Highlights
► There are numerous sources of samples for animal virus discovery. ► Many simple molecular methods exist for the characterization of novel viruses. ► Some human viruses have genetically close animal counterparts. ► Future emerging human viruses may arise from any animal viral families. ► Disease association and causation studies are needed for many new animal viruses.
Abstract
The characterization of viral genomes has accelerated due to improvement in DNA sequencing technology. Sources of animal samples and molecular methods for the identification of novel viral pathogens and steps to determine their pathogenicity are listed. The difficulties for predicting future cross-species transmissions are highlighted by the wide diversity of known viral zoonoses. Recent surveys of viruses in wild and domesticated animals have characterized numerous viruses including some closely related to those infecting humans. The detection of multiple genetic lineages within viral families infecting a single host species, phylogenetically interspersed with viruses found in other host species, reflects past cross-species transmissions. Numerous opportunities for the generation of novel vaccines will arise from a better understanding of animal viromes.
Introduction
The rate of viral discovery has recently increased due to the introduction of next generation sequencing technologies and the analyses of biological samples of diverse geographic origins from multiple host species. By 2006 the number of known human viral species was estimated at approximately 180 [1]. In 2009 the number of all ICTV defined viral species, including both eukaryotic viruses and bacteriophages, stood at approximately 2200 (http://www.ictvonline.org/virusTaxInfo.asp). Compared to the sustained efforts in human virus discovery, viruses infecting other species, including >4200 species of mammals [2], have been greatly under-sampled. While the number of known, globally prevalent human viruses (excluding geographically restricted and emerging viruses), may eventually reach a plateau, the rate of discovery of animal viruses is expected to rapidly increase. The generation of more fully characterized animal viral genomes, from more host species, will improve our understanding of viral evolution, cross species transmissions, and will provide new opportunities for animal vaccine development particularly for domesticated and endangered wild species.
Sources of biological samples for animal virus discovery
Specimens to analyze for new viruses include those from animals with diseases affecting farm productivity or the survival of critically endangered or threatened species. Farmers, with their extensive knowledge and experience of animals, can readily identify new health problems. The health of animals in zoos and aquariums is also of interest given their high level of care, the diverse viral exposures they may experience, and their close proximity to human handlers. Companion animals, due to their extensive health care and close contact with owners may also be a readily accessible source of animal samples for pathogen discovery. Shelters for abandoned or feral animals, because of their crowded conditions and the high susceptibility to infections of their often undernourished and weakened residents also provide a fertile breeding ground for viral epidemics and pathogen discovery (Figure 1 ).
Figure 1.
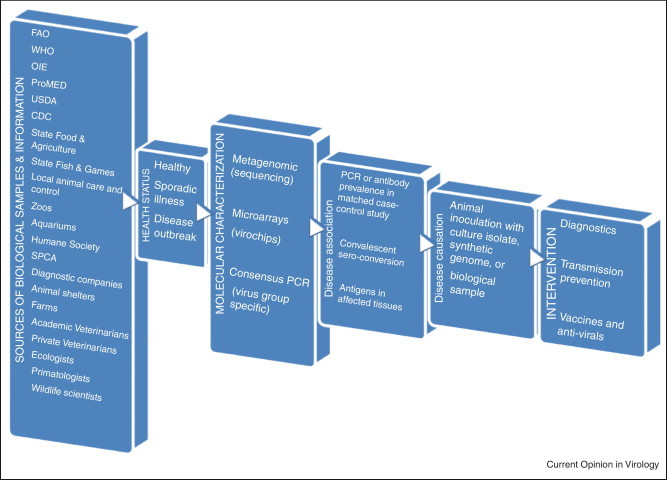
Flow chart of animal virus discovery, pathogenicity determination, and interventions.
Outbreaks of acute disease on farms and in animal shelters greatly facilitate virus-disease association studies if appropriate data and sample collection occurs. The affected animals should all exhibit pathogen-specific markers of infection, such as sero-conversion or the presence of a newly characterized virus. The pathogenicity of a new virus can also be tested in animals following direct inoculation with viral isolates (minimally passaged to prevent attenuation), the original biological samples (if shown to contain no other virus by metagenomics), or by synthesizing the genome and transfecting it in vitro to generate infectious particles. Because of their protected status, such inoculations are not feasible in endangered species where more indirect means of testing disease causation, akin to the situation for novel human viruses, are required [3, 4]. By identifying unusual symptoms or disease outbreaks, both academic and private veterinarians and scientists also contribute to the identification of previously unknown or emerging animal pathogens. Federal departments such as the USDA and CDC, state organizations concerned with fish and game or food and agriculture, and local government groups involved in animal care and control can also identify disease outbreaks in wild, farm or companion animals and collect samples for further studies. The Humane Society and the Society for the Prevention of Cruelty to Animals, by closely monitoring and promoting animal health, may also detect and report early signs of emerging infections. International organizations such as the Food and Agriculture and World Health Organizations of the United Nations, World Organization for Animal Health (i.e. OIE), and ProMED can also assist in the recognition of emerging animal health problems, dissemination of information, and in coordinating international collaborations (Figure 1). A growing realization of animals as the source of most emerging human and animal infections has led to the One Health Initiative to foster collaborations between physicians, veterinarians, and scientists to monitor the exchange of infectious agents between species [5, 6, 7, 8, 9, 10, 11, 12].
Bats, rodents, and primates are notorious sources of zoonotic infections, possibly a result of their very large colony sizes facilitating maintenance of viral transmission chains, frequent association with humans, and their close genetic relatedness to humans respectively. The consumption of wild animals as bush meat, particularly of non-human primates, also provides a portal of entry of animal viruses into human populations [8, 10, 11]. Large unbiased or viral family specific surveys of these and other mammalian groups to characterize their viruses will enhance our understanding of the original animal reservoirs of many current human viruses. Viral infections may be mostly asymptomatic in their long-term hosts, but pathogenic in a new host species. Viral metagenomics and more virus family specific surveys have therefore been used to characterize viral populations in both sick and healthy animals [13, 14, 15•, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28]. The buildup of known animal viral genome sequences will also allow their inclusion in updated high-throughput virus detection assays, such as micro-array ‘virochips,’ able to very sensitively detect known viruses and their close genetic relatives [29, 30, 31•]. Including probes from the growing number of viral genomes on micro-arrays also allows simultaneous disease association studies for multiple viruses using animal (and human) cohorts. The availability of biological samples from large numbers of epidemiologically matched unexplained disease cases and healthy controls is likely to be a major limiting factor for determining which of the rapidly growing number of animal viruses are likely pathogens and therefore targets for transmission control measures or vaccine development.
Molecular methods for viral discovery
Many classical methods of viral discovery such as cell inoculation and monitoring for cytopathic effects can yield pure viral cultures, but are subject to the availability of susceptible cell lines and infectious inoculums. The introduction of molecular methods has greatly simplified the genome characterization of both known and emerging or previously unrecognized viruses. Consensus PCR, targeting conserved viral genome regions, [32, 33] can be used to rapidly screen large numbers of samples for any group of related viruses such as herpes viruses [34], astroviruses [35, 36], and enteroviruses [37]. The downside of this sensitive method is the requirement for a priori knowledge of which viral family is likely to be present in order to avoid the need for numerous PCR primer sets targeting a large number of different viral families. Rolling circle amplification preferentially amplifies circular DNA viral genomes and has greatly enhanced their discovery but is less efficient for linear DNA or RNA genomes [38, 39, 40]. Microarrays spotted with oligonucleotides of the most conserved viral regions have also been highly successful but are limited by the amount of mismatch they can tolerate such that highly divergent species (relative to those previously known and spotted on arrays) may not hybridize [29, 30, 31•, 41, 42, 43, 44]. Random nucleic acid amplification with or without prior enrichment for viral particles [45], followed by DNA sequencing (including next generation sequencing) and in silico similarity searches for sequence related to those of known viruses has been highly productive [46, 47, 48, 49, 50•, 51, 52, 53•, 54, 55]. This metagenomic approach is limited by the need for novel viral sequences to show detectable protein or nucleic acid sequence similarity to those of the many already sequenced viruses.
Anticipating zoonoses
The sources of many emerging viral diseases are animals in contact with the new viral host or with an intermediate bridge species [5, 6, 7, 8, 9, 10, 11, 12]. Initially, cross-species transmissions are thought to result in weakly adapted viruses that through mutations may evolve to increase their pathogenicity and transmissibility in the new host species. A well understood example of cross-species transmission is of a feline parvovirus adapting to dogs in the late 1970s followed by its global spread and increase in pathogenicity [10, 56]. Mutations in the feline parvovirus surface glycoprotein allowed infection and transmission in dogs [57, 58••]. Further adaptation of the original canine parvovirus may have occurred through intermediate species such as raccoons [59]. The emergence of HIV1 groups M and N from chimpanzees, HIV group P from gorillas and HIV2 from sooty mangabeys, most likely through bush meat hunting, butchering, and consumption is also generally accepted [60, 61••, 62, 63]. SIVcpz, the presumed progenitor of HIV, may itself be a recombinant of two retroviruses from monkeys preyed upon on by chimpanzees [64]. Influenza viruses are especially notorious for their ability to transfer from birds to mammals such as pigs, that can act as intermediate hosts, facilitating recombination with porcine influenza viruses before transmission to humans [56, 65, 66]. Bats and rodents appear to be frequent sources of viral zoonosis but the very high number of these animal species and their global distribution makes a systematic determination of their viromes difficult. The genetic characterization of viruses per se in these frequent virus donor species does not a priori provide information regarding the likelihood of successful transmissions to human. Zoonoses are dependent on complex interactions between viral phenotypes and host genetics (particularly surface receptors and innate immune responses), cross-neutralization by antibodies to related viruses, and epidemiological factors influencing viral exposures. The high mutability of viral genomes indicates that, provided some chronic low-level replication occurs in a new host species, these viruses have the potential to further adapt increasing both their viral load and the possibility of transmission in that species. The diversity of emerging or re-emerging human viruses such as HIV (Retroviridae) and the SARS virus (Coronaviridae) that are transmissible between humans, zoonotically acquired viruses capable of only limited transfer between humans such as the Ebola virus (Filoviridae) and Lassa virus (Arenaviridae), ‘dead end’ zoonoses without the necessary adaptation to facilitate ongoing transmission between humans such as rabies virus (Rhabdoviridae), Hendra virus (Paramyxoviridae), and monkeypox virus (Poxviridae), as well as the arthropod vectored West Nile and Japanese encephalitis viruses (Flaviviridae), Crimean-Congo hemorrhagic fever virus (Bunyaviridae), and Chickungunya virus (Togaviridae), indicate that sequence members of any of the known viral families infecting animals could potentially become epidemic in humans. The recent demonstration of an adenovirus (Adenoviridae) from a titi monkey outbreak of respiratory symptoms infecting a scientist at a primate center and this person transmitting the virus to a human contact, further illustrates the wide range of viral families that can be considered capable of at least some level of replication in multiple host species [67••]. This study exemplified the speed with which an adenovirus could be transmitted from an unknown host to titi monkey, between titi monkeys, from a titi monkey to a human and between at least two humans. Some genotypes of hepatitis E virus (Hepiviridae) are capable of oral-fecal transmission between human while other genotypes are acquired by consuming infected animal meat, but are inefficiently transmitted between human [68, 69]. Simian foamy and T-lymphotropic viruses (Retroviridae) that have infected persons exposed to non-human primates can also be considered as viruses constantly ‘probing’ human populations but that, unlike HIV1 and HIV2, have not adapted sufficiently to be transmitted between humans [70, 71, 72••, 73, 74, 75]. Certain human viruses such as influenza (Orthomyxoviridae) are periodically acquired directly from avian or mammalian hosts. The highly lethal H5N1 influenza circulating in birds is currentlyy poorly transmissible between humans or between other mammals although rapid passage experiments in ferrets or direct mutagenesis have exposed its latent capacity to rapidly increase its pathogenicity and transmissibility [76, 77, 78•].
The wide diversity of viruses capable of switching host species therefore highlights the difficulty in predicting from which viral family will emerge the next human viral pandemic. Because increasing genetic distances between hosts is a significant block to cross species transmission [79••, 80, 81], there has been a focus on identifying viruses and immune response shared between non-human primates and people exposed to them [70, 71, 72••, 73, 74, 82]. Since the frequency and intensity of viral exposure can also be expected to increase the likelihood of cross-species transmission, the study of viruses in farm or companion animals with extensive contact with both humans and wildlife should also uncover viral species of concern for future zoonoses. Sero-surveys for antibodies to these viruses would reveal the extent of their replication in highly exposed humans. Arboviruses also present a growing threat as seen with resurgent West Nile, Dengue, Japanese encephalitis and Chikungunya viruses following introductions in new locales or extension of the range of their insect vectors [83•]. Monitoring for new arboviruses in anthropophilic arthropod vectors may provide novel viral genomes whose capacity to infect humans or other mammals can then be tested serologically. Vaccinating animal reservoirs for some arboviruses could warrant considerations to reduce spill-over infections into humans.
The recent characterization of the closest known genetic relative of the human HCV [84••] and enteric Aichi viruses in canine samples [14, 85] point to dogs as a potential zoonotic origin of these now common human infections. The direction of transmission (dogs to human or human to dogs) cannot be revealed by genetic similarities alone and future viral discoveries may reveal yet closer relatives of these and other human viruses [16]. As sampling of animal viruses increases, a complex network of past cross-species transmission will likely emerge. For example until recently only a single species of astrovirus (HAstV) and of parvovirus (B19) were known to infect humans. Viral survey in human have now shown that multiple genera and species within the Parvoviridae [46, 86, 87, 88, 89] and Astroviridae [90, 91, 92] can infect humans. Multiple lineages of these viral families can also be found in pigs and other animals [15•, 17] (Figure 2 ). Distinct phylogenetic clades within a viral family also include viruses found in different mammal hosts, likely reflecting cross-species transmission of parvoviruses [93, 94, 95, 96] (as was recently documented for the feline to canine CPV2 transfer in the 1970s) and of astroviruses (Figure 2) [90, 91, 92].
Figure 2.
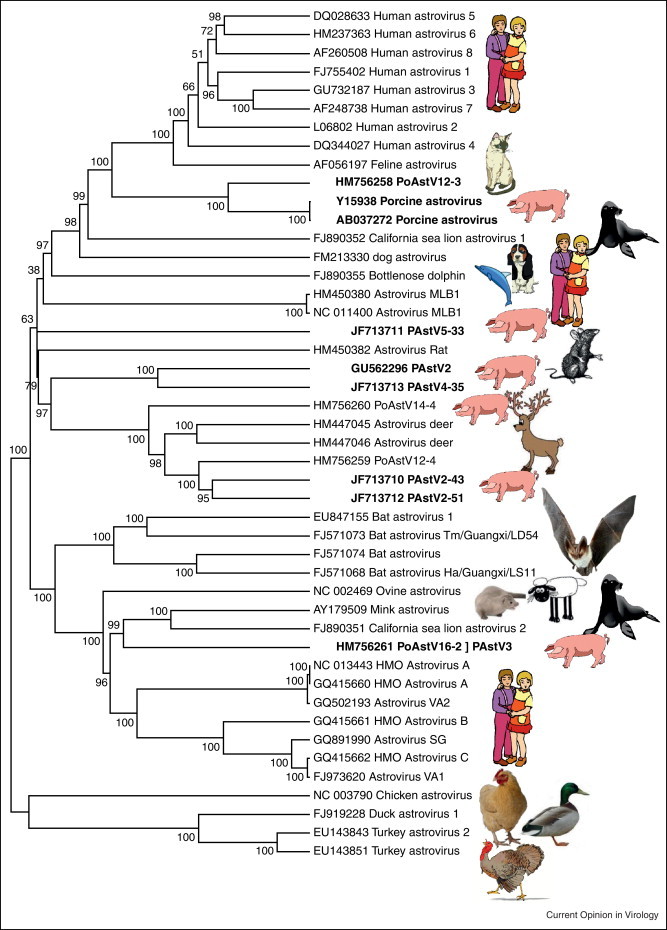
Maximum likelihood phylogenetic analysis of capsid proteins of astroviruses showing that diverse astroviruses infect some mammalian hosts species likely reflecting past cross-species transmissions. Bootstrap values of ≥70% are indicated at each branching point.
While the occurrence of cross-species transmissions is well established the overall frequency of such events is harder to estimate using molecular clocks calibrated based on short-term observations of viral evolution. Estimates of the time to last common ancestor of existing lineages of related viral RNA species yields date of thousands of years which differ greatly from dates derived using molecular clocks based on ancient viral genomes recently found on host chromosomes whose ages since integration are in the millions of years [97, 98, 99••]. In the absence of longitudinally collected epidemiological data confirming the recent emergence of a virus in a new host species, as was possible for HIV1/2, SARS-CoV, and canine parvovirus CPV2, estimating the age and therefore the frequency for other cross-species jumps based on molecular clocks derived from short term viral evolution data, is therefore problematic [100, 101, 102].
Newly characterized animal viruses and disease association
While the rate of viral discovery has greatly accelerated, the epidemiological studies required to associate infections with symptoms has lagged behind due to difficulties in obtaining large numbers of the most appropriate biological samples. In order for human or animal viral vaccine development to proceed, convincing evidence of pathogenicity is required [4, 103]. Genetically characterizing novel viral genomes in diseased animals provides the information required to design high throughput PCR assays with which to compare viral prevalence in epidemiologically matched disease cases versus healthy controls (Figure 1). Matching between cases and controls should optimally include age, sex, location, and type of environment (e.g. high intensity farming or free range). Disease association studies as well as temporal association of symptoms with IgM detection, rising IgG can both provide evidence in support of pathogenicity. Disease induction following animal challenge with the purified virus amplified in cell culture, or other pure virus inoculum can directly demonstrate disease causation (Figure 1). Complicating factors to be considered which may influence clinical outcome include variable host genetics, passive immunity due to maternally acquired antibodies, cross-protection by prior infections with related but less pathogenic viruses, and co-infections with other agents. Repeatability in independent studies can also validate prior conclusions. The severity of symptoms and their frequency must also be onerous enough to justify the cost of vaccine development, efficacy testing under realistic conditions, and ultimately large-scale vaccination.
Recent successes in identifying animal viruses and associating them with disease include the piscine reovirus (PRV) associated with heart and skeletal muscle inflammation in farmed salmon, where viral prevalence and viral loads were higher in affected than in healthy fish and viral expression was detected in affected tissues [50•]. A new bornavirus was also detected and associated with proventricular dilatation disease in psittacine birds [44, 49, 104].
Animal viral vaccines
Once pathogenicity has been established, the efficacy of vaccination must be shown to provide cross-protection against genetically diverse viral ‘field’ strains. In situations where the challenge viruses are highly diverse, the use of multiple viral strains as vaccine antigens may be considered to widen the breadth of cross-protection.
Given the rapid rate of animal virus genome characterization using deep sequencing and other molecular approaches and the ever wider surveys of domesticated and exotic animal populations, it can be anticipated that a subset of the newly identified viruses will be shown to be pathogenic [15•, 17, 18, 36, 54, 55, 93, 96, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119]. The decision to develop animal viral vaccines will depend largely on economic calculations and/or the need to protect animal and/or human health. The ease of developing vaccines for animals, relative to human vaccines, including direct viral challenges and the requirement for only short term protection, will facilitate the rapid manufacture and testing of novel attenuated, inactivated, or subunit animal viral vaccines. Opportunities for vaccine development protecting farm, companion, and endangered animals should therefore rapidly expand in the near future.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Funding was received from NHLBI R01 HLO83254 and BSRI for this study.
References
- 1.Woolhouse M.E., Howey R., Gaunt E., Reilly L., Chase-Topping M., Savill N. Temporal trends in the discovery of human viruses. Proc Biol Sci. 2008;275:2111–2115. doi: 10.1098/rspb.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson D.E., Reeder D.M. Johns Hopkins University Press; 2005. Mammal Species of the World: A Taxonomic and Geographic Reference. [Google Scholar]
- 3.Fredericks D.N., Relman D.A. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Delwart E. From orphan virus to pathogen: the path to the clinical lab. Curr Opin Virol. 2011;1:282–288. doi: 10.1016/j.coviro.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pike B.L., Saylors K.E., Fair J.N., Lebreton M., Tamoufe U., Djoko C.F., Rimoin A.W., Wolfe N.D. The origin and prevention of pandemics. Clin Infect Dis. 2010;50:1636–1640. doi: 10.1086/652860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coker R., Rushton J., Mounier-Jack S., Karimuribo E., Lutumba P., Kambarage D., Pfeiffer D.U., Stärk K., Rweyemamu M. Towards a conceptual framework to support one-health research for policy on emerging zoonoses. Lancet Infect Dis. 2011;11:326–331. doi: 10.1016/S1473-3099(10)70312-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day M.J. One health: the importance of companion animal vector-borne diseases. Parasit Vectors. 2011;4:49. doi: 10.1186/1756-3305-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daszak P., Epstein J.H., Kilpatrick A.M., Aguirre A.A., Karesh W.B., Cunningham A.A. Collaborative research approaches to the role of wildlife in zoonotic disease emergence. Curr Top Microbiol Immunol. 2007;315:463–475. doi: 10.1007/978-3-540-70962-6_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfe N.D., Daszak P., Kilpatrick A.M., Burke D.S. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg Infect Dis. 2005;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parrish C.R., Holmes E.C., Morens D.M., Park E.C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauck M., Hyeroba D., Tumukunde A., Weny G., Lank S.M., Chapman C.A., O’Connor D.H., Friedrich T.C., Goldberg T.L. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS ONE. 2011;6:e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Pesavento P.A., Shan T., Leutenegger C.M., Wang C., Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Shan T., Li L., Simmonds P., Wang C., Moeser A., Delwart E. The fecal virome of pigs on a high-density farm. J Virol. 2011;85:11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Metagenomics characterization of pig enteric viruses showing very high level of co-infections in healthy and diarrhetic piglets.
- 16.Phan T.G.A.K., Wang B.A., Rose C.A., Lipton R.K.A., Delwart H.L.A., Eric E. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Shan T., Wang C., Côté C., Kolman J., Onions D., Gulland F.M., Delwart E. The fecal viral flora of California sea lions. J Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H., Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Runckel C., Flenniken M.L., Engel J.C., Ruby J.G., Ganem D., Andino R., DeRisi J.L. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS ONE. 2011;6:e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng T.F., Willner D.L., Lim Y.W., Schmieder R., Chau B., Nilsson C., Anthony S., Ruan Y., Rohwer F., Breitbart M. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE. 2011;6:e20579. doi: 10.1371/journal.pone.0020579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J Virol. 2010;84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blomström A.L. Viral metagenomics as an emerging and powerful tool in veterinary medicine. Vet Q. 2011;31:107–114. doi: 10.1080/01652176.2011.604971. [DOI] [PubMed] [Google Scholar]
- 23.van den Brand J.M., van Leeuwen M., Schapendonk C.M., Simon J.H., Haagmans B.L., Osterhaus A.D., Smits S.L. Metagenomic analysis of the viral flora of pine marten and European badger feces. J Virol. 2011;86:2360–2365. doi: 10.1128/JVI.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng T.F., Wheeler E., Greig D., Waltzek T.B., Gulland F., Breitbart M. Metagenomic identification of a novel anellovirus in harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J Gen Virol. 2011;92:1318–1323. doi: 10.1099/vir.0.029678-0. [DOI] [PubMed] [Google Scholar]
- 25.Day J.M., Ballard L.L., Duke M.V., Scheffler B.E., Zsak L. Metagenomic analysis of the turkey gut RNA virus community. Virol J. 2010;7:313. doi: 10.1186/1743-422X-7-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L., Kapoor A., Slikas B., Bamidele O.S., Wang C., Shaukat S., Masroor M.A., Wilson M.L., Ndjango J.B., Peeters M. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blinkova O., Victoria J., Li Y., Keele B.F., Sanz C., Ndjango J.B., Peeters M., Travis D., Lonsdorf E.V., Wilson M.L. Novel circular DNA viruses in stool samples of wild-living chimpanzees. J Gen Virol. 2010;91:74–86. doi: 10.1099/vir.0.015446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge X., Li J., Peng C., Wu L., Yang X., Wu Y., Zhang Y., Shi Z. Genetic diversity of novel circular ssDNA viruses in bats in China. J Gen Virol. 2011;92:2646–2653. doi: 10.1099/vir.0.034108-0. [DOI] [PubMed] [Google Scholar]
- 29.Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D., DeRisi J.L. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S., Schnurr D., Ganem D., Derisi J.L., Boushey H.A. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Greninger A.L., Chen E.C., Sittler T., Scheinerman A., Roubinian N., Yu G., Kim E., Pillai D.R., Guyard C., Mazzulli T. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS ONE. 2010;5:e13381. doi: 10.1371/journal.pone.0013381. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sensitivity of detection of influenza RNA by deep sequencing equal to PCR.
- 32.Rose T.M., Henikoff J.G., Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose T.M., Schultz E.R., Henikoff J.G., Pietrokovski S., McCallum C.M., Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanDevanter D.R., Warrener P., Bennett L., Schultz E.R., Coulter S., Garber R.L., Rose T.M. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu D.K., Poon L.L., Guan Y., Peiris J.S. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H.C., Chu D.K., Liu W., Dong B.Q., Zhang S.Y., Zhang J.X., Li L.F., Vijaykrishna D., Smith G.J., Chen H.L. Detection of diverse astroviruses from bats in China. J Gen Virol. 2009;90:883–887. doi: 10.1099/vir.0.007732-0. [DOI] [PubMed] [Google Scholar]
- 37.Oberste M.S., Maher K., Pallansch M.A. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J Virol. 2004;78:855–867. doi: 10.1128/JVI.78.2.855-867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haible D., Kober S., Jeske H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J Virol Methods. 2006;135:9–16. doi: 10.1016/j.jviromet.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Rector A., Tachezy R., Van Ranst M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J Virol. 2004;78:4993–4998. doi: 10.1128/JVI.78.10.4993-4998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schowalter R.M., Pastrana D.V., Pumphrey K.A., Moyer A.L., Buck C.B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palacios G., Quan P.L., Jabado O.J., Conlan S., Hirschberg D.L., Liu Y., Zhai J., Renwick N., Hui J., Hegyi H. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner S.N., Jaing C.J., McLoughlin K.S., Slezak T.R. A microbial detection array (MDA) for viral and bacterial detection. BMC Genomics. 2010;11:668. doi: 10.1186/1471-2164-11-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu C.Y., Greninger A.L., Kanada K., Kwok T., Fischer K.F., Runckel C., Louie J.K., Glaser C.A., Yagi S., Schnurr D.P. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A. 2008;105:14124–14129. doi: 10.1073/pnas.0805968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kistler A.L., Gancz A., Clubb S., Skewes-Cox P., Fischer K., Sorber K., Chiu C.Y., Lublin A., Mechani S., Farnoushi Y. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol J. 2008;5:88. doi: 10.1186/1743-422X-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A. 2001;98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allander T., Andreasson K., Gupta S., Bjerkner A., Bogdanovic G., Persson M.A., Dalianis T., Ramqvist T., Andersson B. Identification of a third human polyomavirus. J Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greninger A.L., Runckel C., Chiu C.Y., Haggerty T., Parsonnet J., Ganem D., Derisi J.L. The complete genome of klassevirus — a novel picornavirus in pediatric stool. Virol J. 2009;6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honkavuori K.S., Shivaprasad H.L., Williams B.L., Quan P.L., Hornig M., Street C., Palacios G., Hutchison S.K., Franca M., Egholm M. Novel borna virus in psittacine birds with proventricular dilatation disease. Emerg Infect Dis. 2008;14:1883–1886. doi: 10.3201/eid1412.080984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Palacios G., Lovoll M., Tengs T., Hornig M., Hutchison S., Hui J., Kongtorp R.T., Savji N., Bussetti A.V., Solovyov A. Heart and skeletal muscle inflammation of farmed salmon is associated with infection with a novel reovirus. PLoS ONE. 2010;5:e11487. doi: 10.1371/journal.pone.0011487. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fine example of associating a novel virus with a disease outbreak using case/control prevalence and detection of antigens in affected tissues.
- 51.Tang P., Chiu C. Metagenomics for the discovery of novel human viruses. Future Microbiol. 2010;5:177–189. doi: 10.2217/fmb.09.120. [DOI] [PubMed] [Google Scholar]
- 52.Delwart E.L. Viral metagenomics. Rev Med Virol. 2007;17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Bexfield N., Kellam P. Metagenomics and the molecular identification of novel viruses. Vet J. 2010 doi: 10.1016/j.tvjl.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of molecular methods for viral discovery.
- 54.Blomström A.L., Widén F., Hammer A.S., Belák S., Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010;48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blomström A.L., Belák S., Fossum C., McKillen J., Allan G., Wallgren P., Berg M. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009;146:125–129. doi: 10.1016/j.virusres.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Parrish C.R., Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol. 2005;59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 57.Hoelzer K., Shackelton L.A., Parrish C.R., Holmes E.C. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J Gen Virol. 2008;89:2280–2289. doi: 10.1099/vir.0.2008/002055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Hoelzer K., Parrish C.R. The emergence of parvoviruses of carnivores. Vet Res. 2010;41:39. doi: 10.1051/vetres/2010011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed review of the emergence of a mammalian pathogen following cross-species transmission.
- 59.Allison A.B., Harbison C.E., Pagan I., Stucker K.M., Kaelber J.T., Brown J.D., Ruder M.G., Keel M.K., Dubovi E.J., Holmes E.C., Parrish C.R. Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus. J Virol. 2012;86:865–872. doi: 10.1128/JVI.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keele B.F., Van Heuverswyn F., Li Y., Bailes E., Takehisa J., Santiago M.L., Bibollet-Ruche F., Chen Y., Wain L.V., Liegeois F. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Sharp P.M., Hahn B.H. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B: Biol Sci. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed review of the emergence of HIV1 and HIV2 by cross-species transmission.
- 62.Plantier J.C., Leoz M., Dickerson J.E., De Oliveira F., Cordonnier F., Lemée V., Damond F., Robertson D.L., Simon F. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 63.Neel C., Etienne L., Li Y., Takehisa J., Rudicell R.S., Bass I.N., Moudindo J., Mebenga A., Esteban A., Van Heuverswyn F. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J Virol. 2010;84:1464–1476. doi: 10.1128/JVI.02129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharp P.M., Shaw G.M., Hahn B.H. Simian immunodeficiency virus infection of chimpanzees. J Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webby R.J., Webster R.G. Emergence of influenza A viruses. Philos Trans R Soc Lond B: Biol Sci. 2001;356:1817–1828. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster R.G. The importance of animal influenza for human disease. Vaccine. 2002;20(Suppl. 2):S16–S20. doi: 10.1016/s0264-410x(02)00123-8. [DOI] [PubMed] [Google Scholar]
- 67••.Chen E.C., Yagi S., Kelly K.R., Mendoza S.P., Tarara R.P., Canfield D.R., Maninger N., Rosenthal A., Spinner A., Bales K.L. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]; Recent example of a cross-species transmission of a virus into monkeys and humans.
- 68.Purdy M.A., Khudyakov Y.E. The molecular epidemiology of hepatitis E virus infection. Virus Res. 2011;161:31–39. doi: 10.1016/j.virusres.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 69.Meng X.J. From barnyard to food table: The omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011;161:23–30. doi: 10.1016/j.virusres.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolfe N.D., Heneine W., Carr J.K., Garcia A.D., Shanmugam V., Tamoufe U., Torimiro J.N., Prosser A.T., Lebreton M., Mpoudi-Ngole E. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci U S A. 2005;102:7994–7999. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Switzer W.M., Bhullar V., Shanmugam V., Cong M.E., Parekh B., Lerche N.W., Yee J.L., Ely J.J., Boneva R., Chapman L.E. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J Virol. 2004;78:2780–2789. doi: 10.1128/JVI.78.6.2780-2789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Betsem E., Rua R., Tortevoye P., Froment A., Gessain A. Frequent and recent human acquisition of simian foamy viruses through apes’ bites in central Africa. PLoS Pathog. 2011;7:e1002306. doi: 10.1371/journal.ppat.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]; Frequent detection of non-human primate foamy retroviruses nucleic acids and antibodies in Cameroonian hunters.
- 73.Mouinga-Ondémé A., Caron M., Nkoghé D., Telfer P., Marx P., Saïb A., Leroy E., Gonzalez J.P., Gessain A., Kazanji M. Cross-species transmission of simian foamy virus to humans in rural Gabon, central Africa. J Virol. 2012;86:1255–1260. doi: 10.1128/JVI.06016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolfe N.D., Switzer W.M., Carr J.K., Bhullar V.B., Shanmugam V., Tamoufe U., Prosser A.T., Torimiro J.N., Wright A., Mpoudi-Ngole E. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- 75.Calattini S., Betsem E.B., Froment A., Mauclere P., Tortevoye P., Schmitt C., Njouom R., Saib A., Gessain A. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg Infect Dis. 2007;13:1314–1320. doi: 10.3201/eid1309.061162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pavia A.T. Laboratory creation of a highly transmissible H5N1 influenza virus: balancing substantial risks and real benefits. Ann Intern Med. 2012 doi: 10.7326/0003-4819-156-6-201203200-00386. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 77.Suguitan A.L., Matsuoka Y., Lau Y.F., Santos C.P., Vogel L., Cheng L.I., Orandle M., Subbarao K. The multi-basic cleavage site of the hemagglutinin of the highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J Virol. 2011 doi: 10.1128/JVI.05546-11. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78•.Schrauwen E.J., Herfst S., Leijten L.M., van Run P., Bestebroer T.M., Linster M., Bodewes R., Kreijtz J.H., Rimmelzwaan G.F., Osterhaus A.D. The multi basic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. J Virol. 2012 doi: 10.1128/JVI.06828-11. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]; Study showing that mutations at the HA cleavage site can affect in vivo distribution of virus in ferrets.
- 79••.Streicker D.G., Turmelle A.S., Vonhof M.J., Kuzmin I.V., McCracken G.F., Rupprecht C.E. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]; Cross-species rabies transmission more common among closely related bat species.
- 80.Tadmor A.D., Ottesen E.A., Leadbetter J.R., Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Longdon B., Hadfield J.D., Webster C.L., Obbard D.J., Jiggins F.M. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog. 2011;7:e1002260. doi: 10.1371/journal.ppat.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murphy H.W., Miller M., Ramer J., Travis D., Barbiers R., Wolfe N.D., Switzer W.M. Implications of simian retroviruses for captive primate population management and the occupational safety of primate handlers. J Zoo Wildl Med. 2006;37:219–233. doi: 10.1638/05-110.1. [DOI] [PubMed] [Google Scholar]
- 83•.Weaver S.C., Reisen W.K. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of factors influencing arbovirus outbreaks.
- 84••.Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J.A., Firth C., Hirschberg D.L., Rice C.M., Shields S., Lipkin W.I. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome characterization of the flavivirus most closely related to the common human pathogen HCV.
- 85.Kapoor A., Simmonds P., Dubovi E.J., Qaisar N., Henriquez J.A., Medina J., Shields S., Lipkin W.I. Characterization of a canine homolog of human Aichivirus. J Virol. 2011;85:11520–11525. doi: 10.1128/JVI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones M.S., Kapoor A., Lukashov V.V., Simmonds P., Hecht F., Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kapoor A., Simmonds P., Slikas E., Li L., Bodhidatta L., Sethabutr O., Triki H., Bahri O., Oderinde B.S., Baba M.M. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis. 2010;201:1633–1643. doi: 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapoor A., Slikas E., Simmonds P., Chieochansin T., Naeem A., Shaukat S., Alam M.M., Sharif S., Angez M., Zaidi S., Delwart E. A newly identified bocavirus species in human stool. J Infect Dis. 2009;199:196–200. doi: 10.1086/595831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arthur J.L., Higgins G.D., Davidson G.P., Givney R.C., Ratcliff R.M. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 2009;5:e1000391. doi: 10.1371/journal.ppat.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kapoor A., Li L., Victoria J., Oderinde B., Mason C., Pandey P., Zaidi S.Z., Delwart E. Multiple novel astrovirus species in human stool. J Gen Virol. 2009;90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finkbeiner S.R., Li Y., Ruone S., Conrardy C., Gregoricus N., Toney D., Virgin H.W., Anderson L.J., Vinjé J., Wang D., Tong S. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol. 2009;83:10836–10839. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finkbeiner S.R., Kirkwood C.D., Wang D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J. 2008;5:117. doi: 10.1186/1743-422X-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sharp C.P., Lebreton M., Kantola K., Nana A., Diffo J.L., Djoko C.F., Tamoufe U., Kiyang J.A., Babila T.G. Widespread infection of chimpanzees and gorillas with homologues of human parvoviruses B19, PARV4 and human bocavirus in the wild. J Virol. 2010;84:10289–10296. doi: 10.1128/JVI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tse H., Tsoi H.W., Teng J.L., Chen X.C., Liu H., Zhou B., Zheng B.J., Woo P.C., Lau S.K., Yuen K.Y. Discovery and genomic characterization of a novel ovine partetravirus and a new genotype of bovine partetravirus. PLoS ONE. 2011;6:e25619. doi: 10.1371/journal.pone.0025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lau S.K., Woo P.C., Tse H., Fu C.T., Au W.K., Chen X.C., Tsoi H.W., Tsang T.H., Chan J.S., Tsang D.N. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J Gen Virol. 2008;89:1840–1848. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- 96.Kapoor A., Mehta N., Esper F., Poljsak-Prijatelj M., Quan P.L., Qaisar N., Delwart E., Lipkin W.I. Identification and characterization of a new bocavirus species in gorillas. PLoS ONE. 2010;5:e11948. doi: 10.1371/journal.pone.0011948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Belyi V.A., Levine A.J., Skalka A.M. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and circoviridae are more than 40 to 50 million years old. J Virol. 2010;84:12458–12462. doi: 10.1128/JVI.01789-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Horie M., Honda T., Suzuki Y., Kobayashi Y., Daito T., Oshida T., Ikuta K., Jern P., Gojobori T., Coffin J.M., Tomonaga K. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99••.Katzourakis A., Gifford R.J. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]; Large-scale detection of RNA and DNA viruses integrated in animal genomes bring doubt to recent viral origins calculations using molecular clocks.
- 100.Holmes E.C. Molecular clocks and the puzzle of RNA virus origins. J Virol. 2003;77:3893–3897. doi: 10.1128/JVI.77.7.3893-3897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holmes E.C. What does virus evolution tell us about virus origins? J Virol. 2011;85:5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Worobey M., Telfer P., Souquière S., Hunter M., Coleman C.A., Metzger M.J., Reed P., Makuwa M., Hearn G., Honarvar S. Island biogeography reveals the deep history of SIV. Science. 2010;329:1487. doi: 10.1126/science.1193550. [DOI] [PubMed] [Google Scholar]
- 103.Haagmans B.L., Andeweg A.C., Osterhaus A.D. The application of genomics to emerging zoonotic viral diseases. PLoS Pathog. 2009;5:e1000557. doi: 10.1371/journal.ppat.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kistler A.L., Smith J.M., Greninger A.L., Derisi J.L., Ganem D. Analysis of naturally occurring avian bornavirus infection and transmission during an outbreak of proventricular dilatation disease among captive psittacine birds. J Virol. 2010;84:2176–2179. doi: 10.1128/JVI.02191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reuter G., Pankovics P., Delwart E., Boros A. Identification of a novel astrovirus in domestic sheep in Hungary. Arch Virol. 2011 doi: 10.1007/s00705-011-1151-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li L., Pesavento P.A., Woods L., Clifford D.L., Luff J., Wang C., Delwart E. Novel amdovirus in gray foxes. Emerg Infect Dis. 2011;17:1876–1878. doi: 10.3201/eid1710.110233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li L., Pesavento P., Shan T., Leutenegger C.M., Wang C., Delwart E. Viruses in diarrhetic dogs include novel kobuviruses and sapoviruses. J Gen Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li L., Shan T., Soji O.B., Alam M.M., Kunz T.H., Zaidi S.Z., Delwart E. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol. 2011;92:768–772. doi: 10.1099/vir.0.028704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reuter G., Pankovics P., Boros A. Identification of a novel astrovirus in a domestic pig in Hungary. Arch Virol. 2010;156:125–128. doi: 10.1007/s00705-010-0827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rivera R., Nollens H.H., Venn-Watson S., Gulland F.M., Wellehan J.F. Characterization of phylogenetically diverse astroviruses of marine mammals. J Gen Virol. 2010;91:166–173. doi: 10.1099/vir.0.015222-0. [DOI] [PubMed] [Google Scholar]
- 111.Smits S.L., van Leeuwen M., Kuiken T., Hammer A.S., Simon J.H., Osterhaus A.D. Identification and characterization of deer astroviruses. J Gen Virol. 2010;91:2719–2722. doi: 10.1099/vir.0.024067-0. [DOI] [PubMed] [Google Scholar]
- 112.Luo Z., Roi S., Dastor M., Gallice E., Laurin M.A., L’homme Y. Multiple novel and prevalent astroviruses in pigs. Vet Microbiol. 2010;149:316–323. doi: 10.1016/j.vetmic.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Toffan A., Jonassen C.M., De Battisti C., Schiavon E., Kofstad T., Capua I., Cattoli G. Genetic characterization of a new astrovirus detected in dogs suffering from diarrhoea. Vet Microbiol. 2009 doi: 10.1016/j.vetmic.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jonassen C.M., Jonassen T.TØ, Sveen T.M., Grinde B. Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res. 2003;91:195–201. doi: 10.1016/s0168-1702(02)00269-1. [DOI] [PubMed] [Google Scholar]
- 115.Kapoor A., Mehta N., Dubovi E.J., Simmonds P., Govindasamy L., Medina J.L., Street C., Shields S., Lipkin W.I. Characterization of novel canine bocaviruses and their association with respiratory disease. J Gen Virol. 2012;93:341–346. doi: 10.1099/vir.0.036624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shan T., Lan D., Li L., Wang C., Cui L., Zhang W., Hua X., Zhu C., Zhao W., Delwart E. Genomic characterization and high prevalence of bocaviruses in swine. PLoS ONE. 2011;6:e17292. doi: 10.1371/journal.pone.0017292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheung A.K., Wu G., Wang D., Bayles D.O., Lager K.M., Vincent A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch Virol. 2010;155:801–806. doi: 10.1007/s00705-010-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng W.X., Li J.S., Huang C.P., Yao D.P., Liu N., Cui S.X., Jin Y., Duan Z.J. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS ONE. 2010;5:e13583. doi: 10.1371/journal.pone.0013583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rijks J.M., Read F.L., van de Bildt M.W., van Bolhuis H.G., Martina B.E., Wagenaar J.A., van der Meulen K., Osterhaus A.D., Kuiken T. Quantitative analysis of the 2002 phocine distemper epidemic in the Netherlands. Vet Pathol. 2008;45:516–530. doi: 10.1354/vp.45-4-516. [DOI] [PubMed] [Google Scholar]


