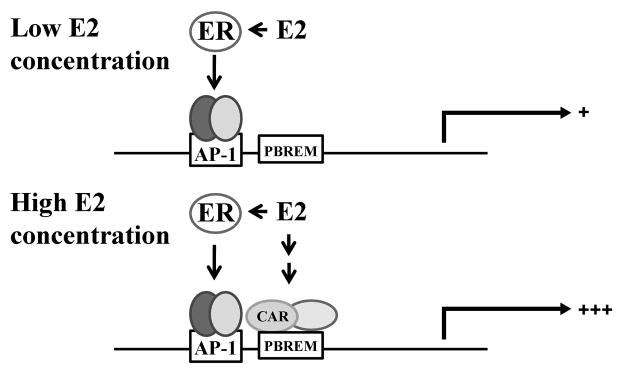
Our results in HepG2 cells indicate the E2 concentration required for CAR activation was high (average concentration ≥ 100 nM), and the activation of CAR by E2 likely occurs only at high E2 concentrations that are reached during pregnancy. These results suggest that E2 differentially activates ER and CAR in a concentration-dependent manner in the liver (Fig. 7). At low plasma concentrations of E2 reached in nonpregnant women or during early stages of pregnancy, E2 likely enhances CYP2B6 expression via activation of ER but not CAR; however, at high concentrations of E2 attained during later stages of pregnancy or in nonpregnant women using oral contraceptives [42], both ER and CAR are activated and synergistically upregulate CYP2B6 expression. This suggests that E2 likely exhibits pregnancy-specific pharmacological activity.
Fig. 7.
Proposed model: ER and CAR that are concurrently activated by E2 at high concentration synergistically enhance CYP2B6 expression. At low concentration (average concentration < 100 nM), E2 upregulates CYP2B6 expression through an ER/AP-1 pathway. At high concentration of E2 (average concentration ≥ 100 nM) (e.g., at full term pregnancy), E2 activates both ER and CAR, which in turn synergistically enhance the promoter activity of CYP2B6.
In conclusion, we demonstrate that E2, at high concentrations reached during pregnancy, enhances expression of CYP2B6 by synergistic actions of ER and CAR. The results imply that elimination of CYP2B6 substrates may increase during pregnancy, potentially requiring a dosage adjustment. These findings should have an important clinical impact on drug therapy in pregnant women as well as on better understanding of the altered physiology during pregnancy.
Supplementary Material
Acknowledgments
We thank Dr. Hyunwoo Lee and Dr. Jonna Frasor for providing critical comments about this manuscript and Ms. Bethany White for providing technical advice in performing ChIP assays. This work was supported by the National Institute of Child Health and Human Development (K12HK055892 and HD065532); and Korean Ministry of Education, Science and Technology. Primary human hepatocytes were obtained through the Liver Tissue Cell Distribution System (NIH Contract #N01-DK-7-0004/HHSN267200700004C).
Abbreviations
- AP-1
activator protein 1
- E2
17β-estradiol
- CYP
Cytochrome P450
- CAR
constitutive androstane receptor
- ER
estrogen receptor
- ERE
estrogen responsive element
- HH
primary human hepatocyte
- PBREM
the phenobarbital-responsive enhancer module
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kwi Hye Koh, Email: khkoh@uic.edu.
Steve Jurkovic, Email: sjurko2@uic.edu.
Kyunghee Yang, Email: khyang1213@gmail.com.
Su-Young Choi, Email: schoi32@uic.edu.
Jin Woo Jung, Email: sildcw@konkuk.ac.kr.
Kwang Pyo Kim, Email: kpkim@konkuk.ac.kr.
Wei Zhang, Email: weizhan1@uic.edu.
Hyunyoung Jeong, Email: yjeong@uic.edu.
References
- 1.Andrade SE, Gurwitz JH, Davis RL, Chan KA, Finkelstein JA, Fortman K, et al. Prescription drug use in pregnancy. Am J Obstet Gynecol. 2004;191:398–407. doi: 10.1016/j.ajog.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 2.Anderson GD. Pregnancy-induced changes in pharmacokinetics: a mechanistic-based approach. Clin Pharmacokinet. 2005;44:989–1008. doi: 10.2165/00003088-200544100-00001. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham FG. Williams obstetrics. New York: McGraw-Hill Medical Publishing Division; 2001. [Google Scholar]
- 4.Sarlis NJ, Gourgiotis L. Hormonal effects on drug metabolism through the CYP system: perspectives on their potential significance in the era of pharmacogenomics. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:439–48. doi: 10.2174/156800805774912971. [DOI] [PubMed] [Google Scholar]
- 5.MGSC Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 7.Shaikh AA. Estrone and estradiol levels in the ovarian venous blood from rats during the estrous cycle and pregnancy. Biol Reprod. 1971;5:297–307. doi: 10.1093/biolreprod/5.3.297. [DOI] [PubMed] [Google Scholar]
- 8.Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, et al. Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos. 2007;35:1935–41. doi: 10.1124/dmd.107.016568. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Yang K, Choi S, Fischer JH, Jeong H. Up-regulation of UDP-glucuronosyltransferase (UGT) 1A4 by 17beta-estradiol: a potential mechanism of increased lamotrigine elimination in pregnancy. Drug Metab Dispos. 2009;37:1841–7. doi: 10.1124/dmd.109.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–70. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 11.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–42. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 12.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J Mol Endocrinol. 2008;41:263–75. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–8. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–31. [PubMed] [Google Scholar]
- 15.Wang H, Tompkins LM. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab. 2008;9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang K, Koh KH, Jeong H. Induction of CYP2B6 and CYP3A4 Expression by 1-Aminobenzotriazole (ABT) in Human Hepatocytes. Drug Metab Lett. 2010 doi: 10.2174/187231210791698410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong H, Choi S, Song JW, Chen H, Fischer JH. Regulation of UDP-glucuronosyltransferase (UGT) 1A1 by progesterone and its impact on labetalol elimination. Xenobiotica. 2008;38:62–75. doi: 10.1080/00498250701744633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi S, Sainz B, Jr, Corcoran P, Uprichard S, Jeong H. Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica; the fate of foreign compounds in biological systems. 2009;39:205–17. doi: 10.1080/00498250802613620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, et al. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–83. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ, Kim KH, Park JS, Jung JW, Kim YH, Kim SK, et al. Comparative analysis of cell surface proteins in chronic and acute leukemia cell lines. Biochemical and biophysical research communications. 2007;357:620–6. doi: 10.1016/j.bbrc.2007.03.191. [DOI] [PubMed] [Google Scholar]
- 21.Gupta MK, Jang JM, Jung JW, Uhm SJ, Kim KP, Lee HT. Proteomic analysis of parthenogenetic and in vitro fertilized porcine embryos. Proteomics. 2009;9:2846–60. doi: 10.1002/pmic.200800700. [DOI] [PubMed] [Google Scholar]
- 22.Kolker E, Hogan JM, Higdon R, Kolker N, Landorf E, Yakunin AF, et al. Development of BIATECH-54 standard mixtures for assessment of protein identification and relative expression. Proteomics. 2007;7:3693–8. doi: 10.1002/pmic.200700088. [DOI] [PubMed] [Google Scholar]
- 23.Swales K, Kakizaki S, Yamamoto Y, Inoue K, Kobayashi K, Negishi M. Novel CAR-mediated mechanism for synergistic activation of two distinct elements within the human cytochrome P450 2B6 gene in HepG2 cells. J Biol Chem. 2005;280:3458–66. doi: 10.1074/jbc.M411318200. [DOI] [PubMed] [Google Scholar]
- 24.Auerbach SS, Stoner MA, Su S, Omiecinski CJ. Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3) Mol Pharmacol. 2005;68:1239–53. doi: 10.1124/mol.105.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–22. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Chen T, Cottrell J, Wang H. Nuclear translocation of adenoviral-enhanced yellow fluorescent protein-tagged-human constitutive androstane receptor (hCAR): a novel tool for screening hCAR activators in human primary hepatocytes. Drug Metab Dispos. 2009;37:1098–106. doi: 10.1124/dmd.108.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–7. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol. 2008;74:443–53. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar SN, Huang RQ, Logan SM, Yi KD, Dillon GH, Simpkins JW. Estrogens directly potentiate neuronal L-type Ca2+ channels. Proc Natl Acad Sci U S A. 2008;105:15148–53. doi: 10.1073/pnas.0802379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborne CK, Wakeling A, Nicholson RI. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. British journal of cancer. 2004;90 (Suppl 1):S2–6. doi: 10.1038/sj.bjc.6601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harnish DC, Evans MJ, Scicchitano MS, Bhat RA, Karathanasis SK. Estrogen regulation of the apolipoprotein AI gene promoter through transcription cofactor sharing. J Biol Chem. 1998;273:9270–8. doi: 10.1074/jbc.273.15.9270. [DOI] [PubMed] [Google Scholar]
- 32.Lo R, Burgoon L, Macpherson L, Ahmed S, Matthews J. Estrogen receptor-dependent regulation of CYP2B6 in human breast cancer cells. Biochim Biophys Acta. 2010;1799:469–79. doi: 10.1016/j.bbagrm.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato S, Tora L, Yamauchi J, Masushige S, Bellard M, Chambon P. A far upstream estrogen response element of the ovalbumin gene contains several half-palindromic 5′-TGACC-3′ motifs acting synergistically. Cell. 1992;68:731–42. doi: 10.1016/0092-8674(92)90148-6. [DOI] [PubMed] [Google Scholar]
- 34.Lambertini E, Tavanti E, Torreggiani E, Penolazzi L, Gambari R, Piva R. ERalpha and AP-1 interact in vivo with a specific sequence of the F promoter of the human ERalpha gene in osteoblasts. J Cell Physiol. 2008;216:101–10. doi: 10.1002/jcp.21379. [DOI] [PubMed] [Google Scholar]
- 35.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–12. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min G, Kim H, Bae Y, Petz L, Kemper JK. Inhibitory cross-talk between estrogen receptor (ER) and constitutively activated androstane receptor (CAR). CAR inhibits ER-mediated signaling pathway by squelching p160 coactivators. J Biol Chem. 2002;277:34626–33. doi: 10.1074/jbc.M205239200. [DOI] [PubMed] [Google Scholar]
- 37.Kawamoto T, Kakizaki S, Yoshinari K, Negishi M. Estrogen activation of the nuclear orphan receptor CAR (constitutive active receptor) in induction of the mouse Cyp2b10 gene. Mol Endocrinol. 2000;14:1897–905. doi: 10.1210/mend.14.11.0547. [DOI] [PubMed] [Google Scholar]
- 38.Gao J, Xie W. Pregnane X receptor and constitutive androstane receptor at the crossroads of drug metabolism and energy metabolism. Drug Metab Dispos. 2010;38:2091–5. doi: 10.1124/dmd.110.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baichwal VR, Tjian R. Control of c-Jun activity by interaction of a cell-specific inhibitor with regulatory domain delta: differences between v- and c-Jun. Cell. 1990;63:815–25. doi: 10.1016/0092-8674(90)90147-7. [DOI] [PubMed] [Google Scholar]
- 40.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–4. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 41.Veitia RA. A sigmoidal transcriptional response: cooperativity, synergy and dosage effects. Biol Rev Camb Philos Soc. 2003;78:149–70. doi: 10.1017/s1464793102006036. [DOI] [PubMed] [Google Scholar]
- 42.Eisenfeld AJ, Aten RF. Estrogen receptors and androgen receptors in the mammalian liver. J Steroid Biochem. 1987;27:1109–18. doi: 10.1016/0022-4731(87)90197-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.