Biochemical form is shaped by biological function. Biochemical investigation of enzymes involved in eukaryotic homologous genetic recombination now is progressing rapidly, having only recently been inaugurated with the study of Rad51 protein, a homolog of the bacterial RecA protein. A significant new front is opened with the first report on the in vitro activities of a second and meiosis-specific RecA homolog, the human Dmc1 protein (1). Results on Rad51 and Dmc1 to date have yielded a number of surprises in the form of some intriguing and perhaps unanticipated differences with respect to the RecA protein. Pondering biochemical differences must inevitably bring one back to a fundamental question: why do cells recombine DNA?
The history of in vitro research into mechanisms of genetic recombination now spans two decades. Until quite recently, bacteria have provided most of the recombination enzymes and most of the biochemical insight. The bacterial RecA protein has been at the center of this activity. Little genetic information is exchanged between bacterial chromosomes without RecA. The RecA protein aligns two DNA molecules, facilitates a strand switch that generates a crossover between them, and promotes migration of the resulting DNA branch. An ever-growing host of additional proteins prepare DNA substrates for RecA action, modulate RecA binding to DNA, or process the branched DNA recombination intermediates created by RecA (2–4).
A parallel universe of recombination enzymes appears to exist in eukaryotes. The curtain opened in 1992 with the demonstration that in Rad51 (5) and Dmc1 (6) yeast possessed at least two RecA homologs. Mammals also possess homologs of both proteins, and their investigation has taken some interesting turns. Targeted disruption of mouse rad51 confers an embryonic lethal phenotype and sensitivity to ionizing radiation (7, 8). Several proteins involved in carcinogenesis interact with Rad51, including p53 (9), BRCA1 (10), and BRCA2 (11). These results have lent some urgency to the in vitro investigation of Rad51. Following the pioneering work of Sung (12), this now is being pursued in more than a dozen laboratories worldwide. The Dmc1 protein of yeast colocalizes with Rad51 on the zygotene chromosomes (13) and also must play an important role in recombination.
The Rad51 proteins of yeast and human appear to be quite similar. They form helical filaments on DNA with a structure similar to that of the RecA protein (14, 15), possess a DNA-dependent ATPase activity (12, 15), promote a DNA strand exchange reaction (12, 16–18), and require a single-strand DNA binding protein (RPA) for optimal strand exchange activity (12, 17, 19). Li et al. (1) demonstrate that the human Dmc1 protein possesses a DNA-dependent ATPase activity and promotes a DNA strand exchange requiring levels of Dmc1 consistent with the formation of a filament as the active form. So far, both Rad51 and Dmc1 sound like RecA. However, the differences between RecA and its eukaryotic counterparts are substantial. The DNA strand exchange reactions promoted by the human Rad51 and Dmc1 are much less robust than those promoted by RecA protein (1, 17, 18). ATP is hydrolyzed by all of the eukaryotic proteins at rates from 1 to 2 orders of magnitude slower than RecA protein. If Rad51 and Dmc1 are the eukaryotic equivalents to RecA, evolution seems to have served up some markedly hobbled proteins for use in eukaryotic recombination.
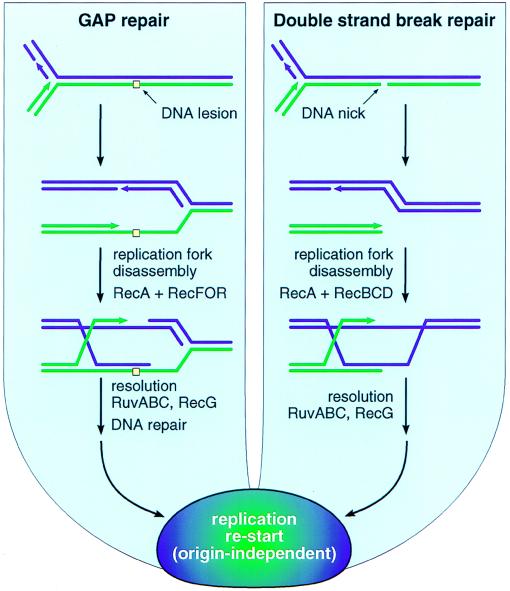
Biological paradigms can both aid and hinder a biochemical investigation. In bacteria, the functional paradigm that has shaped most in vitro investigations involving RecA is centered on conjugational recombination. However, a strong case can be made that recombination evolved in bacteria not as a means to exchange genetic information between cells, but as a DNA repair process (4, 20, 21). Recombinational DNA repair operates in a world of DNA strand breaks and gaps and, in the absence of external factors like ionizing radiation, most of these probably are associated with replication forks. If a fork encounters an unrepaired DNA lesion, the lesion is repaired via a recombinational repair pathway by using the RecF, RecO, RecR, and other functions in addition to RecA protein (Fig. 1). If a fork encounters a DNA strand break (e.g., as might exist at a site undergoing excision or mismatch repair), the resulting double-strand break is repaired via a pathway requiring the RecBCD enzyme, RecA, and other proteins (Fig. 1).
Figure 1.
Major pathways for bacterial recombinational DNA repair.
There are as yet no good estimates as to how often a replication fork is halted by damage, and the probability no doubt varies with growth conditions. However, it almost certainly occurs more often than generally is appreciated. Every Escherichia coli cell suffers 3,000–5,000 DNA lesions per generation, most of which are oxidative lesions (4, 22). Cells lacking RecA exhibit high mortality (which can approach 50% even in the absence of added DNA damaging agents or radiation) (4, 22), much if not all of which may reflect an inability to repair stalled replication forks. Cells lacking RecBCD accumulate high levels of unrepaired double-strand breaks (23), which are likely to represent the normal load of replication-associated breaks handled by the RecBCD repair pathway. These observations alone suggest that every time bidirectional replication is initiated at oriC in a wild-type cell, the chance that one or both replication forks will encounter a situation requiring recombinational DNA repair could be in the range of 10–45%, especially in cells grown aerobically in rich media. A recA culture contains an abundance of dead cells (4), but the fraction of cells with unrepairable stalled replication forks must have an upper limit of 50% in any strain that is capable of increasing cell number with time. If the genome is subjected to an abnormal insult in the form of radiation or an added DNA damaging agent, most cells lacking one of the recombination enzymes simply don’t survive. Recombinational DNA repair provides a highly adaptable and sometimes redundant set of proteins and multiple pathways to deal with the full range of DNA gaps, breaks, and branched structures that might present at a stalled replication fork. The pathways of Fig. 1 represent a crossroads where every aspect of bacterial DNA metabolism comes together. The interface that must exist in these pathways between recombination and replication is particularly interesting and only now being explored. Once halted, the replication forks are likely to disassemble, requiring reassembly after repair in an origin-independent manner.
The study of conjugational recombination has resulted in the identification of numerous recombination enzymes in bacteria and accounts for much of the progress in the field to date; however, not all effects of this paradigm have been positive. Conjugation clearly occurs orders of magnitude less frequently than recombinational DNA repair. The focus on conjugation sometimes has created an unwarranted perception that the RecF pathway for recombination is little more than a minor path observed only in certain mutant backgrounds (24), a perception that may have slowed investigation of RecF pathway biochemistry. However, the sensitivity of bacteria lacking RecF pathway functions to DNA damaging agents (4) indicates that a bacterial cell experiences a different reality. Both RecF and RecBCD pathway functions are important parts of an efficient bacterial DNA repair system. The RecBCD pathway is used almost exclusively during conjugation (3, 24), a fact that simply may reflect the structure of the DNA substrates presented to a bacterial cell on the rare occasions when conjugation occurs.
A paradigm based on recombinational DNA repair offers a potentially insightful perspective and can provide connections to guide in vitro studies of bacterial recombination functions. Recent work linking the RecF and RecR proteins with replication functions (25, 26) highlight an important aspect of the paradigm outlined in Fig. 1. If replication forks often are halted by damage, then an efficient system for the origin-independent reassembly of a replication fork would be essential to the cell. The existence of such a system might have at least two implications. First, it could represent the molecular basis for many of the observations of origin-independent (and recombination-dependent) DNA replication (27, 28). During the SOS response, when elevated levels of DNA breaks and gaps would be present to trigger recombination, a high level of recombination-dependent replication would be almost inevitable. The molecular requirements observed for oriC-independent replication generally fit this scenario quite well (28). Second, replication restart associated with recombinational DNA repair could represent the primary function for the bacterial primosome. Originally characterized in studies of the replication of bacteriophage φX174 DNA (29), it now is clear that most primosome components are not needed for replication initiated at oriC (29, 30). Loss of key primosome proteins such as PriA confer defects not in chromosomal replication but in origin-independent DNA replication and DNA repair (30). The RecF and PriA proteins appear to interact in vivo (25), and the RecF and RecR proteins are required for replication restart after the repair of extensive DNA damage (26). The primosome may not be needed for oriC-initiated replication, but many of the resulting replication forks may be unable to complete their tasks without it.
This scenario also can enlighten a number of underappreciated features of RecA biochemistry that may be relevant to studies of Rad51 and Dmc1. One example is RecA-mediated ATP hydrolysis. RecA-mediated ATP hydrolysis is required to recycle RecA filaments and is coupled to late stages of DNA strand exchange (2, 4, 31). It is not required for DNA pairing and the fundamental process of strand exchange. Instead, it augments that process, rendering strand exchange unidirectional, facilitating the bypass of structural barriers in the DNA and permitting exchange reactions with four DNA strands (4). In effect, the ATP hydrolytic activity of RecA has at least some functions that are best rationalized in the context of DNA repair.
Why do Rad51 and Dmc1 hydrolyze so little ATP? Perhaps the capacity to bypass DNA lesions or promote four-strand exchange reactions no longer is important or is relegated to other enzymatic functions. RecA protein will promote DNA strand exchange with good efficiency if one DNA substrate contains a heterologous insertion of 100 bp or more (4, 32). The DNA strand exchange reaction promoted by yeast Rad51 protein, in contrast, is reduced by a 6-bp insertion and abolished by insertions greater than 14 bp (K. Benjamin, V. Holmes, and N. Cozzarelli, personal communication). Efforts to date to observe a Rad51-promoted four-strand exchange reaction have failed (P. Sung, personal communication).
Of course, Rad51 and Dmc1 are not hobbled proteins, but are just right for their place in the eukaryotic milieu. In eukaryotes, homologous recombination is needed to ensure proper segregation of chromosomes in meiosis I. This and other functions may be as critical or more critical than DNA repair, especially for a meiosis-specific protein such as Dmc1. Rad51 and Dmc1 may act in environments that are even more tightly organized than those seen by RecA. Eukaryotes introduce double-strand breaks into their chromosomes purposefully during meiosis (33), a dangerous thing to do if the stage is not set to initiate recombination. There is no shortage of additional questions raised by the current study of eukaryotic recombination functions. What other proteins (besides RPA) interact with Rad51 and Dmc1, and what are their functions? Why do eukaryotes possess more than one RecA homolog? What do the limitations seen in the strand exchange reactions observed to date with these proteins mean? If Rad51 and Dmc1 do not promote four-strand exchange reactions, how are Holliday intermediates generated in eukaryotes?
Continued biochemical study will provide some answers to the above questions. In yeast, nascent efforts with Rad52, Rad54, Rad55, and Rad57 already have made some progress (34–36). Interestingly, Rad55 and Rad57 may be functional homologs of RecO and RecR (36). Certainly, someone must soon add BRCA1 and BRCA2 to a reaction with human Rad51 and see what happens. It will be apparent to those paying attention that the Radding group is now in the intriguing position of putting human Dmc1 and Rad51 in the same test tube. A careful examination of Rad51 and Dmc1 focusing on those RecA activities known to require ATP hydrolysis also seems to be in order. It is still early, and reactions will get better as each lab gains experience with these new experimental subjects.
Eukaryotic recombination enzymes hold much of the promise for discovery that bacterial enzymes had 20 years ago. No matter what path the biochemistry takes, the answers to some questions still will require a careful consideration of what recombination contributes to DNA metabolism, meiosis, and cell division in a eukaryote. In this effort, the bacterial paradigm established to date will often, but not always, be instructive. The bacterial paradigm, in fact, continues to evolve.
Acknowledgments
I thank Bill Reznikoff, John Petrini, Patrick Sung, and Charles Radding for reading and commenting on this manuscript, and Patrick Sung, Victor Holmes, and Nick Cozzarelli for sharing data before publication. Space limitations required the citation of reviews instead of primary literature wherever possible in this article, and I regret any omissions thereby made necessary. The work described from my laboratory was supported by National Institutes of Health Grants GM32335 and GM52725.
References
- 1.Li Z, Golub E I, Gupta R, Radding C M. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark A J, Sandler S J. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 4.Roca A I, Cox M M. Prog Nucleic Acids Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 5.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 6.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 7.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim D S, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturzbecher H W, Donzelmann B, Henning W, Knippschild U, Buchhop S. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 10.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 11.Sharan S K, Morimatsu M, Albrecht U, Lim D-S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Nature (London) 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 12.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 13.Bishop D K. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 15.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 17.Baumann P, Benson F E, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 18.Gupta R C, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama T, Zaitseva E M, Kowalczykowski S C. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 20.Michod R E, Levin B R, editors. The Evolution of Sex: An Examination of Current Ideas. Sunderland, MA: Sinauer; 1988. [Google Scholar]
- 21.Kuzminov A. Recombinational Repair of DNA Damage. Georgetown, TX: Landes; 1996. [Google Scholar]
- 22.Cox M M. BioEssays. 1993;15:617–623. doi: 10.1002/bies.950150908. [DOI] [PubMed] [Google Scholar]
- 23.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith G R. Cell. 1989;58:807–809. doi: 10.1016/0092-8674(89)90929-x. [DOI] [PubMed] [Google Scholar]
- 25.Sandler S J. Mol Microbiol. 1996;19:871–880. doi: 10.1046/j.1365-2958.1996.429959.x. [DOI] [PubMed] [Google Scholar]
- 26.Courcelle J, Carswell-Crumpton C, Hanawalt P. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogoma T. Proc Natl Acad Sci USA. 1997;94:3483–3484. doi: 10.1073/pnas.94.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 30.Zavitz K H, Marians K J. Mol Microbiol. 1991;5:2869–2873. doi: 10.1111/j.1365-2958.1991.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 31.Bedale W A, Cox M. J Biol Chem. 1996;271:5725–5732. doi: 10.1074/jbc.271.10.5725. [DOI] [PubMed] [Google Scholar]
- 32.Kowalczykowski S C, Eggleston A K. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 33.Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 34.Shen Z, Cloud K G, Chen D J, Park M S. J Biol Chem. 1996;271:148–152. doi: 10.1074/jbc.271.1.148. [DOI] [PubMed] [Google Scholar]
- 35.Jiang H, Xie Y, Houston P, Stemke H K, Mortensen U H, Rothstein R, Kodadek T. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 36.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]