Abstract
Benznidazole is the frontline drug used against Trypanosoma cruzi, the causative agent of Chagas disease. However, treatment failures are often reported. Here, we demonstrate that independently acquired mutations in the gene encoding a mitochondrial nitroreductase (TcNTR) can give rise to distinct drug-resistant clones within a single population. Following selection of benznidazole-resistant parasites, all clones examined had lost one of the chromosomes containing the TcNTR gene. Sequence analysis of the remaining TcNTR allele revealed 3 distinct mutant genes in different resistant clones. Expression studies showed that these mutant proteins were unable to activate benznidazole. This correlated with loss of flavin mononucleotide binding. The drug-resistant phenotype could be reversed by transfection with wild-type TcNTR. These results identify TcNTR as a central player in acquired resistance to benznidazole. They also demonstrate that T. cruzi has a propensity to undergo genetic changes that can lead to drug resistance, a finding that has implications for future therapeutic strategies.
Chagas disease is caused by Trypanosoma cruzi, a flagellated protozoan parasite transmitted by blood-sucking triatomine bugs. In Latin America, 10 million people are infected, with >15 000 deaths annually [1]. Because of migration, the disease is also undergoing globalization. In the United States, there are an estimated 300 000 infected individuals [2]. Chagas disease has 3 phases; acute, indeterminate and chronic. The acute stage is usually asymptomatic, although it can present as a febrile-like illness in children and young adults, with a fatality rate up to 5%. Most symptoms resolve within 4–6 weeks, and patients then enter the indeterminate stage. In the majority of cases, active disease does not proceed further. However, approximately 30% of individuals progress to the chronic phase, a process that can occur many years after the initial infection. This can result in serious cardiac and digestive tract pathologies, where prognosis is poor.
There is no immediate prospect of a Chagas disease vaccine, and infection is lifelong. Chemotherapy is therefore of major importance. For many years, benznidazole and nifurtimox have been the only drugs available [3]. However, their use is characterized by toxicity, and their efficacy against chronic stage disease is unreliable. In addition, cases refractory to treatment are commonly reported [4], and drug-resistant parasites can be selected in the laboratory [5, 6]. Benznidazole and nifurtimox are nitroheterocyclic compounds that contain a nitro group linked, respectively, to an imidazole and furan ring [3]. They are prodrugs and require nitroreductase (NTR)–catalyzed activation within the parasite to have trypanocidal effects. Two classes of NTR have been identified in trypanosomes. Type II NTRs are O2-sensitive flavin-containing enzymes that are capable of 1-electron reduction of nitro drugs to generate an unstable nitro radical [7]. In the presence of O2, this can lead to the production of superoxide anions and regeneration of the parent nitro compound, a process known as redox cycling [8, 9]. Although activation of nitroheterocyclic drugs by T. cruzi has been associated with the formation of reactive oxygen species (ROS) and candidate reductases have been implicated, there is no evidence that enhancing the parasite oxidative defense system has a protective affect [10–15]. Furthermore, addition of benznidazole to T. cruzi extracts does not lead to the generation of ROS [16].
Type I NTRs are O2-insensitive flavin mononucleotide–dependent enzymes that can mediate the 2-electron reduction of nitro drugs through a nitroso, to hydroxylamine derivatives. These can react further to generate nitrenium cations and other highly electrophilic intermediates, which may promote damage to DNA and other macromolecules [17, 18]. Two enzymes with type I activity have been identified in T. cruzi. The first is prostaglandin F2α synthase [19], although this is only capable of mediating 2-electron reduction under anaerobic conditions. The second, for which there is now strong evidence of a central role in activating nitro drugs, is a nicotinamide adenine dinucleotide, reduced (NADH)–dependent mitochondrial type I NTR [5]. In the case of nifurtimox, an active unsaturated open chain nitrile metabolite contributes to the resulting trypanocidal activity [20].
TcNTR can reduce a range of nitroheterocycles, and deletion of the corresponding genes from T. cruzi and Trypanosoma brucei results in loss of sensitivity [5]. Consistent with this, a genome-wide RNA interference screen of T. brucei for genes associated with nifurtimox and benznidazole resistance by loss-of-function mechanisms identified TbNTR as the major candidate [21]. To investigate the capacity of T. cruzi to develop resistance against benznidazole, we generated resistant clones following in vitro selection. Here, we show that distinct drug-resistant clones can arise independently and that, in each case, resistance under selective pressure is associated with loss of TcNTR activity.
MATERIALS AND METHODS
Parasites
T. cruzi MRAT/COL/Gal61 (Table 1) [22] were cultivated in supplemented Roswell Park Memorial Institute (RPMI) 1640 medium at 28°C [23]. Clones were derived by limiting dilution. Transformed T. cruzi were maintained at 10 μg/mL blasticidin or 50 μg/mL G418. Amastigotes were grown in African green monkey kidney (Vero) or rat skeletal myoblast L6 cells cultured in RPMI 1640/10% fetal bovine serum at 37°C in 5% CO2. To generate metacyclic trypomastigotes, epimastigote cultures were grown to stationary phase, at which point they differentiated. These were used to infect monolayers at a ratio of 5 metacyclics per mammalian cell. Following overnight incubation at 37°C, extracellular metacyclics and epimastigotes were removed by several washes. Bloodstream-form trypomastigotes emerged between day 7 and 10, and this homogenous population was used in quantitative infection experiments.
Table 1.
Natural Sensitivity to Benznidazole Is Not Associated With TcNTR Sequence
| Haplotype, Strain | GenBank Accession No. | Biological Origin | Geographical Origin | Phylogenetic Group | IC50, μM |
|---|---|---|---|---|---|
| Haplotype 1 | |||||
| AC17 | JN043349 | Rhodnius pallescens | Chocó | I | 6.53 ± 1.12 |
| AMP07 | JN043351 | Panstrongylus geniculatus | Antioquia | I | 17.6 ± 0.3 |
| B114 | JN043353 | Triatoma dimidiata | Córdoba | I | 18.7 ± 1.4 |
| B138 | JN043352 | T. dimidiata | Córdoba | I | 17.6 ± 0.7 |
| B51 | JN043354 | R. pallescens | Córdoba | I | 20.6 ± 1.1 |
| CAS18 | JN043345 | Didelphis marsupialis | Casanare | I | 3.90 ± 0.78 |
| CG | JN043336 | Homo sapiens | Caquetá | II | 4.61 ± 0.35 |
| GAL52 | JN043347 | D. marsupialis | Sucre | I | 9.07 ± 2.06 |
| GAL61 | JN043346 | Rattus rattus | Sucre | I | 5.85 ± 1.51 |
| HA | JN043337 | H. sapiens | Casanare | I | 4.66 ± 0.61 |
| LB53 | JN043358 | T. dimidiata | Sucre | I | 17.0 ± 0.7 |
| MG | JN043339 | H. sapiens | Arauca | I | 4.90 ± 0.33 |
| MG10 | JN043356 | T. dimidiata | Magdalena | I | 14.9 ± 0.89 |
| OV1 | JN043359 | P. geniculatus | Sucre | I | 17.4 ± 0.76 |
| OV17 | JN043355 | P. geniculatus | Sucre | I | 22.2 ± 2.4 |
| SN3 | JN043361 | Rhodnius prolixus | La Guajira | I | 34.6 ± 1.9 |
| SN5 | JN043360 | R. prolixus | La Guajira | I | 24.2 ± 1.3 |
| SN6 | JN043357 | R. prolixus | La Guajira | I | 16.9 ± 0.9 |
| SP | JN043342 | H. sapiens | Casanare | I | 6.41 ± 0.75 |
| SPR | JN043341 | H. sapiens | Casanare | II | 5.32 ± 1.08 |
| STP33 | JN043350 | R. prolixus | Tolima | I | 11.3 ± 1.0 |
| Haplotype 2 | |||||
| AF1 | JN043348 | P. geniculatus | Antioquia | II | 4.69 ± 1.87 |
| JEM | JN043340 | H. sapiens | Putumayo | I | 5.19 ± 0.65 |
| Distinct haplotypes | |||||
| DA | JN043344 | H. sapiens | Boyacá | I | 32.8 ± 3.3 |
| FCH | JN043334 | H. sapiens | N.de Santander | II | 1.50 ± 0.51 |
| MR | JN043338 | H. sapiens | Cesar | II | 4.71 ± 0.29 |
| W3534 | JN043343 | H. sapiens | Sucre | I | 14.0 ± 1.3 |
| YLY | JN043335 | H. sapiens | Putumayo | I/II | 4.38 ± 0.24 |
TcNTR genes were amplified from DNA of 28 Colombian Trypanosoma cruzi strains and sequenced.
Abbreviation: IC50, median inhibitory concentration.
Intact T. cruzi chromosomes were extracted using an agarose-embedding technique [24] and were fractionated by contour-clamped homogenous field electrophoresis (CHEFE), using a BioRad CHEFE Mapper. For analysis of natural benznidazole sensitivity, TcNTR from 28 T. cruzi strains from different regions of Colombia was amplified and sequenced.
To generate benznidazole resistance, epimastigotes were seeded at the median inhibitory concentration (IC50) and subcultured for several weeks under selective pressure. The drug concentration was then doubled and the process repeated. This was continued until a resistant population was established (61R) at 50 µM, the reported level of therapeutic resistance [25]. IC50 values were determined by an enzymatic micromethod [26]. A total of 2 × 106 epimastigotes/mL were cultured with different drug concentrations for 72 hours at 28°C in 96-well microtiter plates. The plates were then incubated with 10 mg/mL 3(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT) for 90 minutes and MTT reduction to formazan crystals measured at 595 nm.
Construction of Vectors
For expression of TcNTR, a 708–base pair fragment corresponding to the catalytic domain of the protein was amplified using DNA from sensitive and resistant clones [5]. Fragments were digested with BamHI/HindIII and ligated into the vector pTrcHis-C (Invitrogen), and the resulting constructs were used to transform Escherichia coli BL-21. To express active protein in benznidazole-resistant T. cruzi, the full-length TcNTR gene (939 bp) was amplified from 61S DNA and ligated into the BamHI/HindIII site of the vector pTEX [27]. Parasites were electroporated and transformants selected with G418. To generate TcNTR heterozygotes from 61S parasites, we used gene disruption with a construct containing a blasticidin-resistance cassette [5]. All constructs were confirmed by sequencing.
Biochemical Analysis
E. coli transformed with pTrcHis-TcNTR were treated with isopropyl-β-D-thiogalactopyranoside to induce expression of recombinant histidine-tagged proteins, which were purified on nickel–nitriloacetic acid columns [5, 28]. Fractions were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and protein concentrations determined by the BCA assay (Pierce). TcNTR activity was measured by following the changes in absorbance at 340 nm due to NADH oxidation.
The TcNTR flavin cofactor was established by determining the fluorescence spectrum in acidic and neutral buffers [29]. Purified protein (0.5 mg) was desalted and boiled for 5 minutes. Clarified supernatant (90 µL) was then mixed with 10 µL 50 mM NaH2PO4 (pH = 7.6) or 1 M HCl (final pH = 2.2), and the fluorescence profile was measured with a Gemini Fluorescent Plate Reader (Molecular Devices). The mean fluorescence values (excitation λ = 450 nm; emission λ = 535 nm) was determined and compared to flavin mononucleotide and flavin adenine dinucleotide (FAD) standards.
RESULTS
Benznidazole-Resistant T. cruzi Lack One of the Chromosome Bands Containing the TcNTR Gene
To select for benznidazole resistance, T. cruzi GAL61 (Table 1) were submitted to continuously increasing drug pressure until we had established a population (61R) that grew at a comparable rate in the presence or absence of 50 µM benznidazole (Materials and Methods). This population displayed approximately 10-fold resistance. Six clonal lines derived from this population exhibited 3–7-fold resistance, when examined independently (Figure 1A). In the absence of drug, the clones grew slightly slower in culture than the parental cells (doubling times from 28–42 hours, compared with 26 hours) but otherwise displayed no obvious morphological changes. Previously, when we generated nifurtimox-resistant T. cruzi, we found that they were also resistant to other nitroheterocyclic drugs, including benznidazole [5]. A similar cross-resistance phenomenon was observed here, with 2-fold greater resistance to nifurtimox and 4-fold greater resistance to nitrofurazone (Figure 1B).
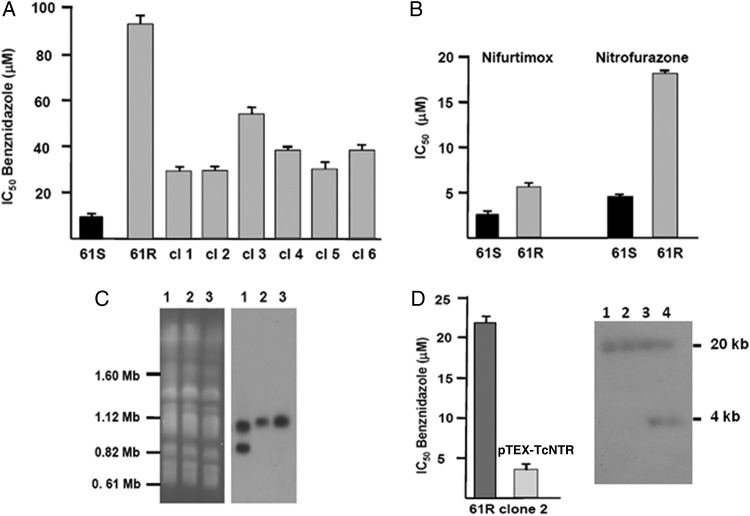
Figure 1.
Properties of Trypanosoma cruzi clones derived by benznidazole selection. A Median inhibitory concentration (IC50) of benznidazole for resistant parasites (61R noncloned population and clones 1–6) and parental (61S) cells (Materials and Methods). B, The 61R cells are cross-resistant to the other nitroheterocycles nifurtimox and nitrofurazone. C, T. cruzi chromosomal DNA separated by contour-clamped homogenous field electrophoresis and hybridized with a TcNTR gene probe. Left panel, ethidium bromide–stained gel; right panel, autoradiograph of gel after Southern blotting. Lane 1, parental 61S; lane 2, 61R clone 3; lane 3, 61R (noncloned population). D, Reintroduction of an active copy of TcNTR into benznidazole-resistant parasites (61R clone 2), using the pTEX vector reverses the drug-resistance phenotype. Growth inhibition data are the mean (±SD) of 3 experiments. An autoradiograph (right) shows BamHI-digested DNA from parental 61R clone 2 (lanes 1 and 2) and pTEX-TcNTR–transformed cells (lanes 3 and 4) hybridized with a TcNTR gene probe.
Nifurtimox resistance in both T. cruzi and T. brucei has been associated with downregulation or loss of a type I NTR gene [5, 21]. We therefore examined the benznidazole-sensitive and -resistant cells for changes in copy number at this locus. In the sensitive parental cells (61S), TcNTR is a single copy gene located on chromosome homologues of 1.1-Mb and 0.85-Mb. With the resistant parasites, however, the 0.85-Mb band was missing in clonal and polyclonal populations (Figure 1C, lanes 2 and 3). There were no other apparent changes to the chromosome profile. To determine whether drug resistance was associated with loss of TcNTR rather than another gene located elsewhere on the missing chromosome, we reintroduced an active copy of TcNTR into 61R clone 2 [27] (Figure 1D). When the transformed cells were assessed, we found that benznidazole sensitivity had been restored.
The Remaining TcNTR Allele in Each Benznidazole-Resistant Clone Encodes an Inactive Protein
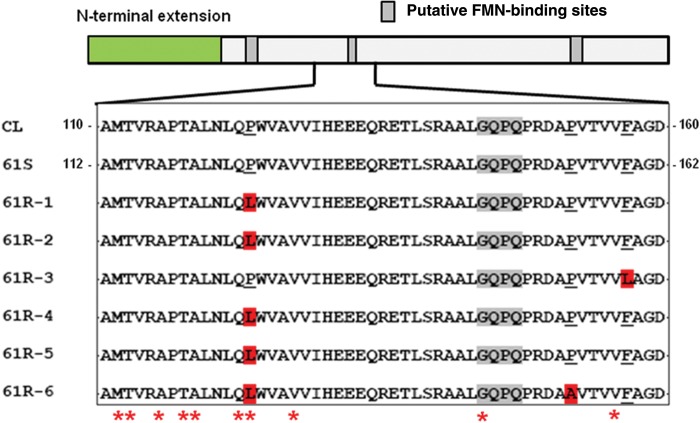
We next investigated whether the remaining chromosomal copy of TcNTR in the benznidazole-resistant 61R parasites had altered. Genes from the 6 resistant clones were amplified and sequenced. Missense mutation(s) were identified in each case. In clones 1, 2, 4, and 5, there was C/T transition at position 374, compared with the TcNTR gene amplified from sensitive clones. In the protein, this would result in replacement of the evolutionarily conserved Pro-125 with leucine (Figure 2). With clone 6, in addition to the mutation at position 374, we also identified a missense mutation at nucleotide 460 (C/G), giving rise to the conversion of Pro-154 to alanine. For clone 3, there was a single missense mutation resulting in C/G transversion at nucleotide 477, leading to the replacement of Phe-159 with leucine. No other mutations were observed in the TcNTR genes isolated from the resistant clones. In the O2-insensitive E. coli nitroreductase nfsB, most mutations associated with nitrofuran resistance are located in the corresponding region to those in TcNTR (Figure 2) [30, 31].
Figure 2.
Mutations in TcNTR from benznidazole-resistant Trypanosoma cruzi. The TcNTR schematic identifies the amino terminal extension (excluded from recombinant proteins) and the location of putative flavin mononucleotide (FMN)–binding regions inferred by analogy with Escherichia coli nfsB [30]. Full-length copies of TcNTR from 61R resistant clones were amplified and sequenced. Differences in the amino acid sequence as compared to the parental TcNTR (61S) were restricted to a single region and are highlighted in red. Several 61S clones were sequenced, but no differences were identified. The sequence in this region of 61S TcNTR (residues 112–162) is identical to that in the genome strain CL Brener (GenBank accession no. XP_810645). The corresponding CL Brener TcNTR residues are numbered 110–160 because of an insertion or deletion in the amino terminal domain. Mutations in the corresponding region of E. coli nfsB that confer nitrofurantoin resistance are indicated by asterisks [31].
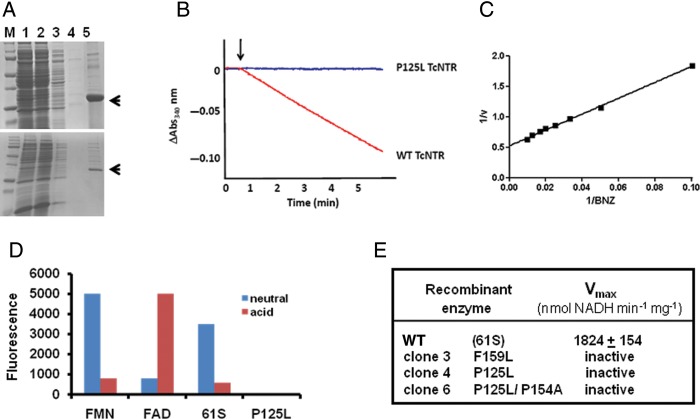
To determine whether the TcNTR mutations had perturbed activity, we amplified a fragment encoding the catalytic region of the enzyme, using DNA from 61S and 61R clones 3, 4, and 6. TcNTR is mitochondrial, and previous attempts to express active full-length enzyme had been unsuccessful [5]. Activity was only detectable when the amino terminal domain was excluded from the recombinant protein (Figure 2). After sequence confirmation, the expressed histidine-tagged proteins were purified on nickel columns (Materials and Methods; Figure 3A). Fractions containing recombinant protein derived from the 61S TcNTR gene were yellow, as expected of a flavoprotein. Those containing enzyme derived from the resistant clones were colorless.
Figure 3.
Biochemical analysis of TcNTR from benznidazole (BNZ)-sensitive and -resistant Trypanosoma cruzi. A, Purification of recombinant TcNTR. Upper image, wild-type (WT; 61S) enzyme; lower, clone 4. Protein expression was induced by isopropyl-β-D-thiogalactopyranoside (Materials and Methods), and a clarified fraction (lane 1) was loaded onto a nickel–nitriloacetic acid column and the flow-through collected (lane 2). The column was washed with 50 mM and then 100 mM imidazole (lanes 3 and 4) and the recombinant protein eluted with 500 mM imidazole, 1% triton X-100 (lane 5). The 32-kDa TcNTR band is highlighted by an arrow. Recombinant protein from each of the resistant clones was purified in a similar manner. B, TcNTR activity was monitored (on the basis of absorbance [Abs] at a wavelength of 340 nm) by following oxidation of nicotinamide adenine dinucleotide, reduced (NADH; 100 μM), in the presence of WT or mutant (P125L, clone 4) enzyme (0.2 μg) and BNZ (100 μM). C, Activity (v) of the WT enzyme (nmol NADH per min per mg) was established by this assay with a fixed concentration of NADH (100 μM) in the presence of different levels of BNZ (10–100 μM). D, Fluorescence (excitation λ = 450 nm; emission λ = 535 nm) of the TcNTR cofactor (WT and P125L) and flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) controls (in arbitrary units) under acidic and neutral conditions (Materials and Methods). E, Activity of WT and mutant TcNTRs.
The capacity of the recombinant enzymes to reduce benznidazole and nifurtimox was established from double reciprocal plots of 1/TcNTR activity against 1/drug concentration, at a fixed NADH concentration (100 μM) (Materials and Methods; Figure 3B and 3C). For the enzyme derived from the sensitive clone, we established apparent Km values (±SD) of 28.0 ± 2.7 μM for benznidazole and 15.5 ± 3.5 μM for nifurtimox. Further analysis gave apparent Vmax values (±SD) of 1824 ± 154 nmol NADH oxidized per minute per milligram for benznidazole and 399 ± 14 nmol NADH oxidized per minute per milligram for nifurtimox. When each of the mutant TcNTRs were analyzed, no activity could be detected, even when 10 times as much recombinant protein was used. We then investigated the mutant proteins for flavin binding (Figure 3D), using fluorescent detection under neutral and acidic conditions (Materials and Methods). At neutral pH and with an excitation wavelength of 450 nm, the flavin mononucleotide standard and 61S TcNTR-derived cofactor both gave a fluorescence profile that peaked at 535 nm, a signal that was quenched under acidic conditions. By contrast with FAD, the 535 nm peak occurs at pH2 and is quenched at pH7. No flavin fluorescence was detected with mutant TcNTR protein (Figure 3D).
Infectivity of Benznidazole-Resistant Parasites
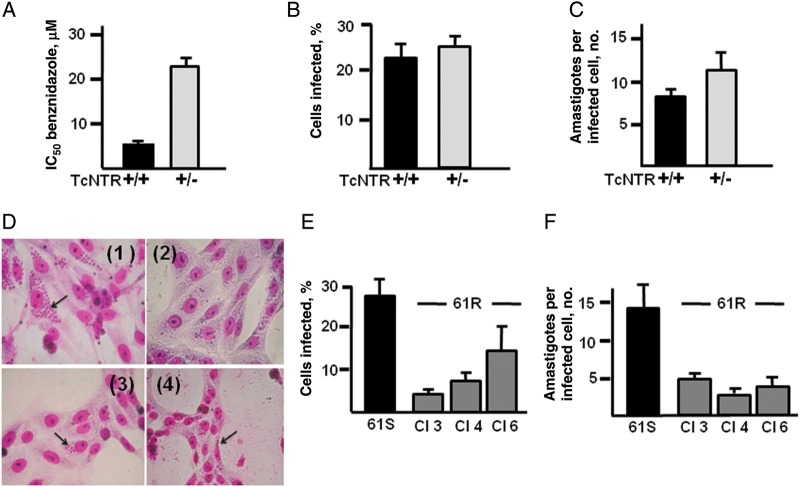
To investigate the scope for drug resistance in the field to result from loss/inactivation of TcNTR genes, we examined the effects of these events on infectivity. First, we generated heterozygous parasites to test for haploid insufficiency. One TcNTR allele in the 61S genome was disrupted by targeted integration (Supplementary Figure 1). The 61S TcNTR+/− epimastigotes grew at the same rate in culture as homozygotes, and to the same density. When these heterozygotes were examined for benznidazole resistance, there had been a 4-fold increase (Figure 4A). These parasites were used to infect rat myoblast L6 cells. No differences were observed in the ability of the heterozygotes to develop into infective metacyclic trypomastigotes, to invade cells (Figure 4B), to grow as intracellular amastigotes (Figure 4C), and subsequently to differentiate into bloodstream trypomastigotes. Therefore, drug resistance that arises through loss of 1 copy of TcNTR is not associated with a reduction in infectivity in vitro.
Figure 4.
Loss of 1 copy of TcNTR does not reduce infectivity. Targeted disruption of TcNTR in 61S epimastigotes was achieved using a construct that confers blasticidin resistance (Supplementary Figure 1). A, TcNTR heterozygotes are benznidazole resistant. 61S TcNTR homozygotes (+/+) and heterozygotes (+/−) were tested to establish their median inhibitory concentration (IC50). Data are the mean (±SD) of 3 experiments. B, Heterozygotes are not deficient in infectivity. Cell-derived trypomastigotes were added to an L6 cell monolayer at a ratio of 1:5 cells to parasites. Infected cells were counted after 72 hours, (experiment performed in triplicate, with data presented mean ± SD). C, Heterozygotes are not deficient in their ability to replicate in mammalian cells. Infections were performed and monitored as described above. D, Infection of Vero cells with 61S sensitive and 61R resistant parasites, stained with Giemsa: (1) 61S; (2) 61R clone 3; (3) 61R clone 4; (4) 61R clone 6. Arrows indicate intracellular amastigotes 48 hours after infection. E, Benznidazole-resistant clones are deficient in their ability to infect Vero cells. Values shown are from 5 experiments (P < .05). (F) Benznidazole-resistant clones are less able to replicate in Vero cells. Values shown are from 5 experiments (P < .05).
The infective phenotype of the 61R resistant clones, which contain a single inactive copy of TcNTR, was also examined. In culture, epimastigotes differentiated into metacyclic trypomastigotes at a level similar to sensitive clones. When culture-derived trypomastigotes were used to initiate infections, all the resistant clones tested (clones 3, 4, and 6) were able to develop through the intracellular cycle as amastigotes and differentiate into bloodstream trypomastigotes, which were released following host cell lysis. At 2 levels, however, we observed a reduction in virulence. When Vero cells were used (Figure 4D), the number infected by resistant clones was significantly less than the level observed with the parental sensitive parasites (Figure 4E), and the average number of amastigotes per infected cell was reduced (Figure 4F). When L6 cells were infected with drug-resistant metacyclics, although released trypomastigotes could be observed, their numbers were too few for a quantifiable infection assay to be performed. This compares to an infection rate of approximately 25% in the case of the 61S TcNTR heterozygotes and homozygotes (Figure 4B). These experiments therefore suggest that functional loss of both TcNTR genes, by the mechanisms identified here, is associated with a reduction in virulence that would reduce the capacity of highly drug-resistant parasites to spread within the population.
TcNTR Diversity and Benznidazole Sensitivity in the Field
To explore possible relationships between natural susceptibility to benznidazole and TcNTR, we sequenced the gene from 28 Colombian strains of different biological and geographical origins and with a range of benznidazole sensitivities (IC50, 1.5–35 μM) (Table 1). TcNTR length varied between 939 and 951 nucleotides in these strains, mainly because of changes in the copy number of a trinucleotide (ATC)5-9 located between residues 210 and 238. This region of the protein is not required for enzyme activity [5]. Excluding this repeat, we identified 42 polymorphisms, 25 of which were nonsynonymous. These amino acid differences were restricted to 7 strains, all but one of human origin. None of the polymorphisms were located in the region of TcNTR where we had identified mutations associated with benznidazole resistance. Most were located in the amino terminal extension (Supplementary Figure 2). The major amino acid haplotype group encompassed 21 strains of various biological and geographical origins. Importantly, these had a wide range of benznidazole sensitivities (IC50, 4–35 μM) (Table 1). This extensive natural variation is therefore independent of TcNTR sequence and must be due to other factors. This suggests that resistance arising from changes to TcNTR is an acquired trait that requires selective pressure.
DISCUSSION
Despite being the frontline drug against T. cruzi infections for >40 years, benznidazole has drawbacks [4, 32]. It can have serious side effects, it requires long-term administration (30–60 days), and its efficacy against chronic stage disease is inconsistent. Treatment failures are widely reported, although the extent to which this is an acquired trait or reflects diversity in the level of susceptibility within natural parasite populations is unknown [33]. As shown here and elsewhere [5, 34, 35], laboratory selection of drug-resistant T. cruzi is readily achievable, but in the case of benznidazole and nifurtimox, it is only recently that a mechanism has been identified [5]. Activation of these prodrugs by the trypanosome type I NTR, an enzyme absent from mammals, is central to their mode of action and explains why they are more toxic to the parasite than to the host. The 61R benznidazole-resistant T. cruzi clones that we investigated were characterized by loss of a 0.85-Mb chromosome band containing TcNTR. Genome plasticity is a common phenomenon in trypanosomes [24]. Confirmation that reduced TcNTR expression caused this resistance was provided by reversion of the phenotype following reintroduction of the gene. Unexpectedly, we also found that in each of the 61R clones examined, the TcNTR gene on the 1.1-Mb chromosome homologue had acquired missense mutation(s) that rendered the expressed product enzymatically inactive (Figures 2 and 3).
The most parsimonious explanation for our data is that drug pressure led initially to selection of benznidazole resistance because of loss of the TcNTR-containing 0.85-Mb chromosome. Continued treatment then resulted in selection, from within this population, of distinct lineages in which mutation(s) had inactivated the remaining TcNTR gene. The acquisition of 2 distinct missense mutations in TcNTR of clone 6 (nucleotides 374 and 460) implies consecutive events. This 2-step process is reminiscent of what happens in E. coli, where increased nitrofuran resistance resulted from consecutive mutations in the type I NTR genes nfsA and nfsB [31]. The mutant TcNTR proteins were found to be deficient in flavin mononucleotide binding. In the NTR group of enzymes, the location of flavin binding is highly conserved within the overall structure [30]. All of the mutations in TcNTR were restricted to a region (residues 125–159; Figure 2) that, in the E. coli enzyme, contains residues that interact with the isoalloxazine O2, N3, O4 face of flavin mononucleotide [30]. The mutation of residue 125 resulted in conversion of an evolutionarily conserved proline to a leucine (clones 1, 2, 4, 5, and 6). At position 154 in clone 6, proline was converted to alanine. Both changes would be expected to perturb structure. In clone 3, the mutation associated with disruption of flavin mononucleotide binding involved conversion of phenylalanine 159 to leucine. Phenylalanine is present at the corresponding position in E. coli and T. cruzi NTRs (Figure 2), suggesting a functionally conserved role.
The ability of distinct TcNTR-deficient T. cruzi clones to arise independently in a single population is strong evidence that the drug-activating properties of this enzyme are central to the trypanocidal mechanism. The TcNTR single knockouts were 4-fold less susceptible to benznidazole (Figure 4), a level of resistance that is significant in the context of this drug, for which the therapeutic window is limited [3]. The virulence properties in vitro were also indistinguishable from TcNTR homozygotes. This potential for benznidazole resistance by a straightforward mechanism, coupled with the absence of haploid insufficiency, may explain some of the observed treatment failures. The inability of the 61S strain to produce a patent infection in mice has restricted us from investigating this further. Complete loss of TcNTR activity in the 61R resistant clones did however have a detrimental effect on infectivity in vitro (Figure 4). This implies that in vivo there will be a limit to the extent of benznidazole resistance achievable by mechanisms involving TcNTR (approximately 4-fold), since parasites need to retain a residual level of enzyme activity. When we investigated possible relationships between susceptibility to benznidazole and TcNTR sequence in a diverse group of parasites (Table 1), we found no correlation. These data suggest that natural variation in sensitivity does not involve mutations in TcNTR and that resistance by this mechanism may be a trait that arises only after selective pressure. Currently, there is no information on the extent to which treatment failures reflect natural or acquired resistance.
An observation, which has wider implications for treatment of Chagas disease, is the ease with which drug resistance can arise. In a single experiment, we identified 2 distinct mechanisms, chromosome loss and point mutation, which acted to reduce TcNTR activity. In the latter case, 3 distinct, independently acquired mutations were identified. T. cruzi is extremely diverse, with a genome characterized by extensive and highly variable surface antigen gene families [36]. This antigenic diversity may have arisen in response to selective immune pressure during evolution, which acted to limit the proofreading ability of DNA polymerase and/or DNA repair mechanisms. As a consequence, the parasite may have acquired an ability to readily develop drug resistance by mutational mechanisms such as those described here. This is an important consideration that should inform drug development strategies for Chagas disease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank INS, Colombia, for providing some of the human T. cruzi strains, and Michael Miles (LSHTM), for critical comments on the manuscript.
Financial support. This work was supported by the Wellcome Trust (grant 084175 to J. M. K. and grant 082342 to S. R. W.), COLCIENCIAS (project 111551929168 to A. M. M. and O. T. C.), and a fellowship (to A. M. M.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Moncayo A, Silveira AC. Current epidemiological trends for Chagas disease in Latin America and future challenges in epidemiology, surveillance and health policy. Mem Inst Oswaldo Cruz. 2009;104:17–30. doi: 10.1590/s0074-02762009000900005. [DOI] [PubMed] [Google Scholar]
- 2.Bern C, Montgomery SP. An estimate of the burden of Chagas disease in the United States. Clin Infect Dis. 2009;49:e52–4. doi: 10.1086/605091. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson SR, Kelly JM. Trypanocidal drugs: mechanisms, resistance and new targets. Expert Rev Mol Med. 2009;11:1–24. doi: 10.1017/S1462399409001252. [DOI] [PubMed] [Google Scholar]
- 4.Castro JA, de Mecca MM, Bartel LC. Toxic side effects of drugs used to treat Chagas’ disease (American trypanosomiasis) Hum Exp Toxicol. 2006;25:471–9. doi: 10.1191/0960327106het653oa. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson SR, Taylor MC, Horn D, Kelly JM, Cheeseman I. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc Natl Acad Sci U S A. 2008;105:5022–7. doi: 10.1073/pnas.0711014105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murta SM, Gazzinelli RT, Brener Z, Romanha AJ. Molecular characterization of susceptible and naturally resistant strains of Trypanosoma cruzi to benznidazole and nifurtimox. Mol Biochem Parasitol. 1998;93:203–14. doi: 10.1016/s0166-6851(98)00037-1. [DOI] [PubMed] [Google Scholar]
- 7.Peterson FJ, Mason RP, Hovsepian J, Holtzman JL. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J Biol Chem. 1979;254:4009–14. [PubMed] [Google Scholar]
- 8.Docampo R, Mason RP, Mottley C, Muniz RP. Generation of free radicals induced by nifurtimox in mammalian tissues. J Biol Chem. 1981;256:10930–3. [PubMed] [Google Scholar]
- 9.Moreno SN, Mason RP, Docampo R. Reduction of nifurtimox and nitrofurantoin to free radical metabolites by rat liver mitochondria. Evidence of an outer membrane-located nitroreductase. J Biol Chem. 1984;259:6298–305. [PubMed] [Google Scholar]
- 10.Kelly JM, Taylor MC, Smith K, Hunter KJ, Fairlamb AH. Phenotype of recombinant Leishmania donovani and Trypanosoma cruzi which over-express trypanothione reductase. Sensitivity towards agents that are thought to induce oxidative stress. Eur J Biochem. 1993;218:29–37. doi: 10.1111/j.1432-1033.1993.tb18348.x. [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson SR, Temperton NJ, Mondragon A, Kelly JM. Distinct mitochondrial and cytosolic enzymes mediate trypanothione-dependent peroxide metabolism in Trypanosoma cruzi. J Biol Chem. 2000;275:8220–5. doi: 10.1074/jbc.275.11.8220. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson SR, Meyer DJ, Taylor MC, Bromley EV, Miles MA, Kelly JM. The Trypanosoma cruzi enzyme TcGPXI is a glycosomal peroxidase and can be linked to trypanothione reduction by glutathione or tryparedoxin. J Biol Chem. 2002;277:17062–71. doi: 10.1074/jbc.M111126200. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson SR, Obado SO, Mauricio IL, Kelly JM. Trypanosoma cruzi expresses a plant-like ascorbate-dependent hemoperoxidase localized to the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2002;99:13453–8. doi: 10.1073/pnas.202422899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson SR, Horn D, Pathalingham R, Kelly JM. RNAi identifies two hydroperoxide metabolising enzymes that are essential to the bloodstream form of the African trypanosome. J Biol Chem. 2003;278:31640–6. doi: 10.1074/jbc.M303035200. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson SR, Prathalingam R, Taylor MC, Ahmed A, Horn D, Kelly JM. Functional characterisation of the iron superoxide dismutase gene repertoire in Trypanosoma brucei. Free Radic Biol Med. 2006;40:198–209. doi: 10.1016/j.freeradbiomed.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 16.Moreno SN, Docampo R, Mason RP, Leon W, Stoppani AO. Different behaviors of benznidazole as free radical generator with mammalian and Trypanosoma cruzi microsomal preparations. Arch Biochem Biophys. 1982;218:585–91. doi: 10.1016/0003-9861(82)90383-6. [DOI] [PubMed] [Google Scholar]
- 17.Streeter AJ, Hoener BA. Evidence for the involvement of a nitrenium ion in the covalent binding of nitrofurazone to DNA. Pharm Res. 1988;5:434–6. doi: 10.1023/a:1015988401601. [DOI] [PubMed] [Google Scholar]
- 18.McCalla DR, Reuvers A, Kaiser C. Breakage of bacterial DNA by nitrofuran derivatives. Cancer Res. 1971;31:2184–8. [PubMed] [Google Scholar]
- 19.Kubata BK, Kabututu Z, Nozaki T, et al. A key role for old yellow enzyme in the metabolism of drugs by Trypanosoma cruzi. J Exp Med. 2002;196:1241–51. doi: 10.1084/jem.20020885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall BS, Bot C, Wilkinson SR. Nifurtimox activation by trypanosomal type I nitroreductases generates cytotoxic nitrile metabolites. J Biol Chem. 2011;286:13088–95. doi: 10.1074/jbc.M111.230847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker N, Alsford S, Horn D. Genome-wide RNAi screens in African trypanosomes identify the nifurtimox activator NTR and the eflornithine transporter AAT6. Mol Biochem Parasitol. 2011;176:55–7. doi: 10.1016/j.molbiopara.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Tropica. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Kendall G, Wilderspin AF, Ashall F, Miles MA, Kelly JM. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase does not conform to the “hotspot” topogenic signal model. EMBO J. 1990;9:2751–8. doi: 10.1002/j.1460-2075.1990.tb07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obado SO, Taylor MC, Wilkinson SR, Bromley EV, Kelly JM. Functional mapping of a trypanosome centromere by chromosome fragmentation identifies a 16 kb GC-rich transcriptional “strand-switch” domain as a major feature. Genome Res. 2005;15:36–43. doi: 10.1101/gr.2895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villarreal D, Nirdé P, Hide M, Barnabé C, Tibayrenc M. Differential gene expression in benznidazole-resistant Trypanosoma cruzi parasites. Antimicrob Agents Chemother. 2005;49:2701–9. doi: 10.1128/AAC.49.7.2701-2709.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 27.Kelly JM, Ward HM, Miles MA, Kendall G. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucl Acid Res. 1992;20:3963–9. doi: 10.1093/nar/20.15.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall BS, Wu X, Hu L, Wilkinson SR. Exploiting the drug-activating properties of a novel trypanosomal nitroreductase. Antimicrob Agents Chemother. 2010;54:1193–9. doi: 10.1128/AAC.01213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faeder EJ, Siegel LM. A rapid micromethod for determination of FMN and FAD in mixtures. Anal Biochem. 1973;53:332–6. doi: 10.1016/0003-2697(73)90442-9. [DOI] [PubMed] [Google Scholar]
- 30.Parkinson GN, Skelly JV, Neidle S. Crystal structure of FMN-dependent nitroreductase from Escherichia coli B: a prodrug-activating enzyme. J Med Chem. 2000;43:3624–31. doi: 10.1021/jm000159m. [DOI] [PubMed] [Google Scholar]
- 31.Whiteway J, Koziarz P, Veall J, et al. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–39. doi: 10.1128/jb.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinazo MJ, Muñoz J, Posada E, et al. Tolerance of benznidazole in treatment of Chagas’ disease in adults. Antimicrob Agents Chemother. 2010;54:4896–9. doi: 10.1128/AAC.00537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filardi LS, Brener Z. Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Trans R Soc Trop Med Hyg. 1987;81:755–9. doi: 10.1016/0035-9203(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 34.Murta SM, Romanha AJ. In vivo selection of a population of Trypanosoma cruzi and clones resistant to benznidazole. Parasitology. 1998;116:165–71. doi: 10.1017/s0031182097002084. [DOI] [PubMed] [Google Scholar]
- 35.Buckner FS, Wilson AJ, White TC, Van Voorhis WC. Induction of resistance to azole drugs in Trypanosoma cruzi. Antimicrob Agents Chemother. 1998;42:3245–50. doi: 10.1128/aac.42.12.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franzén O, Ochaya S, Sherwood E, et al. Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl Trop Dis. 2011;5:e984. doi: 10.1371/journal.pntd.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.