Abstract
Flap endonucleases (FENs), essential for DNA replication and repair, recognize and remove RNA or DNA 5′-flaps. Related to FEN specificity for substrates with free 5′-ends, but controversial, is the role of the helical arch observed in varying conformations in substrate-free FEN structures. Conflicting models suggest either 5′-flaps thread through the arch, which when structured can only accommodate single-stranded (ss) DNA, or the arch acts as a clamp. Here we show that free 5′-termini are selected using a disorder-thread-order mechanism. Adding short duplexes to 5′-flaps or 3′-streptavidin does not markedly impair the FEN reaction. In contrast, reactions of 5′-streptavidin substrates are drastically slowed. However, when added to premixed FEN and 5′-biotinylated substrate, streptavidin is not inhibitory and complexes persist after challenge with unlabelled competitor substrate, regardless of flap length or the presence of a short duplex. Cross-linked flap duplexes that cannot thread through the structured arch react at modestly reduced rate, ruling out mechanisms involving resolution of secondary structure. Combined results explain how FEN avoids cutting template DNA between Okazaki fragments and link local FEN folding to catalysis and specificity: the arch is disordered when flaps are threaded to confer specificity for free 5′-ends, with subsequent ordering of the arch to catalyze hydrolysis.
INTRODUCTION
Structure sensing 5′-nucleases are vital for DNA replication, repair, and recombination. Operating without regard to sequence, 5′-nucleases recognize defined nucleic acid junctions and catalyze the hydrolysis of specific phosphate diester bonds (1–3). Exemplary junctions for 5′-nuclease cleavage are formed during lagging strand DNA replication and long-patch base excision repair (lpBER), where 5′-extensions (flaps) occur at adjacent duplexes (Okazaki fragments and lpBER intermediates) as a consequence of polymerase strand displacement synthesis. Divalent metal ion-dependent flap endonucleases (FENs), the prototypical 5′-nuclease family members, are the enzymes that catalyze removal of 5′-flaps. This hydrolytic processing yields 5′-phosphorylated-nicked DNAs for subsequent ligation and during human replication must take place at least 50 million times per cell cycle. The importance of 5′-flap elimination is demonstrated by the lethality of fen1(−/−) knockouts in mammals (4). FENs endonucleolytically remove 5′-flaps, thereby avoiding repetitive exonucleolytic processing. Even before structures of FEN proteins became available, it was suggested that FEN specificity for junctions with free 5′-termini, and discrimination against other junctions lacking this feature that occur at replication forks, could be achieved by threading the 5′-flap DNA through a hole in the protein (5). Yet this proposal has remained controversial, and the basis for end specificity has remained enigmatic.
Subsequent structural studies did indeed reveal a hole in FEN proteins formed by helices linking the main DNA-binding domains straddling the active site (Figure 1A) (6,7). Known as the helical arch, this subdomain is partially disordered in some X-ray structures (Figure 1B and Supplementary Figure S1A) (8–10). In structured form, the arch is only large enough to accommodate single- but not double-stranded (ds) DNA, appearing to account for FEN specificity. Support for a threading mechanism for end specificity came from biochemical experiments that suggested that forming a duplex within the 5′-single-stranded (ss) flap or binding of proteins to this region of substrates prevented the FEN reaction (11,12). Structural studies of bacteriophage T4FEN bound to a pseudo-Y (pY) DNA substrate did show a 5′-flap within the arch region (9). However, in this complex, the DNA did not occupy the divalent metal ion-free active site and one helix of the arch was disordered (Figure 1C and Supplementary Figure S1B). Nevertheless, models can be created using this structure by overlay with FENs crystallized with ordered arches showing the flap DNA passing through, although not yet positioned in the active site for reaction (Figure 1D), furthering controversy regarding a possible threading mechanism for specificity.
Figure 1.
Structures of T5, T4 and human FENs with and without DNA. (A) Structure of T5FEN (1UT5) with transparent surface to highlight the helical arch and resulting hole above the active site bound divalent metals (black spheres). (B) Structure of hFEN1 (1UL1, X chain; pink) with transparent surface representations showing the disordered arch (missing arch residues, dotted lines; active site metal ions, black). (C) T4FEN structure in complex with a pseudo-Y (pY) substrate (2IHN) without metal ions. Based on alignment with a substrate-free T4FEN structure (1TFR), the location of active site divalent metal ions (black spheres) is shown along with template (brown) and 5′-flap strands (yellow) of the pY and disordered residues (dotted lines). (D) Model of T5FEN (1UT5) in complex with a pY substrate with active site divalent metals, protein and DNA colored as in (A) and (C) based on alignment with the T4FEN-DNA structure (2IHN) shows that the 5′-flap could go through the helical arch. Some steric clashes are observed suggesting conformational changes. (E) Structure of hFEN1 in complex with the product of reaction of a double-flap substrate (3Q8K). Template DNA (brown), the cleaved 5′-flap DNA strand (yellow), and 3′-flap DNA (purple) are shown with active site metal ions (gray) and a K+ ion (purple). (F) Active site of the hFEN1-product DNA complex (3Q8K) showing the 5′-phosphate monoester product interacting with active site divalent metal ions (black spheres). Note, this nucleobase is not paired with the template. 5′-Nuclease superfamily conserved active site carboxylates (red) and helical arch α4 Lys93 and Arg100 (blue) are shown. A tyrosine residue (Tyr40) from α2 stacks upon the unpaired nucleobase.
In contrast, several studies have challenged the hypothesis that the helical arch enforces FEN specificity. Apparently in conflict with earlier literature, human FEN1 (hFEN1) has been demonstrated to endonucleolytically process so-called gapped flap substrates (13). Gapped flaps contain a short 5′-region of duplex that cannot pass through a structured helical arch. Other biochemical studies on the question of FEN 5′-flap accommodation have also produced results that are apparently at odds with a threading model (14,15). Thus, as an alternative to passage of substrate through the arch, this subdomain has been suggested instead to act as a clamp (3,7,14,16). One possible explanation of FEN specificity known as tracking in which FENs were proposed to initially interact with ss flaps either by threading or clamping and slide along these until junctions were encountered has been discredited (2,3,11–13,16–18).
Deciphering the origins of FEN1 specificity for 5′-flaps is made more complex by other 5′-nucleases that are sequence related to FENs but have differing specificities (1–3,19). In humans, EXO1, the mismatch and resection 5′-nuclease is most closely related to FEN1. EXO1 catalyses the processive hydrolyses of the 5′-termini of gapped, nicked and blunt duplex DNAs. Like FEN1, EXO1 can also endonucleolytically remove 5′-flaps (20). Another superfamily member XPG, the 5′-nuclease of nucleotide excision repair (NER), acts upon bubble substrates (21). The major human Holliday junction resolvase is suggested to be GEN1, another superfamily member (22). However, neither NER bubbles nor four-way junctions possess free 5′-termini in vivo. The 5′-portion of these substrates could therefore not be passed through an arch.
Recent structures of hFEN1 and hEXO1 bound to substrates and products in the presence of active site metal ions highlight the similarities between 5′-nuclease superfamily members (Figure 1E) (2,3,23). Despite analogous structures for hFEN1 and hEXO1 complexes with product DNAs, which include conserved contacts between the cleaved 5′-phosphate and helical arch residues (Figure 1F), differing interpretations for the requirement for threading versus clamping indicate that key questions regarding the basis for substrate specificities within the FEN-like 5′-nucleases remain. Here, using functional studies with modified DNAs, we resolve how FENs accommodate the 5′-region of substrates, demonstrate that processing of 5′-gapped flaps is an hFEN1 activity that proceeds by the same mechanism and propose a universal model for departure of DNA from the active sites of 5′-nuclease superfamily members. Moreover, our results explain how FENs can function to rapidly remove flaps during replication without risk of destroying template DNA between Okazaki fragments.
MATERIALS AND METHODS
Over-expression and purification of T5FEN and hFEN1
T5FEN and hFEN1 (wild-type and K93A) were over-expressed and purified as described (2,24).
Synthesis and purification of oligonucleotide substrates
Oligonucleotides (ODNs) were synthesized using an ABI model 394 DNA/RNA synthesizer or by DNA Technology A/S (Risskov, Denmark) using 5′-fluorescein-CE- phosphoramidite (6-FAM) or 3′-(6-FAM)-CPG to incorporate 5′-FAM or 3′-FAM, respectively, and biotin TEG phosphoramidite to add biotin (Link Technologies, Lanarkshire, UK). The long tether 3′-biotin substrate [21 nt pY-3′B] was constructed using 3′-biotin TEG followed by three additions of spacer-CE-phosphoramidite-18. ODNs were purified by reverse-phase (RP) HPLC (Waters × bridge 10 × 250 mm C-18 column) using triethylammonium acetate buffers pH 6.5 with a gradient of acetonitrile. Purified ODNs were desalted using NAP-10 columns and subjected to MALDI-TOF mass spectrometry. Residual divalent metal ion contaminants were removed by treatment with Chelex resin. Experimental MWs were all within 3 Da of calculated. A complete list of ODNs is contained in Supplementary Figure S2.
Determination of the rate of decay of enzyme substrate complexes
Substrates were annealed as described (13,17). Enzyme and substrate were pre-incubated at 20°C (hFEN1) or on ice (T5FEN) for 2 min in 25 mM HEPES, pH 7.5, 50 mM KCl, 2 mM CaCl2, 1 mM DTT and 0.1 mg/ml BSA (hFEN; calcium buffer) or 25 mM HEPES, pH 7.5, 50 mM KCl, 1 mM EDTA, 1 mM DTT and 0.1 mg/ml BSA (T5FEN; EDTA buffer) to form ‘premixed’ complexes. If required, five equivalents (with respect to [S]) of streptavidin (SA) were added before (‘blocked’ reactions) or after (‘trapped’ reactions) addition of enzyme and incubated accordingly for 5 min. Increasing the concentration of streptavidin did not alter the outcome. Samples were warmed at 37°C, and the reaction was initiated by mixing with an equal amount of magnesium buffer as above but containing 16 mM MgCl2 instead of CaCl2, (hFEN1) or 20 mM MgCl2 instead of EDTA (T5FEN). The final concentrations of enzyme and substrate were 500 nM and 5 nM, respectively. For ‘trapped’ and ‘premixed’ reactions, sampling was carried out using quench flow apparatus (RQF-63 quench flow device, Hi-Tech Sci Ltd., Salisbury, UK). After time delays of 6.4 ms to 51.2 s, quench (8 M Urea containing 80 mM EDTA) was added. ‘Blocked’ reactions were sampled manually. Reactions were analyzed by dHPLC equipped with a fluorescence detector (Wave® system, Transgenomic, UK) as described (13,17,25,26). After quenching the presence of SA did not alter the dHPLC retention time with tetrabutyl ammonium bromide as the ion-pairing reagent (Supplementary Figure S3). The formation of product formed over time (Pt) was fitted to Equation (1), to determine the first-order rate constant (k) where P∞ is the amount of product at end point:
| (1) |
Competition experiments
Competitor ODNs were pY or double-flap substrates without FAM label or biotin (Supplementary Figure S2). Enzyme and FAM–biotin–substrate were incubated at 20°C for 2 min (hFEN1) or on ice for 2 min (T5FEN) in either calcium buffer (hFEN1) or EDTA buffer (T5FEN) as above. For ‘trapped’ reactions, five equivalents of SA were added followed by incubation for a further 1 min. Competitor substrate was then added, and the mixtures were incubated for 10 min at 37°C. Increasing this time to 20 min had no impact on the outcome. An equal volume of magnesium buffer (as above) was added to initiate reaction producing final concentrations of enzyme (500 nM), FAM-labeled substrate (5 nM) and competitor (2.5 µM, T5FEN; 5 µM, hFEN1). Reactions were sampled, quenched and the amount of product determined as above. In experiments where streptavidin and/or competitor were not added an equal volume of appropriate buffer was, and all samples underwent identical incubations.
Preparation of azide-alkyne ODN
9 -(5′-O-Dimethoxytrityl-2′-deoxyribofuranosyl)-N2-[(dimethylamino) methylidene]-2-amino-6-methylsulfonylpurine-3′-(2-cyanoethyl-N,N-diisopropyl)-phosphoramidite (27) was used to construct an ODN with 3′-FAM and a 5′-alkyne (6-Hexyn-1-yl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite (Glen Research) using mild/fast deprotection phopshoramidites for dC, dA and dG. Following 1 µmol scale synthesis, the CPG-bound ODN was treated with 200 µl of 11-Azido-3,6,9-trioxaundecan-1-amine:acetonitrile:DBU at a ratio of 9:9:2 at 37°C for 48 h with gentle mixing. Concentrated NH3(aq) (1 ml) was then added and the mixture left for a further 72 h at room temperature. After evaporation to dryness, the residue was suspended in water (150 μl) and extracted with diethyl ether (3 × 500 μl). The aqueous layer was removed and then purified by HPLC as described above. MW (AA-HP) 11 173.56 calculated; 11 176 found.
Preparation of triazole ODN
Reactions contained AA-HP (10 nmol) mixed with CuSO4.5H2O (750 nmol) sodium ascorbate (30 μmol) and tris-3-hydroxypropyltriazolylamine (28) (21 μmol) in a total volume of 1 ml with NaCl (final concentration 0.2 M) and were incubated at room temperature overnight with gentle mixing. The reaction mixture was desalted (NAP-10) and purified by RPHPLC under denaturing conditions (as for other ODNs but at 55°C). The triazole ODN (Z-HP) eluted 3.3 min earlier than AA-HP (Figure 5C).
Figure 5.
Processing of gapped flaps by hFEN1 does not require resolution of secondary structure. (A) Schematic illustration of the procedure used to introduce an azide functionality after ODN synthesis but before removal from CPG. Cleavage from the support and deprotection produced a hairpin containing oligodeoxyribonucleotide (ODN) with a 5′-alkyne, internal azide and 3′-FAM (HP-AA). (B) ‘Click’ reaction using HP-AA ODN produced as in Figure 5A to yield the triazole cross-linked hairpin containing ODN HP-Z. (C) dHPLC traces of the reaction shown in Figure 5B of HP-AA ODN (top) to produce HP-Z ODN (middle). A co-injection of HP-AA and HP-Z ODNs is shown (bottom). (D) A 3 nt DF substrate (3 nt DF Control) used for comparison with gapped DF substrates made with HP-AA and HP-Z. (E) Normalized initial rates of reaction of WT hFEN1 and K93A hFEN1 with substrates as in Figure 5D. Reactions contained 50 nM substrate and either 5pM (WT) or 2.5 nM (K93A) in 50 mM HEPES pH 7.5, 100 mM KCl, 8 mM MgCl2, 1 mM DTT and 0.1 mg/ml BSA.
Determination of the rates of the reaction of triazole cross-linked gapped substrates
Kinetic analysis was carried out using GAP DF-AA and GAP DF-Z substrates at a concentration of 50 nM, with 5 pM WT hFEN1 or 2.5 nM K93A at 37°C in 50 mM HEPES, pH 7.5, 100 mM KCl, 8 mM MgCl2, 1 mM DTT and 0.1 mg/ml BSA. Samples of reaction mixture were quenched in an equal volume of 250 mM EDTA, pH 8.0. Reactions were analyzed as above. Initial rates of reaction were obtained from plots of amount of product versus time.
RESULTS
A 5′-block inhibits FEN catalyzed reactions
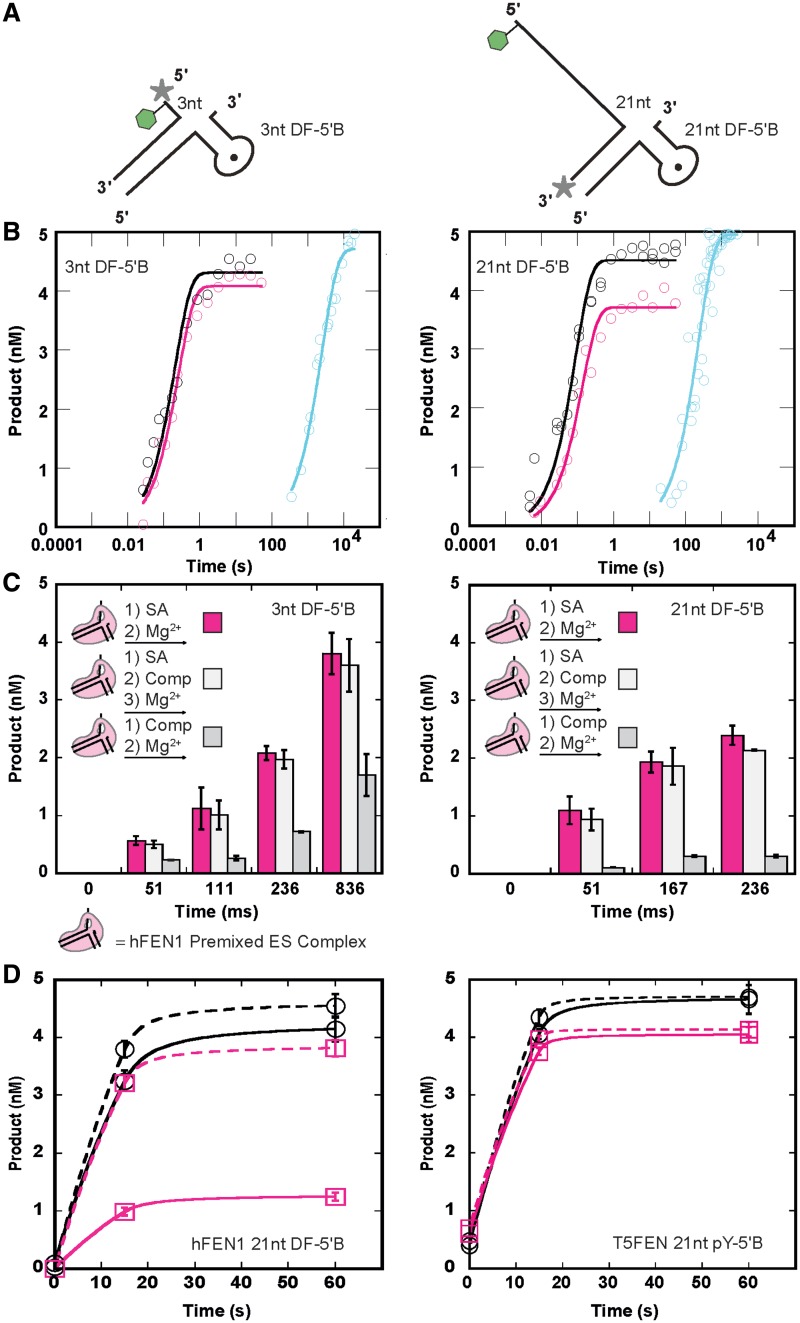
To test whether FENs use a threading or clamping mechanism, we used biotinylated substrates to which streptavidin (SA) could be bound (Figures 2A, 3A and 4A; Supplementary Figure S2). The 53 kDa SA tetramer forms stable complexes with biotin [t1/2 (our reaction conditions) >> 2 h (29)] and is too large to pass through the helical arch even when it is disordered. However, 5′-SA should not prevent clamping at the base of a 5′-flap. The addition of 3′- or 5′-biotin label to T5FEN substrates with 21 nt 5′-flaps did not significantly alter the rate of enzyme catalyzed reactions (Figure 2A and Supplementary Table S1). Neither did adding SA to reactions of non-biotinylated substrates. However, the rates of Mg2+ initiated reaction of T5FEN-substrate ‘blocked’ complexes formed by adding SA-conjugated substrates to enzyme differed by three orders of magnitude depending on the biotinylation site (Figure 2B and C). 5′-Blocked complexes reacted very slowly (Supplementary Figure S3), whereas 3′-SA complexes had a similar rate to those premixed with no block. Similar drastic decreases in the rate of reaction are observed under multiple turnover conditions (Supplementary Table S2).
Figure 2.
The rate of reaction of complexes of T5FEN and streptavidin substrates is dependent on the order of the addition of reagents and the site of substrate biotinylation. (A) Biotinylated substrates for T5FEN with 21 nt 5′-flaps. Green hexagon and gray star indicate biotin and fluorescein (FAM), respectively. (B) Order of the addition of reagents schematic illustrating how premixed, blocked and trapped complexes of FEN and substrates were prepared as detailed in ‘Materials and Methods’ section. (C) The effect of the SA addition on rates of T5FEN reactions containing 5′- (left) and 3′- (right) biotinylated substrates shown as plots of product versus time. Note: the X-axis (time) is in log format. Premixed (black), blocked (cyan, see Supplementary Figure S3) and trapped (pink) reactions were initiated at 37°C at final concentrations 500 nM T5FEN, 5 nM substrate, 10 mM Mg2+, 50 mM KCl, 1 mM EDTA, 1 mM DTT and 0.1 mg/ml BSA at pH 7.5. All data points are the result of three independent experiments (error bars omitted for clarity). Rate constants are summarized in Supplementary Table S3A. (D) The concentrations of product formed at various time intervals after initiation of reactions of trapped and premixed complexes that were challenged with excess competitor (Comp; non-biotinylated and unlabeled 21 nt pY substrate, final concentration 2.5 µM). Trapped and premixed complexes were formed and incubated at 37°C for 10 min with 5-fold excess Comp before initiation of reaction with magnesium buffer. Left, the amounts of product formed from 5′-SA trapped 21 nt pY-5′B complexes with and without the addition of Comp are shown in white and pink, respectively, whereas that formed from premixed 21 nt pY-5′B is shown in gray. Right, the amount of product formed from a 3′-SA trapped 21 nt pY-3′B to which Comp was added is shown in white. All data points are the result of three independent experiments with standard errors shown. See also Supplementary Figure S3.
Figure 3.
The rates of reactions of hFEN1 catalyzed hydrolysis of 5′-biotinylated substrates in the presence of SA are affected by the order of the addition of reagents. (A) Biotinylated DF substrates for hFEN1. A 3 nt 5′-flap DF (left) and 21 nt 5′-flap DF (right) with 5′-biotin and fluorescent label indicated with a green hexagon and gray star, respectively. (B) Product versus time plots of hFEN1 reactions of 3 nt DF-5′B (left) and 21 nt DF-5′B (right) illustrating the effects of order of the SA addition on the rates of reaction. Note: the X-axis (time) is in log format. Premixed (black), blocked (cyan) and trapped (pink) were assembled as in Figure 2B and then reaction initiated at 37°C by addition of Mg2+ at final concentrations 500 nM hFEN1, 5 nM substrate, 8 mM Mg2+, 100 mM KCl, 1 mM EDTA, 1 mM DTT and 0.1 mg/ml BSA at pH 7.5. All data points are the result of three independent experiments (error bars omitted for clarity). Rate constants are summarized in Supplementary Table S3B. (C) The concentrations of product formed at various time intervals after initiation of reactions of trapped and premixed complexes that were challenged with excess competitor (Comp; non-biotinylated and unlabeled 5 nt DF substrate, final concentration 5 µM). (D) Trapped and premixed complexes of hFEN1 and 3 nt DF-5′B (left) and 21 nt DF-5′B (right) were formed and incubated at 37°C for 10 min with 10-fold excess competitor before the addition of magnesium buffer. Concentrations of product formed from trapped complexes with and without the addition of competitor are shown in white and pink, respectively. Concentrations of product formed from premixed complexes challenged with competitor are shown in gray. All data points are the result of three independent experiments with standard errors shown. See also Supplementary Figure S3.
Figure 4.
The effect of short stable hairpins in the 5′-flap on hFEN1 catalyzed reactions. (A) Schematic of the biotinylated gapped DF substrate (GAP DF-5′B) with biotin and a 3′-fluorescent label indicated by the green hexagon and gray star, respectively. (B) Plots of product versus time for wild-type (straight line) and K93A (dashed line) hFEN1 premixed GAP DF-5′B (green) and 21 nt DF-5′B (black) reactions showing the effects of the presence of dsDNA in the 5′-flap and K93A mutation on the rates of reaction. Note: the X-axis (time) is in log format. Curve fits using equation 1 yield first order rate constants 683 ± 56/min (WT hFEN1: 21 nt DF-5′B) 357 ± 34/min, (WT hFEN1:GAP DF-5′B) 0.3 ± 0.02/min (K93A hFEN1: 21 nt DF-5′B), 0.17 ± 0.01/min (K93A hFEN1: GAP DF-5′B). (C) Product versus time plots of hFEN1 reactions containing GAP DF-5′B illustrating the effects of order of the SA addition on the rates of reaction. Note: the X-axis (time) is in log format. Premixed (black), blocked (cyan) and trapped (pink) were assembled, and then the reaction was initiated as in Figure 3B. All data points are the result of three independent experiments (error bars omitted for clarity). Rate constants are summarized in Supplementary Table S3B. (D) The concentrations of product formed at various time intervals after initiation of reactions of trapped and premixed gapped flap complexes that were challenged with excess competitor as in Figure 3C, before addition of magnesium buffer to initiate reaction. Concentrations of product formed from trapped complexes with and without the addition of competitor are shown in white and pink, respectively. Concentration of product formed from premixed complexes challenged with competitor is shown in gray. All data points are the result of three independent experiments with standard errors shown. See also Supplementary Figure S3.
5′-Substrate trapping does not inhibit reaction
To test whether the 5′-block prevented substrates threading through the helical arch and to rule out that 5′-streptavidin itself was inhibitory, we formed 5′-trapped complexes (Figure 2B). A trapped complex contained the same ingredients as a blocked complex, but the order of the addition of reagents differed. Without viable cofactor present, T5FEN and biotinylated substrate were premixed to form a complex, potentially allowing threading to take place and then excess SA was added. Upon initiation of reaction with Mg2+, the rate of decay of 5′-trapped complexes was only 2-fold slower than premixed complexes where no SA was added and was 2000 times faster than 5′-blocked complexes.
5′-Streptavidin addition creates a non-exchangeable-trapped complex
To further test that 5′-SA-trapped complexes were the result of threading, we challenged FAM-labeled substrate complexes with excess unlabeled substrate prior to initiation of reaction. Threaded 5′-trapped complexes would be predicted to have a dissociation rate commensurate with the biotin–streptavidin interaction and should therefore be essentially irreversible and resistant to competition on shorter time scales. Labeled substrate was competed from non-SA premixed complexes; the amount of FAM-product observed upon initiation of reaction with Mg2+ was consistent with ratio of labeled to unlabeled material and the concentration of enzyme (Figure 2C and D; Supplementary Figure S4). However, 5′-SA-trapped complex could not be competed away; the amount of product observed in this case was similar to that produced by the non-challenged trapped complex. In contrast, with the 3′-biotinylated substrate, the SA ‘trapping’ procedure resulted in a readily exchanged complex (Figure 2C and D). Combined blocking, trapping and competition experiments imply the 5′-portion of T5FEN substrates needs to be threaded through the helical arch for optimal enzyme activity.
hFEN1 uses the same mechanism for short and long 5′-flaps
hFEN1 catalyzed reactions have been suggested to proceed by two different mechanisms, dependent on 5′-flap length (16). Longer flaps are posited to thread through the helical arch, whereas substrates with shorter 5′-flaps (<6 nt) are supposed to not thread. Thus, to determine whether hFEN1 does indeed support two different mechanisms, we conducted experiments analogous to those described for T5FEN using 5′-biotinylated double flap (DF) substrates having 3 and 21 nt 5′-flaps (Figure 3A). As with T5FEN, biotinylation of substrates had negligible impact on hFEN1 catalytic parameters (Supplementary Table S1).
Premixed, blocked and trapped hFEN1 substrate complexes were created in the presence of catalytically inert Ca2+ (Figure 2B) (30,31). Results are analogous to those obtained with T5FEN. Rates of reactions of 5′-blocked complexes were decreased by three or four orders of magnitude dependent on flap length (Figure 3B). Similar reductions in reaction rates were also observed under multiple turnover conditions (Supplementary Table S2). In contrast, 5′-SA-trapped hFEN1 complexes behaved like premixed complexes containing non-SA substrates (Figure 3B). When challenged with excess unlabeled substrate prior to initiation of reaction, premixed enzyme–substrate complexes were readily competed (Figure 3C and Supplementary Figure S4). In contrast, 5′-SA-trapped complexes persisted and underwent hydrolysis. Importantly, similar outcomes are observed regardless of the length of the 5′-flap, and all data are in accord with a threading but against a clamping mechanism for all lengths of 5′-flap.
Unlike T5FEN and despite saturating conditions for the hFEN1–substrate interaction, the proportion of complex that was trapped (i.e. cleaved at 60 s, approximately 500 half-lives for trapped substrates) was altered by the conditions of pre-incubation (Figure 3D). Although some hFEN1–substrate complex was trapped in the presence of EDTA (15–20%), a greater proportion of substrate was trapped in the presence of Ca2+ ions (75–80%). Altering the temperature at which SA was added did not significantly alter the outcome. The portion of complex that did not react on a subsecond time scale decayed with a similar rate to 5′-SA blocked species. Thus, in EDTA the hFEN1–substrate complex is in equilibrium between threaded and non-threaded complexes both of which can be captured by 5′-SA. The amount of fast-reacting product reflects the equilibrium state between these two forms. When the trapping procedure is carried out with non-catalytic Ca2+ ions present, this equilibrium was altered in favor of the more catalytically proficient species (Supplementary Figure S5). These differences between higher and lower evolutionary FENs can be accounted for by their respective structures. As revealed by the T4FEN–PsY complex charged and aromatic residues of the partially structured arch interact with the flap in a state where it is not active site positioned due to lack of divalent ions (9). These residues are largely functionally conserved in T5FEN, but not in hFEN1. In hFEN1, the considerable repulsion afforded by the metal-free seven carboxylate active site, together with the possible influence of active site metals on arch conformation, presumably results in a requirement for divalent ions for flap accommodation.
Gap and flap endonuclease activities occur via the same mechanism
Structured helical arches are not large enough for duplex DNAs to pass through. Despite this and somewhat controversially, hFEN1 supports endonucleolytic reactions of gapped flaps containing short duplexes (11,13). To determine whether reactions of gapped substrates are a bona fide activity of hFEN1 instead of a co-purifying activity from Escherichia coli, we compared rates of hydrolysis of gapped and flap substrates with wild-type and K93A mutant hFEN1. Both proteins were rigorously purified using the same procedure. In hFEN1 and hEXO1 structures, this helical arch lysine, known to be important for catalysis in vitro and in vivo, contacts the 5′-phosphate monoester of product DNA positioned on active site metal ions and is suggested to act as an electrostatic catalyst (Figure 1F) (2,3,32,33). The gapped DF substrate contained a short stable hairpin within the 5′-flap (Figure 4A). The rate of reactions with the 21 nt DF and the gapped DF were both severely decreased with the K93A mutant, emphasizing that hFEN1 is responsible for the cleavage and that this reaction is mechanistically similar to the endonucleolytic incision of flaps lacking secondary structure (Figure 4B).
To ascertain whether gapped substrates are passed through the hFEN1 helical arch, a 5′-hairpin DF with a biotin label added to the hairpin turn was used (Figure 4A). Duplex stability was unaffected by biotin addition (Supplementary Figure S6A and S6B). Results were identical to those of 5′-ss-flap substrates. 5′-Blocking the gapped DF with SA severely decreases the rate of reaction (Figure 4C). Similarly, trapped complexes can be formed when SA is added after pre-incubation of biotinylated substrate with hFEN1; the rate of decay of these complexes is analogous to those premixed in the absence of SA. Additionally, although premixed gapped biotin DF–hFEN1 complex was readily competed by unlabeled substrate, it was not competed when 5′-trapped with SA (Figure 4D and Supplementary Figure S4). Thus even a short duplex contained within a 5′-flap is able to pass through the helical arch.
hFEN1 processes gapped flaps without resolving secondary structure
A possible mechanism for accommodation of 5′-gapped-flap substrates within the helical arch involves passage of a transient ss species (13,16). To test if 5′-duplexes must become ss for FEN processing, we formed a hairpin where the secondary structure within the 5′-flap could not be resolved due to a covalent cross-link. The triazole cross-link was formed by Cu(I) catalyzed Huisgen [3 + 2] cycloaddition reaction (‘Click chemistry’) of an alkyne and azide (Figure 5). Introduction of an azide was achieved using the convertible nucleoside sulfone (27) (Figure 5A), which after ODN assembly was reacted with an amino-PEG-azide before removal from the CPG support. The resultant azido functionalized 2,6-diaminopurine derivative base paired with the 5′-terminal dT of the hairpin (Figure 5B). An alkyne was introduced to the 5′-terminus using commercially available 5′-hexynyl phosphoramidite. Formation of the cross-linked hairpin was monitored by reversed-phase HPLC (Figure 5B and C). The resultant cross-linked oligomer, which no longer exhibited a temperature-dependent melting transition (Supplementary Figure S6C and D), was hybridized to a template strand to yield a locked-hairpin gapped DF substrate. hFEN1-catalyzed cleavage of this non-resolvable substrate proceeded with only a modest 2-fold reduction in rate (Figure 5D and E). Thus, accommodation of substrates with short duplex regions within the 5′-flap does not require the resolution of secondary structure.
Implications of threading or clamping models
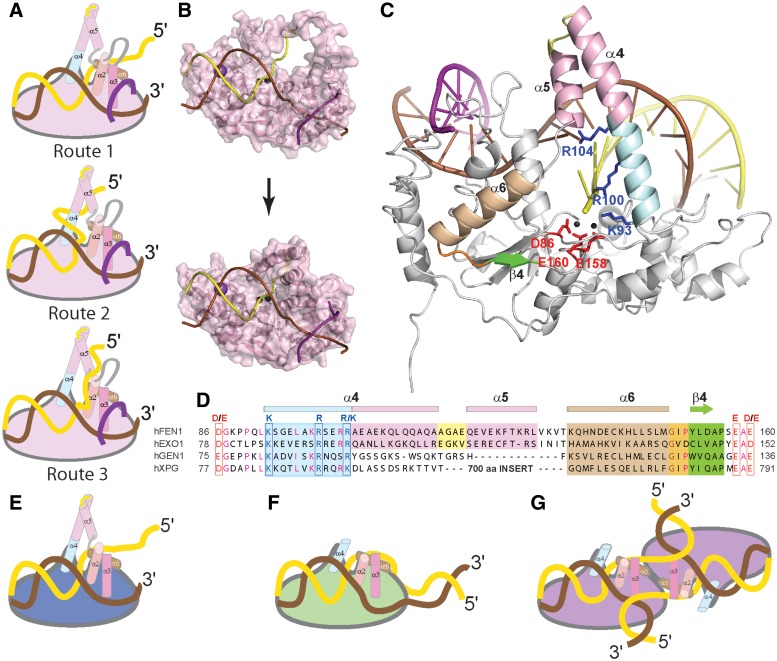
To understand the implications of threading and clamping models for 5′-nucleases, we examined hFEN1 and hEXO1 product DNA structures. Both complexes show the respective product 5′-phosphate monoesters bound by two active site divalent metal ions and superfamily conserved Lys93 and Arg100 residues (hFEN1 numbering) from the first helix (α4) of the helical arch (Figure 1F) (2,3). Three routes are possible for ssDNA extending from the 5′-phosphate, depicted schematically in Figure 6A. Following a simple linear path threads the DNA through the helical arch so that it encloses the first added nucleotide joined to the scissile bond (Figure 6A, route 1). For flapless DNAs that are substrates for both FENs and EXO1, this corresponds to the terminal 5′-nucleotide of the substrate. Introducing a turn at this nucleotide was necessary to create clamping models that allowed substrate to depart either side of the arch. Passing in front of α4 is unhindered but requires the phosphate backbone of the flap to approach the backbone of the substrate duplex (Figure 6A, route 2). Departure of substrate past the other side of the arch involves inserting the ssDNA between α5 and α2 (Figure 6A, route 3) (3). Route 3 is not feasible in hFEN1 or hEXO1 since this path is blocked by interactions between the α5–α6 linker and α2, but has recently been suggested to be the way XPG would accommodate NER bubbles (3). Similar models can also be created with bacteriophage FENs, where arch architecture differs slightly (Supplementary Figure S7). Notably, all clamping models only predict close proximity of the 5′-nuclease and the first 2–4 nt joining the scissile phosphate diester at the base of 5′-flaps.
Figure 6.
Disorder-thread-order model for hFEN1 and a unified model for the accommodation of substrates by the 5′-nuclease superfamily. (A) Schematic models for departure of the 5′-portion of hFEN1 substrates from the active site involving threading (route 1) or clamping (routes 2 and 3). Selected helical elements that define routes 1-3 are represented as cylinders. (B) Proposed disorder-thread-order model for coupling substrate selectivity to catalysis through the helical arch shown for hFEN1. The lower model is based on the structure of the enzyme-product (3Q8K). Bound DF substrate is modeled by extension of product DNA as in Figure 6A route 1. The upper panel is generated from the DNA-free hFEN1 (1UL1, Y chain) with missing regions modeled in with CHARMM and overlaying the substrate DNA from superimposition with the lower panel pdb model. (C) hFEN1-product structure (3Q8K) with secondary structure elements between D86 and D160 including the helical arch (amino acid residues 93–130), highlighted. The superfamily conserved lower part of α4, top of α4 and α5 (helical cap), α6, α6–β4 loop and β4 are colored pale cyan, pink, brown, orange and green, respectively. (D) Sequence alignment of hFEN1, hEXO1, hXPG and hGEN1 between D86 and D160 colored as in (C) showing conservation of catalytic residues and lack of conservation of the helical cap in hGEN1 and hXPG. (E-G) Schematic representations of the proposed unified substrate binding model for the 5′-nuclease superfamily where the 5′-portion of substrates departs from the active site following the same path. (E) EXO1 nuclease domain with pY DNA, (F) XPG nuclease domain with bubble and (G) GEN1 nuclease domain with a four-way junction. In (E) the junction portion of the substrate is positioned as in hEXO1 structures with the added 5′-flap passing through the arch. In (F) and (G), the FEN1/EXO1 helical cap has been removed allowing substrates without free 5′-ends to be accommodated. Reacting duplexes of substrates are positioned as in hFEN1 and hEXO1 structures and the path of 5′-portion of substrate past α4 (blue) is inferred from our data. The positioning of other substrate features is shown for illustrative purposes only.
DISCUSSION
The way FENs accommodate 5′-flaps and whether this in turn confers substrate selectivity have been longstanding questions. This puzzle is further complicated by the 5′-nuclease superfamily, where structurally related proteins with conserved active sites catalyze the same reaction, but preferentially act on different nucleic acid structures (1). Furthermore, despite long-held notions that FEN specificity is mediated by initial recognition of a free 5′-ss flap, analyses show that FENs initially bind the two-way junctions of substrates largely by complementary strand interactions (2,3,11–13,16,17,34). Recent structural analyses imply that double nucleotide unpairing of the reacting duplex is required to form a catalytically competent complex with contact between active site ions and the scissile bond (1–3,30,31). Thus, determining how FENs accommodate the 5′-portion of substrate is essential to understand how the reaction competent complex is formed. Furthermore, this information together with known differences in specific regions of various FEN superfamily members may suggest how this mechanism is adapted by other 5′-nucleases to substrates that lack free 5′-termini.
Our experiments, tested with two FEN family members, were designed to differentiate between threading and clamping models, and functionally determine how FENs accommodate 5′-flaps. When the 53 kDa streptavidin tetramer is bound to the 5′-flap before the addition of enzyme, reactions catalyzed by T5FEN and hFEN1 are inhibited consistent with streptavidin blocking the flap from threading though the arch (Figure 6A, route 1). Inhibition did not occur when streptavidin was added to the preformed enzyme–substrate complex, showing that streptavidin itself is not inhibitory and consistent with the hypothesis that the flap has already passed through the archway. Moreover, inhibition did not occur when streptavidin was added to the 3′-terminus of substrate before interaction with T5FEN, again showing that streptavidin itself is not inhibitory, but that its positioning on substrate is critical to the inhibition. The lack of displacement of labeled substrates by competitor from complexes where T5FEN or hFEN1 is added before 5′-streptavidin, contrasts with the ready competition of non-streptavidin conjugated complexes. This is consistent with a model where the 5′-flap is threaded through the arch and is trapped in this state by streptavidin.
In contrast, it is difficult to reconcile these results with clamping models where the substrate passes on either side of the arch (Figure 6A, routes 2 and 3). 5′-Streptavidin should not interfere with reactions proceeding by a clamping mechanism; indeed measured dissociation constants for hFEN1 5′-biotinylated substrates with long flaps ± streptavidin without cofactor ions are identical (16). The length of the 5′-flap in our substrates, particularly those with 21 nt flaps or gapped flaps, is much greater than the 2-4 nt that would be needed to pass the edge of the arch in either direction. An unlikely scenario where 5′-streptavidin produced non-exchangeable clamped (trapped) species would require an interaction between the tetramer and both T5FEN and hFEN1, regardless of the 5′-flap length. The possibility of a fortuitous interaction with streptavidin is ruled out by the lack of inhibition observed with the 3′-modified substrate where the length of the biotin linker could still allow interaction with the arch region of FENs. Together, our results strongly argue against a clamped structure and favor the threaded structure as the catalytically proficient form of T5FEN and hFEN1 that reacts on a biologically relevant timescale.
Despite the dimensions of structured helical arches that will not permit ds nucleic acid to pass through, our data show that substrates with a short gapped 5′-duplex behave similarly to 5′-ss flaps and must also be passed through the arch. Furthermore, resolution of secondary structure is not required to achieve this. This observation rules out a mechanism where FENs thread DNA through a structured archway and implies a model where the 5′-portion of substrate is initially passed through the unstructured or partially structured arch seen in many FEN structures (Figure 6B) (8,9). Our comparisons of FEN crystal structures, which underscore the flexibility of the arch region even in the crystal environment, are consistent with the opening of the arch rather than the melting of the gapped DNA. Once 5′-flap or 5′-gapped duplex DNA is through the disordered loop, adopting the helical structure correctly positions the catalytic lysine and arginine residues for interaction with the unpaired scissile phosphate diester at the active site metals (Figures 1F and 6B and C), thereby linking reaction site selection to catalysis. Our disorder-thread-order model explains data that appeared at odds with the threading of substrates through a closed aperture, such as the ability of FENs to process 2′-branched flaps or those with moderately bulky modifications such as (cis)-diamine dichloroplatinum (CPPD) adducts and thymine dimers (14,15). Threading though a disordered arch also eliminates concerns with an earlier bind and then thread hypothesis that proposed passing 5′-flaps through a structured archway (18). The act of binding the complementary DNA junction orients the 5′-portion of the substrate into the disordered/partially ordered arch region of the protein, and consequently there is no requirement for an energy source to push or pull a 5′-flap through a small aperture.
Considering the diverse range of substrates that the 5′-nuclease superfamily can process, devising a mechanism that is consistent with all family members and that can readily explain differences in overall specificity is a formidable challenge. Nevertheless, the junction binding abilities of 5′-nucleases moderated by hydrophobic wedges revealed by recent structures of hFEN1 and hEXO1 are consistent with adaptation to recognition of the bubble and four-way junction substrates of XPG and GEN1 (2,3). Similarly, as the site of reaction in all family members is predominantly 1 nt into a duplex, the double nucleotide unpairing mechanism implied by hFEN1 and hEXO1 structures seems likely to apply to all family members (1–3,30,31). As the hEXO1 helical arch so closely resembles that of hFEN1, it seems probable that this too uses a disorder-thread-order mechanism (Figure 6E). However, a considerable problem arises regarding how the 5′-portion of GEN1 and XPG substrates are accommodated. The data presented here do show that catalysis of reactions of blocked substrates can occur, albeit on a non-biologically significant time scale. Analogously, a chimera of the XPG junction binding domain with a FEN helical arch can cleave bubbles, albeit very slowly (2). Thus, if the features that encode specificity for free 5′-termini are removed, processing of bubbles and Holliday junctions could take place. Superfamily sequence comparisons of the arch subdomain and juxtaposed regions reveal that whilst all members conserve the base of α4 that contains conserved basic residues that interact with the scissile phosphate, the latter part of this helix and α5, called the ‘helical cap’, is not conserved by XPG and GEN1 (Figure 6C and D). If in XPG and GEN1 this region of the protein adopts a structure that does not close an aperture around substrates but instead creates a groove or cleft to accommodate a single non-base paired nucleotide, then the departure of all superfamily substrates from the active site can take a similar route past the catalytically relevant α4 residues (Figure 6A, route 1 and Figure 6E–G).
The disorder-thread-order mechanism revealed by the results presented here has implications for the roles of FEN1 and EXO1 during replication and repair and their ability to maintain genome fidelity. Gapped flaps containing short regions of duplex do not drastically alter FEN activity. When such gapped flaps form (e.g. from repeating sequences), they will not severely inhibit FEN1 activity protecting against genome expansions unless flap duplexes are bound stably by proteins (35). However, when duplexes in the 5′-portion of substrates are very long or lack a 5′-terminus, they will inhibit the activity of 5′-nucleases that thread substrates underneath a helical cap. This applies to gapped DNAs occurring between Okazaki fragments during lagging strand replication or as a response to damage. For these gapped DNAs, potentially two junction binding modes of FEN1 and EXO1 could result in reaction at a free 5′-terminus or of the continuous (in replication, parental) strand (Figure 7). The disorder-thread-order model predicts FEN1 or EXO1 catalyzed reaction of the continuous strand is strongly selected against because a long duplex could not pass through the aperture. Thus even when gaps are too small for ss binding proteins to associate with the continuous strand it remains protected from EXO1 and FEN1 action supporting genome fidelity. In contrast, a clamping mechanism would not protect the genome in this way.
Figure 7.
Models of FEN1 and EXO1 substrate interactions and their implications for genome integrity. The gapped junction between Okazaki fragments during lagging strand replication, or an equivalent structure produced as a response to damage, could be bound by FEN1 and EXO1 in two different ways. Both threading and clamping models could lead to reaction at the free 5′-termini after double nucleotide unpairing, with the 5′-nucleotide contained within or clamped by the arch (left). FENs readily process gapped flaps that contain short regions of duplex by threading them through the disordered arch. However, when the duplex is long or lacks 5′-termini, it could not pass through the arch and so reaction on the continuous strand is selected against, thus protecting genome integrity (right). In contrast if the 5′-portion of the substrate were clamped, the reaction of the continuous strand could occur.
The disorder-thread-order mechanism also has implications for targeting FENs for therapeutic intervention and for possible mechanisms for control of activity in other superfamily members including the more distantly related 5′-exoribonucleases. The importance of FEN activity in all organisms (4,33,36,37), differences between higher and lower evolutionary FENs and other superfamily members (1–3,6,9,17,19,30), and the high levels of FEN1 expression in cancer cells (38), all hold promise for possible antibacterial, antiviral or anticancer therapies. Thus, the search for FEN-specific inhibitors has been initiated (39). A requirement for disorder-order transition provides new inhibition strategies directed at preventing structural changes to be explored. Extending the possibility of disorder-order transitions of the base of α4 to other family members, regardless of whether they thread substrates under the helical cap or place them in a groove, suggests the possibility for control of activity of larger family members (EXO1, XPG and GEN1) through intra- and/or inter-molecular protein–protein interactions (3). The 5′-nucleases of RNA degradation Xrn1 and Xrn2 (Rat1 in yeast) conserve the FEN active site including equivalent positioning of lysine and arginine residues on an arch-like domain, seen as partially disordered in current structures (40–42). However, unlike FENs, these enzymes produce only mononucleotide products not flaps from 5′-phosphorylated but not m7GpppN-capped RNAs. It is interesting to note that exit from the back of the ‘arch’ is blocked in XRNs, and extending the threading model, this could explain both their specificity for 5′-phosphorylated species and the size of their products. Multiple rounds of disorder-thread-order transitions may be required for processive hydrolysis by these enzymes.
In contrast to other members of the superfamily, FENs are not activated by other proteins (3,43). Additionally, hFEN1 interaction partners have only modest effects on activity and do not downregulate it (44). Yet, in vitro hFEN1 is an extraordinarily efficient enzyme on DF substrates with second-order rate constants that approach those for the diffusional encounter of biomolecules (107 − 109/M/s) (13). Post-translational modifications may alter activity at certain cell cycle phases (45), but during replication FEN discrimination is paramount. FENs must only process substrates with free 5′-termini and not endanger the genome by cutting other junctions that are formed at replication forks. In particular, FENs must not destroy template DNA between Okazaki fragments. A single nucleotide 3′-flap binding site in hFEN1 affords some preference for the products of displacement synthesis (2,13,46). In concert, passing the 5′-portion of substrates through a disordered aperture, which then orders to position catalytic residues, couples substrate selection to catalysis and provides exquisite specificity for substrates that possess free non-protein bound 5′-termini.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7, Supplementary Tables 1–3.
FUNDING
Biotechnology and Biological Sciences Research Council (grant number BBF0147321 to J.A.G.); Marie Curie International Incoming Fellowship within the 7th European Community Framework Programme (project number PIIF-GA-2009-254386 to L.D.F.); National Cancer Institute (grant numbers RO1 CA081967, P01 CA092584 to J.A.T.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elaine Frary, Robert Hanson and Simon J. Thorpe for expert technical assistance.
REFERENCES
- 1.Tomlinson CG, Atack JM, Chapados B, Tainer JA, Grasby JA. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem. Soc. Trans. 2010;38:433–437. doi: 10.1042/BST0380433. [DOI] [PubMed] [Google Scholar]
- 2.Tsutakawa SE, Classen S, Chapados BR, Arvai A, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyamichev V, Brow MA, Dahlberg JE. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 6.Ceska TA, Sayers JR, Stier G, Suck D. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 7.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 8.Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devos JM, Tomanicek SJ, Jones CE, Nossal NG, Mueser TJ. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates. J. Biol. Chem. 2007;282:31713–31724. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- 10.Mueser TC, Nossal NG, Hyde CC. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the Rad2 family of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 11.Murante RS, Rust L, Bambara RA. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finger LD, Blanchard MS, Theimer CA, Sengerová B, Singh P, Chavez V, Liu F, Grasby JA, Shen B. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J. Biol. Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bornarth CJ, Ranalli TA, Henricksen LA, Wahl AF, Bambara RA. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 1999;38:13347–13354. doi: 10.1021/bi991321u. [DOI] [PubMed] [Google Scholar]
- 15.Barnes CJ, Wahl AF, Shen BH, Park MS, Bambara RA. Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′- to 3′-exonuclease endonuclease RAD2 homologue 1 or flap endonuclease 1. J. Biol. Chem. 1996;271:29624–29631. doi: 10.1074/jbc.271.47.29624. [DOI] [PubMed] [Google Scholar]
- 16.Gloor JW, Balakrishnan L, Bambara RA. Flap Endonuclease 1 Mechanism Analysis Indicates Flap Base Binding Prior to Threading. J. Biol. Chem. 2011;285:34922–34931. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams R, Sengerová B, Osborne S, Syson K, Ault S, Kilgour A, Chapados BR, Tainer JA, Sayers JR, Grasby JA. Comparison of the catalytic parameters and reaction specificities of a phage and an archaeal flap endonuclease. J. Mol. Biol. 2007;371:34–48. doi: 10.1016/j.jmb.2007.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. Contacts between the 5′ nuclease of DNA polymerase I and its substrate DNA. J. Biol. Chem. 2001;276:30167–30177. doi: 10.1074/jbc.M100985200. [DOI] [PubMed] [Google Scholar]
- 19.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 20.Lee BI, Wilson DM. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 21.Hohl M, Thorel F, Clarkson SG, Scharer OD. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 2003;278:19500–19508. doi: 10.1074/jbc.M213155200. [DOI] [PubMed] [Google Scholar]
- 22.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 23.Williams R, Kunkel T. FEN nucleases: bind, bend, fray, cut. Cell. 2011;145:171–172. doi: 10.1016/j.cell.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayers JR, Eckstein F. Properties of overexpressed phage T5 D15 exonuclease. Similarities with Escherichia coli DNA polymerase I 5′-3′ exonuclease. J. Biol. Chem. 1990;265:18311–18317. [PubMed] [Google Scholar]
- 25.Tock MR, Frary E, Sayers JR, Grasby JA. Dynamic evidence for metal ion catalysis in the reaction mediated by a flap endonuclease. EMBO J. 2003;22:995–1004. doi: 10.1093/emboj/cdg098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel D, Tock MR, Frary E, Feng M, Pickering TJ, Grasby JA, Sayers JR. A conserved tyrosine residue aids ternary complex formation, but not catalysis, in phage T5 flap endonuclease. J. Mol. Biol. 2002;320:1025–1035. doi: 10.1016/s0022-2836(02)00547-8. [DOI] [PubMed] [Google Scholar]
- 27.Shibata T, Glynn N, McMurry TB, McElhinney RS, Margison GP, Williams DM. Novel synthesis of O6-alkylguanine containing oligodeoxyribonucleotides as substrates for the human DNA repair protein, O6-methylguanine DNA methyltransferase (MGMT) Nucleic Acids Res. 2006;34:1884–1891. doi: 10.1093/nar/gkl117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocalka P, El-Sagheer A, Brown T. Rapid and efficient DNA strand cross-linking by click chemistry. Chembiochem. 2008;9:1280–1285. doi: 10.1002/cbic.200800006. [DOI] [PubMed] [Google Scholar]
- 29.Chivers C, Crozat E, Chu C, Moy V, Sherratt D, Howarth M. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nature Methods. 2010;7:391–393. doi: 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomlinson C, Syson K, Sengerová B, Atack J, Sayers J, Swanson L, Tainer J, Williams N, Grasby J. Neutralizing mutations of carboxylates that bind metal 2 in T5 flap endonuclease result in an enzyme that still requires two metal ions. J. Biol. Chem. 2011;286:30878–30887. doi: 10.1074/jbc.M111.230391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Syson K, Tomlinson C, Chapados BR, Sayers JR, Tainer JA, Williams NH, Grasby JA. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J. Biol. Chem. 2008;283:28741–28746. doi: 10.1074/jbc.M801264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengerová B, Tomlinson C, Atack JM, Williams R, Sayers JR, Williams NH, Grasby JA. Brønsted analysis and rate-limiting steps for the T5 flap endonuclease catalyzed hydrolysis of exonucleolytic substrates. Biochemistry. 2010;49:8085–8093. doi: 10.1021/bi100895j. [DOI] [PubMed] [Google Scholar]
- 33.Storici F, Henneke G, Ferrari E, Gordenin DA, Hübscher U, Resnick MA. The flexible loop of human FEN1 endonuclease is required for flap cleavage during DNA replication and repair. EMBO J. 2002;21:5930–5942. doi: 10.1093/emboj/cdf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 35.McMurray C. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuzaki Y, Adachi N, Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30:3273–3277. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finger LD, Shen B. FEN1 (flap structure-specific endonuclease 1) Atlas Genet Cytogenet Oncol Haematol. 2010 http://AtlasGeneticsOncology.org/Genes/FEN1ID40543ch11q12.html (2 February 2012, date last accessed) [Google Scholar]
- 39.Tumey LN, Bom D, Huck B, Gleason E, Wang J, Silver D, Brunden K, Boozer S, Rundlett S, Sherf B, et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 2005;15:277–281. doi: 10.1016/j.bmcl.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 40.Chang JH, Xiang S, Xiang K, Manley JL, Tong LA. Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1. Nat. Struct. Mol. Biol. 2011;18:270–276. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinek M, Coyle SM, Doudna JA. Coupled 5′ Nucleotide recognition and processivity in Xrn1-Mediated mRNA decay. Mol. Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Schärer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng L, Jia J, Finger LD, Guo Z, Zer C, Shen BH. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, Shen BH. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 2010;6:766–773. doi: 10.1038/nchembio.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedrich-Heineken E, Hubscher U. The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 2004;32:2520–2528. doi: 10.1093/nar/gkh576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.