Abstract
The Hedgehog pathway is one of the most common signal transduction pathways used by mammalian cells. Most studies have focused on its role during development, primarily of the nervous system, skin, bone and pancreas. Due to the activation of this pathway during proliferation and neoplastic transformation, more recent studies have examined its role in adult tissues. Significant levels of sonic hedgehog are expressed in the gastric mucosa, which has served to direct analysis of its role during organogenesis, gastric acid secretion and neoplastic transformation. Therefore the goal of this review is to apply current knowledge of this pathway to further our understanding of gastrointestinal physiology and neoplasia, using the stomach as a prototype.
Juanita L. Merchant holds faculty appointments in the Departments of Internal Medicine and Integrative & Molecular Physiology at the University of Michigan. She received her BS in Biology from Stanford University and received both her MD and PhD (Cell Biology) from Yale University. She completed Internal Medicine residency and a research fellowship in Gastroenterology at the Massachusetts General Hospital in Boston and her clinical fellowship in Gastroenterology at UCLA. Her primary research interests include transcriptional control mechanisms regulating cell growth and differentiation and microbial–host interactions in the luminal GI tract and their application to cancer pathogenesis. She is known for her work on how regulatory hormones in the stomach, e.g. gastrin and Shh, are regulated by the immune system. Her studies combine a number of modalities including molecular and cell biology, mouse genetics and translation to human biology.
 |
Introduction
This review summarizes the basic components of the Hedgehog (Hh) signalling pathway and considers our current understanding of this pathway in the luminal gastrointestinal tract with a focus on gastric physiology and tumorigenesis. Hh signalling in the pancreas and hepatobiliary systems has been recently reviewed elsewhere (Omenetti & Diehl, 2008; Teglund & Toftgard, 2010; Omenetti et al. 2011)).
Overview of hedgehog pathway
The Hh signalling pathway was initially identified during a mutagenesis screen conducted by the fly geneticists Eric Wieschaus and Christiane Nusslein-Volhard in 1978 (Nusslein-Volhard & Wieschaus, 1980)). The mutated gene corresponding to abnormal denticle formation (hair-like projections) resulted in flies that had the appearance of a hedgehog. They ultimately received the Nobel prize in 1995 for discovering several signalling pathways, including the Hh pathway, which control the segmentation patterning in Drosophila (Roush, 1995)). In flies, the pathway is initiated by a 471-residue protein called Hedgehog (hh). However, its prototypical mammalian counterpart was named Sonic Hedgehog (Shh) after the Sega video game character. Eventually, three mammalian hh genes activating the same signalling pathway were identified (Katoh & Katoh, 2005; Merchant et al. 2010b)). The other two gene products in addition to Shh were named Indian (Ihh) and Desert (Dhh) Hedgehog (Adolphe et al. 2004)) (Shimeld, 1999; Ramalho-Santos et al. 2000)). Since the major genes expressed in the luminal gastrointestinal tract are Shh and Ihh, this review will primarily refer to these two ligands. In general, the three ligands bind the same receptor Patched (Ptch1 and 2) and activate a signal transduction pathway resulting in the proteolytic processing of glioma-associated oncogene (Gli) transcription factor family members (Carpenter et al. 1998)). Ultimately, the processed Gli proteins bind the same DNA consensus binding site (5′-GACCACCCA-3′) (Kinzler & Vogelstein, 1990; Winklmayr et al. 2010)).
Although there are distinct differences in the tissue specific expression of the mammalian ligands, all three bind the Ptch receptors with the same affinity and activate identical signalling cascades. Therefore the difference between the three ligands appears to be their tissue specificity and the level of expression in each tissue (Pathi et al. 2001)). For example, Shh expression is highest in the developing organs of the foregut, which include lung, liver, pancreas, oesophagus and the proximal stomach (corpus) (Litingtung et al. 1998; Motoyama et al. 1998)). By contrast, Ihh expression emerges in hindgut-derived tissues, specifically the distal stomach (antrum), intestine and colon (Kolterud et al. 2009; van Dop et al. 2009; Zacharias et al. 2010)).
Hh ligand processing
Like many signal transduction pathways, there are multiple steps prior to activation of specific target genes. Both Shh and Ihh undergo proteolytic processing from the ∼45 kDa precursor to the ∼19 kDa cleaved ligand (Roelink et al. 1995; Valentini et al. 1997)). Since the steps for processing Shh have been the most intensively studied, generation of this ligand will be described in greater detail. Initially, the 24-amino acid signal peptide is cleaved from the amino terminus of the nascent precursor polypeptide (Lee et al. 1994; Bumcrot et al. 1995; Hammerschmidt et al. 1997; Goetz et al. 2002; Wendler et al. 2006)) then further processed within the endoplasmic reticulum (Chen et al. 2011)). The peptide undergoes cholesterol modification at cysteine 199 followed by autocatalysis mediated by serine protease-like activity residing within the C-terminal domain (Porter et al. 1995, 1996a,b)). The resulting 19 kDa-processed peptide is palmitoylated at cysteine 25 (cysteine 24 in human Shh) (Pepinsky et al. 1998; Buglino & Resh, 2010)) (Fig. 1)). Recent studies show that the palmitoylation step is required for the peptide to be tethered to the plasma membrane in position to be clipped by a member of the ADAM protease family (Dierker et al. 2009; Ohlig et al. 2011)) (Fig. 1)). The cholesterol-modified truncated Shh ligand then forms micelles with the protein Dispatched rendering the micelles capable of diffusion away from the cell of origin (Caspary et al. 2002; Callejo et al. 2011)).
Figure 1. Sonic Hedgehog processing.
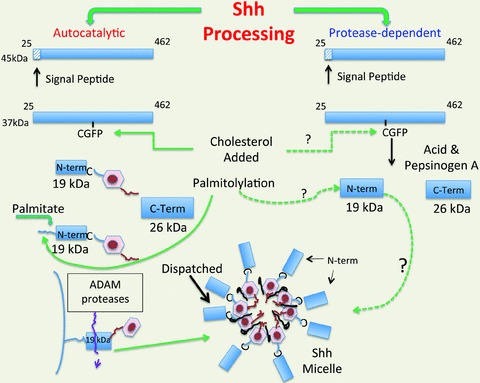
Two mechanisms of Shh processing have been reported. The best known is the autocatalytic mechanism of Shh processing in which the C-terminus functions as a cholesterol esterase adding the sterol to cysteine 199 followed by palmitoylation at residue 25. The fatty acid permits Shh to be tethered to the plasma membrane until it is cleaved by an ADAM protease. The cleaved Shh molecules form miscelles in the presence of the protein Dispatched (Disp). This mechanism has been described for Drosophila cells and cells derived from mesenchyme. By contrast, gastric acid producing parietal cells cleave Shh in a protease (pepsin)-dependent manner. It is not known whether Shh produced from the parietal cell is post-translationally modified with cholesterol or a fatty acid.
By contrast, we have observed in primary parietal cells that Shh processing is acid dependent (Zavros et al. 2007)) (Fig. 1)). Specifically, the consensus site for the aspartic protease pepsin A is GCFP residing at residues 198–201 within the Shh polypeptide. Thus pepsin cleavage is expected to occur on the C-terminal side of the phenylalanine residue (Kageyama, 2002)). Since spontaneous processing of the endogenous ligand to the 19 kDa form was not observed in primary gastric cell cultures, we concluded that Shh processing in the normal stomach requires acid to generate pepsin from its zymogen pepsinogen. It has not been determined whether the N-terminal form generated by gastric parietal cells is lipid-modified or forms micelles. Pepsin is a member of the aspartic protease family that includes both intracellular and transmembrane proteases activated at low pH, e.g. cathepsins and β-secretases, respectively (Vassar et al. 1999; Ivanova et al. 2008)). Thus it is possible that Shh might undergo proteolysis in the vesicular compartments of other cell types, e.g. neurons (Beug et al. 2011)). At present, it is difficult to reconcile whether autocatalytic processing of Hh ligands occurs in the mature gut epithelium. The studies establishing the autocatalytic mechanism for processing Drosophila hh and Shh were performed using transgenes transfected into Drosophila cells, embryos or mesenchymally derived mammalian cell lines, e.g. COS, BOSC, CHO, NIH3T3, HEK293, which typically do not express Hh ligands (Lee et al. 1994; Bumcrot et al. 1995; Porter et al. 1995; Chen et al. 2011)). Recent studies using in situ hybridization and LacZ reporter mice have established that both Shh and Ihh mRNAs are expressed exclusively in the intestinal and colonic epithelium and not in the mesenchyme (Kolterud et al. 2009; Varnat et al. 2010)). However, studies are needed to compare the endogenous peptide forms generated by other epithelium outside of the stomach.
One might speculate that the two different mechanisms of processing Shh ligand reflect tissue specific differences, i.e. one mesenchymal and the other epithelial. Since gastric acid secretion occurs at the apical surface, pepsin-mediated processing implies that Shh produced by the parietal cell could be secreted luminally or basolaterally creating a diffusion gradient. The role of Shh ligand in the lumen has yet to be defined since the Hh target cell typically resides on the basolateral surface in the mesenchyme. Mesenchymal cells express both the receptor Ptch and the Hh signalling apparatus, e.g. Gli1 (Kolterud et al. 2009; Varnat et al. 2010)). However, without direct evidence, one cannot exclude the possibility that Shh ligands secreted apically move via transcellular or paracellular mechanisms to the basolateral surface (Rogers & Schier, 2011)). Therefore, the current dogma based on these observations is that Hh signalling is unidirectional (paracrine) and proceeds from the gut epithelium to the mesenchyme.
Shh gradients and gene targets
As a morphogen, Shh forms a gradient such that the concentration of the active ligand inversely correlates with its diffusion distance (Rogers & Schier, 2011)). It is well established during embryonic development of the neural tube and limb bud that different subgroups of genes become activated or repressed depending on the concentration of Shh binding to the cell (Table 1)). Thus a different group of genes becomes activated or repressed by high concentrations of Shh (close to the cell of origin) compared to those activated or repressed by low concentrations of Shh (far from the cell of origin) (Table 1)). For example in the developing neural tube, the Shh gradient determines polarity along the ventral–dorsal axis, whereas in the developing limb bud, the Hh gradient determines digit formation (Wong & Reiter, 2008)). In this way, distinct gene clusters and subsequent cell fate can be regulated by the morphogen in a time-dependent and concentration-dependent manner (Rogers & Schier, 2011)).
Table 1.
Shh concentration gradient and target genes
| ShhHigh | ShhMed | ShhLow | |
|---|---|---|---|
| Gene A | On | Off | Off |
| Gene B | Off | On | On |
| Gene C | Off | On | Off |
| Neural tube | Ventral | → | Dorsal |
| Limb buds | 5th digit | 3rd digit | Thumb |
Hh signalling and primary cilia
The Drosophila gene encoding the transcription factor targeted by Hh signalling is called Cubitus interruptus (Ci) (Hui & Angers, 2011)) (Fig. 2)). However in mammals, there are three Ci isoforms designated Gli1, Gli2 and Gli3 that exhibit different functions (Kinzler & Vogelstein, 1990; Zhu & Lo, 2010)) (Fig. 2)). The N-terminus contains a repressor domain and binding site for a component of the repressor complex Suppressor of Fused (SuFu); while the C-terminus contains a transcription activation domain (TAD). The five C2–H2 zinc finger DNA binding domain (Znf) lies adjacent to the N-terminal repressor domain (Sasaki et al. 1999)). Centrally located nuclear localization and export motifs between the zinc finger and C-terminal domains direct the cellular location of the Gli factors (Sheng et al. 2006)). However, only Gli 2 and 3 possess an N-terminal repressor and C-terminal activator domain, whereas Gli1 does not contain a repressor domain (Hui & Angers, 2011)). Therefore masking or unmasking these two domains by proteolysis (via the cleavage domain) regulates whether these two transcription factors activate or repress Hh target genes (Wong & Reiter, 2008)).
Figure 2. Schematic diagram of Gli transcription factors.
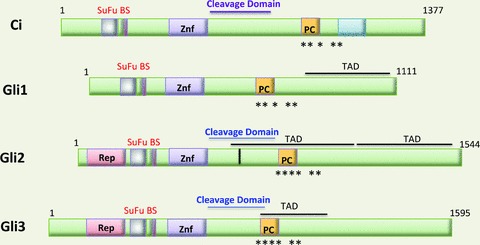
Gli1, Gli2 and Gli3 are zinc finger homologues of Drosophila Cubitus interruptus (Ci). The 4 transcription factors are aligned to demonstrate the homology between the various isoforms. Various functional domains of these proteins are indicated. Znf, zinc finger domain; TAD, transcription activation domain; Rep, repressor domain; SuFu BS, suppressor of fused binding site; PC, phosphorylation cluster; *protein kinase A phosphorylation site; cleavage domain: site of cleavage by proteasome. Modified from Hui & Angers, 2011 with permission.
Components of the Hh signalling pathway accumulate within specific organelles called primary cilia (Pazour & Witman, 2003; Haycraft et al. 2005; Kim et al. 2009)). Initially identified in Chlamydamonas, these plasma membrane protrusions serve both motile and sensory functions. The Hh transmembrane transducer Smoothened (Smo) accumulates in primary cilia upon ligand binding to Ptch1 (Kim et al. 2009)). There is only one primary cilium per cell and it is composed of acetylated α-tubulin condensed into nine microtubule arms arranged around the internal perimeter of the cilium. Since there is no central microtubule, the primary cilia are described as forming a 9 + 0 configuration (Pedersen et al. 2008)).
In the unstimulated state Smo, a 7-pass transmembrane receptor related to the G-protein coupled receptor (GPCR) family, resides in vesicles just below the plasma membrane surface. The repressor complex composed of the serine/threonine kinase Fused, adapter SuFu and kinesin-like Kif7 sequesters Gli 2 and 3 in the cilium tip (Fig. 3A)). Sequential phosphorylation of Gli2 and 3 by protein kinase A (PKA), glycogen 3b (GSK3b) and casein kinase 1 (CK1) create a phophopeptide motif on Gli 2 and 3 that is recognized by β-TrCP (β-transducin repeat-containing protein). β-TrCP binding subsequently recruits the Skip/Cullin1/F-box (SCF) E3 ubiquitin ligase. Addition of ubiquitin then directs the Gli proteins to the proteasome for limited (Gli3) or complete (Gli2) degradation by the proteasome. This in turn regulates the abundance of the Gli2 and Gli3 repressor forms that ultimately inhibit Hh target genes in the nucleus. In the nucleus, the Gli3 repressor inhibits Hh target genes by forming complexes with co-repressors such as Sin3A-HDAC or Ski-HDAC (Fernandez-Zapico, 2008)) (Fig. 3A)).
Figure 3. Hedgehog signalling in primary cilia.
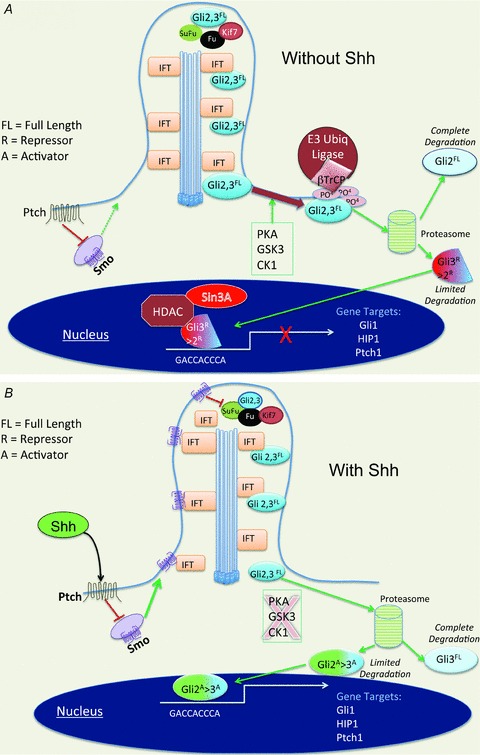
A, in the absence of Shh ligand, Smo-laden vesicles reside in the cytoplasm. Full-length (FL) Gli2 and Gli3 reside at the cilia tip with repressor complex proteins Fused, SuFu and Kif7. The intraflagellar protein (IFT) moves into the cilium with cargo proteins. If Gli2 and 3 are phosphorylated sequentially by PKA, CK or GSK3β, the docking protein βTrCP recognizes the phosphopeptide motif and recruits an E3 ligase to ubiquitinate (Ub) the protein. Ub Gli proteins undergo degradation by the proteasome. The repressor Gli2 protein moves to the nucleus and inhibits genes with co-repressor complexes. B, Hh signalling triggers Smo-laden vesicles to fuse with the plasma membrane once relieved from the inhibitory effects of the receptor Ptch. IFTs carry Smo antegrade to the tip of the cilium. Fused phosphorylates SuFu, Gli2 and Gli3 are released to the cytoplasm, phosphorylation is inhibited and Gli3 repressor is degraded. Gli2 migrates to the nucleus to bind Hh target genes.
Upon Hh ligand binding to its 12-pass transmembrane receptor Ptch, the Smo-containing vesicles fuse with the plasma membrane (Fig. 3B)). ‘Cargo’ proteins such as the intraflagellar proteins (IFTs) latch onto the microtubular tracks and move protein (e.g. Smo) up to the cilium tip where it encounters the Hh repressor complex (Haycraft et al. 2005; Milenkovic & Scott, 2010)). Once Smo interacts with the suppressor complex, Fu phosphorylates SuFu releasing the Gli2 and Gli3 transcription factors. This initiates retrograde migration from the tip of primary cilia to the cytoplasm where phosphopeptide formation and ubiquitination are blocked, preventing complete (Gli2) and limited (Gli3) degradation by the proteasome (Zhu & Lo, 2010)) (Fig. 3B)). Another scenario suggests that the absence of sequential phosphorylation results in limited degradation of Gli2 that removes the N-terminal repressor domain completely unmasking the activator domains (Hui & Angers, 2011)). The resulting ratio of activator to repressor Gli forms then shuttle to the nucleus to bind specific gene targets. Thus, it is the ratio of the Gli 2>3 activator to Gli 3>2 repressor proteins that regulates whether the cell response will be transcriptional induction or repression (Wong & Reiter, 2008)) (Fig. 3B)).
Since Ptch1, Hedgehog interacting protein (Hip1) and Gli1 are all transcriptional targets of Hh signalling, the pathway is capable of activating negative (Ptch1, Hip1) or positive (Gli1) feedback mechanisms that further modify the intensity and duration of the signal (Stecca & Ruiz, 2010)). Hh targets include genes regulating proliferation and differentiation, e.g. cyclin D1, D2, N-myc, Wnts, PdgfRα, Igf2, FoxM1, Hes1; cell survival (Bcl2); self-renewal (Bmi1, Nanog); angiogenesis (Vegf); and epithelial-mesenchymal transition (Snail1, Sip1, Elk1, Msx2). Moreover, crosstalk between the TGFβ-Smad3 (Johnson et al. 2011)) and receptor tyrosine kinase–Ras–Erk pathways through phosphorylation or protein interactions with Gli proteins (Lauth & Toftgard, 2007; Dennler et al. 2009; Chang et al. 2010; Fan et al. 2010)) implicate these two signalling pathways in Hh ligand-independent regulation.
Hedgehog signalling in gastric physiology
Ramalho-Santos and colleagues reported abnormal stomachs in day old Shh null mice (Ramalho-Santos et al. 2000)). Since that fortuitous observation, there has been a flurry of interest in understanding the role of Hh signalling in the luminal gastrointestinal tract (van den Brink, 2007)). The gut tube forms at embryonic day 9.5. During development, Shh gene expression remains high in the anterior gut (destined eventually to become oesophagus and gastric corpus) until embryonic day 15.5. At the pylorus, which demarcates the stomach from the duodenum, Shh expression is repressed and Ihh becomes the predominant ligand in the small and large intestine (Kolterud et al. 2009)). Reports on Shh expression in the adult mouse colon have varied according to the detection method. Normal human colon expresses Hh ligand and signalling components and expression increases with transformation (Oniscu et al. 2004)). Using in situ hybridization, van Dop et al. (2009) showed that Ihh mRNA is the predominant ligand in the adult colon and Shh expression was not detectable. Other reports indicate that Shh expression level is higher than observed in the small intestine, but is still significantly lower than observed in the stomach (Saqui-Salces & Merchant, 2010; Varnat et al. 2010)). Interestingly, Ptch1 and Gli1 (indicators of Hh signalling) overlap with the epithelial expression of Shh until E18.5 when these two Hh target genes become restricted to the mesenchyme. Thus during the early stages of embryonic development, Hh signalling is potentially autocrine, but switches to the epithelial to mesenchymal paracrine pattern just prior to birth.
In the adult corpus (anterior stomach), the stem cell gives rise to four major cell lineages, the acid-secreting parietal cell, mucous producing neck and surface pit cells (Fig. 4)), and the endocrine lineages (not shown) (Karam & Leblond, 1992)). The mucous neck lineage then gives rise to the zymogenic lineage as it migrates to the base of the gland (Ramsey et al. 2007)) Recent studies show that the TFF2-positive metaplasia that develops with gastritis arises from the zymogenic lineage via transdifferentiation (Nozaki et al. 2008; Nam et al. 2010)). Shh expression occurs in all gastric epithelial cells (Waghray et al. 2010)). Stimulation of acid secretion, e.g. with gastrin or histamine, induces Shh gene expression (Zavros et al. 2007)). Todisco and coworkers demonstrated that Shh stimulates H+,K+-ATPase gene expression in primary parietal cells (Stepan et al. 2005)). Moreover, we have shown that acid is required to process the 45 kDa Shh precursor in the stomach (Zavros et al. 2007)). Agents that inhibit acid, e.g. omeprazole, IL-1β and TNFα, block parietal cell Shh expression in primary cell cultures (Waghray et al. 2010)). Thus clearly Hh signalling is essential to the physiology of the stomach and specifically parietal cell acid secretion. The latter point was recently underscored by Zavros and coworkers who conditionally deleted the Shh locus in parietal cells (H+,K+-ATPase Cre mice bred to ShhFL/FL mice) (Xiao et al. 2010)). These mice were hypochlorhydric, hypergastrinaemic and developed foveolar hyperplasia (expansion of the surface mucous pit cell layer). This result suggests that the high levels of Shh produced by the parietal cells restrict expansion of the surface pit compartment (Fig. 4)). We also found that transgenic expression of the transcriptional target and Hh ligand inhibitor called Hip1 from the H+,K+-ATPase promoter inhibits acid secretion and increases plasma gastrin levels in 2-month-old mice (El-Zaatari et al. 2010)).
Figure 4. Gastric Shh inversely correlated with mucous cell compartment expansion.
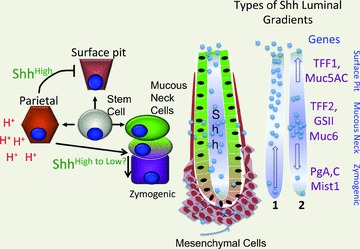
A significant amount of Shh in the stomach is generated by the acid-producing parietal cells. Conditionally null Shh mice expand their surface pit mucous cell compartment (foveolar hyperplasia). Shh processing in stomach requires acid secretion and is stimulated by gastrin. Shh is known to form a concentration gradient and in turn exert a differential effect on genes as a result. Thus there is a sliding scale of Shh concentrations and subsequent effects on gene expression. Therefore gastric Shh might form a gradient with increasing Shh concentration towards the gastric gland lumen (scenario no. 1) or maintain a gradient in which the concentrations are elevated near the mid-gastric gland then diffuse toward the lumen and gland base (scenario no. 2). Examples of potential gene targets expressed by specific cell types are listed on the right.
Under normal conditions, the mucous neck population appears to function as the precursor cell population that gives rise to the pepsinogen producing zymogenic cell lineage (Ramsey et al. 2007)). Thus the high levels of Shh produced by the parietal cells in the central portion of the gland appear to support this transition from mucous neck to zymogenic lineage (Fig. 4)). However, there are no transgenic studies demonstrating a role for Shh on other gastric epithelial cell populations. Nevertheless, the cells at the gland base are likely to be exposed to lower concentrations of Shh secreted from the parietal cells, raising the possibility that it is a gradient from high to low concentrations of Shh that correlates with the lineage shift (Fig. 4)). In gradient scenario no. 1, Shh diffuses from the central portion of the gland populated by parietal and mucous neck cells to the luminal surface (with acid).
By contrast in gradient scenario no. 2, there is bidirectional diffusion of Shh to both the gastric lumen and gland base. Typical lineage markers for these cell types are indicated to infer that the Shh concentration gradient established might affect these gene targets indirectly once canonical gene targets become activated in the mesenchyme (Fig. 4)). Examples of gene targets influenced by a Shh gradient are shown in Table 2 for the longitudinal gut axis and in Table 3 predicted for the gastric corpus gland. Therefore Shh ligand secreted specifically from parietal cells is required to maintain parietal cell homeostasis and secretory function in vivo.
Table 2.
Shh gradients and gut development
| ShhHigh | ShhMed | ShhLow | |
|---|---|---|---|
| H+,K+-ATPase | On | On | Off |
| Gastrin | Off | On | Off |
| Sucrase | Off | Off | Off |
| Gut polarity | Corpus | Antrum | Intestine/Colon |
Table 3.
Model for Shh gradients and gastric cell fate
| ShhHigh | ShhMed | ShhLow | |
|---|---|---|---|
| TFFl | Off | Off | On |
| TFF2 | On | Off | Off |
| Mist1 | Off | On | On |
| Gastric lineages | Mucous neck | Mucous neck to zymogenic | Zymogenic surface pit |
An important exception to the paracrine Hh signalling route is the response of myofibroblasts to tissue injury of which there are several examples that will be highlighted. Normally, the adult liver does not produce Hh ligands. However, the hepatic stellate cell initiates a tissue repair programme in which this myofibroblastic cell type both produces and responds to Shh by expressing collagen (Syn et al. 2009; Choi et al. 2010)). Persistent tissue injury produces liver fibrosis (cirrhosis) a preneoplastic change that can ultimately lead to hepatocellular cancer (Omenetti & Diehl, 2008)). Recently, it has been shown that infiltrating mesenchymal stem cells (MSCs) from the bone marrow will produce Shh once recruited to the stomach during Helicobacter-induced chronic gastritis (Quante et al. 2011)). This example suggests that bone marrow-derived cells recruited to the stomach exhibit a Hh ligand-signal transduction loop in response to tissue injury and inflammation. Finally in a recent study, it has been shown that Hh signalling is required for haematopoietic stem cell (HSC) proliferation and differentiation (Merchant et al. 2010a)). In that study, Gli1 null mice (deficient Hh signalling) showed defective myeloid differentiation and stress-induced haematopoiesis but no effect on T or B cell development. Thus the bone marrow-derived lineages appear to be especially sensitive to deficient Hh signalling.
The best-known example of regulated Shh expression in the setting of tissue injury and neoplastic transformation occurs in the development of cerebellar tumours (medulloblastoma). Specifically, the pro-inflammatory cytokine interferon γ (IFNγ) induces Shh gene expression in cerebellar granule neurons through a STAT1-dependent mechanism. A common theme among the examples in which Hh signalling exhibits an autocrine loop is that all four cell types (hepatic stellate, MSC, HSC, cerebellar granule cell) are a type of stroma-derived progenitor or stem cell (Silbereis et al. 2010; Quante et al. 2011; Reister et al. 2011)). We have confirmed that pro-inflammatory cytokines, e.g. IFNγ, stimulate Shh gene expression in primary parietal cell cultures (El-Zaatari et al. 2010)). Thus a plausible scenario to consider and further examine is the ability of pro-inflammatory cytokines to initiate carcinogenesis through chronic stimulation of Shh ligand production in stromal cells triggering their proliferation (fibrosis) and subsequent secretion of factors (Wnts, TGFβ) that stimulate specific epithelial cell populations, e.g. gastric mucous cell populations (Fig. 5)).
Figure 5. Pro-inflammatory cytokine initiation of Shh feedforward loop.
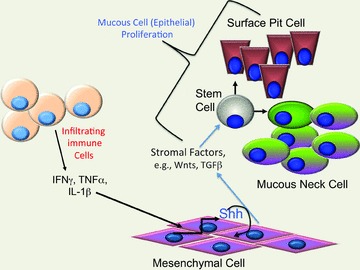
During chronic gastric inflammation, pro-inflammatory cytokines (Th1) are able to stimulate Shh gene expression in stromal cells. Inflammation in the stomach inhibits parietal cell acid secretion and eventually triggers loss of the parietal and zymogenic cell lineages (atrophy). This potentially results in expansion of the mesenchymal cell population and production of pro-proliferative stromal factors, e.g. Wnts, TGFβ. Wnt signalling is then available to induce mucous cell proliferation (metaplasia) and subsequently dysplasia/cancer.
In summary, Hh ligands typically produced by the epithelium target mesenchymal cells in the gut stroma (Kolterud et al. 2009)). This has been described as the classic ‘paracrine’ Hh pathway observed in multiple tissues. By contrast, other ligand-dependent mechanisms observed in cancers include ‘autocrine’ and ‘reverse’ paracrine in which the ligand is generated in stroma in addition to ligand-independent mechansims (cell autonomous) (Scales & de Sauvage, 2009; Marini et al. 2011)). In addition, regulation and tissue specific differences in Hh ligand processing might complicate our understanding of Hh signalling, but this will require analysis of the native protein in other epithelia. During chronic inflammation, pro-inflammatory cytokines create an environment supporting autocrine regulation of Hh signalling in MSCs (Quante et al. 2011)). This mechanism would imply a type of feedforward pathway initiated by inflammation. In the gut, infiltrating MSCs responding to cell autonomous Hh signalling might be an important contributor to gastrointestinal tumorigenesis.
Hedgehog signalling in gastrointestinal cancer
There are two schools of thought with respect to the role of the Hh pathway in gastric tumorigenesis. The first scenario is related to immunohistochemical staining of Hh pathway components in human gastric cancer showing Shh ligand and signalling components expressed in supporting mesenchymal cells (Fukaya et al. 2006; Teglund & Toftgard, 2010)). This theory would suggest that Hh mediates autocrine signalling in the mesenchyme that either initiates or supports neoplastic transformation of the epithelium. However, it is difficult if not impossible to conclude causation by Hh signalling without functional testing by knocking down various components of the pathway in cell lines. The second scenario is based on emerging transgenic mouse studies in which conditional deletion of Shh or Hh signal transduction components (Ptch1, Smo, Gli1) would prevent transformation. As previously discussed, Xiao et al. (2010) performed conditional deletion of Shh from the parietal cells and observed hyperplastic changes. The proliferative phenotype was attributed to activation of epithelial to mesenchymal transition regulators, e.g. Snail.
In the small intestine, inhibition of Hh signalling by transgenic expression of Hip1 results in expansion of the mesenchyme and abnormal villus formation (Madison et al. 2005)). Overexpression of Ihh from the intestinal epithelium stimulates proliferation of the smooth muscle cells and over time epithelial expansion of the crypt compartment (Zacharias et al. 2011)). By contrast, Kaestner and co-workers found using a cell line based approach that mesenchymal targets of the Hh pathway FoxF1 and FoxL1 suppress Wnt signalling (Madison et al. 2009)). Thus when Hh ligands are deleted, specific Fox homeobox genes are not activated and Wnt signalling becomes de-repressed. This result suggests that Hh signalling initiated from the epithelium exerts an anti-proliferative effect on targets in the mesenchyme. Thus loss of Hh ligand from the intestinal epithelium appears to produce similar outcomes. In the stomach, loss of Shh with parietal cell atrophy appears to create an environment of epithelial hyperplasia whereas in the small intestine, there is smooth muscle atrophy (mesenchyme) but subsequent epithelial hyperplasia with loss of Ihh. The stronger mesenchymal response in the intestine might reflect subtle differences between Shh and Ihh.
Several studies have examined the expression of Shh and Hh signalling proteins in human colon cancer by immunohistochemistry (Oniscu et al. 2004; Alinger et al. 2009; Yoshikawa et al. 2009; Saqui-Salces & Merchant, 2010)), but few have demonstrated a causative role. Deletion of the Hh receptor Ptch1 increases the accumulation of myofibroblasts but reduced epithelial proliferation (van Dop et al. 2009)). However, Varnat et al. (2010) showed strong expression of the Shh promoter in normal mouse colon by crossing ShhCre mice to the LacZ indicator mouse line ROSA26. In addition, the group showed that conditional deletion of Smo (decreased Hh signalling) decreases polyp formation in APC mutant mice (APCFl/Fl× CreERT2). Collectively, these initial genetic studies suggest that Hh signalling in the intestine appears to be required for epithelial proliferation and neoplastic transformation.
Future directions
In summary, understanding Hh signalling in the luminal GI tract is still in its infancy but is poised to be of critical importance due to the high levels of expression in the normal adult stomach and to a lesser extent in the colon. Hh signalling is clearly important in the mesenchyme, but might be driven by ligand-independent mechanisms. In this review, the role of Hh signalling in normal gastric physiology, specifically its requirement for the stomach to secrete acid was highlighted due to a number of functional studies that currently exists. Although Shh levels have been strongly implicated in a number of epithelial cancers, e.g. skin, brain, pancreas, the direct role of Hh ligands in gastrointestinal cancers is not yet certain. The current evidence is most convincing for Hh signalling (Ptch1, Smo) inducing transformation, but this might be due to ligand-independent mechanisms. These issues will likely be sorted out as more genetic reagents become available for the luminal GI tract.
Acknowledgments
This work was supported by NIH grant P01-DK62041 to J.L.M. The author wishes to thank Gail Kelsey for the illustrations. The author has no competing financial interests.
References
- Adolphe C, Narang M, Ellis T, Wicking C, Kaur P, Wainwright B. An in vivo comparative study of sonic, desert and Indian hedgehog reveals that hedgehog pathway activity regulates epidermal stem cell homeostasis. Development. 2004;131:5009–5019. doi: 10.1242/dev.01367. [DOI] [PubMed] [Google Scholar]
- Alinger B, Kiesslich T, Datz C, Aberger F, Strasser F, Berr F, Dietze O, Kaserer K, Hauser-Kronberger C. Hedgehog signaling is involved in differentiation of normal colonic tissue rather than in tumor proliferation. Virchows Arch. 2009;454:369–379. doi: 10.1007/s00428-009-0753-7. [DOI] [PubMed] [Google Scholar]
- Beug ST, Parks RJ, McBride HM, Wallace VA. Processing-dependent trafficking of Sonic hedgehog to the regulated secretory pathway in neurons. Mol Cell Neurosci. 2011;46:583–596. doi: 10.1016/j.mcn.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Buglino JA, Resh MD. Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog. PLoS One. 2010;5:e11195. doi: 10.1371/journal.pone.0011195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumcrot DA, Takada R, McMahon AP. Proteolytic processing yields two secreted forms of sonic hedgehog. Mol Cell Biol. 1995;15:2294–2303. doi: 10.1128/mcb.15.4.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejo A, Bilioni A, Mollica E, Gorfinkiel N, Andres G, Ibanez C, Torroja C, Doglio L, Sierra J, Guerrero I. Dispatched mediates Hedgehog basolateral release to form the long-range morphogenetic gradient in the Drosophila wing disk epithelium. Proc Natl Acad Sci U S A. 2011;108:12591–12598. doi: 10.1073/pnas.1106881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D, Stone DM, Brush J, Ryan A, Armanini M, Frantz G, Rosenthal A, de Sauvage FJ. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc Natl Acad Sci U S A. 1998;95:13630–13634. doi: 10.1073/pnas.95.23.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Garcia-Garcia MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, Alcorn HL, Anderson KV. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr Biol. 2002;12:1628–1632. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- Chang H, Li Q, Moraes RC, Lewis MT, Hamel PA. Activation of Erk by sonic hedgehog independent of canonical hedgehog signalling. Int J Biochem Cell Biol. 2010;42:1462–1471. doi: 10.1016/j.biocel.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tukachinsky H, Huang CH, Jao C, Chu YR, Tang HY, Mueller B, Schulman S, Rapoport TA, Salic A. Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J Cell Biol. 2011;192:825–838. doi: 10.1083/jcb.201008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SS, Syn WK, Karaca GF, Omenetti A, Moylan CA, Witek RP, Agboola KM, Jung Y, Michelotti GA, Diehl AM. Leptin promotes the myofibroblastic phenotype in hepatic stellate cells by activating the hedgehog pathway. J Biol Chem. 2010;285:36551–36560. doi: 10.1074/jbc.M110.168542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S, Andre J, Verrecchia F, Mauviel A. Cloning of the human GLI2 Promoter: transcriptional activation by transforming growth factor-beta via SMAD3/beta-catenin cooperation. J Biol Chem. 2009;284:31523–31531. doi: 10.1074/jbc.M109.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierker T, Dreier R, Petersen A, Bordych C, Grobe K. Heparan sulfate-modulated, metalloprotease-mediated sonic hedgehog release from producing cells. J Biol Chem. 2009;284:8013–8022. doi: 10.1074/jbc.M806838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Zaatari M, Zavros Y, Tessier A, Waghray M, Lentz S, Gumucio D, Todisco A, Merchant JL. Intracellular calcium release and protein kinase C activation stimulate sonic hedgehog gene expression during gastric acid secretion. Gastroenterology. 2010;139:2061–2071. doi: 10.1053/j.gastro.2010.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, He M, Sheng T, Zhang X, Sinha M, Luxon B, Zhao X, Xie J. Requirement of TGFβ signaling for SMO-mediated carcinogenesis. J Biol Chem. 2010;285:36570–36576. doi: 10.1074/jbc.C110.164442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Zapico ME. Primers on molecular pathways GLI: more than just Hedgehog? Pancreatology. 2008;8:227–229. doi: 10.1159/000134271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Isohata N, Ohta H, Aoyagi K, Ochiya T, Saeki N, Yanagihara K, Nakanishi Y, Taniguchi H, Sakamoto H, Shimoda T, Nimura Y, Yoshida T, Sasaki H. Hedgehog signal activation in gastric pit cell and in diffuse-type gastric cancer. Gastroenterology. 2006;131:14–29. doi: 10.1053/j.gastro.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Goetz JA, Suber LM, Zeng X, Robbins DJ. Sonic Hedgehog as a mediator of long-range signaling. Bioessays. 2002;24:157–165. doi: 10.1002/bies.10056. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Brook A, McMahon AP. The world according to hedgehog. Trends Genet. 1997;13:14–21. doi: 10.1016/s0168-9525(96)10051-2. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Ivanova S, Repnik U, Bojic L, Petelin A, Turk V, Turk B. Lysosomes in apoptosis. Methods Enzymol. 2008;442:183–199. doi: 10.1016/S0076-6879(08)01409-2. [DOI] [PubMed] [Google Scholar]
- Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, Matrisian LM, Mundy GR, Sterling JA. TGF-beta promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res. 2011;71:822–831. doi: 10.1158/0008-5472.CAN-10-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T. Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell Mol Life Sci. 2002;59:288–306. doi: 10.1007/s00018-002-8423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam SM, Leblond CP. Identifying and counting epithelial cell types in the ‘corpus’ of the mouse stomach. Anat Rec. 1992;232:231–246. doi: 10.1002/ar.1092320208. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Comparative genomics on Sonic hedgehog orthologs. Oncol Rep. 2005;14:1087–1090. [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterud A, Grosse AS, Zacharias WJ, Walton KD, Kretovich KE, Madison BB, Waghray M, Ferris JE, Hu C, Merchant JL, Dlugosz AA, Kottmann AH, Gumucio DL. Paracrine Hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology. 2009;137:618–628. doi: 10.1053/j.gastro.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M, Toftgard R. Non-Canonical Activation of GLI Transcription Factors: Implications for Targeted Anti-Cancer Therapy. Cell Cycle. 2007;6:2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in hedgehog protein biogenesis. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development. 2005;132:279–289. doi: 10.1242/dev.01576. [DOI] [PubMed] [Google Scholar]
- Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J Biol Chem. 2009;284:5936–5944. doi: 10.1074/jbc.M808103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini KD, Payne BJ, Watkins DN, Martelotto LG. Mechanisms of Hedgehog signalling in cancer. Growth Factors. 2011;29:221–234. doi: 10.3109/08977194.2011.610756. [DOI] [PubMed] [Google Scholar]
- Merchant A, Joseph G, Wang Q, Brennan S, Matsui W. Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood. 2010a;115:2391–2396. doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant JL, Saqui-Salces M, El-Zaatari M. Hedgehog signaling in gastric physiology and cancer. Prog Mol Biol Transl Sci. 2010b;96:133–156. doi: 10.1016/B978-0-12-381280-3.00006-3. [DOI] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP. Not lost in space: trafficking in the hedgehog signaling pathway. Sci Signal. 2010;3:pe14. doi: 10.1126/scisignal.3117pe14. [DOI] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- Nam KT, Lee HJ, Sousa JF, Weis VG, O’Neal RL, Finke PE, Romero-Gallo J, Shi G, Mills JC, Peek RM, Jr, Konieczny SF, Goldenring JR. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139:2028–2037. doi: 10.1053/j.gastro.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki K, Ogawa M, Williams JA, Lafleur BJ, Ng V, Drapkin RI, Mills JC, Konieczny SF, Nomura S, Goldenring JR. A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology. 2008;134:511–522. doi: 10.1053/j.gastro.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Ohlig S, Farshi P, Pickhinke U, van den Boom J, Hoing S, Jakuschev S, Hoffmann D, Dreier R, Scholer HR, Dierker T, Bordych C, Grobe K. Sonic hedgehog shedding results in functional activation of the solubilized protein. Dev Cell. 2011;20:764–774. doi: 10.1016/j.devcel.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Omenetti A, Choi S, Michelotti G, Diehl AM. Hedgehog signaling in the liver. J Hepatol. 2011;54:366–373. doi: 10.1016/j.jhep.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenetti A, Diehl AM. The adventures of sonic hedgehog in development and repair. II. Sonic hedgehog and liver development, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol. 2008;294:G595–598. doi: 10.1152/ajpgi.00543.2007. [DOI] [PubMed] [Google Scholar]
- Oniscu A, James RM, Morris RG, Bader S, Malcomson RD, Harrison DJ. Expression of Sonic hedgehog pathway genes is altered in colonic neoplasia. J Pathol. 2004;203:909–917. doi: 10.1002/path.1591. [DOI] [PubMed] [Google Scholar]
- Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, Williams KP. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev. 2001;106:107–117. doi: 10.1016/s0925-4773(01)00427-0. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Witman GB. The vertebrate primary cilium is a sensory organelle. Curr Opin Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Veland IR, Schroder JM, Christensen ST. Assembly of primary cilia. Dev Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski K, Taylor FR, Wang EA, Galdes A. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273:14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, Beachy PA. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996a;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, Beachy PA. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996b;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, Friedman R, Varro A, Tycko B, Wang TC. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, Konieczny SF, Mills JC. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- Reister S, Kordes C, Sawitza I, Haussinger D. The epigenetic regulation of stem cell factors in hepatic stellate cells. Stem Cells Dev. 2011;20:1687–1699. doi: 10.1089/scd.2010.0418. [DOI] [PubMed] [Google Scholar]
- Roelink H, Porter JA, Chiang C, Tanabe Y, Chang DT, Beachy PA, Jessell TM. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- Rogers KW, Schier AF. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol. 2011;27:377–407. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- Roush W. Nobel prizes: fly development work bears prize-winning fruit. Science. 1995;270:380–381. doi: 10.1126/science.270.5235.380. [DOI] [PubMed] [Google Scholar]
- Saqui-Salces M, Merchant JL. Hedgehog signaling and gastrointestinal cancer. Biochim Biophys Acta. 2010;1803:786–795. doi: 10.1016/j.bbamcr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Sheng T, Chi S, Zhang X, Xie J. Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. J Biol Chem. 2006;281:9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- Shimeld SM. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev Genes Evol. 1999;209:40–47. doi: 10.1007/s004270050225. [DOI] [PubMed] [Google Scholar]
- Silbereis J, Heintz T, Taylor MM, Ganat Y, Ment LR, Bordey A, Vaccarino F. Astroglial cells in the external granular layer are precursors of cerebellar granule neurons in neonates. Mol Cell Neurosci. 2010;44:362–373. doi: 10.1016/j.mcn.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz IAA. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepan V, Ramamoorthy S, Nitsche H, Zavros Y, Merchant JL, Todisco A. Regulation and function of the sonic hedgehog signal transduction pathway in isolated gastric parietal cells. J Biol Chem. 2005;280:15700–15708. doi: 10.1074/jbc.M413037200. [DOI] [PubMed] [Google Scholar]
- Syn WK, Jung Y, Omenetti A, Abdelmalek M, Guy CD, Yang L, Wang J, Witek RP, Fearing CM, Pereira TA, Teaberry V, Choi SS, Conde-Vancells J, Karaca GF, Diehl AM. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137:1478–1488. doi: 10.1053/j.gastro.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Valentini RP, Brookhiser WT, Park J, Yang T, Briggs J, Dressler G, Holzman LB. Post-translational processing and renal expression of mouse Indian hedgehog. J Biol Chem. 1997;272:8466–8473. doi: 10.1074/jbc.272.13.8466. [DOI] [PubMed] [Google Scholar]
- van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- van Dop WA, Uhmann A, Wijgerde M, Sleddens-Linkels E, Heijmans J, Offerhaus GJ, van den Bergh Weerman MA, Boeckxstaens GE, Hommes DW, Hardwick JC, Hahn H, van den Brink GR. Depletion of the colonic epithelial precursor cell compartment upon conditional activation of the hedgehog pathway. Gastroenterology. 2009;136:2195–2203. doi: 10.1053/j.gastro.2009.02.068. [DOI] [PubMed] [Google Scholar]
- Varnat F, Zacchetti G, Ruiz i Altaba A. Hedgehog pathway activity is required for the lethality and intestinal phenotypes of mice with hyperactive Wnt signaling. Mech Dev. 2010;127:73–81. doi: 10.1016/j.mod.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. β-Secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Waghray M, Zavros Y, Saqui-Salces M, El-Zaatari M, Alamelumangapuram CB, Todisco A, Eaton KA, Merchant JL. Interleukin-1β promotes gastric atrophy through suppression of Sonic Hedgehog. Gastroenterology. 2010;138:562–572. doi: 10.1053/j.gastro.2009.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler F, Franch-Marro X, Vincent JP. How does cholesterol affect the way Hedgehog works? Development. 2006;133:3055–3061. doi: 10.1242/dev.02472. [DOI] [PubMed] [Google Scholar]
- Winklmayr M, Schmid C, Laner-Plamberger S, Kaser A, Aberger F, Eichberger T, Frischauf AM. Non-consensus GLI binding sites in Hedgehog target gene regulation. BMC Mol Biol. 2010;11:2. doi: 10.1186/1471-2199-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SY, Reiter JF. The primary cilium at the crossroads of mammalian hedgehog signaling. Curr Top Dev Biol. 2008;85:225–260. doi: 10.1016/S0070-2153(08)00809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Ogle SA, Schumacher MA, Orr-Asman MA, Miller ML, Lertkowit N, Varro A, Hollande F, Zavros Y. Loss of parietal cell expression of Sonic hedgehog induces hypergastrinemia and hyperproliferation of surface mucous cells. Gastroenterology. 2010;138:550–561. doi: 10.1053/j.gastro.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Shimada M, Miyamoto H, Higashijima J, Miyatani T, Nishioka M, Kurita N, Iwata T, Uehara H. Sonic hedgehog relates to colorectal carcinogenesis. J Gastroenterol. 2009;44:1113–1117. doi: 10.1007/s00535-009-0110-2. [DOI] [PubMed] [Google Scholar]
- Zacharias WJ, Li X, Madison BB, Kretovich K, Kao JY, Merchant JL, Gumucio DL. Hedgehog is an anti-inflammatory epithelial signal for the intestinal lamina propria. Gastroenterology. 2010;138:2368–2377. doi: 10.1053/j.gastro.2010.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Madison BB, Kretovich KE, Walton KD, Richards N, Udager AM, Li X, Gumucio DL. Hedgehog signaling controls homeostasis of adult intestinal smooth muscle. Dev Biol. 2011;355:152–162. doi: 10.1016/j.ydbio.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavros Y, Waghray M, Tessier A, Bai L, Todisco A, Gumucio DL, Samuelson LC, Dlugosz A, Merchant JL. Reduced pepsin a processing of sonic hedgehog in parietal cells precedes gastric atrophy and transformation. J Biol Chem. 2007;282:33265–33274. doi: 10.1074/jbc.M707090200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Lo HW. The human glioma-associated oncogene homolog 1 (GLI1) family of transcription factors in gene regulation and diseases. Curr Genomics. 2010;11:238–245. doi: 10.2174/138920210791233108. [DOI] [PMC free article] [PubMed] [Google Scholar]


