Eukaryotic viruses and their multicellular hosts have coevolved complex interrelationships to permit virus reproduction without destruction of the host. The IFN-induced cellular antiviral response is the first line of defense against viral infection within an animal host. On viral infection, the expression of IFN is induced at the transcriptional level. The IFN is secreted to protect adjacent cells from secondary infection, thereby limiting viral spread. Type I IFNs are composed of the different types of IFN-α that are produced in leukocytes and the IFN-β that is produced in fibroblasts and epithelial cells. Type I IFNs bind to their receptors and activate a signaling cascade that culminates in the transcriptional induction of at least 30 genes (1). Two of these genes encode latent enzymes requiring activation by binding to double-stranded (ds)RNA that is produced as a replication intermediate in the viral life cycle. First, latent 2′-5′-oligoadenylate synthetase is activated by dsRNA to increase synthesis of 2′-5′ oligoadenylates that are required to activate 2′-5′-A-dependent RNase L (2). Activated RNase L nonspecifically degrades single-stranded RNAs and thus limits virus production. Second, the dsRNA-activated protein kinase (PKR), the most well characterized IFN-induced gene product, mediates the antiviral actions of type I IFNs. In studies reported in this issue of PNAS, Lau and coworkers (3) show that reduction in PKR level acts to delay cell death and thereby converts a lytic infection by encephalomyocarditis virus (EMCV) into a persistent infection. These studies as well as those from other laboratories support the idea that viral pathogenesis may be mediated by PKR-induced cell death and suggest that modification of PKR function in vivo may be a feasible approach to influence viral pathogenesis. In addition, the study presents a model system that may be applied to study mechanisms responsible for the establishment of persistent infections for other viruses such as HIV and hepatitis C virus.
PKR is activated by dsRNA binding to two dsRNA binding motifs in its amino terminus to promote dimerization that subsequently stimulates trans-autophosphorylation (4). Phosphorylated dimeric PKR is the activated form that is able to recognize and phosphorylate the eukaryotic translation initiation factor-2 on Ser-51 of its α-subunit (eIF-2α; Fig. 1; ref. 5). eIF-2 is a heterotrimeric protein that binds GTP and initiator tRNAmet. This ternary complex binds to the small 40S ribosomal subunit to form a 43S complex. The 43S complex binds mRNA and then the 60S ribosomal subunit joins with concomitant hydrolysis of GTP. To promote another round of initiation, GDP bound to eIF-2 must be exchanged for GTP in a reaction catalyzed by eIF-2B. Phosphorylation of eIF-2α on Ser-51 increases the affinity of GDP for eIF-2 by 100-fold, such that GDP–GTP exchange is inhibited. As a consequence, eIF-2–GDP forms a stable complex with eIF-2B. Because eIF-2 is in excess over eIF-2B in the cell, phosphorylation of only a small percentage of eIF-2α can sequester all the cellular eIF-2B activity to prevent further translation initiation events. In such a manner, PKR restricts cellular as well as viral protein synthesis.
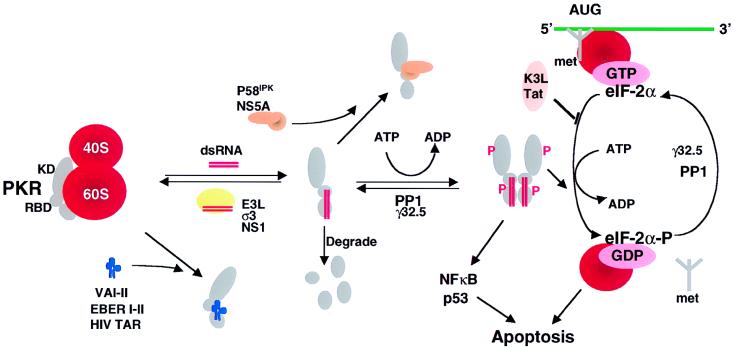
Figure 1.
Viral inhibitors of the PKR pathway. PKR is depicted as a monomer with a kinase domain (KD) and two dsRNA binding domains (RBD) bound to the 60S ribosomal subunit. The binding of dsRNA induces a conformational change to promote PKR dimerization, autophosphorylation, and activation of the eIF-2α kinase activity. Activated PKR also leads to activation of NF-κB, p53, and IFN regulatory factor 1 (IRF1). Viral inhibitors that act through different mechanisms are depicted. Adenovirus VA RNAs, Epstein–Barr virus EBER RNAs, and HIV transactivator responsive region TAR RNA bind and inhibit PKR and presumably displace PKR from the ribosome. Numerous viral proteins, such as vaccinia virus E3L, influenza virus NS1, and reovirus σ3 bind and sequester dsRNA, thereby preventing activation by dsRNA. Vaccinia virus K3L and HIV trans-activating transcriptional activator Tat act to inhibit PKR binding to eIF-2α. Protein phosphatase PP1 dephosphorylates phosphorylated eIF-2α as well as phosphorylated PKR. Herpes simplex virus 1 encodes a protein γ32.5 that facilitates activation of PP1. Hepatitis C virus nonstructural protein NS5A prevents PKR dimerization. In addition, influenza virus activates a cellular inhibitor P58IPK that also inhibits PKR dimerization. Poliovirus induces PKR degradation. (See ref. 5 and references therein). met represents initiator methionyl tRNA, and AUG represents the initiator codon.
To establish a productive infection, viruses must first overcome the IFN-induced blockades imposed on viral replication. As a consequence, IFN-resistant viruses have evolved many diverse mechanisms to inhibit PKR function (Fig. 1; ref. 6). These mechanisms include (i) production of RNAs or proteins that act as dsRNA antagonists to prevent PKR activation; (ii) synthesis of proteins that bind and sequester dsRNA; (iii) synthesis of proteins that prevent PKR dimerization; (iv) production of proteins that interfere with PKR binding to substrate eIF-2α; (v) activation of phosphatases that dephosphorylate eIF-2α and PKR; and (vi) inhibition of PKR expression or induction of PKR degradation. The role of PKR in viral infection is underscored by the finding that viruses, such as hepatitis C virus (7–9), vaccinia virus (10–12), and possibly HIV (13–15), have individually evolved multiple mechanisms to ensure inactivation of the PKR pathway.
To elucidate the role of PKR in viral infection, investigators have employed multiple strategies to interfere with the PKR pathway; these include expression of trans-dominant negative mutants of PKR, use of PKR antisense oligonucleotides, expression of viral inhibitors of PKR, and PKR gene deletion in mice. Results from these studies support the hypothesis that PKR plays a fundamental role in inhibiting protein synthesis and limiting viral replication and pathogenesis. However, recent studies also revealed that PKR plays a more pivotal role in this process by inducing apoptosis (16–18). Apoptosis is a genetically programmed event whereby cells undergo systematic self-destruction in response to a variety of stimuli. Recently, it was shown that increased PKR activity induces apoptosis in response to numerous stimuli, which include tumor necrosis factor-α, serum deprivation, dsRNA, and viral infection (16, 18–20). These recent observations on the ability of PKR to induce apoptosis support a model whereby PKR limits viral replication, not simply by inhibiting protein synthesis, but by actively inducing apoptosis.
Previous studies with Epstein–Barr virus and α-virus infection suggested that preventing apoptosis can convert a lytic infection into a persistent one (21, 22). In the studies of Lau and coworkers (3), PKR expression was reduced by antisense PKR RNA expression in the promonocytic cell line U937. On infection with EMCV, a cytolytic virus that is sensitive to IFN, cells with reduced PKR were resistant to lysis and became persistently infected with continual shedding of virus. Establishment of persistent infection correlated with reduced apoptosis that the authors attribute to reduced PKR activity. Although these results are consistent with a role of PKR in establishing persistent infection, this role cannot be confirmed from the antisense PKR experiments alone. Reduction of PKR levels could alter the expression of many genes that may contribute significantly to inhibiting apoptosis and/or establishment of persistence. For example, PKR induces transcription of genes that are responsive to dsRNA, which include the IFNs. It is known that EMCV is more sensitive to the IFN-induced 2′-5′-oligoadenylate synthetase pathway than to the PKR pathway (23, 24). EMCV is resistant to the inhibitory effects of IFN in cells that are defective in the 2′-5′ A pathway (25–27). In addition, 2′-5′-A-RNase L is also known to promote apoptosis (26, 28).
Another interesting observation of the present study was the coevolution of both virus and host that occurred with time. Over time, the cells assumed a slower growth rate, a reduced expression of IFN-α, an increased expression of Fas receptor and p53, and a resistance to superinfection with EMCV. At the same time, the EMCV evolved to become dramatically attenuated, although EMCV RNA was expressed at the same level. The continual evolution of both cells and virus is reminiscent of observations of EMCV persistent infection of K562 cells (29). The cells that are resistant to infection are selected to grow and eventually take over the population. Meanwhile, the virus also evolves such that variants are produced that can infect the resistant cells effectively. As a consequence, periodic fluctuations are observed in the viability of the coevolving cultures (29). This type of coevolving persistent infection in culture provides a useful model to study viral evolution.
What is the mechanism by which PKR expression activates apoptosis? Evidence supports the idea that PKR activates apoptosis upstream of the antiapoptotic function of Bcl-2 and involves activation of the death-executing enzyme caspase 3 (30). To date, studies have invoked phosphorylation of eIF-2α as well as activation of NF-κB or p53 in PKR-mediated apoptosis. Expression of a Ser51Ala mutant eIF-2α induced transformation of NIH 3T3 cells (31) and protected them from serum deprivation and tumor necrosis factor-α-induced apoptosis (20). It also protected from apoptosis induced by PKR overexpression in HeLa cells (32). In addition, expression of a Ser51Asp eIF-2α mutant, which functions to mimic phosphorylated eIF-2α, was sufficient to induce apoptosis in COS-1 cells (20). The findings provide evidence that eIF-2α phosphorylation may induce apoptosis directly. How might eIF-2α phosphorylation influence the apoptotic pathway? It was proposed that eIF-2α phosphorylation may induce preferential translation, by a mechanism that may be similar to the amino acid deprivation response in Saccharomyces cerevisiae, of selective mRNAs that provide apoptotic functions. Amino acid starvation in yeast activates the eIF-2α kinase Gcn2p to stimulate translation GCN4 mRNA encoding a transcription factor required to induce expression of the amino acid biosynthetic genes (33). GCN4 mRNA contains multiple short ORFs upstream from the AUG initiation codon. These reading frames are not recognized when eIF-2 is limiting, thereby allowing ribosomes to scan through and initiate at the authentic AUG initiation codon. It has been suggested that translation of some mRNAs encoding proapoptotic functions, such as the Bcl-2 family member Bax, may be controlled in this manner (34). An alternative hypothesis is that translation inhibition may deplete the cell of short-lived proteins that provide antiapoptotic functions, such as the inhibitors of apoptosis (35).
In addition to eIF-2α phosphorylation, PKR can lead to activation of the transcription factors NF-κB (32, 36, 37), IRF1 (37), and p53 (38), which are known to promote proinflammatory and proapoptotic responses. The NF-κB heterodimer is held in an inactive complex with its inhibitor IκB. In response to activators, Iκb is phosphorylated on residues 32 and 36 to induce its ubiquitin-dependent proteasome-mediated degradation, allowing NF-κB to translocate to the nucleus. Activation of PKR in cells leads to phosphorylation of IκB and activation of NF-κB (36), possibly by directly phosphorylating the IκB kinase complex (ref. 39 and B. R. G. Williams, personal communication). Cells that are defective in PKR are also defective in activation of NF-κB in response to dsRNA (37, 40). Although NF-κB provides primarily an antiapoptotic function, numerous reports have correlated NF-κB activation with apoptosis (ref. 32 and references therein). NF-κB induces transcription of several death-promoting transcription factors including p53 and cMyc as well as other death-promoting genes, e.g., Fas, Fas ligand, IRF1, and caspase-1. Therefore, under certain conditions, activation of NF-κB can lead to apoptosis. In the context of EMCV infection, Schwarz et al. (41) showed that NF-κB potentiates virulence of EMCV.
In addition, PKR can activate p53, although the mechanism by which this activation occurs is not understood. The tumor suppressor p53 is a transcription factor that regulates the cell cycle on genotoxic shock and is known to activate apoptosis (42). Phosphorylation of amino-terminal serine residues in p53 potentiates p53 function. PKR can phosphorylate p53 at Ser-392, although whether this phosphorylation activates p53 function is not known (43). Recently, it was shown that p53 function is impaired in PKR-deficient cells (38).
Given the fundamental role of PKR in apoptosis, it was surprising that mice with an amino-terminal or carboxyl-terminal disruption of the PKR gene had no evidence of increased tumorigenesis (40, 44). It is possible that another PKR-related gene may be responsible for the lack of tumorigenesis. It was interesting that mouse embryo fibroblasts derived from the amino-terminal PKR-deleted mice were defective in both induction of type I IFNs and activation of NF-κB in response to poly(I)⋅poly(C). However, this defect was restored by pretreatment with IFN. Although it is possible that poly(I)⋅poly(C) increased levels of the amino-terminal-deleted PKR gene product that could provide functional kinase activity, it is also possible that another IFN-inducible gene product was supplying the PKR function. Recently, a vertebrate homologue of yeast GCN2 was identified (45). Because yeast Gcn2p responds to amino acid deprivation, it is conceivable that this vertebrate gene product provides a PKR-like function in metabolically disrupted cells, such as may occur on transformation and/or viral infection. It is also important to note that another eIF-2α kinase, termed PKR-related endoplasmic reticulum kinase (PERK) or pancreatic eIF-2α kinase (PEK) was identified recently as a transmembrane protein localized to the endoplasmic reticulum (46, 47). PERK is activated on endoplasmic reticulum stress to phosphorylate eIF-2α. In the context of virus infection, it is interesting to consider that overexpression of viral glycoproteins may activate PERK, possibly duplicating the role of PKR in response to enveloped virus infection.
To date, studies indicate that the status of PKR activation in the cell may have several outcomes that depend on the virus and the host. In some viral infections, PKR activation will elicit early apoptosis, which limits viral spread. Viruses that have mechanisms to counteract PKR activation may establish persistent or latent infections that eventually will be detrimental to the host. Alternatively, some viruses may destroy host tissue by virus-induced PKR-mediated apoptosis and thereby contribute to the pathogenesis of viral disease. Certainly, the ability to alter the pathogenesis of viral infection by modulation of the PKR pathway would seem to be an attractive therapeutic approach. Presently, there are three known targets that may mediate PKR-induced apoptosis; eIF-2α, NF-κB, and p53. Future experiments need to identify which, if any, of these are important in viral pathogenesis. As we learn more about the mechanisms that viruses use to evade the PKR pathway, the feasibility of developing therapeutic strategies to interfere in this process increases and may be one step toward a cure for some viral diseases.
Footnotes
See companion on page 11860.
References
- 1.Sen G C, Ransohoff R M. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 2.Player M R, Torrence P F. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeung M C, Chang D L, Camanitigue R E, Lau A L. Proc Natl Acad Sci USA. 1999;96:11860–11865. doi: 10.1073/pnas.96.21.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, Kaufman R J. J Biol Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 5.Pain V M. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 6.Gale M, Jr, Katze M G. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 7.Gale M, Jr, Kwieciszewski B, Dossett M, Nakao H, Katze M G. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 9.Hatada E, Saito S, Fukuda R. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies M V, Chang H-W, Jacobs B L, Kaufman R J. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp T V, Witzel J E, Jagus R. Eur J Biochem. 1997;250:85–91. doi: 10.1111/j.1432-1033.1997.00085.x. [DOI] [PubMed] [Google Scholar]
- 12.Sharp T V, Moonan F, Romashko A, Joshi B, Barber G N, Jagus R. Virology. 1998;250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 13.Brand S R, Kobayashi R, Mathews M B. J Biol Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 14.Gunnery S, Rice A P, Robertson H D, Mathews M B. Proc Natl Acad Sci USA. 1990;87:8687–8691. doi: 10.1073/pnas.87.22.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunnery S, Green S R, Mathews M B. Proc Natl Acad Sci USA. 1992;89:11557–11561. doi: 10.1073/pnas.89.23.11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S B, Esteban M. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 17.Yeung M C, Liu J, Lau A S. Proc Natl Acad Sci USA. 1996;93:12451–12455. doi: 10.1073/pnas.93.22.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Der S D, Yang Y L, Weissmann C, Williams B R. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibler K V, Shors T, Perkins K B, Zeman C C, Banaszak M P, Biesterfeldt J, Langland J O, Jacobs B L. J Virol. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava S P, Kumar K U, Kaufman R J. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 21.Gregory C D, Dive C, Henderson S, Smith C A, Williams G T, Gordon J, Rickinson A B. Nature (London) 1991;349:612–614. doi: 10.1038/349612a0. [DOI] [PubMed] [Google Scholar]
- 22.Levine B, Huang Q, Isaacs J T, Reed J C, Griffin D E, Hardwick J M. Nature (London) 1993;361:739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 23.Chebath J, Benech P, Revel M, Vigneron M. Nature (London) 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 24.Rysiecki G, Gewert D R, Williams B R. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 25.Kumar R, Choubey D, Lengyel P, Sen G C. J Virol. 1988;62:3175–3181. doi: 10.1128/jvi.62.9.3175-3181.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou A, Paranjape J, Brown T L, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman R H. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassel B A, Zhou A, Sotomayor C, Maran A, Silverman R H. EMBO J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz-Guerra M, Rivas C, Esteban M. Virology. 1997;236:354–363. doi: 10.1006/viro.1997.8719. [DOI] [PubMed] [Google Scholar]
- 29.Pardoe I U, Grewal K K, Baldeh M P, Hamid J, Burness A T. J Virol. 1990;64:6040–6044. doi: 10.1128/jvi.64.12.6040-6044.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S B, Rodriguez D, Rodriguez J R, Esteban M. Virology. 1997;231:81–88. doi: 10.1006/viro.1997.8494. [DOI] [PubMed] [Google Scholar]
- 31.Donze O, Jagus R, Koromilas A E, Hershey J W, Sonenberg N. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gil J, Alcami J, Esteban M. Mol Cell Biol. 1999;19:4653–4663. doi: 10.1128/mcb.19.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinnebusch A. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 199–244. [Google Scholar]
- 34.Jagus R, Joshi B, Barber G N. Int J Biochem Cell Biol. 1999;31:123–138. doi: 10.1016/s1357-2725(98)00136-8. [DOI] [PubMed] [Google Scholar]
- 35.Deveraux Q L, Reed J C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 36.Kumar A, Hague J, Lacoste J, Hiscott J, Williams B R G. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuddihy A R, Li S, Tam N W, Wong A H, Taya Y, Abraham N, Bell J C, Koromilas A E. Mol Cell Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y-L, Reis L F, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams B R G, Aguet M, Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz E M, Badorff C, Hiura T S, Wessely R, Badorff A, Verma I M, Knowlton K U. J Virol. 1998;72:5654–5660. doi: 10.1128/jvi.72.7.5654-5660.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 43.Cuddihy A R, Wong A H, Tam N W, Li S, Koromilas A E. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- 44.Abraham N, Stojdl D F, Duncan P I, Methot N, Ishii T, Dube M, Vanderhyden B C, Atkins H L, Gray D A, McBurney M W, et al. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 45.Olsen D S, Jordan B, Chen D, Wek R C, Cavener D R. Genetics. 1998;149:1495–1509. doi: 10.1093/genetics/149.3.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X Z, Harding H P, Zhang Y, Jolicoeur E M, Kuroda M, Ron D. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Vattem K M, Sood R, An J, Liang J, Stramm L, Wek R C. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]