SUMMARY
How and why the brain reversibly switches from a waking to a sleep state remain among the most intriguing questions in biology. We show that CyclinA (CycA) and Regulator of Cyclin A1 (Rca1), essential cell cycle factors, function in post-mitotic neurons to promote sleep in Drosophila melanogaster. Reducing the abundance of CycA in neurons delayed the wake-sleep transition, caused multiple arousals from sleep and reduced the homeostatic response to sleep deprivation. CycA is expressed in ~40–50 neurons in the adult brain, most of which are intermingled with circadian clock neurons, suggesting functional interactions among neurons controlling sleep and circadian behavior.
Rest in Drosophila shares many features with mammalian sleep (1, 2). Flies spend a large portion of their waking time moving, and the locomotor assay developed for circadian behavior (3) can therefore be used to screen for sleep mutants in this animal. A bout of inactivity lasting five minutes or longer is associated with an increased arousal threshold (1, 2), as well as changes in neural activity (4), and is used to define sleep in the fly. Since the discovery of sleep in Drosophila a decade ago (1, 2), the results of several genetic screens for sleep factors have been reported, and many of the genes identified have conserved roles across species [reviewed in (5)]. A surprisingly large fraction of these genes are ion channels or their regulators, or are involved with broadly acting neurotransmitter pathways. These findings led to the hypothesis that there are no dedicated sleep genes but rather that sleep is regulated by genes that control normal neuronal function [reviewed in (5)].
We decided to screen for sleep genes solely within the nervous system, bypassing the potential confounding effects of these genes in other tissues. We screened ~4,000 UAS-RNA interference (UAS-RNAi) lines using the pan-neuronal driver elavGal4 to knock down gene expression, while simultaneously driving the expression of UAS-Dicer2 to enhance RNAi efficacy (6). After three retesting rounds, we identified ~20 genes whose RNAi-mediated neuronal depletion led to reproducible changes in sleep pattern, quantity, or both. One sleep-promoting factor we isolated was Rca1 (Regulator of Cyclin A1), a conserved, essential cell cycle gene (7–10).
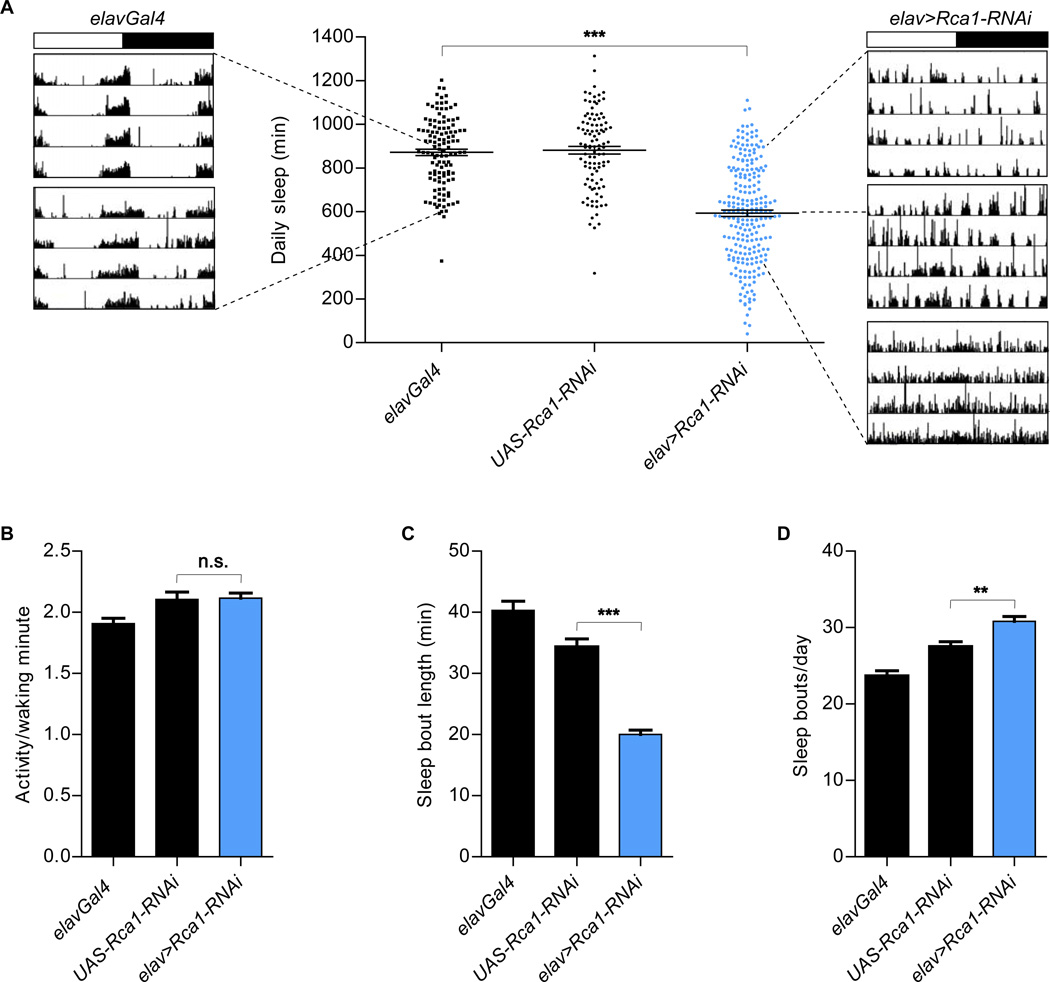
Depletion of Rca1 from neurons (elavGal4 + UAS-Rca1-RNAi, denoted “elav>Rca1-RNAi”) decreased the amount of total daily sleep (Figs. 1A and 2F), without affecting the levels of activity per waking minute (Fig. 1B). It did so by decreasing the duration of individual sleep episodes [(sleep bouts), Fig. 1C]. The number of sleep episodes was slightly increased in elav>Rca1-RNAi animals (Fig. 1D), likely because of the highly fragmented nature of their sleep. The net reduction in sleep duration understates the severity of the elav>Rca1-RNAi phenotype. For example, elav>Rca1-RNAi flies that showed no net change in daily sleep often failed to display behavior that is organized into prolonged intervals of rest and activity seen in the control flies (see examples of locomotor traces in Fig. 1A).
Figure 1. Reduced sleep in animals expressing neuronal Rca1-RNAi.
(A) Pan-neuronal Rca1-RNAi (elav>Rca1-RNAi, Number of animals (n) =235) reduced the duration of daily sleep compared to that of the parental control animals tested as hemizygotes [elavGal4 (n=121), UAS-Rca1-RNAi (n=103)]. Shown are example actograms for elavGal4 and elav>Rca1-RNAi flies that have average or extreme phenotypes. The top actogram for elav>Rca1-RNAi shows an example of a fly that is a “normal” sleeper (800+ minutes of sleep), but that did not show consolidation of rest and activity states (phenotype seen in 18 out 49 “normal” elav>Rca1-RNAi sleepers). Dashed lines point from the actograms to the amount of sleep seen in these individual flies. The white and black portions of the bar above the actograms indicate the light and dark periods of the experiment, respectively. (B) Comparable activity per waking minute in elav>Rca1-RNAi flies and controls. (C, D) Decreased sleep bout length (C) and increased number of daily sleep bouts (D) in elav>Rca1-RNAi flies, compared to the parental controls. Statistical analysis performed using One-way ANOVAs with Tukey post-test. Shown is statistical significance for experimental genotypes compared to controls with the most similar values. In all figures, error bars represent SEM; in all figures *P<0.05, **P <0.01, ***P<0.001. In all experiments described in this figure, elavGal4 drives the expression of UAS-Dicer2.
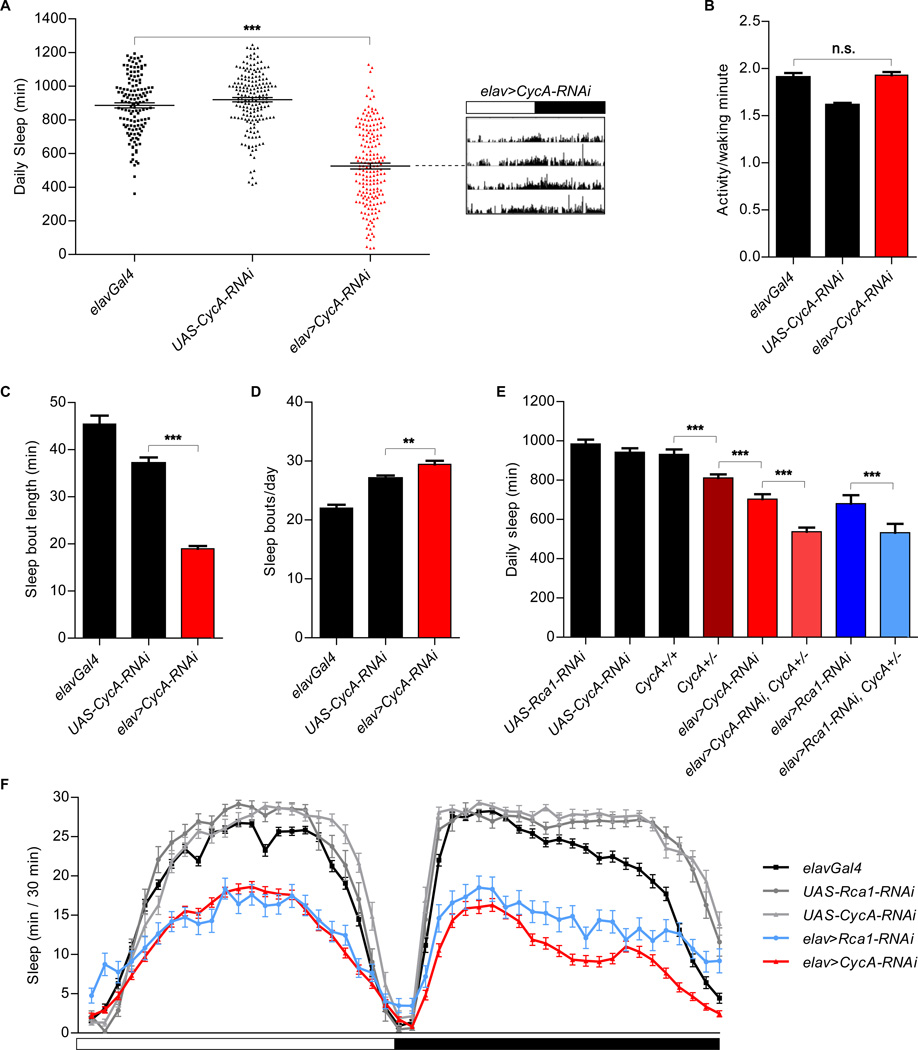
Figure 2. Reduced sleep in animals expressing neuronal CycA-RNAi.
(A) Reduced sleep in animals expressing pan-neuronal CycA-RNAi [elav>CycA-RNAi (n=185)] compared to that of the parental controls [elavGal4 (n=136), UAS-CycA-RNAi (n=178)]. Shown is an actogram of an elav>CycA-RNAi with an average phenotype. The white and black portions of the bar above the actogram indicate the light and dark periods of the experiment, respectively. (B) Normal levels of activity per waking minute in animals expressing CycA-RNAi in neurons. (C, D) Average sleep bout length was decreased (C), while the number of daily sleep bouts was increased (D), in elav>CycA-RNAi flies. (E) Shorter total sleep of CycA null heterozygous flies [CycA+/− (n=136)] than that of flies with both functional copies of the gene [CycA+/+ (n= 82)]. Introducing RNAi against either CycA or Rca1 into the CycA heterozygous background enhanced the sleep phenotypes [elav>CycA-RNAi, CycA+/− (n=99); elav>Rca1-RNAi, CycA+/− (n=35)]. Other controls shown are UAS-Rca1-RNAi (n=33), UAS-CycA-RNAi (n=56), elav>CycA-RNAi (n=78) and elav>Rca1-RNAi (n=22). (F) Sleep profile of elav>CycA-RNAi (n=126), elav>Rca1-RNAi (n=58), and control [elavGal4 (n=88), UAS-Rca1-RNAi (n=24), UAS-CycA-RNAi (n=36)] flies. Statistical analysis performed using One-way ANOVAs with Tukey post-test. In all experiments described in this figure, except that shown in panel (E), elavGal4 drives the expression of UAS-Dicer2.
Drosophila Rca1 is homologous to the mammalian early mitotic inhibitor (Emi1) (9, 10), and plays a critical role in preventing premature cyclin degradation (8). Cyclins regulate cell cycle progression through activation of specific Cyclin-Dependent Kinases (CDKs). The main target of Drosophila Rca1 regulation is Cyclin A (CycA) (7), so we tested whether depletion of CycA might affect sleep. Indeed, expression of CycA-RNAi in neurons (elavGal4 + UAS-CycA-RNAi, denoted “elav>CycA-RNAi”) caused a decrease in the time animals spent sleeping (Fig. 2A and F) without significantly affecting levels of activity per minute while awake (Fig. 2B). This reduction was a consequence of decreased sleep bout duration (Fig. 2C), whereas the number of sleep episodes was slightly increased (Fig. 2D).
We noticed that the ectopic over-expression of Rca1 or CycA in neurons produced a small and rough eye phenotype. A similar eye phenotype is associated with a mutation in a negative regulator of CycA and was used in a modifier screen to identify new genes involved in cell cycle regulation (7). We used this morphological readout of CycA levels to confirm that the UAS-Rca1-RNAi and UAS-CycA-RNAi lines were effective, by showing that they suppressed eye phenotypes associated with Rca1 or CycA over-expression, respectively (fig. S1A and S1B). Furthermore, UAS-Rca1-RNAi also suppressed the eye phenotypes resulting from CycA over-expression (fig. S1C), arguing that Rca1 can stabilize CycA protein in neurons.
Flies homozygous for CycA null alleles (CycA−) do not survive and could not be examined. However, CycA− heterozygotes are viable and showed a decrease in total sleep amounts, a phenotype further enhanced by expression of CycA-RNAi in this background (Fig. 2E), even in the absence of Dicer2 over-expression. We observed an additive effect between the heterozygous CycA− mutation and neuronal Rca1-RNAi as well (Fig. 2E). The reduction in total sleep seen in these experiments may not fully reflect the influence of CycA/Rca1 in sleep control, as CycA protein was still detectable in the brains of all of our experimental genotypes.
As CycA is thought to be the main target of Rca1 regulation during the cell cycle, and our behavioral and genetic analyses indicated a similar relationship with respect to the effects of these genes on sleep, we focused our subsequent studies on CycA.
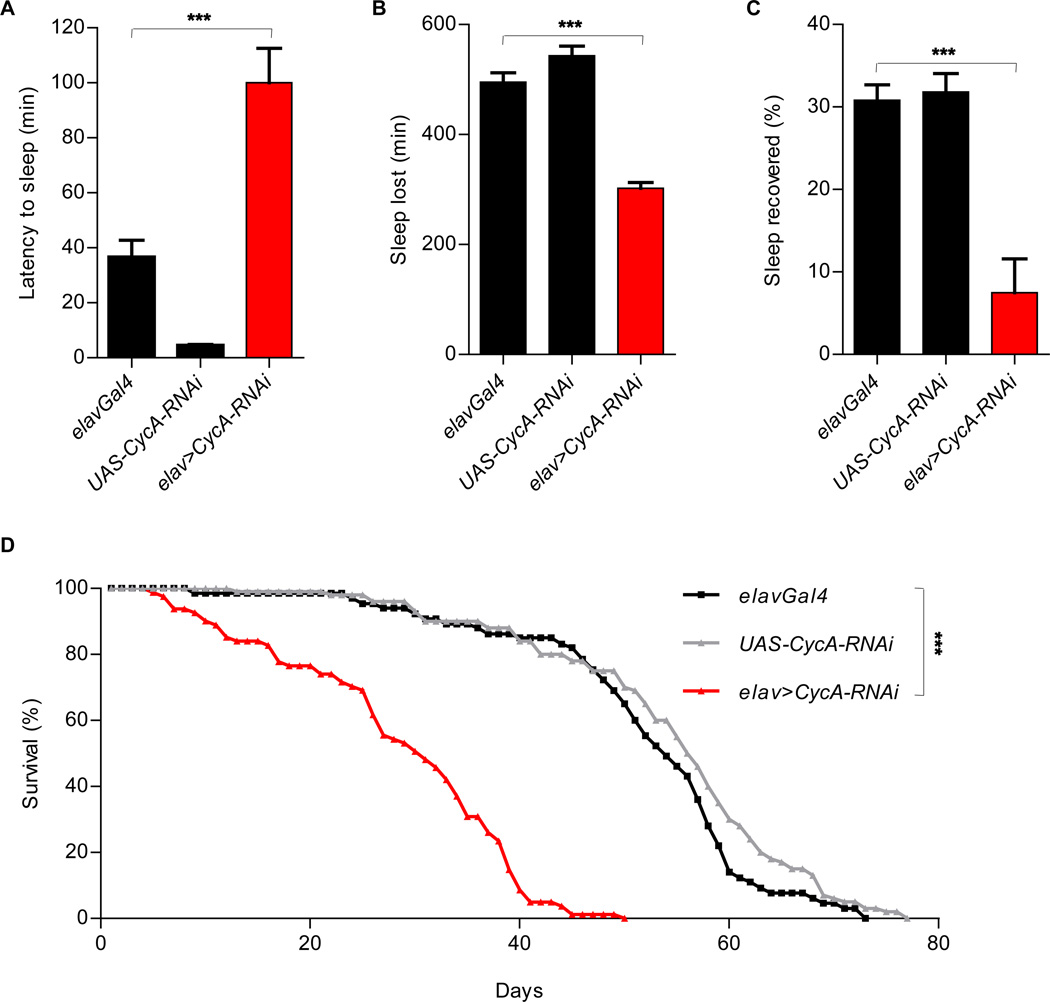
It took significantly longer for elav>CycA-RNAi flies to fall asleep after lights went off than it did for the controls (Fig. 3A). This was not the case for all genes that were identified in our screen. This increased latency to sleep suggests that a mechanism regulating wake-sleep transition is disrupted in elav>CycA-RNAi flies. To examine if sleep can be homeostatically controlled in these animals, we examined their response to sleep deprivation. We mechanically stimulated the flies during the night and examined their sleep pattern during the subsequent morning, when “rebound” sleep is normally seen in wild-type flies that have been sleep-deprived the night before (1, 2). Although we saw some increase in rebound sleep in flies of all the genotypes tested, this response was reduced in elav>CycA-RNAi flies (Fig. 3 B and C), suggesting a defective sleep homeostat. In accordance with the idea that sleep serves an essential function, neuronal depletion of CycA shortens lifespan (Fig. 3D), presumably due to chronic sleep deprivation experienced by these animals. However, it is also possible that CycA contributes to lifespan through other functions in post-mitotic neurons.
Figure 3. Reduced homeostasis and decreased lifespan in elav>CycA-RNAi animals.
(A) Delayed sleep onset in elav>CycA-RNAi animals (n=80) compared to controls [elavGal4 (n=64), UAS-CycA-RNAi (n=36)] after lights went off. (B, C) Decreased recovery of sleep after overnight sleep deprivation in elav>CycA-RNAi animals (n=27), compared to controls [elavGal4 (n=52), UAS-CycA-RNAi (n=10)]. Sleep lost (B) was calculated by subtracting sleep on the night of the deprivation from undisturbed sleep the night before. All animals analyzed lost at least 95% of their sleep. To account for the differences in baseline sleep between the genotypes, recovery data (C) were normalized. (D) Reduced lifespan in elav>CycA-RNAi (n=113) animals compared to controls [elavGal4 (n=102), UAS-CycA-RNAi (n=100)]. Statistical analysis of sleep was performed using One-way ANOVAs with Tukey post-test, and statistical analysis of longevity was performed using Log-rank tests. In all experiments described in this figure, elavGal4 drives the expression of UAS-Dicer2.
Sleep is regulated in a circadian fashion, so we wondered whether some of the sleep defects we observed in elav>CycA-RNAi animals resulted from a disrupted circadian clock. Most elav>CycA-RNAi flies showed robust rhythms in constant darkness, with normal periods (figs. S2A and S3A). Circadian rhythmicity was observed even in animals with severe sleep defects (fig. S2B through D). Furthermore, elav>CycA-RNAi flies eclosed with strong circadian rhythms (fig. S3B). Finally, Period (PER), a core component of the circadian clock (11), accumulated rhythmically in the pacemaker neurons of elav>CycA-RNAi animals (fig. S3C). Thus, the sleep defects observed in elav>CycA-RNAi animals appear not to reflect circadian clock anomalies.
Staining with two different antibodies to CycA showed a restricted expression pattern in adult brains (fig. S4). Both antibodies detected the same ~ 12 dorsal neurons, and one of the antibodies labeled cells in several other brain regions, including the pars intercerebralis, a known fly sleep center (12, 13) (fig. S4A and S4B). The antibody that detected CycA in more cells was raised against the whole CycA protein, whereas the one that labeled fewer cells was raised against the CycA N-terminus. Both appear to be specific to CycA, because their staining was strongly reduced in embryos in which CycA-RNAi was driven by a strong, ubiquitous Gal4 driver (fig. S5). The broader CycA expression pattern in the brain was supported by co-localization with CycAGal4-driven GFP, although GFP was detected in additional cells in all the clusters (fig. S4A). Overall, CycA protein was detected in ~40–50 neurons in the brain (fig. S4C).
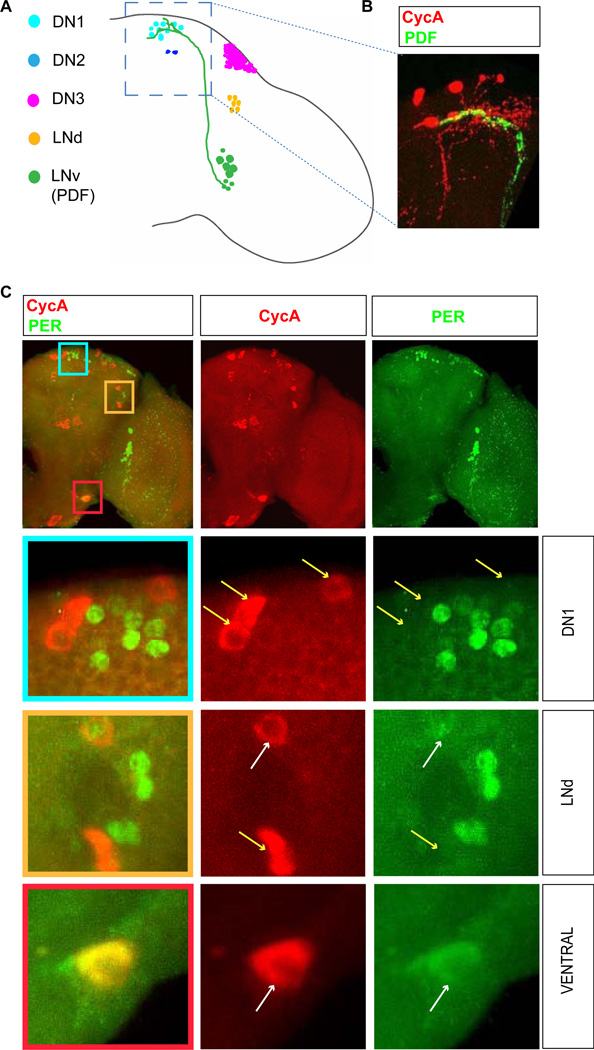
In the adult fly brain, ~150 clock neurons are organized into six major clusters: three dorsal clusters (DN1, DN2 and DN3), a dorsal-lateral cluster (LNd) and two groups of ventral-lateral neurons - small (s-LNv) and large (l-LNv). The latter two include neurons that produce the neuropeptide PDF (Pigment Dispersing Factor), a major regulator of clock output (14) (Fig. 4A). s-LNvs are the major pacemaker neurons (15, 16), which also regulate sleep (17). Co-staining of brains for CycA and PDF revealed that termini of PDF-producing s-LNvs project near the dorsal CycA neurons (Fig. 4B). Indeed, as judged by PDFrGal4-driven GFP, which marks presumptive PDF receptor-producing neurons, these CycA cells appear to express the PDF receptor (fig. S6A), (18). Such co-expression was found in all CycA neurons (examples shown in fig. S6A), and suggests a molecular and neuronal connection between circadian and sleep systems. Co-expression with PDFr also provides an opportunity to knock down CycA levels in a more limited set of neurons than our pan-neuronal driver had afforded. PDFrGal4-driven CycARNAi resulted in a severe sleep reduction (fig. S6B).
Figure 4. Association of CycA neurons and circadian clock neurons in the brain.
(A) A schematic representation of an adult brain with known groups of clock neurons indicated by different colors. LNv neurons project to the dorsal area of the brain where DN1 clock neurons are found. (B) The bodies and arborizations of the dorsal CycA neurons (red) are found in the vicinity of the LNv neuronal termini (green, marked by the expression of neuropeptide PDF). (C) Immunostaining for CycA (red) and a core circadian clock component Period which marks circadian clock neurons (PER, green). The bottom three panels show higher magnification images indicated by different color squares in the upper panel. The labels “DN1” and “LNd” refer to the types of circadian neurons found in these respective areas, while “ventral” denotes an area that has no known clock neurons. In different regions of the brain, cells labeled for CycA and PER cluster (DN1 region), cluster and partially overlap (LNd region), or overlap (ventral region). White and yellow arrows point to overlapping and non-overlapping cells, respectively. For simplicity, only one brain hemisphere is shown. The brains shown were fixed at Zeitgeber time 2 (ZT2, the time point at which levels of PER are high).
To determine if dorsal CycA neurons are in fact DN1 circadian neurons, we asked whether they express per. A subset of the dorsal CycA-containing neurons expressed perGal4-driven GFP (fig. S6C). A similar partial overlap was seen in all other CycA clusters, with the exception of the pars intercerebralis (fig. S6C). As perGal4 was expressed in a smaller subset of CycA neurons than PDFrGal4, we used the former to knock down CycA in a more limited neuronal subpopulation. In these experiments, sleep was consistently lower than in control animals, although the extent of sleep loss was less extreme than with pan-neuronal CycA knockdown (fig. S6D). Interestingly, CycA-containing dorsal neurons that expressed perGal4 were never stained by an antibody to PER, and thus do not detectably express PER protein. Therefore, dorsal CycA neurons are closely associated with the DN1 cluster (Fig. 4), but do not appear to be clock neurons themselves. A similar relationship exists between CycA-expressing and DN3 or LNv neurons (fig. S7). In contrast, we occasionally found CycA and PER proteins in the same cell within a subset of dorsal lateral neurons (LNds), a group of clock neurons that regulate the evening peak of activity (Fig. 4) (15, 16). Finally, a bilaterally symmetric pair of large ventral neurons expressed both CycA and PER proteins (Fig. 4). These ventral neurons do not belong to any known clock neuronal clusters. In summary, CycA-containing neurons are often intermingled with circadian pacemaker neurons. As CycA neurons appear to express the receptor for the circadian neuropeptide PDF, such an arrangement suggests that signals produced by neurons regulating circadian rhythmicity may be directed to neurons which control sleep.
There are other known instances of the repurposing of cell cycle machinery in neurons. Examples include the recent observations that two Cyclin-dependent kinase pathways are essential for proper axonal targeting of presynaptic components in C. elegans (19), and that Cyclin E regulates synaptic plasticity and memory formation in mice (20). We observed no obvious defects in the gross morphology or number of CycA-containing neurons in elav>CycA-RNAi flies, and have not detected any obvious rhythmic patterns of CycA protein expression. Nevertheless, such features might become evident in studies focused on specific cells or specific subcellular regions, or in developing or aging flies. Future understanding of the role CycA plays in determining the structure or activity of neurons specialized for the regulation of sleep seems likely to shed light on the molecular mechanisms underlying this enigmatic behavior. As the role of CycA in cell cycle regulation is conserved throughout metazoans we can speculate that its role in sleep regulation might be conserved as well.
Supplementary Material
Acknowledgments
We thank R. Allada, B. Dickson, J. Hall, C. Lehner, A. Sehgal, F. Sprenger and NIG-Fly Stock Center for reagents; Nicholas Stavropoulos for custom MatLab software; C. Bargmann, M. Crickmore, C. Desplan, A. Keene, S. Shaham, L. Vosshall, and members of the Young lab for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health (NS053087 and GM054339) to MWY. DR was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research and by a Merck fellowship.
Footnotes
Supporting Online Material
Materials and Methods
Figures S1, S2, S3, S4, S5, S6, S7
REFERENCES
- 1.Hendricks JC, et al. Neuron. 2000 Jan;25:129. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 2.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Science. 2000 Mar 10;287:1834. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 3.Rothenfluh A, Abodeely M, Price JL, Young MW. Genetics. 2000 Oct;156:665. doi: 10.1093/genetics/156.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Curr Biol. 2002 Nov 19;12:1934. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- 5.Sehgal A, Mignot E. Cell. Jul 22;146:194. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzl G, et al. Nature. 2007 Jul 12;448:151. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 7.Dong X, et al. Genes Dev. 1997 Jan 1;11:94. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- 8.Grosskortenhaus R, Sprenger F. Dev Cell. 2002 Jan;2:29. doi: 10.1016/s1534-5807(01)00104-6. [DOI] [PubMed] [Google Scholar]
- 9.Zielke N, Querings S, Grosskortenhaus R, Reis T, Sprenger F. EMBO Rep. 2006 Dec;7:1266. doi: 10.1038/sj.embor.7400851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reimann JD, et al. Cell. 2001 Jun 1;105:645. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 11.Boothroyd CE, Young MW. Ann N Y Acad Sci. 2008;1129:350. doi: 10.1196/annals.1417.006. [DOI] [PubMed] [Google Scholar]
- 12.Foltenyi K, Greenspan RJ, Newport JW. Nat Neurosci. 2007 Sep;10:1160. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 13.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Neuron. Mar 11;65:670. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helfrich-Forster C. Genes Brain Behav. 2005 Mar;4:65. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- 15.Grima B, Chelot E, Xia R, Rouyer F. Nature. 2004 Oct 14;431:869. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 16.Stoleru D, Peng Y, Agosto J, Rosbash M. Nature. 2004 Oct 14;431:862. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 17.Parisky KM, et al. Neuron. 2008 Nov 26;60:672. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lear BC, Zhang L, Allada R. PLoS Biol. 2009 Jul;7:e1000154. doi: 10.1371/journal.pbio.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou CY, et al. Cell. May 28;141:846. [Google Scholar]
- 20.Odajima J, et al. Dev Cell. Oct 18;21:655. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.