Abstract
Slug (Snai2), a member of the Snail family of zinc finger transcription factors, plays a role in the epithelial-to-mesenchymal transformation (EMT) that occurs during melanocyte emigration from the neural crest. A role for Slug in the EMT-like loss of cell adhesion and increased cell motility exhibited during melanoma progression has also been proposed. Our immunohistochemical studies of melanoma arrays, however, revealed that Slug expression was actually higher in nevi than in primary or metastatic melanomas. Moreover, Slug expression in melanomas was not associated with decreased expression of E-cadherin, the canonical Slug target in EMT. Comparisons of endogenous Slug and E-cadherin expression in cultured normal human melanocytes and melanoma cell lines supported our immunohistochemical findings. Expression of exogenous Slug in melanocytes and melanoma cells in vitro, however, suppressed E-cadherin expression, enhanced N-cadherin expression, and stimulated cell migration and invasion. Interestingly, both in tumors and cultured cell lines, there was a clear relationship between expression of Slug and MITF, a transcription factor known to regulate Slug expression during development. Taken together, our findings suggest that Slug expression during melanomagenesis is highest early in the process and that persistent Slug expression is not required for melanoma progression. The precise role of Slug in melanomagenesis remains to be elucidated and may be related to its interactions with other drivers of EMT, such as Snail.
Snail (Snai1) and Slug (Snai2), evolutionarily conserved members of the Snail family of zinc finger transcription factors, play an important role in embryonic development.1–4 They regulate the process of epithelial-mesenchymal transformation (EMT), characterized by loss of intercellular adhesions and acquisition of a migratory phenotype.5,6 EMT is essential for a variety of developmental processes in vertebrates, including mesoderm formation during gastrulation and neural crest cell migration.7 Slug is expressed in both premigratory and migratory neural crest cells in the chick, but only in migratory neural crest cells in the mouse. Conversely, Snail is expressed only in migratory neural crest cells in the chick, but in both premigratory and migratory cell populations in the mouse. These findings suggest that the site and timing of expression of the two Snail family members have been exchanged in the avian compared to the mammalian lineage. Overexpressing Slug and Snail in the chick embryo hindbrain demonstrates that Slug and Snail drive very similar events in neural crest formation, despite these differences in expression patterns.8
Snail appears to be essential for normal mesoderm formation in many species, and Snail knockout mice die early during gestation.9–11 Treatment of the chick embryo with Slug antisense oligonucleotides impairs both mesoderm and neural crest delamination, thus Slug is essential for normal embryogenesis in this species.9 Slug knockout mice, on the other hand, are viable and undergo relatively normal gastrulation and neural crest cell differentiation, although it appears that melanocyte migration and maturation are compromised in the absence of Slug.1,12 Slug knockout mice exhibit hypopigmentation at their extremities and on their heads reminiscent of Waardenberg syndrome in humans, a condition characterized by the absence of melanocytes leading to heterochromia irides, hypopigmentation, and hearing loss.12–14 In humans, deletions of Slug have been associated with Waardenberg syndrome.14 Mutations in the MITF, endothelin 3, or its receptor EDNRB genes also produce Waardenberg syndrome.15,16 Slug has been shown to be a target of MITF regulation in Xenopus, and Slug may also reciprocally regulate MITF in this species.17 Piebaldism, a heritable human condition characterized by patchy depigmented areas of skin is usually due to KIT gene mutations, but it has also been linked to Slug deletion.18 Studies in mice have shown that Slug is a downstream target of Kit activation.12 A further link between Slug and melanocytes is provided by the observation that overexpression of Slug in Xenopus embryos results in increased numbers of melanocytes.19
The Snail family also appears to play a role in melanocyte transformation. Slug and Snail are expressed at high levels in a variety of tumor types, including melanoma. In melanoma cells in vitro, Slug and Snail drive EMT-like processes that would be expected to enhance tumor progression, invasion, and metastasis in vivo.20–23 Expression of Snail has been reported to be elevated in melanoma cells compared to untransformed melanocytes.24,25 In melanoma cells, expression of Snail is associated with loss of cell-cell adhesion mediated by E-cadherin, increased motility, and enhanced expression of proteases.24–28 Slug is reportedly expressed at comparable levels in melanocytes and in melanoma cells in vitro, although the expression of Slug is essential for metastasis of murine melanoma allografts in nude mice.23,29 Moreover, expression of Slug also increases resistance of human melanoma cells to chemotherapeutic drugs.22,30 It has been suggested that the role of Slug in melanocyte migration during development predisposes melanomas to metastasize via a Slug-dependent mechanism.29
In the present studies, we examined the expression of Slug protein by immunohistochemistry in nevi, primary melanomas, and metastatic melanomas. Slug expression was significantly higher in nevi compared to primary and metastatic melanomas. Expression of Slug was not correlated with loss of protein expression of its best known target gene E-cadherin, but was positively associated with protein expression of MITF, a known upstream regulator of Slug expression. In vitro studies showed that Slug mRNA and protein expression was higher in primary melanocytes than in melanoma cell lines. These findings suggest that Slug expression during melanomagenesis is highest early during the process and that persistent Slug expression is not required for the maintenance of invasive or metastatic behavior in melanomas.
Materials and Methods
Tissue Samples
A commercial tissue array (commercial array) consisting of triplicate samples of 30 cases of primary melanoma (10 acral, 17 nonacral, 1 mucosal, and 2 from unspecified sites) and 30 cases of metastatic melanoma was purchased (ME207) (US Biomax, Rockville, MD). A second, previously described, custom melanocytic tumor progression array (custom array) was composed of 36 nevi, 57 primary melanomas, and 75 melanoma metastases (two to six tissue cores per case).31 This array was prepared as a collaboration of Skin SPOREs sponsored by the Organ Systems Branch of the National Cancer Institute. Tumor samples consisting of melanomas with residual nevus cells were obtained from the Tissue Resource and Pathology Core (Melanoma Tumor Bank) of the MD Anderson Melanoma Specialized Program of Research Excellence (SPORE) project. This study was approved by the University of Texas MD Anderson Cancer Center Institutional Review Board (protocol no. LAB11-0081) and was conducted according to the Health Insurance and Portability and Accountability Act guidelines. A written informed consent was obtained from all subjects.
Immunohistochemistry
Both arrays were stained for Slug (#9585; Cell Signaling Technology, Danvers, MA), E-cadherin (#4065; Cell Signaling Technology), and MITF (D5) (Thermo Fisher Scientific, Rockford, IL). For Slug and E-cadherin immunohistochemistry, slides were deparaffinized and rehydrated, endogenous peroxidase activity was blocked with 3% H2O2 in water, antigen retrieval was performed in 10 mmol/L citrate buffer, and nonspecific binding was blocked with Biocare Blocking Reagent (Concord, CA). Slug antibody was applied at a dilution of 1:50 for an overnight incubation at 4°C, followed by 30-minutes incubation at room temperature with Biocare Rabbit on Rodent HRP-Polymer. E-cadherin antibody was diluted 1:50 and applied for 1 hour at room temperature; Envision Plus labeled anti-rabbit HRP polymer (Dako, Carpinteria, CA) was then applied for 30 minutes at room temperature. The chromagen used was 3,3′-diaminobenzidine. Immunohistochemistry for MITF on the commercial array was performed at a primary antibody dilution of 1:40. The Bond polymer detection system and an automated stainer (Leica Microsystems, Newcastle upon Tyne, UK) were used to detect immunohistochemical reactivity, following the manufacturer's guidelines. Immunohistochemistry for MITF on the custom array was performed as previously described.31
Scoring
Each tissue array was examined by two pathologists: one American Board of Pathology certified physician (VG, LD, or CTC) and one American College of Veterinary Pathology certified veterinary pathologist (DK). All cases stained by single immunohistochemical stain were evaluated by a single primary pathologist; scores assigned by the primary examiner were verified by a second pathologist and a single final score was obtained for each tumor. Tissues were scored on a scale of 0 to 3 based on the combined extent and intensity of staining. The final score for a case represented the predominant staining pattern in all tissue cores for that case. E-cadherin staining intensity was based on total E-cadherin staining, including both cytoplasmic and membrane localized staining. In addition, the predominant pattern of E-cadherin staining, whether localized to cell membranes or intracytoplasmic, was determined for each tumor scored as E-cadherin positive.
Cell Culture, Adenoviral Transduction, and Immunofluorescence
Normal human melanocytes (NHM) of neonatal origin were obtained from the ATCC (Manassas, VA) and were maintained in Dermal Basal Medium supplemented with a melanocyte growth kit (ATCC). Melanoma cell lines (WC62 and WM115) were obtained from the Coriell Institute (Camden, NJ) and maintained in Eagle's Modified Essential Medium (ATCC) supplemented with 10% normal calf serum (Thermo Scientific, Hudson, NJ). Cells were grown to 75% confluence then incubated with a Slug-expressing adenovirus Slug (Ad-Slug) or a control adenovirus (Ad-Null from Vector Biolabs, Philadelphia, PA).32 For transduction, 10 μl of adenoviral stock (viral titer 108 ifu/mL) mixed with 1 mL of medium was added to each well of a 6-well plate. Cells were incubated with the adenovirus for 24 hours before the medium was changed. After cell harvest, mRNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) and protein was isolated using radioimmunoprecipitation assay buffer supplemented with Halt protease inhibitor cocktail (Thermo Scientific). For immunofluorescence studies, NHM transduced with Ad-Null or Ad-Slug were grown in chamber slides, fixed briefly with neutral buffered formalin, and thoroughly rinsed with PBS. After blocking for an hour with 5% normal goat serum and 0.3% Triton X-100 in PBS, E-cadherin antibody (#4065) (Cell Signaling Technology) diluted 1:200 in 1% bovine serum albumin and 0.3% Triton X-100 in PBS was applied for overnight incubation at 4°C. The next day, cells were rinsed and secondary antibody (goat anti-rabbit IgG) (H+L), F(ab′)2 fragment, Alexa Fluor 488 Conjugate (#4412; Cell Signaling Technology) diluted 1:1000 in 1% bovine serum albumin and 0.3% Triton X-100 in PBS was applied for an hour. Slides were rinsed and coverslipped using Vectashield mounting medium (Vector Laboratories, Burlingame, CA). All incubations were performed in the dark.
Western Blot
Whole cell lysates in RIPA buffer plus Halt protease inhibitor cocktail (Thermo Scientific) were cleared by centrifuging at 10,000 rpm for 10 minutes at 4°C. Forty μg of protein was loaded on 4% to 20% Tris-HCL gel (BioRad, Hercules, CA). Following SDS-PAGE, protein was transferred to polyvinylidene difluoride membrane (Sigma-Aldrich, St. Louis, MO) and probed for Slug (#9585; Cell Signaling Technology), Snail (#3895; Cell Signaling Technology), E-cadherin (#4065; Cell Signaling Technology), N-cadherin (antibody 610920; BD Biosciences, San Jose, CA), MITF (ab73930; Abcam, Cambridge, MA), or GAPDH (ab9485; Abcam). The secondary antibody used for Slug, E-cadherin, MITF, and GAPDH detection was anti-rabbit IgG horseradish peroxidase (NA934V; GE Healthcare Life Sciences, Piscataway, NJ). The secondary antibody used for N-cadherin and Snail was anti-mouse IgG horseradish peroxidase (#7076; Cell Signaling Technology). Primary antibodies were used at 1:1000 dilutions, whereas secondary antibody concentrations dilutions were 1:5000. Blots were incubated overnight at 4°C with primary antibody and 2 hours at room temperature with secondary antibodies. Membranes were incubated in Super Signal West ECL (Thermo Fisher Scientific) and then exposed to film.
Quantitative RT-qPCR
For mRNA analysis, RNA was isolated using the RNeasy kit from Qiagen and cDNA was generated using the High Capacity cDNA RT kit from Applied Biosystems (Foster City, CA). Following reverse transcription, qPCR was performed using TaqMan gene expression assays for Snai1 (Hs00195591), Snai2 (Hs00950344), Cdh1 (E-cadherin; Hs01023814), Cdh2 (N-cadherin; Hs00362037), and RN18S1 (18S; Hs03928990) together with TaqMan Universal Master Mix (4304437; Applied Biosystems). Amplifications were performed on a 7900HT Real-Time PCR Analyzer (Applied Biosystems) using the standard curve method for quantitation.33
Proliferation Assays
Cells were seeded in 96 well dishes and transduced with Ad-Slug or Ad-Null 24 hours after plating. On successive days, 3-[4,5-dimethylthiazol-2-yl]−2,5-diphenyl tetrazolium bromide (MTT) (Sigma-Aldrich) was added and cells were incubated for 3 hours. MTT crystals were dissolved in MTT solvent (0.1 N HCl in anhydrous isopropanol) and absorbance was measured at 570 nm. Concurrently, cell numbers were counted manually by the Trypan Blue exclusion method.
Migration Assays
NHM were transduced with Ad-Slug or Ad-Null and plated into uncoated or Matrigel-coated (Becton, Dickinson and Company Biosystems, Bedford, MA) transwell chambers containing complete medium 48 hours later. After 24 hours, upper chambers were cleaned of remaining cells and membranes were fixed and stained to visualize migrating cells (DiffQuick Kit, Sigma-Aldrich). Dye was extracted from transwell membranes using methanol and absorbance measured with a spectrophotometer at 550 nm. For cell migration assays, cells were transduced with Ad-Slug or Ad-Null; 24 hours after infection, a linear defect was generated in cell monolayers using a 10 μL pipette tip. Images were captured on days 0, 1 and 3 to monitor cell migration. All migration assays were conducted in the presence of 10 mg/mL mitomycin C (Sigma-Aldrich) to block proliferation.
Statistical Analysis
For statistical comparison of tumor immunohistochemistry, cases were classified as negative (scores of 0 or 1) or positive (scores of 2 or 3). Pairwise associations between positive staining and tumor stage and between positive staining for different markers were determined by Fisher's exact test. Values for RT-qPCR and migration and invasion assays are the average of two to three independent studies, each performed in triplicate; groups were compared using Student's t-test. Oncomine (Compendia Bioscience, Ann Arbor, MI) was used for analysis of previously published and publically available RNA microarray studies.
Results
Progression-Related Slug Expression in Melanocytic Tumors
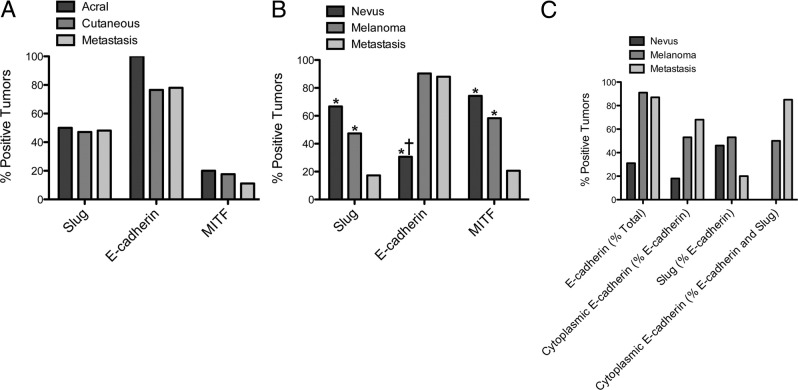
As shown in Figure 1A, approximately 50% of all melanomas in the commercial array were Slug positive. There was no difference between acral and nonacral melanomas or between primary and metastatic melanomas. This commercial array contained a fairly small number of nonacral melanomas and no samples of nevi, and therefore we examined a more comprehensive custom array. When this custom array was examined, there was a clear difference in Slug expression in nevi, primary melanomas, and metastatic melanomas. Sixty-seven percent of nevi were Slug positive, compared to 47% of primary cutaneous melanomas and 17% of metastatic melanomas (Figure 1B). Differences in Slug protein expression between nevi and metastatic melanomas (P ≤ 0.0001) and between primary and metastatic melanomas (P ≤ 0.0002) were highly significant, and the difference between nevi and primary melanomas (P = 0.0534) approached significance. In this regard, it is important to note that immunostaining for Slug in the present studies was performed using a highly specific antibody, whereas previous studies often used less specific antibodies that give spurious cytoplasmic staining.20,34
Figure 1.
Immunohistochemistry demonstrates that expression of Slug, E-cadherin, and MITF protein in melanomas is altered during tumor progression. A: Significant differences were not found in expression of Slug, E-cadherin, and MITF in primary acral, primary nonacral, and metastatic melanomas when a commercial array was evaluated. B: In a custom array, there were significant differences among nevi, primary cutaneous melanomas, and metastatic melanomas in expression of Slug, E-cadherin, and MITF protein. For both tissue arrays, immunohistochemistry was performed, and tumors were scored as negative (0 or + 1 staining) or positive (+2 or +3 staining), and results were compared in a pairwise fashion by Fisher's exact test *P < 0.05 for difference from metastatic melanoma; †P < 0.05 for difference from primary melanoma). C: E-cadherin protein localization was evaluated in the custom array. The percentage of all tumors that were E-cadherin positive was lowest in nevi. The percentage of E-cadherin-positive tumors with predominantly membrane E-cadherin localization decreased as a function of tumor progression. The percentage of E-cadherin-positive tumors that were also Slug positive was lowest in metastatic melanomas. In tumors that were both Slug and E-cadherin positive, membrane localization of E-cadherin decreased with tumor progression. None of these associations was statistically significant.
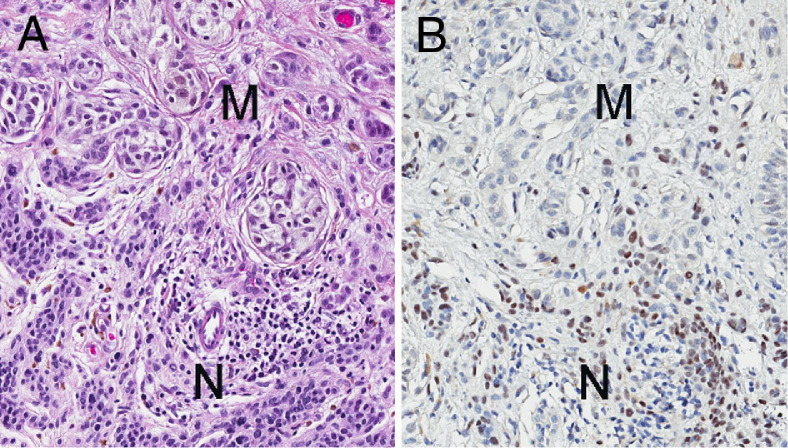
To further investigate the relationship between Slug protein expression in nevi versus primary melanomas, expression of Slug was compared in four different melanomas that arose within nevi. For these tumors, slides were available that included both melanoma and residual nevus cells, allowing direct comparison of Slug staining in the two cell populations. In three of these tumors, Slug staining was clearly more intense in nevus cells compared to melanoma cells (Figure 2); in one of the four tumors, melanoma cells stained more strongly for Slug than residual nevus cells. These findings suggest that Slug protein expression in nevi is often lost or reduced as the nevi undergo malignant transformation.
Figure 2.
In most melanomas with residual nevus cells, Slug immunoreactivity is strongest in nevus cells. A: In this H&E-stained section of cutaneous melanoma, both melanoma (M) and nevus (N) cells can be identified. B: Immunohistochemical staining of an adjacent section of the same tumor shows stronger Slug immunoreactivity in nevus than in melanoma cells. The small lymphocytes admixed with nevus cells are Slug-negative. A similar pattern of staining was seen in three of four melanomas containing residual nevus cells.
Expression of Slug-Related Genes in Melanocytic Tumors
E-cadherin (Cdh1) is perhaps the best studied Slug target gene. Slug directly represses the E-cadherin promoter in a variety of cell types and elevated Slug expression has been linked to decreased E-cadherin expression and tumor progression in several tumor types.6,35–41 Moreover, loss of E-cadherin is a critical hallmark of EMT.42 In the commercial array, 77% to 100% of tumors expressed E-cadherin protein (Figure 1A), and there was no difference in E-cadherin expression among the different tumor types. Moreover, there was no significant association between Slug and E-cadherin protein expression in any tumor type. In the custom array, there were significantly fewer E-cadherin expressing nevi than primary (P ≤ 0.0001) or metastatic melanomas (P ≤ 0.0002), but there was no difference between primary and metastatic melanomas (Figure 1B). Surprisingly, there was no significant association between Slug and E-cadherin protein expression at any stage of tumor progression, in either the commercial or custom array.
Localization of E-cadherin in E-cadherin-positive tumors was also evaluated (Figure 1C). The number of melanomas that could be evaluated in the commercial array was too small to permit analysis; however, when the larger custom array was examined, clear trends of E-cadherin localization emerged. The proportion of tumors, with cytoplasmic as opposed to membrane localization of E-cadherin, increased with tumor stage from 18% in nevi to 55% in primary melanomas and 68% in metastatic melanomas. This was consistent with previous reports that loss of membrane-bound E-cadherin is associated with invasiveness in melanomas.43 Among the E-cadherin-positive tumors, approximately 50% of nevi and primary melanomas were Slug positive, but only 20% of metastatic melanomas were Slug positive, suggesting that co-expression of E-cadherin and Slug protein was lost as tumors progressed. In addition, the proportion of Slug-positive tumors with cytoplasmic E-cadherin localization increased dramatically with tumor progression from 0% in nevi to 50% in primary melanomas to 85% in metastatic melanomas. This was consistent with previous reports of Slug-dependent E-cadherin internalization in other cell types.44,45 These associations between Slug expression and E-cadherin localization, although suggestive, were not statistically significant. Taken together, our results indicated that there was no simple relationship between Slug protein expression and E-cadherin protein expression or localization in human melanocytic tumors.
MITF, the master regulator of melanogenesis, has been reported both to transcriptionally regulate Slug expression and to be a potential target of Slug regulation.14 In the present study, MITF protein was expressed in 11% to 20% of tumors on the commercial array, with expression highest in acral melanomas and lowest in metastatic melanomas; differences, however were not statistically significant. There was no significant relationship between Slug and MITF protein expression. MITF positivity was higher in the custom array, ranging from 21% of metastatic melanomas to 58% of primary melanomas to 77% of nevi (Figure 1B), and differences between expression in nevi versus metastatic melanomas (P ≤ 0.0001) and primary melanomas versus metastatic melanomas (P ≤ 0.0001) were significant. MITF expression paralleled Slug expression (Figure 1B), and there was a significant positive association between Slug and MITF protein in nevi (P = 0.0239) and primary melanomas (P = 0.0038).
Slug Expression in Melanocytes and Melanoma Cells
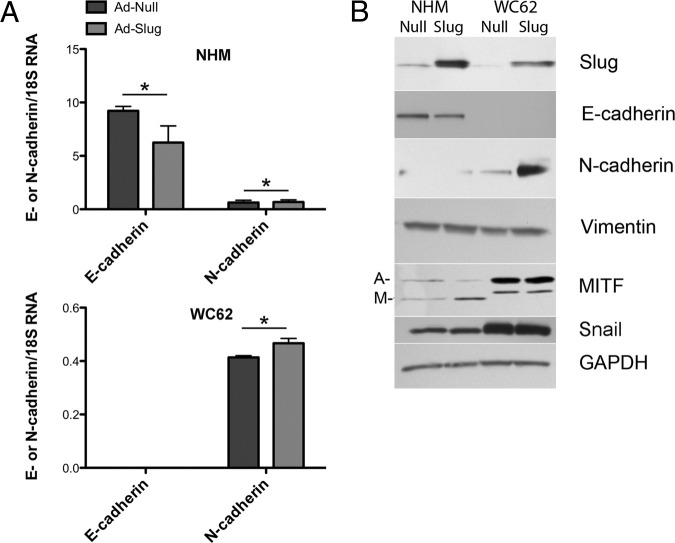
The finding that Slug protein expression was actually higher in nevi than melanomas was somewhat unexpected, although few nevi have been previously examined for Slug expression.46 This suggested that Slug might play its most important role early in melanoma progression (ie, in the emergence of nevi rather than in the acquisition of invasive and metastatic characteristics). Thus, Slug expression was compared in NHM of neonatal origin and primary (WM115) and metastatic (WC62) human melanoma cell lines. Primary and metastatic cell lines were originally obtained from the same patient (Coriell, Camden, NJ). All studies were performed in confluent cells, as cell density dramatically influences basal Slug expression in some cell lines,34,47 and expression was compared by RT-qPCR (Figure 3A) and Western blot (Figure 3B). Endogenous Slug mRNA and protein expression was highest in NHM and lowest in metastatic melanoma cells. In contrast, Snail expression was higher in both melanoma cell lines as compared to NHM. Snail mRNA expression was higher in metastatic compared to primary melanomas, whereas Snail protein levels were higher in primary than in metastatic melanomas. This observation underscored the fact that mRNA levels of Snail and Slug might not have accurately reflected their protein levels. Differences in Slug and Snail mRNA expression were statistically significant (P < 0.01) for NHM compared to both primary and metastatic melanoma cells (Figure 3B). Thus, mRNA and protein expression of these two highly related Snail family genes was inversely related, an unexpected finding based on previous reports of increased Snail expression in melanomas and the fact that Slug and Snail are believed to have overlapping functions.21,24,26,28
Figure 3.
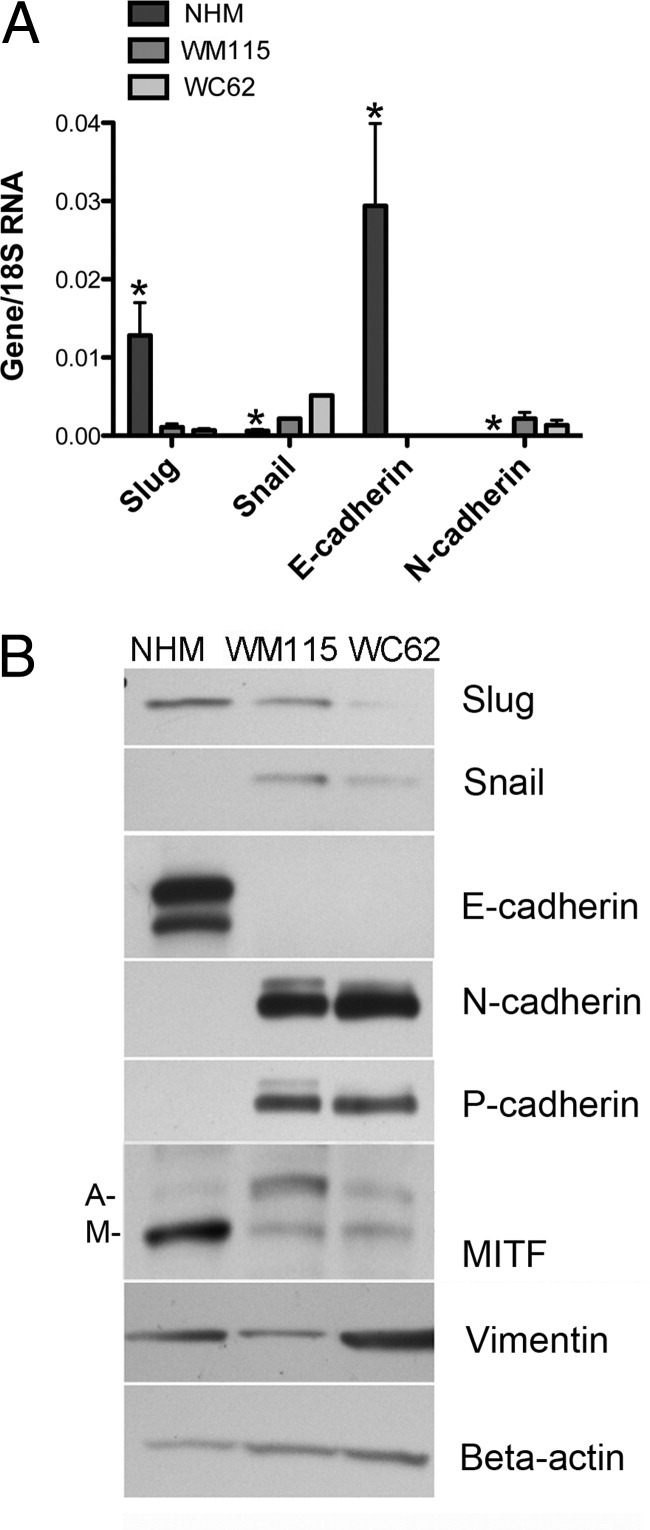
Endogenous Slug expression is higher at both mRNA and protein levels in NHM than in primary (WM115) or metastatic (WC62) melanoma cell lines. A: RT-qPCR revealed significantly higher expression of Slug and E-cadherin mRNAs in NHM than in primary or metastatic melanoma cells, whereas Snail and N-cadherin expression were significantly lower. Error bars indicate SDs. *P < 0.01 compared to both primary and metastatic melanoma cells. B: Western blots confirmed that differences in mRNA levels were reflected in differences in protein expression in NHM versus primary and metastatic melanoma cell lines. Both mRNA and protein were extracted from confluent cell cultures to eliminate density-dependent variations in expression of Slug and its putative target genes. MITF isoforms include both melanocyte-specific (M) and ubiquitous (A) isoforms.
In NHM expressing high levels of endogenous Slug, E-cadherin mRNA, and protein expression were also high, whereas E-cadherin mRNA was undetectable in melanoma cell lines (WM115 and WC62) that expressed low levels of Slug (Figures 3, A and B). The high levels of Slug mRNA and protein expression in NHM compared to melanoma cell lines was somewhat unexpected, given that the analysis of custom melanoma array indicated an increase in E-cadherin expression during melanoma progression. The two datasets, however, were not strictly comparable. E-cadherin expression was not evaluated immunohistochemically in normal human melanocytes in tissue sections, nor were nevus cell lines compared with NHM and melanoma cell lines. Taken together, however, tissue immunohistochemistry and cell culture results highlighted the fact that there was no simple relationship between Slug and E-cadherin protein expression.
N-cadherin mRNA and protein were expressed at very low levels in NHM, but at significantly (P < 0.01) higher levels in both melanoma cell lines. A shift from E-cadherin to N-cadherin expression is a well-known feature of melanoma progression,43 but our findings suggested that this shift was not related to increased Slug expression. P-cadherin and vimentin protein expression was higher in metastatic melanoma cells than in normal melanocytes, as previously reported.26,48,49 Multiple isoforms of MITF, however, were observed on Western blots; there was higher expression of the melanocyte-specific isoform of MITF in NHM compared to melanoma cells (Figure 3B).
Effects of Exogenous Slug on Melanocytes and Melanoma Cells
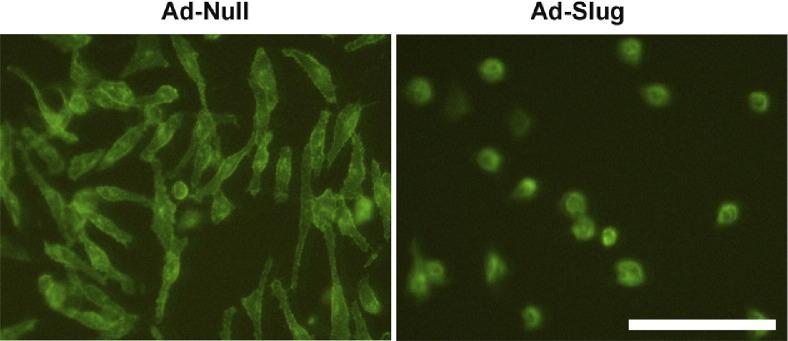
NHM and metastatic melanoma cells were infected with adenoviral vectors inducing high levels of exogenous Slug (Ad-Slug) to determine whether Slug had differential effects on the two cell types. Expression levels were determined 72 hours after transduction by Western blot (Figure 4, A and B). High levels of Slug protein expression were obtained in both NHM and metastatic melanoma cells (WC62). Expression of exogenous Slug significantly (P < 0.0001) reduced expression of E-cadherin mRNA in NHM, but had no effect on E-cadherin expression in melanoma cells, which did not express detectable E-cadherin mRNA either before or after Slug transduction. Moreover, Slug overexpression caused a dramatic internalization of E-cadherin protein in NHM (Figure 5). Levels of N-cadherin mRNA and protein expression were low in both NHM and melanoma cells, but Slug significantly (P < 0.01) increased N-cadherin mRNA expression in both cell types. The effect of Slug on cadherin expression in the two cell lines, although modest, was consistent with previous reports.5,41,46,50 Enhanced Slug expression appeared to have little effect on vimentin protein expression in either cell type (Figure 4B).
Figure 4.
NHM and melanoma cells transduced with Slug-expressing adenovirus (Ad-Slug) express less E-cadherin and more N-cadherin mRNA and protein than cells transduced with a control adenovirus (Ad-null). A: RT-qPCR revealed significantly lower E-cadherin expression in NHM transduced with Ad-Slug compared to Ad-Null-transduced cells. Neither Ad-Slug nor Ad-Null-transfected WC62 cells expressed detectable E-cadherin mRNA. N-cadherin levels were significantly increased in Ad-Slug-transduced NHM and melanoma cells compared to Ad-Null-transduced cells, although basal levels of expression were low and increases were modest. Note the difference in scale for the two graphs. Error bars indicate SEMs. *P < 0.03. B: Western blot results confirmed that Ad-Slug enhanced Slug protein expression in NHM and melanoma cells and that the differences seen in mRNA levels for E-cadherin in NHM and N-cadherin in melanoma cells were reflected in differences in protein expression. In addition, Ad-Slug transduction also enhanced expression of melanocyte-specific MITF (M) in NHM but not in melanoma cells. Melanoma cells expressed higher levels of the MITF isoform constitutively expressed in many tissues (A) and expression of this isoform was not altered by Ad-Slug transduction. Ad-Slug transduction did not alter levels of Snail expression in either cell type.
Figure 5.
Slug-expressing adenovirus (Ad-Slug) causes relocalization of E-cadherin protein from the cell membrane of NHM. NHM transduced with a control adenovirus (Ad-null) expressed abundant E-cadherin on the cell surface, whereas Ad-Slug-transduced NHM exhibited little or no membrane localization of E-cadherin.
MITF is a recognized marker of melanocyte lineage. MITF expression promotes melanoma cell survival and stimulates melanoma cell migration.51,52 In NHM expressing high levels of exogenous Slug, there was an increase in expression of the M isoform of MITF, the isoform reported to be relatively melanocyte-specific;53 however, expression of exogenous Slug in melanoma cells did not enhance production of the MITF M isoform. Slug overexpression did not alter Snail expression in either cell type (Figure 4B), suggesting that Slug did not directly regulate Snail expression.
There were no differences in proliferation in Slug-overexpressing cells compared to control cells as determined by MTT and standard cell counting methods, and neither Slug-transduced NHM nor melanoma cells acquired the ability to grow in soft agar (data not shown). Slug substantially enhanced cell migration in both NHM and metastatic melanoma (WC62) cells, as measured by migration to cover a cell layer defect (Figure 6A), and significantly (P < 0.05) enhanced their migration through an uncoated Boyden chamber (Figure 6B). Slug also enhanced melanoma cell invasiveness, as indicated by significantly (P < 0.05) increased migration through a Matrigel-coated Boyden chamber (Figure 6C); however, Slug had little effect on NHM invasiveness. The pronounced effect of Slug overexpression on cell migration appeared to be relatively independent of the effect of Slug on E-cadherin expression. Slug overexpression only modestly reduced total E-cadherin protein expression in NHM; however, Slug overexpression did cause E-cadherin protein to be relocalized from the melanocyte cell membrane to the cytoplasm, which might have accounted for the effect of Slug on the motility of these cells. Melanoma cells, in contrast, expressed miniscule amounts of E-cadherin, either with or without Slug overexpression, yet Slug overexpression significantly enhanced their motility. In addition, while increased expression of melanocyte-specific MITF might have contributed to the enhanced motility of Slug-overexpressing melanocytes, a similar increase in MITF expression was not seen in Slug-overexpressing melanoma cells with increased motility.
Figure 6.
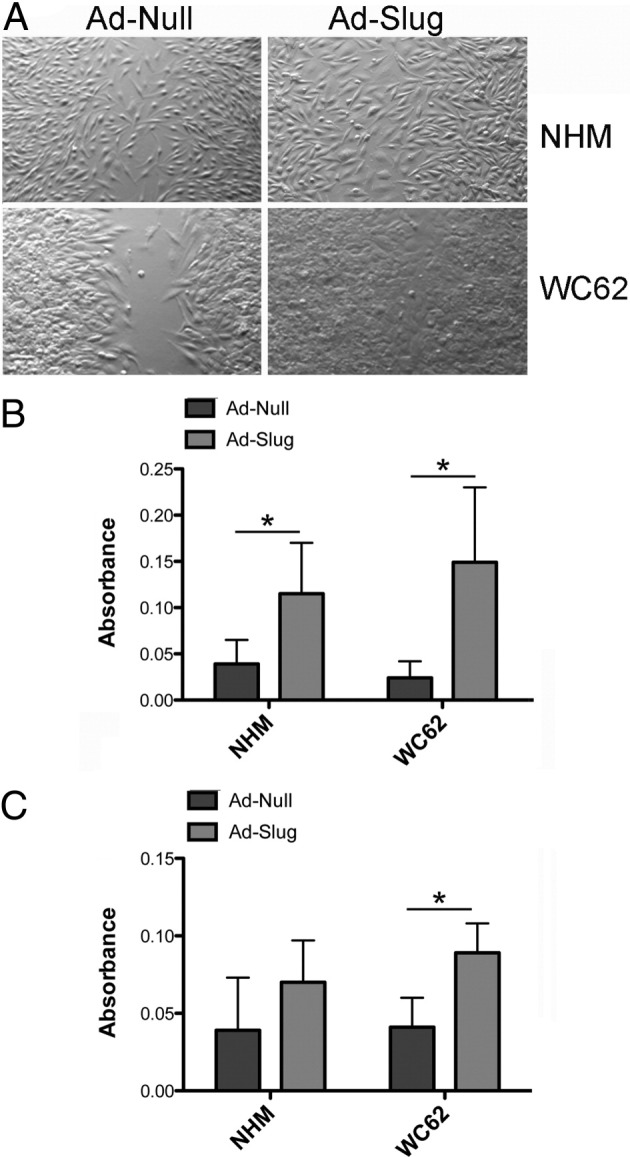
Slug-expressing adenovirus (Ad-Slug) enhances migration and invasion in NHM and melanoma cells (WC62). A: NHM and melanoma cells expressing exogenous Slug (Ad-Slug) migrated more rapidly to cover a defect in the confluent cell layer than cells expressing a control adenovirus (Ad-Null). Cultures were photographed 3 days after the introduction of the defect. B: Ad-Slug-transduced NHM and melanoma cells had significantly enhanced motility compared to Ad-Null-transduced cells. Migration of cells through an uncoated membrane measured motility. Cells were transduced with Ad-Slug or Ad-Null; after 72 hours, cells were placed in the upper portion of the cell culture chamber and allowed to migrate for 24 hours. Cell migration was quantified by removing cells from the upper surface of the membrane, staining cells on the lower surface of the membrane, extracting the retained dye, and determining the absorbance of the extract. Migration of cells through an uncoated membrane measured motility. C: Ad-Slug significantly enhanced the invasiveness of melanoma cells, but not NHM. Migration of cells through a Matrigel-coated membrane measured invasiveness. Error bars indicate SDs. *P < 0.05.
Discussion
The role of Slug in driving EMT during melanocyte differentiation suggests that Slug may also play an important role in melanoma progression by enhancing motility and invasiveness. Thus Slug expression would be expected to increase as melanomas become more invasive and metastatic. RNA microarray studies of melanomas and melanoma cell lines have generally compared Slug expression in melanomas to expression of internal standards or to Slug expression in normal skin. In melanoma cell lines, Slug expression has been reported to be increased by 8- to 156-fold, depending on the cell lines and standards used.54–56 These reports should be interpreted with caution, however, as our previous studies of Slug expression in keratinocytes have shown that Slug expression is highly dependent on cell density.34 Examining Slug expression in nonconfluent melanoma cell lines may thus provide artificially high estimates of basal Slug expression. RNA microarray studies using tumor samples suggest a somewhat lower level of Slug expression in nevi and melanomas, with Slug mRNA expression being elevated only 2.8- to 3.3-fold in nevi and 2- to 4.5-fold in melanomas compared to normal skin.57,58 Indeed, one study failed to detect any difference between Slug mRNA expression in skin versus melanomas.59 It is important to note that Slug is expressed in the basal keratinocytes of normal epidermis,60 thus normal skin may not be the best choice as a standard for comparison. In addition, a significant limitation of all RNA microarray studies is that Slug mRNA levels in cell lines or in tumors may not correlate well with Slug protein levels, as protein levels are modulated post-translationally.61
In contrast to expectations, our immunohistochemistry studies examining a large number of melanocytic tumors in a custom array clearly showed that Slug protein expression was significantly higher in nevi and primary melanomas than in metastatic melanomas (P < 0.05) and higher in nevi than in primary melanomas (P = 0.0534). Moreover, our in vitro studies revealed that confluent NHM expressed higher levels of Slug mRNA and protein than cells derived from primary or metastatic melanomas. Taken together, these findings suggest a role for Slug early during melanoma development, rather than during later stages of melanoma invasion and metastasis. This conclusion is consistent with the findings of Koefinger et al,46 who reported higher expression of Slug protein in nevi than in primary or metastatic melanomas. However, these investigators also reported that cytoplasmic Slug staining was stronger in primary melanomas and metastases than in nevi, whereas we have never observed cytoplasmic Slug immunoreactivity. Cytoplasmic staining observed in previous studies is likely to be artifactual, as the lack of specificity of many antibodies raised against Slug is well known.20,34
Although a role for Slug in melanoma metastasis has been postulated, we show here and a number of RNA microarray studies have shown that Slug is not a predictor of melanoma metastasis.58,59,62–64 Critical changes in expression of a variety of oncogenes and tumor suppressor genes may take place relatively early during melanoma progression;59,64,65 perhaps Slug expression is similarly required only early in melanomagenesis. An early role for Slug is consistent with the inverse relationship between Slug and Snail expression we observed in NHM and melanoma cells. In this regard, it has been suggested that Slug expression precedes Snail expression during breast cancer development and that Slug sets the stage for Snail-mediated tumor progression.36 In melanomas, Slug, but not Snail or Twist cooperates with ZEB1 to repress E-cadherin expression, indicating differential, perhaps sequential expression of EMT drivers.50
E-cadherin is the canonical target of Slug and Snail modulation during EMT.6,35,41 Slug and Snail bind to E-boxes in the E-cadherin (Cdh1) promoter to suppress its expression.5,50 Studies in a variety of cell types, including melanoma cell lines, have demonstrated that expression of exogenous Slug enhances motility and reduces the expression of adherens junctions.7 On the other hand, knocking down Slug expression renders cells resistant to growth factor-induced EMT and inhibits the switch from E- to N-cadherin characteristic of EMT.44,46 In our in vitro studies, exogenous Slug expression enhanced motility in NHM and melanoma cells, increased invasiveness in melanoma cells, suppressed E-cadherin expression in NHM, and enhanced N-cadherin expression in NHM and melanoma cells. Overall, these effects were consistent with previous reports.5,41,46,50
A number of immunohistochemical studies of human tumors have shown the expected inverse relationship between Slug and E-cadherin expression.6,11,35–41 In the present study, however, we were unable to validate a relationship between Slug and E-cadherin protein expression by immunohistochemistry in tumor microarrays. Other studies have also failed to demonstrate an association between Slug and E-cadherin expression in tumors.66–70 Thus, the relationship between Slug and E-cadherin expression in tumors remains somewhat unclear, and studies in cell lines may not accurately predict findings in primary tumors. Previous immunohistochemical studies performed with poorly characterized antibodies and without careful assessment of nuclear versus cytoplasmic staining may not be reliable.20 For the present studies, we used a validated antibody that revealed Slug immunopositivity only in nuclei.34
We did not see a clear relationship between expression of Slug and its target gene E-cadherin (Cdh1), although we did find an association between expression of MITF and Slug. MITF binds to and activates the Slug promoter in vitro.14 Interestingly, overexpression of Slug in Xenopus embryos dramatically increases MITF expression, suggesting that Slug and MITF may reciprocally regulate one another;17 however, Slug expression is not required for MITF expression, as MITF is detected in Slug null cells.14 Multiple observations support a role for Slug in melanogenesis. Slug deficient mice display hypopigmentation at the extremities and on the head and Slug mutations are found in the hereditary pigmentary defects Waardenberg syndrome type II and piebaldism.14,71 The pathway by which Slug regulates pigmentation is unknown, although MITF and SCF/c-kit signaling pathways likely play critical roles.
Studies to date suggest that Slug plays a role in melanomagenesis and that this role is related to the function of Slug in EMT during development. Although Slug appears to enhance melanocyte motility both in emigrating neural crest cells and in normal melanocytes and melanoma cells, presumably by down-regulating E-cadherin expression, it remains to be seen whether the two processes are strictly comparable. Our studies highlight the fact that there is no simple relationship between Slug and E-cadherin expression either in cultured melanocytes and melanoma cells or in primary human melanocytic tumors. Moreover, the effect of Slug on melanocyte and melanoma cell motility is relatively independent of the effect of Slug on E-cadherin expression or localization. Surprisingly, Slug and Snail, two closely related genes, both of which promote cell motility, appear to be differentially regulated in melanocytes and melanoma cells.
The mechanisms by which Slug affects melanoma progression and the interactions of Slug with other EMT drivers may have important implications for intervention in melanoma progression. Our studies suggest a role for Slug early in the transformation of melanocytes to melanoma cells, although the timing, consequences, and importance of Slug expression during melanoma progression remain unclear.
Acknowledgments
We thank the Integrated Imaging, Pathology, and Histology Facility Core at the Science Park Research Division of the University of Texas MD Anderson Cancer Center for their outstanding histology support.
Footnotes
Supported by grants from the University of Texas MD Anderson Cancer Center SPORE in Melanoma (P50 CA093459), University of Texas MD Anderson Cancer Center Support Grant (P30 CA016672), the Center for Research in Environmental Disease (P30 ES007784), the Research Training in Carcinogenesis and Mutagenesis (T32 CA09480), and a Baker Foundation Fellowship (to S.H.S.).
References
- 1.Jiang R., Lan Y., Norton C.R., Sundberg J.P., Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- 2.Locascio A., Manzanares M., Blanco M.J., Nieto M.A. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc Natl Acad Sci USA. 2002;99:16841–16846. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sechrist J., Nieto M.A., Zamanian R.T., Bronner-Fraser M. Regulative response of the cranial neural tube after neural fold ablation: spatiotemporal nature of neural crest regeneration and up-regulation of Slug. Development. 1995;121:4103–4115. doi: 10.1242/dev.121.12.4103. [DOI] [PubMed] [Google Scholar]
- 4.Sefton M., Sanchez S., Nieto M.A. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- 5.Bolos V., Peinado H., Perez-Moreno M.A., Fraga M.F., Esteller M., Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 6.Cano A., Perez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G., Portillo F., Nieto M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 7.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–923. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 8.del Barrio M.G., Nieto M.A. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129:1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- 9.Nieto M.A., Sargent M.G., Wilkinson D.G., Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 10.Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 11.Carver E.A., Jiang R., Lan Y., Oram K.F., Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Losada J., Sanchez-Martin M., Rodriguez-Garcia A., Sanchez M.L., Orfao A., Flores T., Sanchez-Garcia I. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood. 2002;100:1274–1286. [PubMed] [Google Scholar]
- 13.Read A.P., Newton V.E. Waardenburg syndrome. J Med Genet. 1997;34:656–665. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Martin M., Rodriguez-Garcia A., Perez-Losada J., Sagrera A., Read A.P., Sanchez-Garcia I. SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet. 2002;11:3231–3236. doi: 10.1093/hmg/11.25.3231. [DOI] [PubMed] [Google Scholar]
- 15.Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 16.Tachibana M., Kobayashi Y., Matsushima Y. Mouse models for four types of Waardenburg syndrome. Pigment Cell Res. 2003;16:448–454. doi: 10.1034/j.1600-0749.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 17.Kumasaka M., Sato S., Yajima I., Goding C.R., Yamamoto H. Regulation of melanoblast and retinal pigment epithelium development by Xenopus laevis Mitf. Dev Dyn. 2005;234:523–534. doi: 10.1002/dvdy.20505. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Martin M., Perez-Losada J., Rodriguez-Garcia A., Gonzalez-Sanchez B., Korf B.R., Kuster W., Moss C., Spritz R.A., Sanchez-Garcia I. Deletion of the SLUG (SNAI2) gene results in human piebaldism. Am J Med Genet A. 2003;122A:125–132. doi: 10.1002/ajmg.a.20345. [DOI] [PubMed] [Google Scholar]
- 19.LaBonne C., Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- 20.Peinado H., Olmeda D., Cano A. Snail: Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 21.Hemavathy K., Ashraf S.I., Ip Y.T. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 22.Becker K.F., Rosivatz E., Blechschmidt K., Kremmer E., Sarbia M., Hofler H. Analysis of the E-cadherin repressor Snail in primary human cancers. Cells Tissues Organs. 2007;185:204–212. doi: 10.1159/000101321. [DOI] [PubMed] [Google Scholar]
- 23.Bastid J., Bouchet B.P., Ciancia C., Pourchet J., Audoynaud C., Grelier G., Puisieux A., Ansieau S. The SNAIL family member SCRATCH1 is not expressed in human tumors. Oncol Rep. 2010;23:523–529. [PubMed] [Google Scholar]
- 24.Poser I., Dominguez D., de Herreros A.G., Varnai A., Buettner R., Bosserhoff A.K. Loss of E-cadherin expression in melanoma cells involves up-regulation of the transcriptional repressor Snail. J Biol Chem. 2001;276:24661–24666. doi: 10.1074/jbc.M011224200. [DOI] [PubMed] [Google Scholar]
- 25.Kuphal S., Palm H.G., Poser I., Bosserhoff A.K. Snail-regulated genes in malignant melanoma. Melanoma Res. 2005;15:305–313. doi: 10.1097/00008390-200508000-00012. [DOI] [PubMed] [Google Scholar]
- 26.Poser I., Bosserhoff A.K. Transcription factors involved in development and progression of malignant melanoma. Histol Histopathol. 2004;19:173–188. doi: 10.14670/HH-19.173. [DOI] [PubMed] [Google Scholar]
- 27.Dissanayake S.K., Wade M., Johnson C.E., O'Connell M.P., Leotlela P.D., French A.D., Shah K.V., Hewitt K.J., Rosenthal D.T., Indig F.E., Jiang Y., Nickoloff B.J., Taub D.D., Trent J.M., Moon R.T., Bittner M., Weeraratna A.T. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyormoi O., Bar-Eli M. Transcriptional regulation of metastasis-related genes in human melanoma. Clin Exp Metastasis. 2003;20:251–263. doi: 10.1023/a:1022991302172. [DOI] [PubMed] [Google Scholar]
- 29.Gupta P.B., Kuperwasser C., Brunet J.P., Ramaswamy S., Kuo W.L., Gray J.W., Naber S.P., Weinberg R.A. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vannini I., Bonafe M., Tesei A., Rosetti M., Fabbri F., Storci G., Ulivi P., Brigliadori G., Amadori D., Zoli W. Short interfering RNA directed against the SLUG gene increases cell death induction in human melanoma cell lines exposed to cisplatin and fotemustine. Cell Oncol. 2007;29:279–287. doi: 10.1155/2007/540821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazarian R.M., Prieto V.G., Elder D.E., Duncan L.M. Melanoma biomarker expression in melanocytic tumor progression: a tissue microarray study. J Cutan Pathol. 2010;37(Suppl 1):41–47. doi: 10.1111/j.1600-0560.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 32.Chandler H.L., Colitz C.M., Lu P., Saville W.J., Kusewitt D.F. The role of the slug transcription factor in cell migration during corneal re-epithelialization in the dog. Exp Eye Res. 2007;84:400–411. doi: 10.1016/j.exer.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Chen C.Y., Shyu A.B. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirley S.H., Hudson L.G., He J., Kusewitt D.F. The skinny on Slug. Mol Carcinog. 2010;49:851–861. doi: 10.1002/mc.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batlle E., Sancho E., Franci C., Dominguez D., Monfar M., Baulida J., Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 36.Come C., Magnino F., Bibeau F., De Santa Barbara P., Becker K.F., Theillet C., Savagner P. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12:5395–5402. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- 37.Uchikado Y., Natsugoe S., Okumura H., Setoyama T., Matsumoto M., Ishigami S., Aikou T. Slug Expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1174–1180. [PubMed] [Google Scholar]
- 38.Giannelli G., Bergamini C., Fransvea E., Sgarra C., Antonaci S. Laminin-5 with transforming growth factor-beta1 induces epithelial to mesenchymal transition in hepatocellular carcinoma. Gastroenterology. 2005;129:1375–1383. doi: 10.1053/j.gastro.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 39.Elloul S., Silins I., Trope C.G., Benshushan A., Davidson B., Reich R. Expression of E-cadherin transcriptional regulators in ovarian carcinoma. Virchows Arch. 2006;449:520–528. doi: 10.1007/s00428-006-0274-6. [DOI] [PubMed] [Google Scholar]
- 40.Alves C.C., Carneiro F., Hoefler H., Becker K.F. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front Biosci. 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]
- 41.Hajra K.M., Chen D.Y., Fearon E.R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 42.Hay E.D. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 43.Haass N.K., Smalley K.S., Herlyn M. The role of altered cell-cell communication in melanoma progression. J Mol Histol. 2004;35:309–318. doi: 10.1023/b:hijo.0000032362.35354.bb. [DOI] [PubMed] [Google Scholar]
- 44.Savagner P., Yamada K.M., Thiery J.P. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zavadil J., Bitzer M., Liang D., Yang Y.C., Massimi A., Kneitz S., Piek E., Bottinger E.P. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98:6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koefinger P., Wels C., Joshi S., Damm S., Steinbauer E., Beham-Schmid C., Frank S., Bergler H., Schaider H. The cadherin switch in melanoma instigated by HGF is mediated through epithelial-mesenchymal transition regulators. Pigment Cell Melanoma Res. 2011;24:382–385. doi: 10.1111/j.1755-148X.2010.00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conacci-Sorrell M., Simcha I., Ben-Yedidia T., Blechman J., Savagner P., Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauer R., Hein R., Bosserhoff A.K. A secreted form of P-cadherin is expressed in malignant melanoma. Exp Cell Res. 2005;305:418–426. doi: 10.1016/j.yexcr.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 49.Li M., Zhang B., Sun B., Wang X., Ban X., Sun T., Liu Z., Zhao X. A novel function for vimentin: the potential biomarker for predicting melanoma hematogenous metastasis. J Exp Clin Cancer Res. 2010;29:109. doi: 10.1186/1756-9966-29-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wels C., Joshi S., Koefinger P., Bergler H., Schaider H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J Invest Dermatol. 2011;131:1877–1885. doi: 10.1038/jid.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beuret L., Flori E., Denoyelle C., Bille K., Busca R., Picardo M., Bertolotto C., Ballotti R. Up-regulation of MET expression by alpha-melanocyte-stimulating hormone and MITF allows hepatocyte growth factor to protect melanocytes and melanoma cells from apoptosis. J Biol Chem. 2007;282:14140–14147. doi: 10.1074/jbc.M611563200. [DOI] [PubMed] [Google Scholar]
- 52.Gentile A., Trusolino L., Comoglio P.M. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 53.Amae S., Fuse N., Yasumoto K., Sato S., Yajima I., Yamamoto H., Udono T., Durlu Y.K., Tamai M., Takahashi K., Shibahara S. Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem Biophys Res Commun. 1998;247:710–715. doi: 10.1006/bbrc.1998.8838. [DOI] [PubMed] [Google Scholar]
- 54.Pratilas C.A., Taylor B.S., Ye Q., Viale A., Sander C., Solit D.B., Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shankavaram U.T., Reinhold W.C., Nishizuka S., Major S., Morita D., Chary K.K., Reimers M.A., Scherf U., Kahn A., Dolginow D., Cossman J., Kaldjian E.P., Scudiero D.A., Petricoin E., Liotta L., Lee J.K., Weinstein J.N. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 56.Wagner K.W., Punnoose E.A., Januario T., Lawrence D.A., Pitti R.M., Lancaster K., Lee D., von Goetz M., Yee S.F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S.G., Amler L., Ashkenazi A. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 57.Haqq C., Nosrati M., Sudilovsky D., Crothers J., Khodabakhsh D., Pulliam B.L., Federman S., Miller J.R., 3rd, Allen R.E., Singer M.I., Leong S.P., Ljung B.M., Sagebiel R.W., Kashani-Sabet M. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talantov D., Mazumder A., Yu J.X., Briggs T., Jiang Y., Backus J., Atkins D., Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 59.Riker A.I., Enkemann S.A., Fodstad O., Liu S., Ren S., Morris C., Xi Y., Howell P., Metge B., Samant R.S., Shevde L.A., Li W., Eschrich S., Daud A., Ju J., Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parent A.E., Choi C., Caudy K., Gridley T., Kusewitt D.F. The developmental transcription factor slug is widely expressed in tissues of adult mice. J Histochem Cytochem. 2004;52:959–965. doi: 10.1369/jhc.4A6277.2004. [DOI] [PubMed] [Google Scholar]
- 61.Shih J.Y., Yang P.C. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 2011;32:1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 62.Bittner M., Meltzer P., Chen Y., Jiang Y., Seftor E., Hendrix M., Radmacher M., Simon R., Yakhini Z., Ben-Dor A., Sampas N., Dougherty E., Wang E., Marincola F., Gooden C., Lueders J., Glatfelter A., Pollock P., Carpten J., Gillanders E., Leja D., Dietrich K., Beaudry C., Berens M., Alberts D., Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 63.Hoek K.S., Schlegel N.C., Brafford P., Sucker A., Ugurel S., Kumar R., Weber B.L., Nathanson K.L., Phillips D.J., Herlyn M., Schadendorf D., Dummer R. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 64.Smith A.P., Hoek K., Becker D. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol Ther. 2005;4:1018–1029. doi: 10.4161/cbt.4.9.2165. [DOI] [PubMed] [Google Scholar]
- 65.Zaidi M.R., Day C.P., Merlino G. From UVs to metastases: modeling melanoma initiation and progression in the mouse. J Invest Dermatol. 2008;128:2381–2391. doi: 10.1038/jid.2008.177. [DOI] [PubMed] [Google Scholar]
- 66.Martin T.A., Goyal A., Watkins G., Jiang W.G. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488–496. doi: 10.1245/ASO.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 67.Sivertsen S., Hadar R., Elloul S., Vintman L., Bedrossian C., Reich R., Davidson B. Expression of Snail: Slug and Sip1 in malignant mesothelioma effusions is associated with matrix metalloproteinase, but not with cadherin expression. Lung Cancer. 2006;54:309–317. doi: 10.1016/j.lungcan.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 68.Joseph M.J., Dangi-Garimella S., Shields M.A., Diamond M.E., Sun L., Koblinski J.E., Munshi H.G. Slug is a downstream mediator of transforming growth factor-beta1-induced matrix metalloproteinase-9 expression and invasion of oral cancer cells. J Cell Biochem. 2009;108:726–736. doi: 10.1002/jcb.22309. [DOI] [PubMed] [Google Scholar]
- 69.Shioiri M., Shida T., Koda K., Oda K., Seike K., Nishimura M., Takano S., Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li G., Satyamoorthy K., Meier F., Berking C., Bogenrieder T., Herlyn M. Function and regulation of melanoma-stromal fibroblast interactions: when seeds meet soil. Oncogene. 2003;22:3162–3171. doi: 10.1038/sj.onc.1206455. [DOI] [PubMed] [Google Scholar]
- 71.Jiang R., Norton C.R., Copeland N.G., Gilbert D.J., Jenkins N.A., Gridley T. Genomic organization, expression and chromosomal localization of the mouse Slug (Slugh) gene. Biochim Biophys Acta. 1998;1443:251–254. doi: 10.1016/s0167-4781(98)00225-5. [DOI] [PubMed] [Google Scholar]