Abstract
Ataxia-telangiectasia (A-T) is a neurodegenerative disease caused by mutation of the Atm gene. Here we report that ATM-deficiency causes nuclear accumulation of histone deacetylase 4 (HDAC4) in neurons and promotes neurodegeneration. Nuclear HDAC4 binds to chromatin as well as to MEF2A and CREB, leading to histone de-acetylation and altered neuronal gene expression. Blocking either HDAC4 activity or its nuclear accumulation blunts the neurodegenerative changes and rescues several behavioral abnormalities of Atm mutants. Full rescue, however, also requires HDAC4 in the cytoplasm, suggesting that the A-T phenotype results both from a loss of cytoplasmic HDAC4 and its nuclear accumulation. To remain cytoplasmic, HDAC4 must be phosphorylated. The HDAC4 phosphatase, PP2A, is down regulated by ATM-mediated phosphorylation. In ATM deficiency, enhanced PP2A activity leads to HDAC4 dephosphorylation and nuclear accumulation. Our results define a crucial role of nuclear accumulation and cytoplasmic depletion of HDAC4 in the events leading to A-T neurodegeneration.
Ataxia telangiectasia (A-T) is a hereditary multisystemic disease resulting from mutations in the ATM gene, which encodes a 370kD member of the PI3-kinase family of kinases1-4. A-T symptoms include non-neurological phenotypes such as immune system defects, germ cell defects, hypersensitivity to ionizing radiation and increased susceptibility to cancer2,3,5. Yet it is the neuronal cell loss, the most direct cause of the devastating ataxia, which is arguably the least understood phenotype. We report on a newly discovered role for HDAC4 in this process. Class I and class IIa histone deacetylases (HDACs) play important roles in brain development and neuron survival6-10. HDAC4 (Class IIa) is abundant in neurons where it is predominantly cytoplasmic11-13. Relevant to the phenotype of A-T, HDAC4 deficiency in mouse is marked by a postnatal atrophy of the cerebellum with surviving Purkinje cells notably reduced in dendritic complexity14. HDAC4 is normally phosphorylated by calcium/calmodulin-dependent kinases (CaMKs) enabling its binding to the 14-3-3 family of protein chaperones7,15-17. HDAC4, like other class IIa HDACs, associates with the prosurvival transcription factors MEF2 (myocyte enhancer factor 2) and CREB (cAMP response element-binding protein), repressing theirtranscriptional activity18-20. Though well studied in other cell types, the relationship between HDAC4 and MEF2- or CREB-dependent gene expression in neurons remains largely unexplored.
Results
Nuclear accumulation of HDAC4 suppresses MEF2A- and CREB-dependent transcription
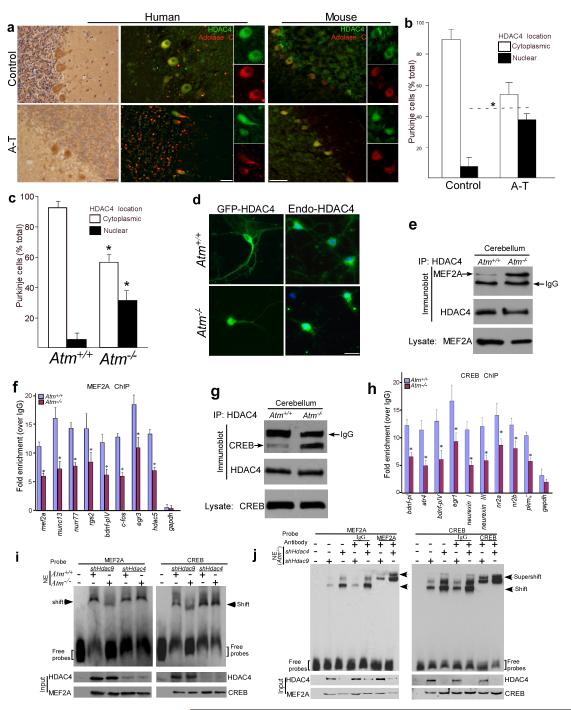
The analogies of neuron death and dendritic atrophy in the cerebella of ATM- and HDAC4-deficient mice9,18,21,22 prompted us to examine HDAC4 in human A-T cerebella. Normally, HDAC4 immunoreactivity is found in Purkinje cell cytoplasm (Fig.1a). In A-T samples by contrast Purkinje cell nuclei had strong HDAC4 staining (Fig. 1a, b). The nuclear accumulation of HDAC4 was specific; despite structural and functional similarities23,24, HDAC5 and HDAC9 showed little nuclear accumulation (Supplementary Fig.1a). These observations were replicated in mice. HDAC4, but not HDAC5 or HDAC9 (Supplementary Fig.1b), showed significant nuclear accumulation in Atm−/− but not in wild-type mouse Purkinje cells (Fig.1a, c). Similar shifts in HDAC4 localization were found in other mouse brain regions including neocortex and brain stem (Supplementary Fig.1c). The effect was direct; nuclear accumulation of GFP-HDAC4 was found in Atm−/− neurons (Fig. 1d). Further, after exposure of cultured cortical neurons to siRNA against Atm, HDAC9 remained in the cytoplasm while endogenous HDAC4 shifted to the nucleus (Supplementary Fig. 1d).
Figure 1.
Nuclear accumulation of HDAC4 in ATM-deficient neurons leads to suppression of MEF2- and CREB-related transcriptional activities
a) Paraffin sections of human cerebellar cortex from controls and A-T patients and cryostat sections of Atm+/+ and Atm−/− mouse cerebellum were immunostained with HDAC4 antibody using either HRP immunocytochemistry (brown) or immunofluorescence (green). Aldolase C (red) immunostaining was used as a cytoplasmic marker of Purkinje cells. Scale bar, 50μm.
b-c) The percentage of Purkinje cells with nuclear accumulation of HDAC4 were shown from A-T samples (b) and Atm−/− mice (c). Values represent the percentage of the total Purkinje cell population (Aldolase C counts). Each bar represents the average of three independent experiments; error bars denote SEM. (* = p < 0.05).
d) Images of endogenous and exogenous HDAC4 traffic in cultured neocortical neurons from both Atm+/+ and Atm−/− embryos. Scale bar, 20μm.
e, g) Protein extracts from Atm+/+ and Atm−/− mouse cerebella were immunoprecipitated with HDAC4 and blotted with MEF2A (e) or CREB (g) antibodies.
f - h) Following ChIP with MEF2A or CREB antibody from Atm+/+ and Atm−/− cerebellum, quantitative real-time PCR analysis was performed for the presence of specific MEF2A (f) or CREB (h) target genes (* = p < 0.01). Gapdh was used as a control. All q-PCR primers are listed in Table S1.
i, j) Validation of the effect of nuclear HDAC4 on MEF2A-DNA and CREB-DNA interaction in Atm−/− neurons. Nuclear extracts (NE) from Atm+/+and Atm−/− neurons with lentiviral shHdac9 and shHdac4 infection were incubated with biotin-labeled probes as indicated.
We next assayed the interaction of MEF2A and CREB proteins with HDAC4, and found it to be dramatically increased in Atm−/− cerebellar extracts as well as after overexpression of nuclear HDAC4 mutants (Fig. 1e, g, Supplementary Fig. 2a). That this served to inactivate MEF2A and CREB was suggested by their reduced promoter occupancy at known target genes in Atm−/− animals. We performed chromatin immunoprecipitation (ChIP) and assayed by qPCR the extent to which various promoter regions were occupied – Egr3, Fos and Mef2a for MEF2A; Atf, Nrxn and Erg-1 for CREB. In each case we found decreased levels of promoter occupancy in the Atm−/− samples (Fig. 1f, h). We next performed electrophoretic mobility shift assays with MEF2A and CREB DNA-binding consensus sequences. Nuclear extracts from Atm−/− mouse brains showed reduced binding to these probes compared to wild-type (Fig. 1i). Further, the DNA-binding of MEF2A and CREB as well as their supershifts could be restored in Atm−/− cells by knocking down HDAC4 but not HDAC9 (Fig. 1i, j). The specificities of the probes were documented (Supplementary Fig. 2b), as were the efficacies of the shRNAs (Supplementary Fig. 2c).
Nuclear HDAC4 suppresses neuronal gene expression driving neurodegenerative events
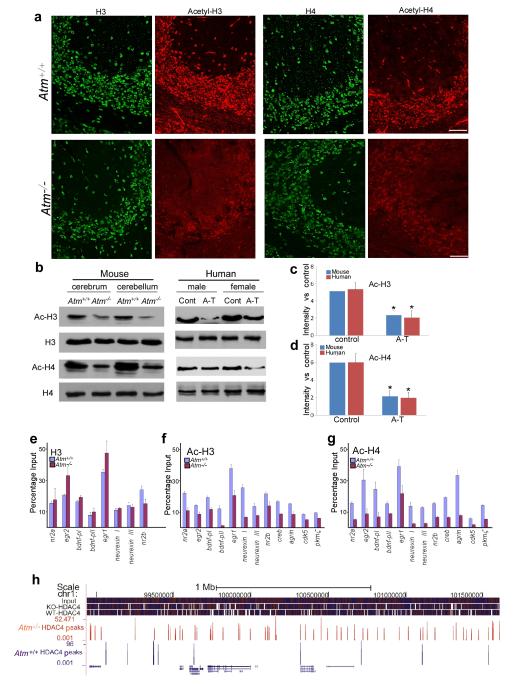
Enhanced HDAC4 in Atm−/− neuronal nuclei suggests a reduction of histone acetylation in these cells. Immunostaining for total histone 3 (H3) and 4 (H4) of Atm−/− cerebellum was similar to that of wild-type (Fig. 2a). By contrast, reduced immunostaining for acetylated histone 3 (AcH3) and 4 (AcH4) were found in Atm−/− neocortex and hippocampus (data not shown). These findings were confirmed by Western blots (Fig. 2b-d). We then examined the association of AcH3 and AcH4 with specific gene promoters. ChIP followed by qPCR assays was performed with extracts of wild-type and Atm−/− mouse cerebellum (Fig. 2e-g). H3 ChIP revealed no consistent difference between wild-type and Atm−/− samples in chromatin association at the promoters we examined (Fig. 2e). By contrast, the association of AcH3 and AcH4 with multiple neuronal growth genes was consistently decreased in Atm−/−. The loss of histone acetylation in a promoter region suggests a closed chromatin configuration and reduced transcription. As predicted, in Atm−/− samples, we observed decreased transcription of multiple neuronal genes at which promoter occupancy by acetylated histones was reduced, e.g., Bdnf, NR2a and Nrxn (Fig. 2f-g). Their occupancy by H3 (Fig. 2e) and H4 (not shown) were equivalent to wild-type.
Figure 2.
Nuclear accumulation HDAC4 leads to global effects on histone acetylation and neuronal gene expression
a) Fluorescent images of Atm+/+ and Atm−/− cerebellum sections immunostained for different histones and acetylated histones as indicated.
b) Protein extracts of cortex and cerebellum from wild-type and Atm−/− mice were probed with various histone antibodies as labeled to the left of the gels.
c-d) Quantification of 3 repetitions of the experiment illustrated in panel I. Error bars denotes standard deviations. * = p<0.05 (by Student’s T-test).
e-g) Fragmented chromatin was immunoprecipitated with the antibodies indicated and quantified with real-time PCR. The primers used for q-PCR are listed in Table S1. Statistical analysis was carried out using Student’s t test. Error bars represent SEM
h) An illustration of the HDAC4 ChIP-seq alignment and peaks. A 2.7 Mb sample region of chromosome 1 shows the density of coverage of 35 nt sequencing tags from input DNA or ChIP from wild type (blue) or Atm−/− (red) mouse brain.
We next performed ChIP with HDAC4 and found that it directly associates with chromatin (Supplementary Fig. 3a, b). We analyzed the HDAC4-precipitated DNA with ChIP-sequencing (ChIPseq) using a SOLiD™ DNA fragment library platform. The aligned ChIP sequence tags revealed patterns of HDAC4 chromatin-binding differed substantially between wild-type and Atm−/− (Fig. 2h and Supplementary Fig. 3c). To validate the ChIPseq results, ChIP-qPCR of selected genes was performed using primers designed within the ChIPseq-identified Atm−/− peak as well as in two sites flanking the peak by approximately 900 bp (Supplementary Fig. 3d-g lower panels). All four qPCR assays displayed increased HDAC4 binding only within the Atm−/− peak predicted by sequencing and peakfinding.
Inhibition of nuclear HDAC4 activity prevents neuronal dysfunction and behavioral abnormalities of Atm−/− mice
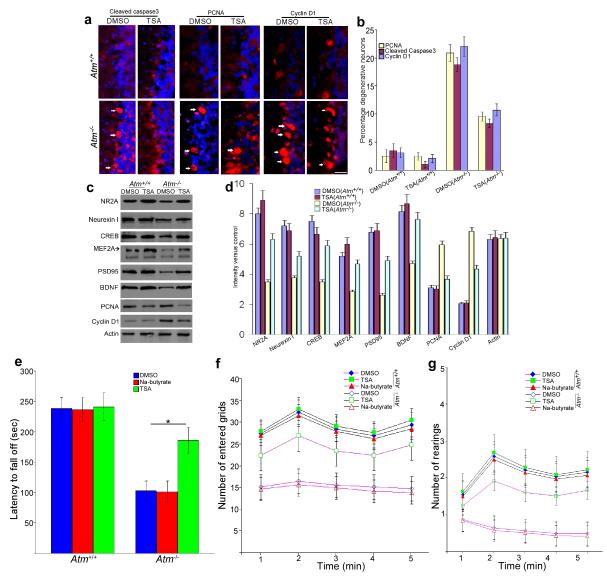
Our data indicate that shifting the location of HDAC4 from cytoplasm to nucleus is neurotoxic. Curiously, HDAC4 knockout mice have smaller brains, which suggests that genetic deletion of HDAC4 is also neurotoxic14. To explore this potential discrepancy, we treated Atm−/− and wild-type mice with trichostatin A (TSA), a classic HDAC inhibitor25-27. We used Na-butyrate, a Class I HDAC inhibitor, as a control. After seven days of TSA treatment, the levels of markers of degeneration such as cleaved caspase-3 and the cell cycle markers PCNA and cyclin D1 were substantially reduced from those normally found in Atm−/− cerebellum (Fig. 3a, b); neuronal survival proteins, by contrast were increased (Fig. 3c, d). Na-butyrate had little effect (Supplementary Fig. 4a, b).
Figure 3.
Inhibition of HDAC4 and blocking its nuclear accumulation partially reverses the A-T phenotype
a) TSA injection reverses neuronal degeneration markers in the Atm−/− cerebellum. Fluorescent images of Atm−/− brain sections immunostained for cleaved caspase-3 as well as PCNA and cyclin D1. White arrows indicate the labeled Purkinje cells. Scale bar, 25μm.
b) Quantification of the degeneration markers for the experiment illustrated in (a). Each bar represents the average of three independent experiments; error bars denote SEM.
c) Immunoblot assays of neuronal and cell cycle genes in mice cerebella lysates prepared from DMSO- or TSA-injected wild-type and Atm−/− mice)
d) Quantification of western blot bands illustrated in panel (c). Error bars denotes standard deviations.
e) Effects of TSA on the motor function of Atm−/− and wild-type animals. Motor performance measured as the average latency before falling from a rota-rod. Each treatment group consisted of 4-6 animals.
f-g) Effects of TSA on the spontaneous locomotor activities (f) and the exploratory activities (g) in Atm−/−mice were observed by open-field test. Data are presented as mean values ± SEM.
Atm−/− mice have motor abnormalities consistent with cerebellar dysfunction28, and the data in Fig. 3a-d predict that TSA should prevent them. Rota-rod and open-field performance of wild-type and Atm−/− mice were evaluated after a 3-week course of either TSA or Na-butyrate. After one additional week of training, both genotypes performed equally well at slow rotation speeds (10 rpm – data not shown). At more strenuous speeds (16 rpm), the performance of Atm−/− animals was significantly worse; TSA-treatment prevented this deficit (p < 0.05 – Fig. 3e). A similar effect was seen on spontaneous locomotor activity (Fig. 3f), as measured by open-field behavior. Exploratory activity (rearing – Fig. 3g) and total distance traveled (data not shown) of TSA-treated Atm−/− mice was significantly greater than that of untreated mutants (p < 0.05) and approached the levels found for wild-type mice. Na-butyrate had little effect on mutant performance in any behavioral tests (Fig. 3e-g).
HDAC4 and the DNA damage response
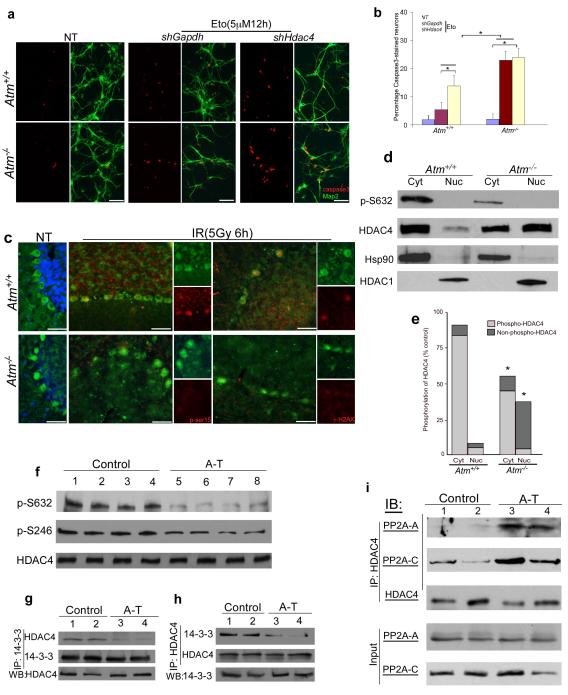
To determine whether HDAC4 played a role in the neuronal DNA damage response, we exposed cultures of wild-type neurons to low doses of etoposide. This enhanced the activation of caspase-3, but the effect was 10-fold greater in Atm−/− neurons (Fig. 4a, b). When shRNA against HDAC4 was used, however, the cell death induced by etoposide increased in wild-type with little effect on Atm−/− cells (Fig. 4a, b). Thus loss of cytoplasmic HDAC4 makes neurons more sensitive to DNA damage. Yet HDAC4 itself does not respond to DNA damage. We exposed mice to 5 Gy of whole body irradiation, activating ATM in wild-type, but not in Atmtm1Bal mutants (Fig. 4c). The cytoplasmic location of HDAC4 nonetheless remained unchanged in both genotypes (Fig. 4c).
Figure 4.
HDAC4 cytoplasmic localization requires its phosphorylation and is independent of DNA damage.
a) Effect of shHdac4 on caspase3 activation in Atm−/− neurons. Activation of caspase3 (red) was used as an index of impending neurodegeneration; Map2 (green) was used as a neuronal marker. Scale bar, 50 μm.
b) Cell death was quantified by counting the number of activated caspase3 immunostained cells and expressing these numbers as a percentage of the total Map2-stained neurons (* = p < 0.05).
c) Mice were treated with or without 5 Gy whole-body irradiation. Cryostat sections of Atm+/+ and Atm−/− cerebellum were immunostained for HDAC4 (green) and γ–H2AX or phospho-S15 of p53 (both in red). Scale bar, 50 μm.At least three-pair of age-matched animals were used for each experiment.
d) Immunoblot assays of HDAC4 and phospho-S632-HDAC4 in nuclear or cytoplasmic extracts prepared from Atm+/+ and Atm−/− mouse cerebellum. Hsp90 and HDAC1 were used as cytoplasmic and nuclear marker respectively.
e) Quantification of the bands shown in (d) reveals a significant decrease in the ratio of phosphorylated to non-phosphorylated HDAC4 in Atm−/− mouse cerebellum (* = p < 0.05).
f) ) Immunoblot assays of HDAC4 and phospho-HDAC4 in protein extracts prepared from frozen cerebellar samples of 4 human controls and 4 individuals with A-T.
g-h) Co-immunoprecipitations show the interaction between HDAC4 and 14-3-3 in lysates of cerebellar tissue from human control and A-T brain.
i) Co-immunoprecipitations show the association of HDAC4 with PP2A subunits in lysates of cerebellar tissue from human control and A-T brain.
Hypophosphorylation of HDAC4 induces its nuclear accumulation
Cytoplasmic and nuclear fractions were prepared from wild-type and Atm−/− mouse cerebella, and Western blotted with an antibody against phospho-S632-HDAC4 or total HDAC4. In wild-type, most HDAC4 is cytoplasmic and phosphorylated (Fig. 4d, e). In Atm−/− mouse cerebellum, however, a large part of HDAC4 is nuclear where it is almost entirely unphosphorylated. The levels of total HDAC4 were largely unchanged (Fig. 4d, e). Homogenates of human A-T cerebellar samples behaved similarly (Fig. 4f), and a second HDAC4 phosphospecific antibody (against phospho-S246) confirmed the phospho-S632 results (Fig. 4f). The reduced level of HDAC4 phosphorylation led to a reduced association with the 14-3-3 protein as assayed by co-immunoprecipitation from human A-T cerebellum (Fig. 4g, h) or Atm−/− mouse brain (Supplementary Fig. 5a, b).
We could find no canonical [S/T]Q ATM target site on HDAC4 itself, nor on the kinases responsible for its phosphorylation, CaMKII and CaMKIV7,17,29. Therefore, we asked whether the HDAC4 protein phosphatase, PP2A30, was involved. PP2A is a heterotrimeric protein31 that must associate with its enzymatic target for full phosphatase activity. It consists of a structural A subunit (PR65), a tissue specificity regulatory B subunit (PR55) and a catalytic C subunit (pp2ca). Immunoprecipitations of HDAC4 from controls revealed little interaction with either the PP2A-A or PP2A-C subunits. In the A-T patients, by contrast, a robust association was observed with both (Fig. 4i). The increased HDAC4/PP2A association was also found in Atm−/− mouse brain (Supplementary Fig. 5c). Thus, the decreased phosphorylation of HDAC4 in ATM-deficiency is due to an increased association between HDAC4 and its phosphatase, PP2A.
PP2A mediates HDAC4 nuclear translocation in an ATM-dependent manner
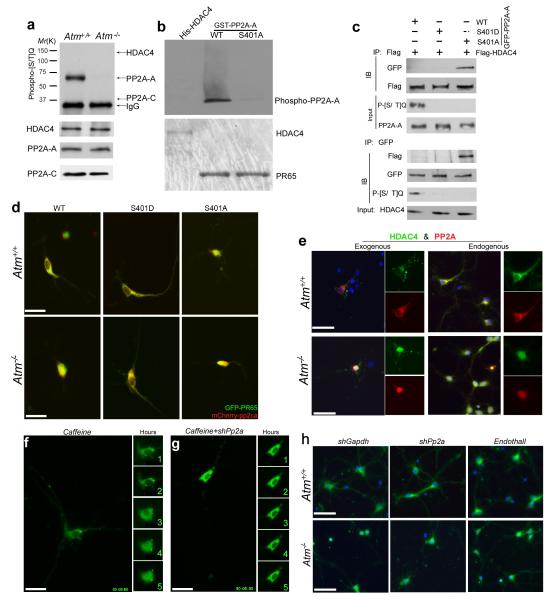
Serine 401 of PP2A-A is a highly probable site of ATM phosphorylation32. We therefore performed immunoprecipitation with PP2A-A, PP2A-C and HDAC4 antibodies and probed the resulting Western blots with an antibody against phosphorylated serine or threonine preceding a glutamine residue (P-[S/T]Q) – the canonical ATM/ATR target site. A strong P-[S/T]Q signal was found on the PP2A-A band from wild-type but not from Atm−/− mice (Fig. 5a). No P-[S/T]Q signal was found in either genotype with PP2A-C or HDAC4 immunoprecipitates (Fig. 5a). As further proof that HDAC4 itself is not an ATM substrate, no P-[S/T]Q signal was also found in N2a cells with overexpressing Flag-HDAC4 (Supplementary Fig. 6a). We verified that S401 is the predominant ATM phosphorylation site on PP2A by in vitro kinase assays (Fig. 5b); an S401A PP2A-A mutant could not be phosphorylated by ATM. We overexpressed Flag-tagged HDAC4 with GFP-tagged isoforms of PP2A-A, and analyzed the Flag-HDAC4 immunoprecipitates for PP2A-A. We found a strong HDAC4-PP2A association with the non-phosphorylatable (S401A) PP2A-A isoform (Fig. 5c), but not with wild-type or the phosphomimetic (S401D) isoform (Fig. 5c). This was confirmed by probing PP2A-A immunoprecipitates for Flag-HDAC4 (Fig. 5c).
Figure 5.
The PP2A-A subunit, PR65, is a novel ATM target and mediates nuclear accumulation of HDAC4 in ATM-deficient neurons
a) Protein extracts from Atm+/+ and Atm−/− mouse cerebellum were immunoprecipitated with the PR65, PP2A-C or HDAC4 and blotted with phospho-[S/T]Q antibody.
b) In vitro ATM kinase assays of His-tagged HDAC4, GST-tagged PR65 or PR65S401A were performed with N2a cell extract.
c) Co-immunoprecipitation assays of PP2A-A and HDAC4 in lysates prepared from N2a cells with overexpression of GFP-PP2A-A (WT, S401A or S401D) and Flag-HDAC4. Lysates were immunoprecipitated with anti-Flag or anti-GFP antibodies and blotted with phospho-[S/T]Q antibody.
d) Representative images of PP2A distribution in Atm+/+ and Atm−/− cultured neurons with co-expression of GFP-PP2A-A, wild-type, S401A or S401Dand mCherry-PP2A-C. Scale bar, 20 μm.
e) Immunofluorescent images of endogenous or exogenous HDAC4 (green) and PP2A (red) at DIV14 in wild-type and Atm−/− primary neurons. Scale bar, 25 μm.
f) Effect of inhibition of ATM activity by caffeine on the localization of GFP-HDAC4 in wild-type E16.5 cortical neurons. The five small panels to the right are isolated images of the cell body. The numeral in each of the small panels represents the time elapsed (hours) since the addition of ATM inhibitor. Scale bar, 20 μm.
g) Effect of knocking down PP2A on ATM-deficient GFP-HDAC4 nuclear accumulation in neurons. Scale bar, 20 μm. Small panels as in (f).
h) Immunofluorescent images of HDAC4 (green) at DIV7 in either shGapdh- or shPp2a-infected wild-type and Atm−/− primary neurons with one hour pretreatment with the PP2A-specific inhibitor, Endothall (5 μM). Scale bar, 25 μm.
ATM-dependent phosphorylation of PP2A-A also alters the localization of the PP2A holoenzyme itself. Endogenous PP2A-A was predominantly cytoplasmic in wild-type neurons, but predominantly nuclear in Atm−/− mutant neurons (Fig. 5e). In wild-type primary neurons, when we co-expressed mutant or wild-type GFP-PP2A-A with mCherry-PP2A-C both subunits distributed primarily in the cytoplasm (Fig. 5d). In Atm−/− neurons, however, unmodified PP2A-A and C translocated to the nucleus while the S401D mutant was predominantly cytoplasmic , even in ATM-deficient neurons . The S401A mutation was nuclear in both genotypes (Fig. 5d). In distinct contrast, the location of the HDAC4 kinases, CaMKII and CaMKIV, were unaffected by ATM-deficiency (data not shown).
We inhibited ATM activity with 2 mM caffeine or 10 μM KU55933. Either inhibitor caused HDAC4 to move from cytoplasm to nucleus within 3 hours (Fig. 5f and Supplementary Fig. 6b). This translocation of HDAC4 could be blocked by pretreating the cells with the PP2A inhibitor, endothall (Fig. 5h and Supplementary Fig.6c) or simultaneously knocking down PP2A (Supplementary Fig. 6d). The genotype-dependent nuclear translocation of endogenous HDAC4 could also be blocked by endothall or by infecting Atm−/− neurons with pr65 and pp2ca shRNAs (Fig. 5h).
Cytoplasmic HDAC4 improves the neurological phenotypes of A-T
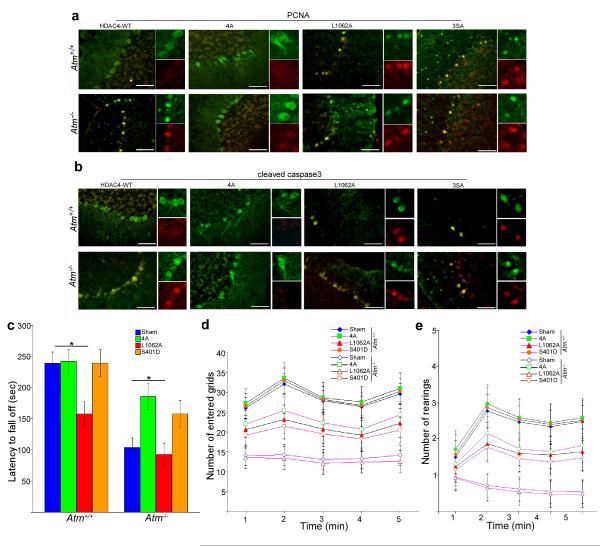
To verify that cytoplasmic HDAC4 prevents cell cycle re-entry and other degenerative changes in Atm−/− mice, we co-injected lentiviral particles encoding human wild-type HDAC4, a nuclear localization HDAC4 mutant (4A – R269A/R280A/K280A/R281A), a nuclear export mutant (L1062A) and a non-phosphorylatable mutant (3SA) together with shHdac4 (specific for the mouse Hdac4 message – Supplementary Fig.7a, b) into the cerebella of wild-type and Atm−/− mice. One week after injection of the two lentiviruses, gene transfer was monitored by immunohistochemistry using a human-specific HDAC4 antibody (Supplementary Fig.7c, d). In infected Atm−/− cerebellar neurons, cytoplasmic HDAC4 (4A) prevented cell cycle reentry and caspase-3 activation (Fig. 6a, b). By contrast, nuclear HDAC4 (L1062A and 3SA) resulted in cell cycle reentry and caspase-3 activation in both Atm−/− and wild-type mice (Fig. 6a, b). Reduced histone acetylation accompanied the increase in nuclear HDAC4, as confirmed by immunocytochemistry of Ac-H3 (Supplementary Fig.7e). Overexpression of unmodified human HDAC4 had little effect on either wild-type or Atm−/− neurons.
Figure 6.
a-b) Representative images of PCNA- and cleaved caspase3-stained Purkinje cells show the effects of lentiviral delivery of different HDAC4 mutants on degenerative progression in Atm−/− mouse cerebellum.. NLS-HDAC4 (cytoplasmic) = 4A; nuclear export mutant HDAC4 (nuclear) = L1062A; Non-phosphorylatable HDAC4 (nuclear) = 3SA. Scale bar, 50 μm.
c) Rota-rod tests show average latency to fall for wild type (+/+) and Atm−/− animals after injection of different HDAC4 as well as S401D lentiviral particles.
d-e) Open–field tests show effects of different lentiviral HDAC4 as well as S401D on the spontaneous locomotor activity (d) and exploratory activity (e) in Atm−/−mice. Each treatment group consisted of 4-6 animals. Data are presented as mean values ± SEM.
In primary neurons, overexpression of HDAC4 demonstrated that neuronal cell cycle activity was tightly correlated with the presence of HDAC4 nuclear localization (Supplementary Fig.6e-f). Cytoplasmic HDAC4 (4A) was protective and prevented Atm−/− neuronal cell cycle reentry. This offered us the opportunity to separately test the functions of nuclear and cytoplasmic HDAC4 and relate them to the presence or absence of behavioral abnormalities in Atm−/− mice. We co-injected lentiviruses encoding shHdac4 (against the mouse message) together with viruses encoding exclusively cytoplasmic (4A) HDAC4 into mouse cerebellum at postnatal day 18 (P18). At this age, the area infected by virus should be larger than following a comparable injection into an adult. At P39, when transgene expression is maximal, we subjected the mice to the same training and trials described above. Cytoplasmic 4A-injected Atm−/− mice were able to stay on the rota-rod significantly longer (p < 0.05) than controls (Fig. 6c), and the results of spontaneous locomotor activity observed in the open-field test were consistent with this difference (Fig. 6d). Exploratory activity (Fig. 6e) of 4A-injected Atm−/− mice were significantly greater (p < 0.05) than those of nuclear L1062A-injected ones (Fig. 6d, e) and 3SA-injected ones (data not shown). Note that L1062A-injected wild-type mice performed substantially worse than controls (Fig. 6c-e). To test the involvement of PP2A, we injected lentivirus carrying wild-type PP2A-A or the S401A or S401D isoform into mouse cerebella. The S401D mutant blocked HDAC4 nuclear translocation and inhibited cell cycle and capase-3 activation; wild-type PP2A-A and the S401A mutant had little effect (Supplementary Fig.8 a, b). The impact of the PP2A S401D mutant on the Atm−/− behavioral abnormalities, however, was modest (Fig. 6c-e). This might be due to the broad range of substrates whose phosphorylation would be changed by PP2A activity as well as to the indirect nature of the effect of PP2A on ATM.
Discussion
Our data provide new insights into the functions of both ATM and HDAC4 in the health and survival of CNS neurons. HDAC4 is normally maintained in the neuronal cytoplasm through its interaction with the 14-3-3 protein. This interaction requires CaMKII or CaMKIV-mediated HDAC4 phosphorylation. ATM plays an essential part in the cytoplasmic retention of HDAC4, but its action is indirect. It targets neither the CaMK kinases nor HDAC4 itself. Rather, it suppresses the HDAC4 phosphatase, PP2A, by phosphorylating serine 401 of its A subunit (diagrammed in Supplementary Fig.8c-e). In ATM deficiency, hypophosphorylated HDAC4 translocates to the nucleus where it triggers a significant decrease in histone H3 and H4 acetylation and large scale changes in the pattern of gene expression.
This study uncovers several significant new features of the epigenetic landscape of the Atm−/− neuronal genome. First, our ChIP analysis has shown genome-wide changes in the location of AcH3 and AcH4 in Atm−/− mice compared to wild-type in contrast to unmodified histone H3 or H4. Second, by performing ChIPseq with HDAC4 antibody, we provide data in support of the idea that HDAC4 is itself associated with specific genomic locations and these locations differ depending on the Atm genotype of the neuron. Most models of HDAC4 action are based on its ‘buffering’ proteins such as MEF2A thus decreasing their effective concentration at the promoters of key target genes33,34. Our ChIP and HDAC4 ChIPseq data suggest that in the absence of ATM, HDAC4 accumulates in the nucleus where it directly associates with chromatin. While it may remain associated with MEF2A and/or CREB, which is suggested by the increased binding of nuclear HDAC4 (3SA and L1062A) to MEF2A and CREB (Supplementary Fig. 3a), it can be described as a full participant in transcriptional suppression. This is emphasized by comparing HDAC4 chromatin-binding patterns with those of AcH3 and AcH4 or MEF2A and CREB alone. Several neuronal genes such as Creb, Mef2A and Bdnf have reduced AcH3 and AcH4 or MEF2A and CREB association yet show high levels of association with HDAC4 itself (Supplementary Fig. 3a, b). The third new epigenetic insight is that, in the promoter region of each genetic locus we have examined, the changes in gene transcription predicted by a more open conformation in the presence of acetylated histone proteins are born out by both RT-PCR and Western blot analysis. This suggests that important aspects of the neuronal phenotypes observed in A-T and in Atm−/− mice are caused not only by the absence of the ATM kinase activity itself, but by the indirect epigenetic changes in the pattern of histone acetylation and transcription factor buffering that occur as a result of nuclear HDAC4.
This concept suggests several new targets for pharmacological intervention in the treatment of A-T. For example, inhibiting HDAC4 activity or blocking its nuclear accumulation both reverse the altered histone acetylation pattern and rescue various aspects of the neurodegenerative phenotype of mouse ATM-deficient neurons. Note in particular, the strong phenotypic rescue of both the neurodegenerative changes of Atm−/− cerebellar neurons in vivo as well as the improvements in motor behavior when HDAC4 nuclear activity is inhibited or cytoplasmic HDAC4 is overexpressed (Fig. 3e-g and Fig. 6c-e). The lentivirus injection studies illustrate this concept on a cell-by-cell basis; the TSA injections make the same point at the level of the whole brain. TSA is admittedly a very broad spectrum HDAC inhibitor; yet the reversal of cell cycle protein expression is dramatic. In addition, the failure of Na-butyrate to elicit comparable effects suggests no involvement of Class I HDAC activities. This picture of a two-edged sword – neuroprotection of HDAC4 in the cytoplasm but neurotoxicity of HDAC4 in the nucleus – suggests that it may be more strategic to focus on HDAC4 location rather than activity as the most useful therapeutic target.
Methods
ChIP sequencing analysis
ChIP sequencing libraries were constructed from Anti-HDAC4-immunoprecipitated chromatin using the SOLiD™ DNA fragment library kit (Invitrogen) following the standard ChIP-Seq protocol. Libraries containing Atm+/+ or Atm−/− samples were bar-coded, applied to beads with emulsion PCR, enriched, and sequenced using the SOLiD™ System v3.5 at the Waksman Genomics Laboratory of Rutgers University. A sample prepared without immunoprecipitation from Atm+/+ was used as the input control. The resulting sequence files and quality scores are available from the NIH SRA, accession number SRA023500. Sequenced reads and quality strings were aligned to the mouse genome (mm9) using Bowtie 0.12.535 to identify the single, best-quality match location. Results were converted to BAM format using Samtools36. Peaks were selected, comparing Atm+/+ or Atm−/− against the input control, using Find Peaks 4.037. Peak tracks were created and visualized using the UCSC Genome Browser where subsets of peaks were chosen for qPCR validation.
Administration of TSA
Trichostatin A (TSA) and Na-butyrate were from Sigma. The inhibitors were administered by a single intraperitoneal (i.p.) injection to two-month old wild-type and Atm−/− mice twice a week at doses of 10 mg/kg (TSA) or 50 mg/kg (Na-butyrate). Seven days after the injection, the mice were killed and samples were collected. For the behavioral experiments, TSA and Na-butyrate were administered i.p. for 3 weeks, beginning at 4 weeks of age. Doses given were 10 mg/kg for TSA and 50mg/kg for Na-butyrate.
Lentivirus production and cerebellar injections
The hairpin sequences for MISSION® shRNA Lentiviral Transduction Particles used in cerebellum infection including shHdac4-3 and -4 (Table S2) were from Sigma. Human HDAC4 and PR65 lentiviral constructs were from GeneCopoeia. Site-direct mutation of lentiviral PR65 to S401A and S401D and lentiviral HDAC4 to 4A (R269A/R280A/K280A/R281A) and L1062A and 3SA (S246A/S467A/S632A) were performed by QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA). The high lentiviral particle titers were prepared with Lenti-Pac™ Expression Packaging Kits (GeneCopoeia) in HEK 293T cells and purified by Ultra-Pure Lentivirus purification Kits (Applied Biological Material Inc. Vancouver). Stereotaxic intra-cerebellar infusions were delivered to wild-type and Atm−/− mice (8 weeks of age)38,39 under isoflurane anesthesia. Mice were positioned in a Kopf stereotaxic apparatus and burr holes drilled into the skull and a 5μl Hamilton syringe fitted with a 33-gauge needle was lowered into the cerebellum for vector delivery. The coordinates of the burr hole and ventral location for infusion were: −7.2 mm from bregma, 1.0 mm lateral from the midline, and 3 mm ventral. For each cerebellum, slow infusion over 15 minutes of 3-5μl of lentiviral particles (1-5×109 IU/ml), was performed, with the needle withdrawn 5 min after completing infusion. For knocking down endogenous HDAC4 and overexpressing human HDAC4, we delivered shhdac4: human HDAC4 lentiviruses as 1:1 ratio. After surgery, animals were injected subcutaneously with 0.3 ml pre-warmed saline to avoid dehydration, and were allowed 7 days recovery prior to tissue collection. For behavioral experiments, lentiviruses were injected into mice cerebellum at postnatal 18 days (P18). 3 weeks after injection, mice were trained and for behavioral tests.
Rota-rod test
For the rotating rod test, age-matched animals were given two training trials (intertrial interval, 2 hr) with the rota-rod adjusted to accelerate from 6 rpm to 40 rpm over a 5 min period each day. Latency to fall off was measured. After 1 week, mice were tested using the rota-rod adjusted to maintain a constant speed for the entire 5 min test period. Each mouse was tested with the rota-rod set at 10 rpm and 16 rpm.
Open-Field Test
The open-field test was conducted as previously described40. Briefly, a floor open field (63 × 57 × 28 cm) was used to assess general motor activity. The floor of the arena was divided into a grid of 30 squares, each ~11 cm on a side. After placing a mouse at the center of the arena, locomotor activity, measured by the number of crossed grids, and exploratory activity, measured by the number of rearings on the hind feet, were recorded during a 5 min period. Total distance traveled was measured from recorded videotapes using video-tracking software (Spontaneous Motor Activity Recording and Tracking (SMART), San Diego instruments, San Diego, CA).
Statistical analyses
All data are presented as the means ± standard errors of the mean (SEM) of a minimum of 3 replicates. For most analyses, we evaluated statistical differences by Student’s t-test. For all analyses, we considered p < 0.05 to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Yang Xu for providing AtmtmBal/+ mutant strain. We thank Dr. Richard Gatti for sharing human A-T paraffin-fixed samples. Human frozen tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders (NICHD Contract #N01-HD-4-3368 and N01-HD-4-3383). We thank Drs. Michael B. Kastan, Tso-pang Yao and Stuart Schreiber for providing plasmids: Flag-ATM wild and Kinase dead (KD), GFP-HDAC4 and Flag-HADC4. Mavis Swerdel prepared the SOLiD ChIP-seq libraries. The long-term support of the A-T Children’s Project to K.H. is gratefully acknowledged. This work was also supported by grants from the NIH (NS20591 and NS71022) to K.H. and RPH (R21 MH085088). CLR is an NSF IGERT fellow.
Footnotes
Contributions J.L. and K.H. designed the experiments, analyzed data and wrote the manuscript. C.L.R. and R.P.H. developed, carried out and analyzed data for ChIP-seq analyses. J.L. and J.C. carried out the immunocytochemistry experiments. J.C. performed all of the q PCR experiments. J.L. and A.K. carried out the mouse cerebellar lentiviral injections. M.S.S and J.L and A.K. carried out behavioral tests.
References
- 1.Barzilai A, Biton S, Shiloh Y. The role of the DNA damage response in neuronal development, organization and maintenance. DNA Repair (Amst) 2008;7:1010–1027. doi: 10.1016/j.dnarep.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Bundey S. Clinical and genetic features of ataxia-telangiectasia. Int J Radiat Biol. 1994;66:S23–29. [PubMed] [Google Scholar]
- 3.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 4.Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 5.Sedgwick RP, Boder E. Progressive ataxia in childhood with particular reference to ataxia-telangiectasia. Neurology. 1960;10:705–715. doi: 10.1212/wnl.10.7.705. [DOI] [PubMed] [Google Scholar]
- 6.D’Mello SR. Histone deacetylases as targets for the treatment of human neurodegenerative diseases. Drug News Perspect. 2009;22:513–524. doi: 10.1358/dnp.2009.9.1428871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majdzadeh N, Morrison BE, D’Mello SR. Class IIA HDACs in the regulation of neurodegeneration. Front Biosci. 2008;13:1072–1082. doi: 10.2741/2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darcy MJ, Calvin K, Cavnar K, Ouimet CC. Regional and subcellular distribution of HDAC4 in mouse brain. J Comp Neurol. 2010;518:722–740. doi: 10.1002/cne.22241. [DOI] [PubMed] [Google Scholar]
- 12.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang AH, et al. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majdzadeh N, et al. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev Neurobiol. 2008;68:1076–1092. doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang AH, et al. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, et al. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J Biol Chem. 2001;276:35042–35048. doi: 10.1074/jbc.M105086200. [DOI] [PubMed] [Google Scholar]
- 18.Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youn HD, Grozinger CM, Liu JO. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J Biol Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]
- 21.Kuljis RO, Xu Y, Aguila MC, Baltimore D. Degeneration of neurons, synapses, and neuropil and glial activation in a murine Atm knockout model of ataxia-telangiectasia. Proc Natl Acad Sci U S A. 1997;94:12688–12693. doi: 10.1073/pnas.94.23.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 23.Backs J, Backs T, Bezprozvannaya S, McKinsey TA, Olson EN. Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol Cell Biol. 2008;28:3437–3445. doi: 10.1128/MCB.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mejat A, et al. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- 25.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nervi C, et al. Inhibition of histone deacetylase activity by trichostatin A modulates gene expression during mouse embryogenesis without apparent toxicity. Cancer Res. 2001;61:1247–1249. [PubMed] [Google Scholar]
- 27.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 28.Barlow C, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Randall WR, Schneider MF. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J Cell Biol. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paroni G, et al. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008;19:655–667. doi: 10.1091/mbc.E07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 33.Chan JK, Sun L, Yang XJ, Zhu G, Wu Z. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J Biol Chem. 2003;278:23515–23521. doi: 10.1074/jbc.M301922200. [DOI] [PubMed] [Google Scholar]
- 34.Miska EA, et al. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [PMID: 19505943] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fejes AP, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaemmerer WF, et al. In vivo transduction of cerebellar Purkinje cells using adeno-associated virus vectors. Mol Ther. 2000;2:446–457. doi: 10.1006/mthe.2000.0134. [DOI] [PubMed] [Google Scholar]
- 39.Cortes ML, Oehmig A, Perry KF, Sanford JD, Breakefield XO. Expression of human ATM cDNA in Atm-deficient mouse brain mediated by HSV-1 amplicon vector. Neuroscience. 2006;141:1247–1256. doi: 10.1016/j.neuroscience.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 40.Cooper JF, Kusnecov AW. Methylmercuric chloride induces activation of neuronal stress circuitry and alters exploratory behavior in the mouse. Neuroscience. 2007;148:1048–1064. doi: 10.1016/j.neuroscience.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.