Abstract
Nitric oxide participates in cellular signal transduction largely through S-nitrosylation of allosteric and active-site cysteine thiols within proteins, forming S-nitrosoproteins (SNO-proteins). S-nitrosylation of proteins has been demonstrated to affect a broad range of functional parameters including enzymatic activity, subcellular localization, protein–protein interactions, and protein stability. Analogous to other ubiquitous posttranslational modifications that are regulated enzymatically, including phosphorylation and ubiquitinylation, accumulating evidence suggests the existence of enzymatic mechanisms for regulating protein S-nitrosylation. In particular, studies have led to the identification of multiple enzymes (nitrosylases and denitrosylases) that participate in targeted S-nitrosylation or denitrosylation of proteins in physiological settings. Nitrosylases are best characterized in the context of transnitrosylation in which a SNO-protein transfers an NO group to an acceptor protein (Cys-to-Cys transfer), but examples of transnitrosylation catalyzed by metalloproteins (Metal-to-Cys transfer) also exist. By contrast, denitrosylases remove the NO group from SNO-proteins, ultimately using reducing equivalents derived from NADH or NADPH. Here, we focus on the recent discoveries of nitrosylases and denitrosylases and the notion that their aberrant activities may play roles in health and disease.
Keywords: S-nitrosylation, SNO-proteins, GSNO, Nitric oxide, Active-site cysteine thiols, Nitrosylases, Denitrosylase
Introduction
Background
S-nitrosylation, the reversible, covalent addition of a nitrogen monoxide (NO) moiety to the thiol side chain of cysteine (Cys), has emerged as an important regulatory mechanism in nitric oxide-related signaling. Both proteins and low-molecular-weight thiols, including in particular glutathione, are subject to S-nitrosylation, generating S-nitrosoproteins (SNO-proteins) and S-nitrosoglutathione (GSNO), respectively [1]. Initial studies of the function of nitric oxide as a signaling molecule in smooth muscle demonstrated a role for cGMP as a second messenger [2]. However, evidence accumulated over the past two decades has demonstrated that nitric oxide exerts its ubiquitous influence on signal transduction and other aspects of cellular function largely through cGMP-independent S-nitrosylation of proteins [3]. In mammals, cellular S-nitrosylation is coupled to nitric oxide synthesis carried out principally by three isoforms of nitric oxide synthase (NOS): neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3). S-nitrosylation results from the reaction of Cys thiols with nitric oxide-derived species, such as N2O3, by oxidation of a SNO radical anion (RSNO•−) or by transnitrosylation, which is the transfer of the NO group from an S-nitrosothiol (SNO) to an acceptor Cys thiol [3, 4]. A growing body of research indicates an essential role for homeostatic regulation of cellular SNOs in normal physiology, which is maintained, in part, by the opposing actions of enzymes involved in either addition or abstraction of NO from SNOs. Dysregulated S-nitrosylation has been implicated as a cause or consequence of a broad range of diseases, including asthma, cystic fibrosis, Parkinson disease, heart failure, and stroke (Table 1), and the role of nitrosylases and denitrosylases in governing levels of S-nitrosylation under both physiological and pathophysiological conditions is increasingly appreciated.
Table 1.
Dysregulated S-nitrosylation of proteins associated with pathophysiology
| Pathophysiology | SNO-proteins | Reference |
|---|---|---|
| Neurological | ||
| Alzheimer disease | Dynamin-related protein 1 | [44] |
| Cyclin-dependent kinase 5 | [43] | |
| Protein disulfide isomerase | [86] | |
| X-linked inhibitor of apoptosis | [40] | |
| Apolipoprotein E | [87] | |
| Parkinson disease | Parkin | [88, 89] |
| Peroxiredoxin-2 | [90] | |
| Protein disulfide isomerase | [86] | |
| X-linked inhibitor of apoptosis | [91] | |
| Stroke | Matrix metalloproteinase 9 | [92] |
| Cardiovascular | ||
| Heart failure | Ryanodine receptor 2 | [93] |
| Long Q/T syndrome | Cardiac sodium channel SN5Ca | [94] |
| Preeclampsia | Serum albumin | [95, 96] |
| SNO-proteins | [97] | |
| Pulmonary arterial hypertension | Hemoglobin | [29] |
| HIF1α | [98] | |
| NSF | [99] | |
| eNOS | [99] | |
| Caveolin 1 | [99] | |
| Clathrin heavy chain | [99] | |
| VHL | [98] | |
| Septic shock | Hemoglobin | [30] |
| CD40 | [100] | |
| Arterial fibrillation/arrhythmia | L-type Ca2+ channel (α1C subunit) | [101–103] |
| Slowly activating delayed- rectifier K+ channel | [104, 105] | |
| Diabetes (type 1) | Hemoglobin | [106, 107] |
| Glucokinase | [108] | |
| Diabetes (type 2) | Insulin receptor β | [109, 110] |
| Insulin receptor substrate 1 | [109, 110] | |
| Ischemic coronary syndrome | Albumin | [111] |
| Hematological | ||
| Blood transfusion: storage defect | Hemoglobin | [28] |
| Sickle cell anemia | Hemoglobin | [27, 32] |
| AE1 | [27] | |
| Pulmonary | ||
| Asthma | SNO-proteins, GSNO | [65] |
| Cystic fibrosis | Hsp70/Hsp90 organizing protein | [112] |
| Lung inflammation | Surfactant protein B | [113] |
| COPD | HDAC2a | [120] |
| Cancer | ||
| Liver cancer | O6-alkylguanine-DNA alkyl transferase | [68] |
| Tumor maintenance | Ras | [114] |
| Tumor radiosensitivity | HIF-1α | [115] |
| Skeletal muscle | ||
| Duchenne muscular dystrophy | Ryanodine receptor 1 | [116] |
| Malignant hyperthermia | Ryanodine receptor 1 | [117] |
Implicated in steroid insensitivity
Specificity determinants
In multiple analyses of the role of protein S-nitrosylation in the context of cellular signal transduction, it has emerged that this posttranslational modification exhibits a high degree of spatiotemporal precision [3]. Selectivity is conferred in part by the interaction of substrates with sources of NO groups including NOSs and NO donors (including GSNO and SNO-proteins) and through SNO motifs that facilitate in vivo S-nitrosylation [5, 6] of only a small subset of cysteines within proteins [7].
Initial analyses of mammalian SNO-hemoglobin suggested an acid–base SNO motif whereby S-nitrosylation is targeted by charged (acidic and basic) side chains within 6 Å of the modified thiol (Cysβ93) [8]. Hemoglobin also contains features of a hydrophobic motif that is characterized by signature aromatic residues (Tyr, Trp) that are thought to facilitate S-nitrosylation by a variety of mechanisms [3, 7], including micellar catalysis (that may explain nitrosylation of solvent-inaccessible Cys, such as that in Cox2 [9]) and transnitrosation, as exemplified by NO group exchange between Trp and Cys in albumin [10]. Tyr and Cys coupling, well known to increase Cys reactivity by lowering thiol pKa, may also facilitate S-nitrosylation by radical routes, catalyzed by a Tyr cation radical that will extract an electron from an adjacent Cys (which then reacts rapidly with NO•). A microarray analysis of S-nitrosylated yeast proteins showed that the SNO site within S-nitrosylated glutamine amidotransferases is situated in a “nucleophilic elbow” at the N terminus of a helix that increases thiol nucleophilicity (as do proximate His and Tyr residues that are features of acid–base and hydrophobic motifs) [11], and additional structural analyses of endogenously nitrosylated mouse proteins revealed over-representation of SNO sites in helices and under-representation in coils relative to unmodified cysteines [12]. The nature of the side chain and stereochemistry of the SNO donor has also been shown to provide additional specificity for protein S-nitrosylation [11]. Subsequent analysis of the acid–base motif has suggested a more distal (8 Å) influence of charged side chains. Importantly, charged residues were suggested to play a role in protein–protein interactions that may promote site-specific S-nitrosylation [13]. In addition, S-nitrosylation of proteins itself has been proposed to generate novel protein–protein interactions by changing the surrounding surface charge distribution and by allosterically generating solvent-exposed binding sites [13]. The potential importance of protein–protein interactions in targeting S-nitrosylation is consistent with the emerging evidence for a major role of transnitrosylative transfer of NO groups from SNO-proteins (Fig. 1b).
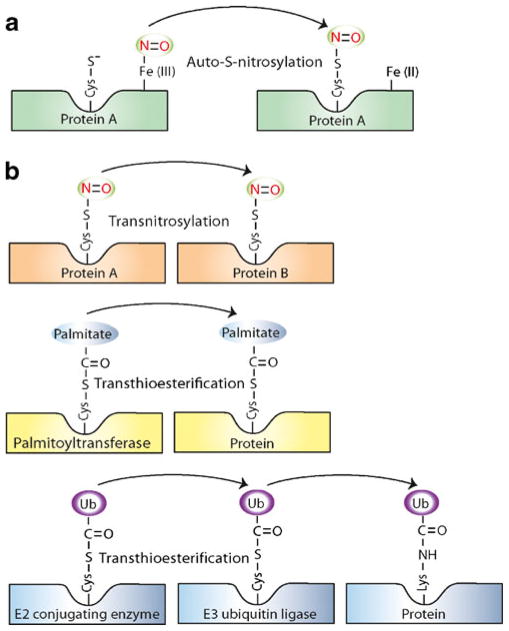
Fig. 1.
Metal-to-Cys and Cys-to-Cys nitrosylases. a The mechanism of action of Metal-to-Cys transnitrosylases may entail intramolecular NO group transfer (auto-S-nitrosylation) or intermolecular reactions with glutathione (not shown) to form SNO-proteins or GSNO, respectively. b The mechanism of action of Cys-to-Cys transnitrosylases exhibits similarities with enzymes involved in other ubiquitous posttranslational modifications. Transnitrosylases, palmitoyltransferases, and E2-conjugating enzymes participate in Cys-to-Cys transfer of NO, palmitate, or ubiquitin, respectively. E3 ubiquitin ligases transfer ubiquitin (ligated to cysteine) to target proteins
Nitrosylases
Metal-to-Cys
Nitrosylases are proteins involved in either Metal-to-Cys (Fig. 1a) or Cys-to-Cys (Fig. 1b) transfer of the NO group. Metal-to-Cys nitrosylases are proteins that mediate the transfer of NO from an intrinsically bound heme iron or other transition metal (e.g., Fe2+, Cu2+) complex to cysteine thiol. It is increasingly appreciated that, in Metal-to-Cys nitrosylases (which include globins and, potentially, dinitrosyl iron complexes ([X2Fe(NO)2] where X is S or N), transition metals can fulfill the redox requirement for SNO synthesis and may do so without a chemical role for molecular oxygen [14–16]. For instance, in the case of mammalian hemoglobin (Hb), an allosterically coupled intramolecular transfer of NO from heme iron (iron nitrosyl, HbFeNO) to Cysβ93 results in auto-S-nitrosylation (SNO-Hb) [17, 18]. Additionally, transfer of metal-coordinated NO from cytochrome c [19] or ceruloplasmin [20] to glutathione is employed in the synthesis of GSNO. It has also been reported that the redox cycling of ceruloplasmin-bound copper supports S-nitrosylation and denitrosylation of glypican-1 [21]. Other examples of Metal-to-Cys nitrosylases that catalyze auto-S-nitrosylation include neuroglobin [22], cytoglobin [22], and possibly nitrophorin [23] (Table 2). In spite of the paucity of freely available cellular pools of redox-active metals, chelation of divalent metal ions results in decreased cellular S-nitrosylation in yeast grown under both aerobic and anaerobic conditions [16], highlighting the general importance of metals in physiological SNO formation.
Table 2.
Nitrosylases and their substrates
| Nitrosylases | Substrates | Reference | |
|---|---|---|---|
| Metal-to-Cys | |||
| Hemoglobin |
|
Auto-S-nitrosylation | [17, 18] |
| Cytoglobina | [22] | ||
| Neuroglobina | [22] | ||
| Cytochrome ca | Glutathione | [19] | |
| Ceruloplasmin | Glutathione | [20] | |
| Glypican-1 | [21] | ||
| Cys-to-Cys | |||
| Hemoglobin | AE1 | [27] | |
| Glutathione | [30] | ||
| Thioredoxin | Caspase-3 | [46–48] | |
| ~ 50 proteins | [48] | ||
| GAPDH | HDAC | [37] | |
| SIRT-1 | [37] | ||
| DNA-PK | [37] | ||
| Caspase-3 | Thioredoxin | [50] | |
| X-linked inhibitor of apoptosis | [40] | ||
| Cyclin-dependent kinase 5 | Dynamin-related protein 1 | [44] | |
Indicates in vitro reaction of unknown physiological relevance
Cys-to-Cys
Cys-to-Cys transnitrosylases (referred to hereinafter as transnitrosylases) are typically SNO-proteins involved in Cys-to-Cys transfer of the NO group from donor to acceptor protein [24]. Mechanistically, the operation of transnitrosylases can be considered analogous to palmitoyltransferases that add palmitate to thiol groups in proteins by transthioes-terification or to E2-conjugating enzymes, which transfer ubiquitin to E3 ubiquitin ligases by transthioesterification; E3 ubiquitin ligases then transfer ubiquitin to protein substrates (Fig. 1b). To date, a number of transnitrosylases have been shown to function in physiological contexts, as described below (Table 2).
SNO-hemoglobin
SNO-hemoglobin has served as a classical model for allosteric control of protein S-nitrosylation [17, 18, 25]. The relaxed (R), oxygenated state of Hb promotes S-nitrosylation of its highly conserved Cysβ93 residue by intramolecular transfer of NO from heme iron, as described above. Deoxygenation of Hb induces an allosteric transition to the tense (T) structure, promoting transnitrosylative transfer of the NO group to the cytosolic domain of the red blood cell (RBC) membrane protein anion exchanger-1 (AE1) and to glutathione. This transnitrosylative transfer subserves export of vasodilatory NO bioactivity [17, 26]. Therefore, SNO-Hb can be functionally classified as an AE1 (and glutathione) nitrosylase. As discussed above, hemoglobin also exhibits an auto-S-nitrosylation activity in which NO bound to heme within the β-subunit can exchange with the Cysβ93 residue (in the R structure). Thus, hemoglobin can both synthesize SNO (Fig. 1a) and nitrosylate its cognate targets (Table 2).
Accumulating evidence suggests a role of aberrant transnitrosylation by SNO-Hb in numerous pathologies. Generally speaking, conditions that disfavor the allosteric transition in Hb from T to R state (hemoglobinopathies, including sickle cell disease) or that promote deoxygenation of Hb (hypoxemia, acidosis) will be associated with reduced synthesis of SNO-Hb. In sickle cell anemia, sickle hemoglobin (HbS; Glu6Val Hb) is impaired both in its ability to form SNO-Hb (Metal-to-Cys transfer of the NO group) and to transnitrosylate AE1 (Cys-to-Cys transfer of the NO group) (Fig. 2) [27]. The resultant decrease in levels of RBC membrane SNOs is correlated with the severity of impaired vasodilation by sickle RBCs demonstrated in vitro [27]. In addition, it has been demonstrated that banked blood (stored for transfusion) exhibits a rapid decrease in levels of SNO-Hb and a corresponding impairment of hypoxic vasodilatory function of RBCs, both of which are ameliorated by restoration of SNO-Hb levels (SNO repletion) [28]. Furthermore, RBCs exposed to sustained hypoxia exhibit impaired pO2-coupled vasodilation in the lungs and are deficient in increasing blood oxygenation; importantly, these parameters change in concert with alterations in SNO-Hb levels within RBCs and are ameliorated by SNO repletion using the inhaled S-nitrosylating gas ethyl nitrite (ENO) [29]. Moreover, hypoxemic patients with pulmonary arterial hypertension exhibit a decrease in SNO-Hb levels and reduced hypoxic vasodilation, which are reversed by ENO, further establishing a cause-and-effect relationship between SNO-Hb and improvements in pulmonary parameters [29]. Finally, SNO-Hb levels are markedly increased in sepsis [30–32] and at altitude [33, 34]. Both septic and hypoxemic conditions are characterized by decreased O2 utilization at the tissue level and increases in SNO-Hb may be viewed as means to normalize O2 delivery by increasing blood flow. Indeed, blood concentrations of SNO-Hb are a positive predictor of aerobic exercise capacity [34].
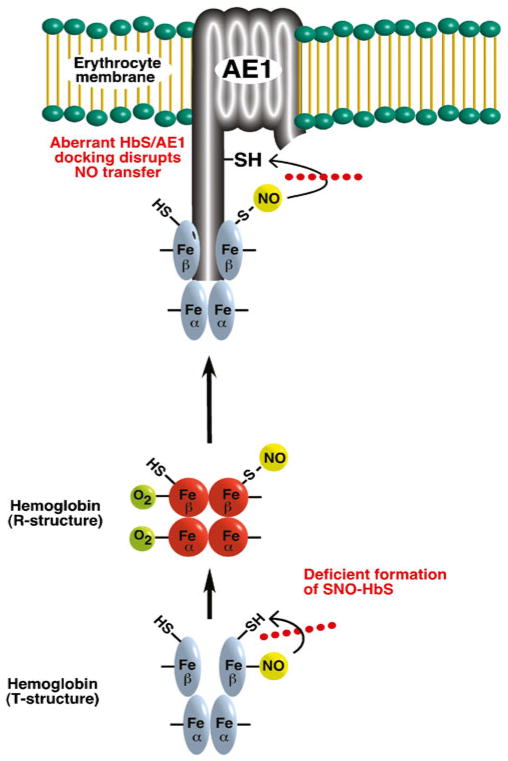
Fig. 2.
Dysregulation of hemoglobin’s nitrosylase activity in sickle cell anemia. In RBCs from patients with sickle cell anemia, aberrant intramolecular transfer of NO from heme iron nitrosyl to Cys (impaired Metal-to-Cys nitrosylase activity) results in deficient formation of SNO-sickle hemoglobin (SNO-HbS). Additionally, aberrant docking of S-nitrosylated SNO-HbS to the membrane protein AE1 disrupts transnitrosylative transfer of the NO group to the membrane (impaired Cys-to-Cys nitrosylase activity). Decreased levels of membrane SNO are associated with decreased ability of RBCs to effect hypoxic vasodilation, which may contribute to the vaso-occlusive crisis of sickle cell anemia [27]
SNO-GAPDH
In addition to its role as a glycolytic enzyme, there is a growing appreciation of the role GAPDH plays in extraglycolytic pathways in the nucleus, including DNA repair, telomere binding, and facilitation of apoptosis [35]. Hara et al. [36] demonstrated that, upon apoptotic stimulation, GAPDH is nitrosylated at its active-site Cys150, which promotes its binding to the E3 ubiquitin ligase Siah1 [36]. This results in the cotranslocation to the nucleus of SNO-GAPDH and Siah1 and in the stabilization of Siah1 that contributes to cell death. More recently, Kornberg et al. [37] showed that SNO-GAPDH binds to and transnitrosylates a number of nuclear proteins including DNA-PK, SIRT1, and HDAC2 and that S-nitrosylation of SIRT1 inhibits its downstream effects on transcription. Additionally, the degree of S-nitrosylation of these nuclear proteins in situ is correlated with levels of expression of GAPDH, as analyzed by over-expression or knockdown of GAPDH in mammalian cells [37]. Therefore, SNO-GAPDH can be classified as a physiological nitrosylase for a subset of nuclear proteins, thereby functioning to implement apoptotic and transcriptional programs.
SNO-caspase 3
The caspase family of proteases plays a central role in apoptosis. Caspase 3, which cleaves a number of target proteins, is the major executioner of apoptosis [38]. Inhibitors of apoptosis (IAPs), which confer protection, contain a baculovirus IAP repeat (BIR) and a RING domain [39]. The BIR domain interacts directly with a caspase, whereas the RING domain has E3 ubiquitin ligase activity [39]. The IAP-designated X-linked inhibitor of apoptosis protein (XIAP) interacts with active caspases 3/7/9 in the cytosol and is considered to be the most potent endogenous caspase inhibitor [40]. XIAP also ubiquitinylates caspases and thereby targets them for proteasomal degradation. XIAP is nitrosylated at its RING domain by either exogenous or endogenous sources of NO in vitro or in vivo; S-nitrosylation of XIAP inhibits its E3 ubiquitin ligase activity, reflected in the stabilization of caspase 3 [40]. Consistent with these findings, knockdown of XIAP in primary cerebrocortical neurons (nNOS-expressing cells) leads to an increase in caspase 3 activity and facilitation of cell death induced by NMDA (an agonist of the NMDA-type glutamate receptor) [40]. Furthermore, SNO-XIAP levels are increased in patients with neurodegenerative conditions including Alzheimer disease and diffuse Lewy body disease, implicating S-nitrosylation of XIAP in the etiology of neuronal damage [40]. Notably, SNO-caspase 3 functions as a specific transnitrosylase for XIAP in vitro, and this reaction is subserved by a physical interaction between SNO-caspase 3 and XIAP [40]. Transnitrosylation of XIAP by SNO-caspase 3 has been implicated in the pathophysiology of cell death, and thus, may be construed as aberrant nitrosylase activity [40].
SNO-cyclin-dependent kinase 5
Cyclin-dependent kinase 5 (Cdk5), which is predominantly a neuron-specific kinase, plays an important role in neuronal development, including cell survival, axon guidance, neuronal migration, and regulation of synaptic spine plasticity [41]. Dysregulation of Cdk5 has been implicated in the pathogenesis of several neurological disorders, including Alzheimer disease and Parkinson disease [42]. A recent study by Lipton and colleagues [43] demonstrated that exposure of primary cortical neurons (nNOS-expressing cells) to NMDA results in the S-nitrosylation of Cdk5 and that SNO-Cdk5 exhibits enhanced kinase activity. Importantly, SNO-Cdk5 can transnitrosylate dynamin-related protein 1 (Drp1) [43]. S-nitrosylation of Drp1 following exposure of cells to amyloid protein leads to its dimerization and enhanced GTPase activity [44]. Consequently, SNO-Drp1 promotes mitochondrial fission with an increase in neuronal damage. Finally, both SNO-Drp1 and SNO-Cdk5 levels are enhanced in patients with Alzheimer disease [43, 44], pointing toward SNO-Cdk5-mediated transnitrosylation of Drp1 as a potential therapeutic target.
SNO-Trx
The Trx system, which consists of redox-coupled thioredoxin (Trx) and thioredoxin reductase (TR), is well characterized as a principal protein disulfide reductase that maintains cellular redox equilibrium. The two redox-active, catalytic Cys in the active site of Trx (Cys32-X-X-Cys35) participate in the reversible reduction of disulfides in substrate proteins, producing oxidized Trx (Cys32-Cys35 disulfide linkage). Oxidized Trx is reduced by the NADPH-dependent Trx reductase (TrxR) [45]. In addition to the active site, human Trx has three additional cysteines (Cys62, Cys69, and Cys73) [45]. Cys73 has emerged as a physiological target of S-nitrosylation that participates in transnitrosylation reactions with other proteins [46–48]. Initial analysis of the function of Trx as a nitrosylase showed that SNO-Trx, S-nitrosylated at Cys73, transnitrosylated caspase 3 (forming SNO-caspase) at a rate approximately 100 times faster than GSNO in vitro [46]. Importantly, in vivo, this transnitrosylation reaction requires the interaction between SNO-Trx and casapse-3, as evidenced by a loss of transnitrosylase activity of a Trx mutant missing the local contact residues (Trx-E70A/K72A) [47]. Subsequent analysis indicated that oxidation of the active-site cysteines within Trx promotes S-nitrosylation of Cys73, which subserves transnitrosylation of target proteins including peroxiredoxin 1 and caspase 3 [48]. Additionally, it was found that SNO-Trx does not serve as a substrate for TR [49]. Consistent with these findings, substantial levels of oxidized Trx are detected in several tissues, including the lung and kidney [48], providing support for the idea that Trx can be uncoupled from TR, which promotes its nitrosylase function. Overexpression of a Trx1 C32S/C35S mutant in HeLa cells promoted the S-nitrosylation of Trx1 C32S/C35S compared to wild-type and revealed 47 novel potential substrates for trans-S-nitrosylation by Trx1 [48]. Thus, Trx may operate under some conditions as a multiprotein nitrosylase (Table 2). Importantly, reduced Trx mediates denitrosylation of numerous SNO-proteins, as detailed below.
Denitrosylases
Denitrosylases are enzymes involved in the removal of NO groups from the Cys thiol side chain of proteins or low-molecular-weight thiols (Table 3). By abstracting NO from proteins, denitrosylases may function to ameliorate nitrosative stress and to regulate signal transduction. Denitrosylases also play important roles in setting levels of cellular nitrosylation by analogy to phosphatases, which often set levels of phosphorylation. To date, two enzymatic denitrosylating systems have been demonstrated to function in physiological contexts: the Trx/TR system and the glutathione (GSH)/GSNO reductase (GSNOR) system [1].
Table 3.
Denitrosylases and their substrates
| Denitrosylases | Substrates | Reference |
|---|---|---|
| GSNO reductase | GRK2 | [66] |
| β-arrestin 2 | [67] | |
| HIF1α | [73] | |
| Ras | [118] | |
| Ryanodine receptor 2 | Unpublished | |
| Connexin | [119] | |
| Thioredoxin/thioredoxin reductase | Caspase 3 | [50] |
| Caspase 9 | [50] | |
| PTP-1B | [50] | |
| NSF | [51] | |
| Insulin receptora | [52] | |
| Akta | [52] | |
| PDE3Ba | [52] | |
| ~50 proteins | [54] | |
| Ceruloplasmin | Glypican-1 | [21] |
| Candidate denitrosylases (in vitro) | ||
| Carbonyl reductaseb | GSNO | [59] |
| Protein disulfide isomerasec | GSNO | [85] |
| Xanthine oxidasec | GSNO | [84] |
| CysNO | [84] | |
| Glutathione peroxidasec | GSNO | [83] |
Indirect evidence
Activity shown in cell lysates
Activity in isolated system
Trx system (Trx, TR, NADPH)
Denitrosylation of SNO-caspase 3 is constitutive in the cytosol and stimulus coupled at the mitochondria, the latter in the context of apoptotic stimulation by Fas ligand [50]. The Trx1 and Trx2 systems serve as the cognate denitrosylases for cytosolic and mitochondrial SNO-caspases, respectively [50]. Trx1 has also been reported to denitrosylate N-ethylmaleimide-sensitive factor (NSF) and thereby promote granule exocytosis that has been implicated in vascular inflammation [51]. Decreased levels of TrxR were associated with increased S-nitrosylation of the insulin receptor, Akt kinase, and the phosphodiesterase, PDE3B, in the adipose tissue of obese subjects, which was implicated in the decreased antilipolytic action of insulin associated with obesity [52]. Thioredoxin-interacting protein (Txnip) inhibits the denitrosylase function of Trx, thereby enhancing cellular levels of SNO-protein [53]. Proteomic analyses revealed that Trx likely serves as a major SNO-protein denitrosylase in mammalian cells where it mediates denitrosylation of a broad spectrum of SNO-proteins [54, 55]. Trx-mediated SNO-protein denitrosylation proceeds via the formation of a mixed disulfide intermediate between Trx and its substrate and likely through transnitrosylation to release the reduced NO moiety, nitroxyl (HNO) [50]. Although the Trx system has also been shown to catalyze the reduction of GSNO in vitro, the physiological relevance of this activity is not known [56]. In addition, lipoic acid has been reported to denitrosylate SNO-albumin (as well as GSNO) in vitro (after reduction to dihydrolipoic acid by lipoic acid dehydrogenase), but the physiological relevance of this activity has not been determined [57].
In physiological settings, Trx exists primarily in the reduced state, which facilitates its function as a SNO-protein denitrosylase [58]. As discussed above, uncoupling of Trx from TR allows S-nitrosylation of (oxidized) Trx and may promote the function of SNO-Trx as a transnitrosylase. Given that Trx is thought to play a role in numerous diseases, its transnitrosylase and denitrosylase activities may serve as therapeutic targets, particularly in the context of oxidative and/or nitrosative stress (nitrosoredox imbalance).
GSNO reductase system (GSNOR, GSH, NADH)
Glutathione is the major low-molecular-weight thiol in mammalian cells (up to 10 mM) [59]. GSNO, which was initially identified as a physiological entity in human airways [60], can transnitrosylate proteins, forming SNO-proteins. A GSNO-metabolizing activity was purified that is broadly conserved across phylogeny (bacteria to man) and was identified as class III alcohol dehydrogenase (ADH3) [61, 62]. It was determined that the principal substrate for ADH3 is in fact GSNO, and the enzyme was renamed GSNO reductase (GSNOR) [62]. GSNOR utilizes NADH to carry out a 2e− reduction of GSNO to generate glutathione sulfinamide [63]. GSNOR is the major source of NADH-dependent GSNO-metabolizing activity in mammalian tissues [30]. Strikingly, knockout of GSNOR in yeast and mice was shown to increase cellular levels of both GSNO and SNO-proteins (upon treatment with an SNO donor or lipopolysaccharide (LPS)). Thus, GSNO and at least some SNO-proteins are in an equilibrium governed by Cys-to-Cys transnitrosylation, and GSNOR is an important regulator of SNO-protein denitrosylation [30, 62]. Consistent with these findings, the addition of glutathione to SNO-proteins leads to the rapid denitrosylation of a large subset of proteins in vitro [64] and accumulating data suggest that GSNOR operates in concert with all isoforms of NOS in vivo [30, 65, 66].
The physiological role of GSNOR as a regulator of protein denitrosylation is well established in the context of signal transduction through G protein-coupled receptors (GPCRs) and, in particular, the β2-adrenergic receptor (β2AR). It has been shown that ligand-coupled S-nitrosylation of multiple proteins regulates β2AR desensitization and internalization. Agonist-induced S-nitrosylation of GPCR kinase 2 (GRK2) inhibits its ability to phosphorylate the receptor and thereby prevents binding of the receptor to β-arrestin 2, resulting in attenuated desensitization of G protein-mediated signaling and decreased receptor internalization [66]. In addition, stimulus-coupled S-nitrosylation of β-arrestin 2 promotes its binding to the clathrin/AP2-based internalization machinery and thereby enhances internalization of the receptor–β-arrestin 2 complex [67]. Regulation by GSNOR is demonstrated by the findings that, in the heart and lungs of GSNOR−/− mice, S-nitrosylation of GRK2 and β-arrestin 2 are augmented, and surface expression of the β2AR is enhanced [66, 67].
Initial evidence of aberrant GSNOR-mediated denitrosylation in disease was established in a sepsis model employing GSNOR−/− mice [30]. Mice deficient in GSNOR exhibit substantial increases in mortality and tissue injury after LPS treatment, the degree of which was directly correlated with levels of cellular S-nitrosylation [30]. In addition, Wei et al. [68] demonstrated that GSNOR-deficient mice are more susceptible to spontaneous and carcinogen-induced hepatocellular carcinoma (HCC). Following challenge with diethylnitrosamine (DEN) or LPS, they observed increased S-nitrosylation, proteasomal degradation, and depletion of the DNA repair enzyme, O6-alkylguanine-DNA alkyl transferase (AGT) in the liver of GSNOR−/− mice (Fig. 3). Consequently, levels of O6-ethylydeoxyguanosine, which is a substrate for AGT, are elevated in GSNOR−/− mice after DEN challenge, implicating SNO-AGT as a physiological target of GSNOR [68]. Furthermore, GSNOR−/− mice are more susceptible to DEN-induced hepatic carcinogenesis, which has been suggested to result from a defect in the DNA repair mechanism [68]. Most notably, as much as 50% of human HCC has been attributed to chromosomal deletion of GSNOR [68]. Collectively, these studies suggest a crucial role for GSNOR in ameliorating nitrosative stress.
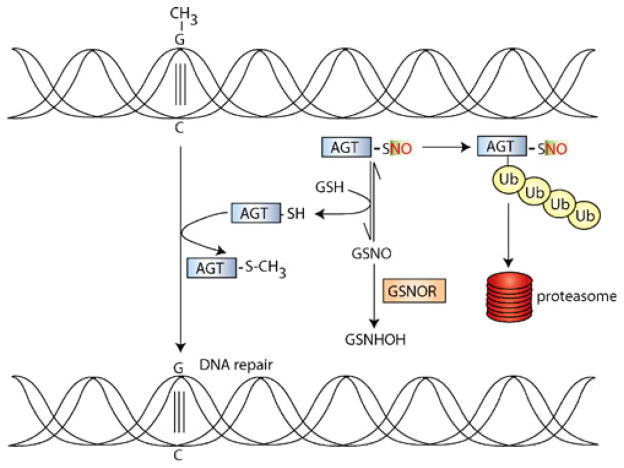
Fig. 3.
Deficiency of cellular GSNOR activity in the etiology of hepatic carcinoma. The O6-alkylguanine-DNA alkyl transferase (AGT) protein transfers the alkyl group (shown as a methyl, CH3, group) from the O6 position of guanine to its active-site cysteine, thereby repairing DNA. However, AGT is S-nitrosylated in inflammatory conditions (in which iNOS is upregulated), and S-nitrosylation promotes ubiquitinylation and subsequently proteasomal degradation. GSNO is in equilibrium with SNO-AGT, and in wild-type mice, GSNOR regulates SNO-AGT denitrosylation by shifting the equilibrium towards GSNO, resulting in decreased SNO-AGT levels. However, in GSNOR−/− mice, the absence of GSNOR activity results in increased S-nitrosylation and degradation of AGT. A chromosomal deletion, inclusive of the GSNOR gene, is found in 50% of patients with HCC. Thus, GSNOR denitrosylase activity may play a tumor suppressor function in the etiology of HCC
Reduced levels of GSNO have also been implicated in the pathogenesis of respiratory and cardiovascular diseases. Mice deficient in GSNOR are protected in an experimental model of asthma, in association with enhanced intracellular SNO levels [65]. Whereas inflammatory responses to ovalbumin treatment are not diminished in GSNOR−/− mice, they are, nonetheless, protected against ovalbumin-induced hyperresponsiveness to methacholine, a cholinergic agonist and bronchoconstrictor [65]. SNPs in GSNOR have been associated in multiple studies with asthma and responsiveness to bronchodilators [69–71] and GSNOR has been strongly linked to asthmatic responsiveness in humans [72]. Furthermore, GSNOR−/− mice display enhanced cardioprotection after myocardial infarction, associated with increased myocardial capillary density [73]. Mechanistically, this increased cardioprotection has been ascribed to enhanced S-nitrosylation of HIF1α, which promotes its stabilization and transcriptional activation of VEGF [73]. Given the critical role of GSNOR in SNO homeostasis, there have been increasing efforts to develop inhibitors targeting GSNOR [74–76]. One inhibitor is in phase II clinical trials for the treatment of acute asthma.
Interestingly, an increasing body of research suggests a critical role for GSNOR in plant pathogenesis. This work has been facilitated by the identification of plant strains with both increased and decreased GSNOR activity [77–79]. For example, GSNOR regulates pathogen-induced increases in cellular S-nitrosylation in Arabidopsis thaliana, and an increase in total SNO levels corresponds with susceptibility to various diseases [77]. In addition, Tada et al. [79] reported that, in Arabidopsis, salicylic acid (SA)-mediated NPR1 oligomer-to-monomer generation is regulated by GSNOR, as evidenced by reduced monomeric NPR1 levels in the nucleus of atgsnor1–3 (Arabidopsis mutant lacking GSNOR activity). Accordingly, SA-mediated NPR1-dependent PR-1 (pathogenesis-related gene 1) expression is inhibited in the null mutant [79]. Furthermore, increased SNO levels as a result of GSNOR deficiency have been shown to regulate programmed cell death and heat shock tolerance in plants [80, 81]. Thus, SNOs, regulated by GSNOR, have important roles in most organisms.
Candidate denitrosylases
On the basis of in vitro analysis, a number of additional mammalian enzymes have been identified as potential denitrosylases for small-molecular-weight SNOs (GSNO and/or CysNO), although their physiological relevance has not been established: superoxide dismutase [82], glutathione peroxidase [83], xanthine oxidase [84], protein disulfide isomerase [85], and carbonyl reductase [59], the latter accounting for ~30% of NADPH-dependent GSNO reductase activity in A549 lung adenocarcinoma cell lysates. Both the identification of additional physiological denitrosylases acting on small-molecular-weight SNOs and/or SNO-proteins and the extent to which denitrosylases operate on distinct sets of substrates (consistent with varying stability of SNO-proteins in vivo as revealed by global kinetic analysis of SNO stability [55]) remain important subjects of experimental analysis.
Conclusion
Protein S-nitrosylation is the prototypic redox-based post-translational modification. Homeostasis of S-nitrosylation is crucial for the maintenance of normal physiology, as evidenced by an increasing number of diseases associated with dysregulated S-nitrosylation. Multiple nitrosylases and denitrosylases function in concert both to maintain homeostasis of protein S-nitrosylation and to dynamically regulate cellular signal transduction. Thus, aberrant nitrosylase and denitrosylase activities may be a cause of disease and may serve as important and novel therapeutic targets.
Acknowledgments
This work was supported by National Institutes of Health Grants R01HL059130, R01HL095463, R01HL091876, and P01HL075443 and by DARPA N66001-10-C-2015. The authors thank Dr. Douglas T. Hess for the discussion and advice.
Contributor Information
Puneet Anand, Institute for Transformative Molecular Medicine and Department of Medicine, Case Western Reserve University and University Hospitals Case Medical Center, Cleveland, OH 44106, USA. Graduate Training Program, Department of Biochemistry, Duke University Medical Center, Durham, NC 27710, USA.
Jonathan S. Stamler, Email: jonathan.stamler@case.edu, Institute for Transformative Molecular Medicine and Department of Medicine, Case Western Reserve University and University Hospitals Case Medical Center, Cleveland, OH 44106, USA.
References
- 1.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 2.Murad F. Cyclic guanosine monophosphate as a mediator of vasodilation. J Clin Invest. 1986;78:1–5. doi: 10.1172/JCI112536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 4.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Mato I, Castro C, Ruiz FA, Corrales FJ, Mato JM. Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J Biol Chem. 1999;274:17075–17079. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 6.Savidge TC, Urvil P, Oezguen N, Ali K, Choudhury A, Acharya V, Pinchuk I, Torres AG, English RD, Wiktorowicz JE, et al. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat Med. 2011;17:1136–1141. doi: 10.1038/nm.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 9.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 10.Nedospasov A, Rafikov R, Beda N, Nudler E. An autocatalytic mechanism of protein nitrosylation. Proc Natl Acad Sci USA. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc Natl Acad Sci USA. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci USA. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of transnitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 15.Bosworth CA, Toledo JC, Jr, Zmijewski JW, Li Q, Lancaster JR., Jr Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci. 2009;106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. A genetic analysis of nitrosative stress. Biochemistry. 2009;48:792–799. doi: 10.1021/bi801813n. [DOI] [PubMed] [Google Scholar]
- 17.Jia L, Bonaventura C, Bonaventura J, Stamler JS. S-nitrosohaemoglobin: a dynamic activity of blood involved in vascular control. Nature. 1996;380:221–226. doi: 10.1038/380221a0. [DOI] [PubMed] [Google Scholar]
- 18.Gow AJ, Stamler JS. Reactions between nitric oxide and haemoglobin under physiological conditions. Nature. 1998;391:169–173. doi: 10.1038/34402. [DOI] [PubMed] [Google Scholar]
- 19.Basu S, Keszler A, Azarova NA, Nwanze N, Perlegas A, Shiva S, Broniowska KA, Hogg N, Kim-Shapiro DB. A novel role for cytochrome c: efficient catalysis of S-nitrosothiol formation. Free Radic Biol Med. 2010;48:255–263. doi: 10.1016/j.freeradbiomed.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue K, Akaike T, Miyamoto Y, Okamoto T, Sawa T, Otagiri M, Suzuki S, Yoshimura T, Maeda H. Nitrosothiol formation catalyzed by ceruloplasmin. Implication for cytoprotective mechanism in vivo. J Biol Chem. 1999;274:27069–27075. doi: 10.1074/jbc.274.38.27069. [DOI] [PubMed] [Google Scholar]
- 21.Mani K, Cheng F, Havsmark B, David S, Fransson LA. Involvement of glycosylphosphatidylinositol-linked ceruloplasmin in the copper/zinc-nitric oxide-dependent degradation of glypican-1 heparan sulfate in rat C6 glioma cells. J Biol Chem. 2004;279:12918–12923. doi: 10.1074/jbc.M313678200. [DOI] [PubMed] [Google Scholar]
- 22.Petersen MG, Dewilde S, Fago A. Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J Inorg Biochem. 2008;102:1777–1782. doi: 10.1016/j.jinorgbio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Weichsel A, Maes EM, Andersen JF, Valenzuela JG, Shokhireva T, Walker FA, Montfort WR. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc Natl Acad Sci USA. 2005;102:594–599. doi: 10.1073/pnas.0406549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamler JS, Hess DT. Nascent nitrosylases. Nat Cell Biol. 2010;12:1024–1026. doi: 10.1038/ncb1110-1024. [DOI] [PubMed] [Google Scholar]
- 25.Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Annu Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 26.Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 27.Pawloski JR, Hess DT, Stamler JS. Impaired vasodilation by red blood cells in sickle cell disease. Proc Natl Acad Sci USA. 2005;102:2531–2536. doi: 10.1073/pnas.0409876102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds JD, Ahearn GS, Angelo M, Zhang J, Cobb F, Stamler JS. S-nitrosohemoglobin deficiency: a mechanism for loss of physiological activity in banked blood. Proc Natl Acad Sci USA. 2007;104:17058–17062. doi: 10.1073/pnas.0707958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon TJ, Ahearn GS, Moya MP, Gow AJ, Huang YC, Luchsinger BP, Nudelman R, Yan Y, Krichman AD, Bashore TM, et al. A nitric oxide processing defect of red blood cells created by hypoxia: deficiency of S-nitrosohemoglobin in pulmonary hypertension. Proc Natl Acad Sci USA. 2005;102:14801–14806. doi: 10.1073/pnas.0506957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 31.Crawford JH, Chacko BK, Pruitt HM, Piknova B, Hogg N, Patel RP. Transduction of NO-bioactivity by the red blood cell in sepsis: novel mechanisms of vasodilation during acute inflammatory disease. Blood. 2004;104:1375–1382. doi: 10.1182/blood-2004-03-0880. [DOI] [PubMed] [Google Scholar]
- 32.Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, et al. Hemoglobin conformation couples erythrocyte S-nitrosothiol content to O2 gradients. Proc Natl Acad Sci USA. 2005;102:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erzurum SC, Ghosh S, Janocha AJ, Xu W, Bauer S, Bryan NS, Tejero J, Hemann C, Hille R, Stuehr DJ, et al. Higher blood flow and circulating NO products offset high-altitude hypoxia among Tibetans. Proc Natl Acad Sci USA. 2007;104:17593–17598. doi: 10.1073/pnas.0707462104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janocha AJ, Koch CD, Tiso M, Ponchia A, Doctor A, Gibbons L, Gaston B, Beall CM, Erzurum SC. Nitric oxide during altitude acclimatization. N Engl J Med. 2011;365:1942–1944. doi: 10.1056/NEJMc1107887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 37.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmgren A. Biochemistry. SNO removal. Science. 2008;320:1019–1020. doi: 10.1126/science.1159246. [DOI] [PubMed] [Google Scholar]
- 39.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 42.Cruz JC, Tsai LH. A Jekyll and Hyde kinase: roles for Cdk5 in brain development and disease. Curr Opin Neurobiol. 2004;14:390–394. doi: 10.1016/j.conb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Qu J, Nakamura T, Cao G, Holland EA, McKercher SR, Lipton SA. S-nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by beta-amyloid peptide. Proc Natl Acad Sci USA. 2011;108:14330–14335. doi: 10.1073/pnas.1105172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lillig CH, Holmgren A. Thioredoxin and related molecules—from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci USA. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, Liu T, Chen W, Oka S, Fu C, Jain MR, Parrott AM, Baykal AT, Sadoshima J, Li H. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol Cell Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashemy SI, Holmgren A. Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S-nitrosylation of cysteine residues. J Biol Chem. 2008;283:21890–21898. doi: 10.1074/jbc.M801047200. [DOI] [PubMed] [Google Scholar]
- 50.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito T, Yamakuchi M, Lowenstein CJ. Thioredoxin increases exocytosis by denitrosylating N-ethylmaleimide-sensitive factor. J Biol Chem. 2011;286:11179–11184. doi: 10.1074/jbc.M110.201780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ovadia H, Haim Y, Nov O, Almog O, Kovsan J, Bashan N, Benhar M, Rudich A. Increased adipocyte S-nitrosylation targets anti-lipolytic action of insulin: relevance to adipose tissue dysfunction in obesity. J Biol Chem. 2011;286:30433–30443. doi: 10.1074/jbc.M111.235945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forrester MT, Seth D, Hausladen A, Eyler CE, Foster MW, Matsumoto A, Benhar M, Marshall HE, Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J Biol Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benhar M, Thompson JW, Moseley MA, Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikitovic D, Holmgren A. S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J Biol Chem. 1996;271:19180–19185. doi: 10.1074/jbc.271.32.19180. [DOI] [PubMed] [Google Scholar]
- 57.Stoyanovsky DA, Tyurina YY, Tyurin VA, Anand D, Mandavia DN, Gius D, Ivanova J, Pitt B, Billiar TR, Kagan VE. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J Am Chem Soc. 2005;127:15815–15823. doi: 10.1021/ja0529135. [DOI] [PubMed] [Google Scholar]
- 58.Wu C, Parrott AM, Fu C, Liu T, Marino SM, Gladyshev VN, Jain MR, Baykal AT, Li Q, Oka S, et al. Thioredoxin 1-mediated post-translational modifications: reduction, transnitrosylation, denitrosylation, and related proteomics methodologies. Antioxid Redox Signal. 2011;15:2565–2604. doi: 10.1089/ars.2010.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bateman RL, Rauh D, Tavshanjian B, Shokat KM. Human carbonyl reductase 1 is an S-nitrosoglutathione reductase. J Biol Chem. 2008;283:35756–35762. doi: 10.1074/jbc.M807125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins ME, Sugarbaker DJ, Chee C, Singel DJ, et al. Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc Natl Acad Sci USA. 1993;90:10957–10961. doi: 10.1073/pnas.90.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen DE, Belka GK, Du Bois GC. S-nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem J. 1998;331(Pt 2):659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 63.Staab CA, Alander J, Brandt M, Lengqvist J, Morgenstern R, Grafst rom RC, Hoog JO. Reduction of S -nitrosoglutathione by alcohol dehydrogenase 3 is facilitated by substrate alcohols via direct cofactor recycling and leads to GSH-controlled formation of glutathione transferase inhibitors. Biochem J. 2008;413:493–504. doi: 10.1042/BJ20071666. [DOI] [PubMed] [Google Scholar]
- 64.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Que LG, Liu L, Yan Y, Whitehead GS, Gavett SH, Schwartz DA, Stamler JS. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, et al. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 67.Ozawa K, Whalen EJ, Nelson CD, Mu Y, Hess DT, Lefkowitz RJ, Stamler JS. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med. 2010;2:19ra13. doi: 10.1126/scitranslmed.3000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choudhry S, Que LG, Yang Z, Liu L, Eng C, Kim SO, Kumar G, Thyne S, Chapela R, Rodriguez-Santana JR, et al. GSNO reductase and beta2-adrenergic receptor gene–gene interaction: bronchodilator responsiveness to albuterol. Pharmacogenet Genomics. 2010;20:351–358. doi: 10.1097/FPC.0b013e328337f992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore PE, Ryckman KK, Williams SM, Patel N, Summar ML, Sheller JR. Genetic variants of GSNOR and ADRB2 influence response to albuterol in African-American children with severe asthma. Pediatr Pulmonol. 2009;44:649–654. doi: 10.1002/ppul.21033. [DOI] [PubMed] [Google Scholar]
- 71.Wu H, Romieu I, Sienra-Monge JJ, Estela Del Rio-Navarro B, Anderson DM, Jenchura CA, Li H, Ramirez-Aguilar M, Del Carmen Lara-Sanchez I, London SJ. Genetic variation in S-nitrosoglutathione reductase (GSNOR) and childhood asthma. J Allergy Clin Immunol. 2007;120:322–328. doi: 10.1016/j.jaci.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Que LG, Yang Z, Stamler JS, Lugogo NL, Kraft M. S-nitrosoglutathione reductase: an important regulator in human asthma. Am J Respir Crit Care Med. 2009;180:226–231. doi: 10.1164/rccm.200901-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, Messina E, Bowles D, Kontos CD, Hare JM, et al. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci USA. 2009;106:6297–6302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun X, Qiu J, Strong SA, Green LS, Wasley JW, Blonder JP, Colagiovanni DB, Mutka SC, Stout AM, Richards JP, et al. Discovery of potent and novel S-nitrosoglutathione reductase inhibitors devoid of cytochrome P450 activities. Bioorg Med Chem Lett. 2011;21:5849–5853. doi: 10.1016/j.bmcl.2011.07.103. [DOI] [PubMed] [Google Scholar]
- 75.Sun X, Qiu J, Strong SA, Green LS, Wasley JW, Colagiovanni DB, Mutka SC, Blonder JP, Stout AM, Richards JP, et al. Structure–activity relationships of pyrrole based S-nitrosoglutathione reductase inhibitors: pyrrole regioisomers and propionic acid replacement. Bioorg Med Chem Lett. 2011;21:3671–3675. doi: 10.1016/j.bmcl.2011.04.086. [DOI] [PubMed] [Google Scholar]
- 76.Colagiovanni DB, Drolet DW, Langlois-Forget E, Piche MP, Looker D, Rosenthal GJ. A nonclinical safety and pharmacokinetic evaluation of N6022: a first-in-class S-nitrosoglutathione reductase inhibitor for the treatment of asthma. Regul Toxicol Pharmacol. 2011;62:115–124. doi: 10.1016/j.yrtph.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 77.Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478:264–268. doi: 10.1038/nature10427. [DOI] [PubMed] [Google Scholar]
- 79.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, et al. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 81.Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jourd’heuil D, Laroux FS, Miles AM, Wink DA, Grisham MB. Effect of superoxide dismutase on the stability of S-nitrosothiols. Arch Biochem Biophys. 1999;361:323–330. doi: 10.1006/abbi.1998.1010. [DOI] [PubMed] [Google Scholar]
- 83.Hou Y, Guo Z, Li J, Wang PG. Seleno compounds and glutathione peroxidase catalyzed decomposition of S-nitrosothiols. Biochem Biophys Res Commun. 1996;228:88–93. doi: 10.1006/bbrc.1996.1620. [DOI] [PubMed] [Google Scholar]
- 84.Trujillo M, Alvarez MN, Peluffo G, Freeman BA, Radi R. Xanthine oxidase-mediated decomposition of S-nitrosothiols. J Biol Chem. 1998;273:7828–7834. doi: 10.1074/jbc.273.14.7828. [DOI] [PubMed] [Google Scholar]
- 85.Sliskovic I, Raturi A, Mutus B. Characterization of the S-denitrosation activity of protein disulfide isomerase. J Biol Chem. 2005;280:8733–8741. doi: 10.1074/jbc.M408080200. [DOI] [PubMed] [Google Scholar]
- 86.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 87.Abrams AJ, Farooq A, Wang G. S-nitrosylation of ApoE in Alzheimer’s disease. Biochemistry. 2011;50:3405–3407. doi: 10.1021/bi200266v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, et al. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 90.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsang AH, Lee YI, Ko HS, Savitt JM, Pletnikova O, Troncoso JC, Dawson V, Dawson TM, Chung KK. S-nitrosylation of XIAP compromises neuronal survival in Parkinson Disease. Proc Natl Acad Sci USA. 2009;106:4900–4905. doi: 10.1073/pnas.0810595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 93.Gonzalez DR, Beigi F, Treuer AV, Hare JM. Deficient ryanodine receptor S-nitrosylation increases sarcoplasmic reticulum calcium leak and arrhythmogenesis in cardiomyocytes. Proc Natl Acad Sci USA. 2007;104:20612–20617. doi: 10.1073/pnas.0706796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ueda K, Valdivia C, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, Ackerman MJ, Makielski JC. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci USA. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gandley RE, Tyurin VA, Huang W, Arroyo A, Daftary A, Harger G, Jiang J, Pitt B, Taylor RN, Hubel CA, et al. S-nitrosoalbumin-mediated relaxation is enhanced by ascorbate and copper: effects in pregnancy and preeclampsia plasma. Hypertension. 2005;45:21–27. doi: 10.1161/01.HYP.0000150158.42620.3e. [DOI] [PubMed] [Google Scholar]
- 96.Tyurin VA, Liu SX, Tyurina YY, Sussman NB, Hubel CA, Roberts JM, Taylor RN, Kagan VE. Elevated levels of S-nitrosoalbumin in preeclampsia plasma. Circ Res. 2001;88:1210–1215. doi: 10.1161/hh1101.092179. [DOI] [PubMed] [Google Scholar]
- 97.Zhang HH, Wang YP, Chen DB. Analysis of nitrosoproteomes in normotensive and severe preeclamptic human placentas. Biol Reprod. 2011;84:966–975. doi: 10.1095/biolreprod.110.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmer LA, Doctor A, Chhabra P, Sheram ML, Laubach VE, Karlinsey MZ, Forbes MS, Macdonald T, Gaston B. S-nitrosothiols signal hypoxia-mimetic vascular pathology. J Clin Invest. 2007;117:2592–2601. doi: 10.1172/JCI29444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mukhopadhyay S, Lee J, Sehgal PB. Depletion of the ATPase NSF from Golgi membranes with hypo-S-nitrosylation of vasorelevant proteins in endothelial cells exposed to monocrotaline pyrrole. Am J Physiol Heart Circ Physiol. 2008;295:H1943–H1955. doi: 10.1152/ajpheart.00642.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Godoy LC, Moretti AI, Jurado MC, Oxer D, Janiszewski M, Ckless K, Velasco IT, Laurindo FR, Souza HP. Loss of CD40 endogenous S-nitrosylation during inflammatory response in endotoxemic mice and patients with sepsis. Shock. 2010;33:626–633. doi: 10.1097/SHK.0b013e3181cb88e6. [DOI] [PubMed] [Google Scholar]
- 101.Sun J, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel alpha1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 102.Burger DE, Lu X, Lei M, Xiang FL, Hammoud L, Jiang M, Wang H, Jones DL, Sims SM, Feng Q. Neuronal nitric oxide synthase protects against myocardial infarction-induced ventricular arrhythmia and mortality in mice. Circulation. 2009;120:1345–1354. doi: 10.1161/CIRCULATIONAHA.108.846402. [DOI] [PubMed] [Google Scholar]
- 103.Carnes CA, Janssen PM, Ruehr ML, Nakayama H, Nakayama T, Haase H, Bauer JA, Chung MK, Fearon IM, Gillinov AM, et al. Atrial glutathione content, calcium current, and contractility. J Biol Chem. 2007;282:28063–28073. doi: 10.1074/jbc.M704893200. [DOI] [PubMed] [Google Scholar]
- 104.Asada K, Kurokawa J, Furukawa T. Redox- and calmodulin-dependent S-nitrosylation of the KCNQ1 channel. J Biol Chem. 2009;284:6014–6020. doi: 10.1074/jbc.M807158200. [DOI] [PubMed] [Google Scholar]
- 105.Bai CX, Namekata I, Kurokawa J, Tanaka H, Shigenobu K, Furukawa T. Role of nitric oxide in Ca2+ sensitivity of the slowly activating delayed rectifier K+ current in cardiac myocytes. Circ Res. 2005;96:64–72. doi: 10.1161/01.RES.0000151846.19788.E0. [DOI] [PubMed] [Google Scholar]
- 106.Milsom AB, Jones CJ, Goodfellow J, Frenneaux MP, Peters JR, James PE. Abnormal metabolic fate of nitric oxide in type I diabetes mellitus. Diabetologia. 2002;45:1515–1522. doi: 10.1007/s00125-002-0956-9. [DOI] [PubMed] [Google Scholar]
- 107.Padron J, Peiro C, Cercas E, Llergo JL, Sanchez-Ferrer CF. Enhancement of S-nitrosylation in glycosylated hemoglobin. Biochem Biophys Res Commun. 2000;271:217–221. doi: 10.1006/bbrc.2000.2617. [DOI] [PubMed] [Google Scholar]
- 108.Ding SY, Tribble ND, Kraft CA, Markwardt M, Gloyn AL, Rizzo MA. Naturally occurring glucokinase mutations are associated with defects in posttranslational S-nitrosylation. Mol Endocrinol. 2010;24:171–177. doi: 10.1210/me.2009-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carvalho-Filho MA, Ueno M, Hirabara SM, Seabra AB, Carvalheira JB, de Oliveira MG, Velloso LA, Curi R, Saad MJ. S-nitrosation of the insulin receptor, insulin receptor substrate 1, and protein kinase B/Akt: a novel mechanism of insulin resistance. Diabetes. 2005;54:959–967. doi: 10.2337/diabetes.54.4.959. [DOI] [PubMed] [Google Scholar]
- 110.Pauli JR, Ropelle ER, Cintra DE, Carvalho-Filho MA, Moraes JC, De Souza CT, Velloso LA, Carvalheira JB, Saad MJ. Acute physical exercise reverses S-nitrosation of the insulin receptor, insulin receptor substrate 1 and protein kinase B/Akt in diet-induced obese Wistar rats. J Physiol. 2008;586:659–671. doi: 10.1113/jphysiol.2007.142414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Massy ZA, Fumeron C, Borderie D, Tuppin P, Nguyen-Khoa T, Benoit MO, Jacquot C, Buisson C, Drueke TB, Ekindjian OG, et al. Increased pasma S-nitrosothiol concentrations predict cardiovascular outcomes among patients with end-stage renal disease: a prospective study. J Am Soc Nephrol. 2004;15:470–476. doi: 10.1097/01.asn.0000106716.22153.bb. [DOI] [PubMed] [Google Scholar]
- 112.Marozkina NV, Yemen S, Borowitz M, Liu L, Plapp M, Sun F, Islam R, Erdmann-Gilmore P, Townsend RR, Lichti CF, et al. Hsp 70/Hsp 90 organizing protein as a nitrosylation target in cystic fibrosis therapy. Proc Natl Acad Sci USA. 2010;107:11393–11398. doi: 10.1073/pnas.0909128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guo CJ, Atochina-Vasserman EN, Abramova E, Foley JP, Zaman A, Crouch E, Beers MF, Savani RC, Gow AJ. S-nitrosylation of surfactant protein-D controls inflammatory function. PLoS Biol. 2008;6:e266. doi: 10.1371/journal.pbio.0060266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bellinger AM, Reiken S, Carlson C, Mongillo M, Liu X, Rothman L, Matecki S, Lacampagne A, Marks AR. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Durham WJ, Aracena-Parks P, Long C, Rossi AE, Goonasekera SA, Boncompagni S, Galvan DL, Gilman CP, Baker MR, Shirokova N, et al. RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell. 2008;133:53–65. doi: 10.1016/j.cell.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marozkina NV, Yemen S, Wei C, Wallrabe H, Nagji AS, Liu L, Morozkina T, Jones DR, Gaston B. S-nitrosoglutathione reductase in human lung cancer. Am J Respir Cell Mol Biol. 2011;46:63–70. doi: 10.1165/rcmb.2011-0147OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Straub AC, Billaud M, Johnstone SR, Best AK, Yemen S, Dwyer ST, Looft-Wilson R, Lysiak JJ, Gaston B, Palmer L, et al. Compartmentalized connexin 43 s-nitrosylation/denitrosylation regulates heterocellular communication in the vessel wall. Arterioscler Thromb Vasc Biol. 2011;31:399–407. doi: 10.1161/ATVBAHA.110.215939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Malhotra D, Thimmulappa RK, Mercado N, Ito K, Kombairaju P, Kumar S, Ma J, Feller-Kopman D, Wise R, Barnes P, et al. Denitrosylation of HDAC2 by targeting Nrf2 restores glucocorticosteroid sensitivity in macrophages from COPD patients. J Clin Invest. 2011;121:4289–4302. doi: 10.1172/JCI45144. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]