Abstract
DNA bending can be promoted by reducing the net negative electrostatic potential around phosphates on one face of the DNA, such that electrostatic repulsion among phosphates on the opposite face drives bending toward the less negative surface. To provide the first assessment of the energetic contribution to DNA bending when electrostatic asymmetry is induced by a site-specific DNA binding protein, we manipulated the electrostatics in the EcoRV endonuclease-DNA complex by mutation of cationic sidechains that contact DNA phosphates and/or by replacing a selected phosphate in each strand with uncharged methylphosphonate. Reducing the net negative charge at two symmetrically located phosphates on the concave DNA face contributes −2.3 to −0.9 kcal/mol (depending on position) to complex formation. In contrast, reducing negative charge on the opposing convex face produces a penalty of +1.3 kcal/mol. Förster resonance energy transfer experiments show that the extent of axial DNA bending (about 50°) is little affected in the modified complexes, implying that modification affects the energetic cost but not the extent of DNA bending. Kinetic studies show that favorable effects of induced electrostatic asymmetry on equilibrium binding derive primarily from a reduced rate of complex dissociation, suggesting stabilization of the specific complex between protein and markedly bent DNA. A smaller increase in the association rate may suggest that the DNA in the initial encounter complex is mildly bent. The data imply that protein-induced electrostatic asymmetry makes a significant contribution to DNA bending, but is not itself sufficient to drive full bending in the specific EcoRV-DNA complex.
Keywords: protein-DNA recognition, electrostatic interactions, restriction endonuclease, asymmetric phosphate neutralization, chiral methylphosphonate
Introduction
Protein-induced DNA bending is important for a variety of cellular processes including the initiation and regulation of transcription1–6, recombination7, DNA compaction8; 9, and correct positioning of elements required for the formation of catalytic sites of DNA endonucleases10; 11. By juxtaposing DNA segments that are separated in primary sequence, DNA bending may allow a single protein to interact with two separate segments of DNA and/or facilitate contacts between proteins bound at distally located DNA sites. Such interactions play crucial roles in regulating replication and transcription. DNA bending also plays both structural and energetic roles in sequence discrimination by site-specific binding proteins. For example, the formation of the specific EcoRV endonuclease-DNA and lac repressor-operator complexes are accompanied by dramatic DNA distortion/bending, whereas in the corresponding non-specific complexes the DNA is not distorted5; 11; 12. DNA bending may be required to increase steric and electrostatic complementarity between protein and DNA surfaces, but bending incurs an energetic cost that must be compensated by favorable free-energy contributions from protein-phosphate interactions, protein-base interactions, and water release from nonpolar surfaces13. For non-specific complexes, the smaller contributions from these favorable factors are insufficient to drive DNA bending13.
Double helical DNA in solution can be described as a relatively rigid elastic rod with a persistence length of ~150 bp14–19. Rigidity derives from both favorable base stacking interactions and repulsive electrostatic forces among uniformly distributed, negatively charged phosphates20. More recent studies on shorter DNA length scales suggest that the double helix may be many times less rigid than classical models have predicted21; 22. In addition, local DNA structure and conformational flexibility are sequence dependent, and significant deviations from the ideal B-form have been observed for free DNA molecules23; 24. In some cases a protein-DNA complex contains a DNA conformation that is not spontaneously accessible to the free DNA; this is equivalent to saying that bending requires the input of energy, primarily required for destacking of DNA bases25; 26.
Proteins are capable of inducing very sharp and localized bends in the DNA axis, generally by altering the roll or tilt angles at specific base-pair steps. The role of the protein appears essential, because most short oligonucleotides, even A-tract sequences, exhibit relatively small deviations from B-form structure23. For example, while A-tracts can be bent by as much as 20°27, the transcription factors TATA binding protein (TBP) and catabolite activator protein (CAP) induce 80° and 90° bends, respectively28; 29. Another striking example is the prokaryotic integration host factor (IHF), which binds in the DNA minor groove and induces a 160° bend by means of two kinks separated by nine base-pairs30.
Some proteins that bend DNA away from their surfaces intercalate aromatic amino acid sidechains between base-pairs, thus replacing some of the lost stacking energy2; 3; 31. Computational studies32 suggest that immersing a section of DNA backbone in a low dielectric protein environment is sufficient to provoke bending away from the protein. On the other hand, proteins that bend DNA toward their surfaces typically form energetically favorable interactions to both bases and phosphates, and if the same set of interactions cannot form with unbent DNA, then these interactions stabilize the bent DNA.
Over 30 years ago, Mirzabekov and Rich33 proposed that DNA bending could also be induced by “asymmetric neutralization of phosphate groups”. By this mechanism, the proximity of a positively charged protein surface to one face of the DNA reduces the net negative electrostatic potential on that face, resulting in an imbalance of interphosphate repulsive forces and a collapse of the DNA by bending of the helix axis toward the protein. The apposition of a positively charged group to the negatively charged DNA backbone is commonly referred to in the literature as “neutralization” of phosphate charge 34–37. This usage can be narrowly construed to mean that the electrostatic potential in the neighborhood is driven from highly negative toward a more “neutral” value; however, the negative charge on the nearest phosphate has not vanished, nor is the less negative electrostatic potential confined to the phosphate nearest the positive charge.
Asymmetric chemical neutralization of phosphates has been shown experimentally to drive DNA bending. For example, neutralization of multiple phosphates on one face of a free DNA molecule using uncharged methylphosphonate (PMe) analogs results in spontaneous bending of 6–24° toward the neutralized surface38. Experimental and computational neutralization of phosphates, corresponding to those forming salt bridges in the E. coli CAP-DNA complex, increased DNA bending in the predicted direction for a free DNA molecule containing the CAP binding site39; 40. Computational neutralization of phosphates contacted by a wide variety of other DNA bending proteins has also been shown to induce DNA bending and increase DNA flexibility41. Furthermore, bending of free DNA duplexes can also be induced by covalent tethering of cations to the DNA backbone42; 43.
Mutational studies of GCN4 and the bZIP family of transcription factors have demonstrated protein-induced DNA bending toward key positively charged amino acid residues on the protein surfaces44–46. However, simple mutational studies are insufficient to demonstrate a specific role of induced electrostatic asymmetry in promoting bending. To isolate and evaluate the free-energy contribution to DNA bending when electrostatic asymmetry is induced by a site-specific DNA binding protein, we have chosen as a model the restriction endonuclease EcoRV. The EcoRV homodimer bends its specific target sequence, GATATC, approximately 50° toward the major groove11, primarily by destacking of the central T-A base-pairs11, where the T-A step is unusually susceptible to bending11; 23; 47; 48. The protein preferentially apposes cationic sidechains to the concave face of the DNA, so an excess of negative potential on the opposite face might provide a driving force for axial bending and provide some of the energy to compensate for unfavorable base-pair destacking.
A detailed model of DNA bending by EcoRV endonuclease has been proposed based on a set of binary enzyme-DNA crystal structures determined in distinct crystal lattices10. These structures showed a series of bent states with increasing surface area burial between the enzyme surface and DNA minor groove. Catalytic activity in the crystalline state was observed only in the lattice providing the most highly bent structure, suggesting that the series of snapshots might correspond to a bending pathway in solution10; 11. According to the model, electrostatic interactions with flanking DNA facilitate initial bending. The remainder of the 50° bend, the required global reorientation of the DNA binding domains and final assembly of the active sites is then driven by the formation of specific protein-DNA interactions and increased burial of nonpolar surface.
To investigate whether and how much the asymmetry of negative electrostatic potential induced in the region of the DNA by EcoRV contributes to DNA bending, we have modified the electrostatic landscape of the EcoRV-DNA interface and measured the thermodynamic, structural and kinetic consequences. We have manipulated the electrostatics both by mutating a Lys or Arg residue in various positions to Ala, and by replacing selected anionic phosphates with uncharged methylphosphonates. The disruption of charge-charge pairing in a protein-DNA interface can affect measured binding free energy by many mechanisms, including effects on direct protein-DNA interaction, effects on intraprotein interactions, effects of unsatisfied charge when one member of an ion pair is eliminated, and electrostatic effects on DNA bending energy. We isolate the electrostatic effect of a phosphate charge by focusing on chemical neutralization in the context of mutant protein lacking the corresponding cationic sidechain. Molecular dynamics simulations show that in this interface, a charged phosphate remains solvated and associated with counterions even when bound to a mutant protein in which its normal charge partner is absent; thus, energetic penalties for desolvating a charged phosphate do not obscure the issue in this context. Most strikingly, our observations strongly support the hypothesis that we are isolating effects on the DNA bending energy: (a) Reducing negative charge on the concave face is favorable to protein-DNA binding, whereas reducing negative charge on the convex face is unfavorable to binding. (b) The favorable effects on the concave DNA face are stronger near the apex of the bend and weaker as interphosphate distance increases farther from the apex. These observations are uniquely consistent with the hypothesis that the protein promotes axial DNA bending by inducing a charge imbalance between the two faces of the DNA. Förster resonance energy transfer (FRET) experiments show that DNA in the modified complexes is bent to the same extent as in the unmodified complex, so the energetic effects of the induced charge imbalance derive from a lower cost of DNA bending. Kinetic experiments show that the favorable effects of induced charge asymmetry on equilibrium binding derive primarily from a reduced rate of complex dissociation, suggesting stabilization of the specific complex between protein and markedly bent DNA. Neutralization with PMe on the concave face slightly accelerates complex association, whereas neutralization on the convex face slightly slows association, consistent with the possibility that the initial encounter complex embodies some degree of DNA bending.
Results and Discussion
Manipulating and dissecting electrostatic effects at the EcoRV-DNA interface
We aim to test the hypothesis that a protein can promote DNA bending by placing positively charged Lys or Arg sidechains in position to reduce net negative potential preferentially on one face of the DNA. In the EcoRV-DNA complex (Fig. 1a), the bent DNA (Fig. 1b) is contacted on its concave face by three positively charged amino acid sidechains per monomer (K119, R221 and R226). These sidechains are expected to reduce net repulsive forces on the concave face of the bound DNA, such that excess interphosphate repulsion on the opposite face will drive bending toward the less negative face. From a thermodynamic perspective, induced electrostatic asymmetry is expected to reduce the energetic cost of bending the helix axis. Conversely, interaction on the convex face by R140 should increase the cost of bending, since interphosphate repulsion on the convex face provides the driving force. Deletion of the 29 amino acid C-terminal subdomain, which contains R221 and R226, results in a significant decrease in equilibrium DNA bending by EcoRV, suggesting that the +4 positive charge contributed by this subdomain plays a key role in the bending process49. This study, however, did not distinguish between (1) stabilization of the bent conformation by electrostatically attractive protein-DNA contacts and (2) a specific favorable role for induced electrostatic asymmetry.
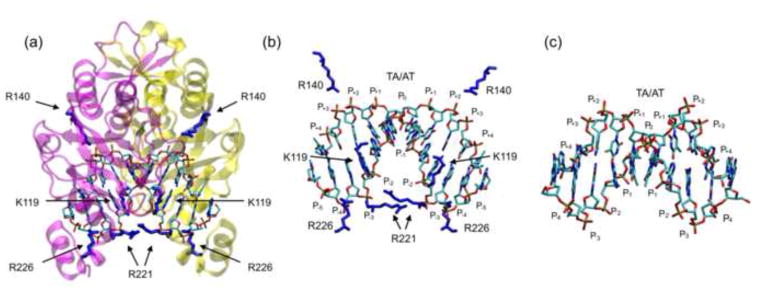
Figure 1. The EcoRV-DNA-Ca2+ complex.
(a) Location of the eight basic residues (4 per monomer) that contact the phosphate backbone in the specific complex (PDB ID 1B94)60. The protein backbone is in magenta and yellow (to distinguish the subunits) and cationic side chains that contact DNA phosphates are in dark blue. (b) Magnification of the DNA from the complex with protein backbone removed for clarity. Basic side chains that contact the phosphate backbone are shown in blue. The DNA bends downward and phosphate positions on each DNA strand are numbered as in Fig. 2a. (c) Conformation of free CCGATATCGG (PDB ID 1ZFC)105 for comparison. Molecular images in all figures were created with the molecular graphics program “VMD”106.
In comparing the free DNA containing an EcoRV site (Fig. 1c) to the protein-bound DNA (Fig. 1d), the protein-induced axial bend is most evident in a positive 50° roll at the central TA base step, causing energetically unfavorable destacking of the central base-pairs10; 11; 50; 51. Milder destacking is associated with negative roll angles (−9° to −12°) at the G-A and T-C steps10. The bend narrows the major groove11 and brings several phosphates on the concave face (notably positions P0, P−1, P−2 and P−3; see Fig. 2a for nomenclature) closer to one another relative to their positions in unbent DNA of similar sequence. Conversely, most interphosphate distances on the convex face (P+1, P+2, P+3, P+4) are greater than in the unbent DNA, thus relaxing the repulsion on this face.
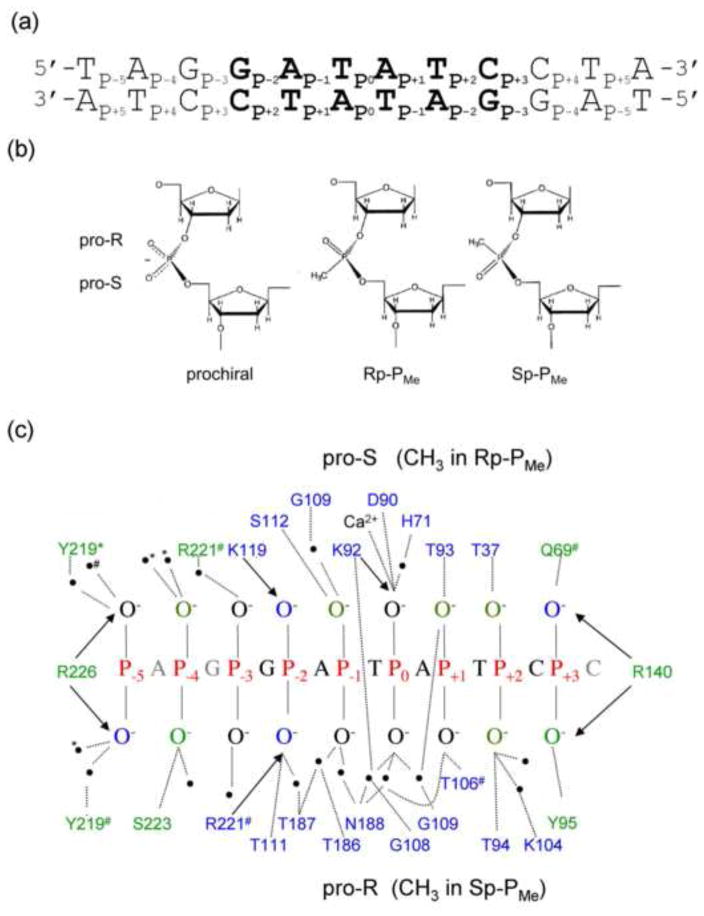
Figure 2. Phosphate contacts made in the specific EcoRV-DNA recognition complex.
(a) Specific DNA substrate used for all thermodynamic experiments. The specific recognition sequence GATATC (bold) is embedded in a 24 bp parent oligonucleotide. All PMe substitutions were made symmetrically in both DNA strands. (b) Comparison of anionic prochiral phosphodiester (left) with uncharged chiral methylphosphonates (PMe). By IUPAC convention, replacement of the pro-S (O1P) non-bridging phosphoryl oxygen with a methyl group results in an Rp-PMe diastereomer; replacement of the pro-R (O2P) phosphoryl oxygen results in an Sp-PMe diastereomer. (c) Phosphate contacts made to one strand of the specific EcoRV-bound DNA, based on 10 crystal structures of EcoRV-DNA ternary complexes. The base sequence is that used in our thermodynamic experiments. Hydrogen bonds (<3.5Å) to the phosphoryl oxygens from polar side chains and water molecules (black circles) are shown as dotted lines; Coulombic interactions (≤4.3Å) with cationic sidechains are denoted by arrows. Most contacts are symmetrical with respect to protein subunits and DNA strands; those contacts made to only one DNA chain are denoted by # or *; these correspond to chains C and D in PDB ID: 1B94. Sidechains that contact phosphoryl oxygens within the GATATC recognition site are in blue; those making contact outside the GATATC site are in green.
We hypothesize that the apposition of a positively charged Lys or Arg sidechain to the concave DNA face makes two favorable contributions to binding free energy: (1) the contribution that arises directly from the short-range interaction of the sidechain with the nearest phosphate, the net of (unfavorable) desolvation of the two charged groups and the formation of the attractive charge-charge interaction, which predominates because the groups lie within hydrogen bonding distance 52; 53; (2) the favorable contribution from decreased net repulsion among phosphates on the concave face, such that excess interphosphate repulsion on the opposing convex face provides a driving force for bending. As a complicating factor, each DNA phosphate may form other energetically favorable interactions, direct or water-mediated, with uncharged groups on the protein, using the same and/or different phosphoryl oxygen. Although our experiments manipulate the DNA phosphates pairwise (e.g., P−2 on both DNA strands), it must be borne in mind that mutual repulsion occurs among all six phosphates on the concave face of the DNA.
Our objective in this work was to isolate and evaluate the contribution of induced electrostatic asymmetry to binding free energy. We used two methods to manipulate the charge landscape at the interface: 1) Removing a cationic Lys or Arg side chain that contacts DNA phosphate by mutating it to Ala, thus affecting both the protein-DNA charge-charge interaction and the reduced-repulsion contributions, further confounded by altered intraprotein interactions. 2) Removing negative charge from a DNA phosphate (chemical neutralization) by substituting uncharged methylphosphonate (PMe) (Fig. 2b), thus affecting not only the protein-DNA charge-charge and reduced-repulsion factors, but also perhaps the favorable contribution(s) from interaction with uncharged protein groups (Fig. 2c). It is therefore the case that neither mutation nor chemical modification alone can extract the contribution from decreased repulsion.
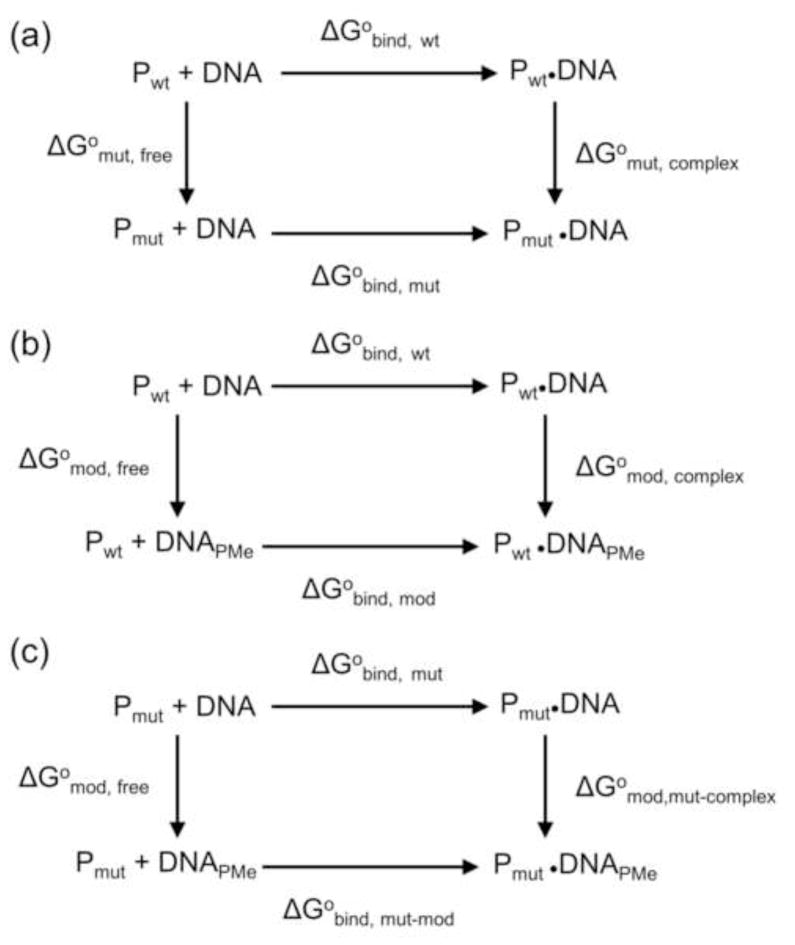
The contribution of protein-induced electrostatic asymmetry can be cleanly extracted by combining thermodynamic pseudocycles (Fig. 3) for the mutational and chemical manipulations in two kinds of cases: (1) Where only one cationic sidechain contacts a single phosphoryl oxygen (e.g., K119 contacts the pro-S oxygen at P−2, Fig. 2c); (2) Where one cationic sidechain lies approximately equidistant between the pro-S and pro-R phosphoryl oxygens (e.g., R226 at P−5, Fig. 2c). First we compare mutant and wild-type proteins with unmodified DNA (Fig. 3a). The difference in observed binding free energy changes ΔΔG°obs,mut is
Figure 3. Thermodynamic pseudocycles.
(a) Equilibrium binding of unmodified DNA to wild type and mutant EcoRV; (b) Equilibrium binding of wild type EcoRV to unmodified and PMe modified DNA; (c) Equilibrium binding of mutant EcoRV to unmodified and PMe modified DNA. The free energy changes (ΔG°) for the reversible processes indicated by horizontal arrows can be determined by equilibrium binding experiments, but the “processes” indicated by vertical arrows represent chemical modifications or protein mutations, with free energy changes that are not experimentally accessible.
| (1) |
Since the sum of free energy changes around the cycle must be zero, it follows that the effect of mutation on the protein-DNA complex ΔG° mut,complex is
| (2) |
Note that it cannot be assumed that ΔG°mut,free is negligible, since the mutation may perturb intraprotein interactions. This factor, often ignored in mutational analysis, is in fact significant in many instances (see below).
Similarly for the DNA modification pseudocycle (Fig. 3b),
| (3) |
where ΔΔG°obs,mod ≡ ΔG°bind,mod − ΔG°bind,wt
These two cycles can be combined as in Fig. 3c:
| (4) |
The first two terms on the right (Fig. 3c; horizontal arrows) are determined by measuring the binding of mutant protein to PMe-modified or unmodified DNA, respectively. The third term on the right, the free-energy effect of single PMe modification of the free DNA (Fig. 3c; left vertical arrow), is assumed to be negligible. (We show below that these single PMe substitutions do not induce any measurable bending of the free DNA.) The term ΔG°mod,mut-complex (Fig. 3c; right vertical arrow) is the free-energy effect of replacing a charged phosphate with an uncharged PMe in the context of mutant protein where both the protein sidechain-phosphate interaction and the protein attenuation of negative electrostatic potential surrounding the phosphate are absent. Thus, this term should closely approximate to the desired relief of interphosphate repulsion, as distinct from any effect of the Coulombic protein-phosphate interaction. If indeed there is a contribution from relief of repulsion, this contribution is expected to be net favorable for a phosphate on the concave face. Neutralization on the convex face should be conversely unfavorable. Note that considering only mutant protein in this pseudocycle avoids the issue of ΔG°mut,free and particularly the issue of contributions from intraprotein interactions formed by the Lys or Arg sidechain (see example of K119, below.)
One important limitation of this strategy is that the PMe substitution must not also perturb interactions of the phosphate with other, uncharged protein groups (e.g., the T111 interaction with P−2, Fig. 2c). This condition can be met approximately at P−2 by an appropriate choice of a chiral RP-PMe, which directs the methyl group toward the cationic K119 sidechain being mutated, thus leaving a nonbridging phosphoryl oxygen to interact with the uncharged T111 (Fig. 2c). This nonbridging oxygen carries a fractional negative charge54; 55, so interaction with T111 should be minimally perturbed. At P−5, R226 places a positive charge approximately equidistant between the two phosphoryl oxygens (this is true in 9 different co-crystal structures), and there is no other sidechain that directly interacts here, so the racemic RP,SP-PMe provides the same effect as either diastereomer. As will be seen below, there are other phosphates in the EcoRV complex for which there are more complicated considerations.
Protein interaction at P−2 on the concave face promotes DNA bending
We first consider the phosphate at P−2, located on the concave DNA face in the complex (Fig. 1). The pro-S phosphoryl oxygen is contacted by K119, while the pro-R phosphoryl oxygen is contacted by R221 and T111, and is part of a network of water-mediated interactions, which bridge all the phosphates in the recognition interface (Fig. 2c). Thus, in the context of wild-type EcoRV, PMe substitution at the P−2 phosphate would not only remove a negative charge from the interface, but would also perturb protein-phosphate interactions, depending on the stereochemistry of the modification. (By IUPAC nomenclature convention, a chiral RP-methylphosphonate replaces the pro-S phosphoryl oxygen with a methyl group and the pro-R phosphoryl oxygen with P=O. The SP-PMe replaces the pro-R oxygen with a methyl group. See Fig. 2b,c.)
Following the strategy outlined above, we measured equilibrium association constants (KA) (see Materials & Methods). Binding assays were done in the presence of saturating amounts of Ca2+, which enhances protein-DNA binding affinity56–58, but does not support DNA hydrolysis under assay conditions. Because two Ca2+ ions in each half-site are chelated by the protein and both phosphoryl oxygens of the two P0 phosphates50; 59; 60, repulsion between the two P0 phosphates and other phosphates is largely excluded from our analysis. Mutant EcoRV proteins lack the targeted cationic sidechain in both subunits of the homodimer. Similarly, we made single PMe substitutions in both strands of the DNA duplex, so modified substrates lack two negative charges on symmetrically disposed phosphates.
We first examine the K119A mutation, the effects of which correspond to the thermodynamic cycle of Fig. 3a. In the wild type EcoRV-DNA complex the K119 side chain interacts with the pro-S phosphoryl oxygen at P−2 and makes an intraprotein interaction with the T111 main chain carbonyl oxygen (Fig. 4a). The K119A mutant protein binds unmodified DNA 6,500-fold worse than does wild type EcoRV (ΔΔG°bind = +5.2 kcal/mol; Table 1). The unfavorable effects of the K119A mutation likely derive from (1) removing the interaction from K119-Nε to phosphate P−2 in each half-site; (2) removing two positive charges (one side chain per monomer), which interact with the concave face of the bent DNA and attenuate the negative electrostatic potential, most strongly nearest P−2; (3) removing the intra-protein interaction K119:Nε-T111:O in each half-site. In two different crystal structures of the free EcoRV protein, the electron density of the K119 side chain is either missing (PDB ID: 1AZ361), or is present, but does not interact with the T111 main chain (PDB ID: 1RVE11). Thus, the intraprotein interaction may contribute favorable binding free energy ΔG°bind, wt (Fig. 3a), and removing this interaction will make an equivalent unfavorable contribution to the K119A-DNA complex (ΔG°mut,complex, Fig. 3a). Note that effects (1) and (3) cannot be separated from each other by any combination of thermodynamic pseudocycles, but the electrostatic effect (2) can be isolated by a combined pseudocycle (Fig. 3c).
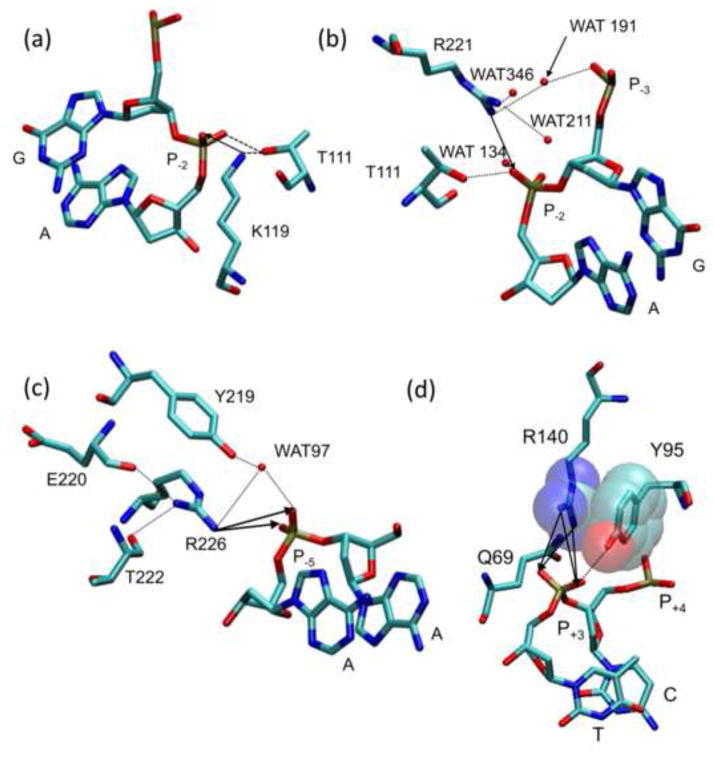
Figure 4. Interactions of cationic sidechains with DNA phosphates.
Contacts made to the (a) K119, (b) R221, (c) R226, and (d) R140 side chains in crystal structure PDB ID 1B94. Neighboring side chains, bases, and water molecules are labeled and atoms are represented by color (C = cyan, O = red, N = blue, and P = brown). Hydrogen bonds and Coulombic interactions are represented by dotted lines and arrows respectively. (d) Van der Waals interactions between the R140 guanidino and the Y95 phenyl are shown in a semi-transparent space fill representation.
Table 1.
Effects of phosphate neutralization on EcoRV-DNA equilibrium binding
| Enzyme | DNAa | KA (M−1)b | KA(ref)/KAd | ΔΔG°bind (kcal/mol)d |
|---|---|---|---|---|
| wild type EcoRV | GATATC | 1.0 (±0.1) ×1011 | 1 | 0 |
| CONCAVE FACE | ||||
| K119A | GATATC | 1.5 (±0.1) ×107 | 6,500 | +5.2 (±0.1) |
| K119A | Rp-PMe-2 | 7.7 (±1.4) ×108 | 133 | +2.9 (±0.2) |
| K119A | PMe-2 | 2.1 (±0.1) ×108 | 476 | +3.7 (±0.1) |
| R226A | GATATC | 7.9 (±0.3) ×106 | 13,000 | +5.5 (±0.1) |
| R226A | PMe-5 | 3.6 (±0.5) ×107 | 2,800 | +4.6 (±0.4) |
| R221A | GATATC | 5.7 (±0.5) × | 180 | +3.1 (±0.1) |
| R221A | PMe-2 | 6.1 (±3.0) ×106 | 16,393 | +5.7 (±0.4) |
| R221A | Sp-PMe-2 | 2.2 (±0.2) ×106 | 45,454 | +6.3 (±0.1) |
| R221A | PMe-3 | 3.7 (±0.2) ×108 | 270 | +3.3 (±0.1) |
| R221A | PMe-5 | 1.2 (±0.2) ×109 | 80 | +2.6 (±0.1) |
| CONVEX FACE | ||||
| R140A | GATATC | 9.3 (±2.0) ×107 | 1100 | +4.2 (±0.2) |
| R140A | PMe+3 | 8.2 (±0.2) ×106 | 12,200 | +5.5 (±0.4) |
The double stranded 24 bp oligodeoxyribonucleotide d(TGTGTTGTAGGATATCCTACAGGT) was used for all equilibrium binding experiments. PMe (n) denotes the position of PMe substitutions (Fig. 2a), which were made symmetrically in both DNA strands. Unless diastereomer is specified, substitution was racemic RP,SP,-PMe.
Equilibrium binding constants were measured at 295K in binding buffer plus 0.24 M K+
KA(ref) is the equilibrium association constant for the unmodified DNA.
For modified complexes with equilibrium association constant KA,mod, the difference in standard binding free energy (ΔΔG°bind) relative to that for a wild type EcoRV complex with unmodified DNA is calculated as ΔΔG°bind = −RTln (KA,mod/KA,unmod).
We next examine the PMe substitution at P−2, corresponding to the thermodynamic cycle of Fig. 3b. The stereospecific RP-PMe-2 substitution replaces only the pro-S phosphoryl oxygen with a methyl group, thereby removing the full formal negative charge on the P−2 phosphate and abolishing the hydrogen bonding interaction (2.7Å) with the K119 side chain. The RP-PMe-2 modification reduces wild type EcoRV binding affinity 420-fold relative to unmodified DNA (ΔΔG°bind = +3.5 kcal/mol; Table 1). This is likely the net of (1) Diminished interphosphate repulsion on the concave face; (2) Disruption of the K119-P−2 interaction; (3) Perturbation of the K119:Nε-T111:O intraprotein interaction (Fig. 4a); (4) Possible unfavorable effects of displacing the K119 side chain (see below). In MD simulations, the contacts (Fig. 2c) from R221 and T111 to the pro-R phosphoryl oxygen are preserved in the wild-type EcoRV-RP-PMe-2 complex, suggesting that the effects of this modification are local. However, the simulations show the K119 sidechain in this complex is solvated and displaced toward the minor groove of the DNA relative to its position in the unmodified complex. Mutating the K119 side chain increases binding affinity 3-fold in the context of the RP-PMe-2 modified DNA (compare binding affinities for wt-RP-PMe-2; Table 2 and K119A-RP-PMe-2; Table 1). This observation suggests that displacing the K119 side chain has other unfavorable consequences beyond disrupting the interaction with T111.
Table 2.
Effect of single PMe substitutions on binding of wild type EcoRV endonuclease
| DNAa | KA(M−1)b | KA(ref)/KAc | ΔΔG°bind (kcal/mol)d |
|---|---|---|---|
| GATATC | 1.0(±0.1) ×1011 | 1 | 0 |
| CONCAVE FACE | |||
| PMe-5 | 9.1(±1.0) ×1010 | 1 | 0 (±0.1) |
| PMe-4 | 6.3 (±1.3) ×108 | 160 | +3.0 (±0.1) |
| PMe-3 | 8.3 (±1.0) ×109 | 12 | +1.5 (±0.1) |
| Rp-PMe-2 | 2.4 (±0.5) ×108 | 420 | +3.5 (±01) |
| Sp-PMe-2 | 3.3 (±0.2) ×107 | 3000 | +4.7(±0.1) |
| PMe-2 | 8.4 (±1.3) ×107 | 1200 | +4.1(±0.1) |
| CONVEX FACE | |||
| PMe+1 | 9.0 (±1.1) ×109 | 11 | +1.4 (±0.1) |
| PMe+2 | 4.3 (±0.3) ×1010 | 2 | +0.5 (±0.1) |
| PMe+3 | 1.5 (±0.35) ×107 | 6600 | +5.1 (±0.1) |
The double stranded 24 bp oligodeoxyribonucleotide d(TGTGTTGTAGGATATCCTACAGGT) was used for all equilibrium binding experiments. PMe(n) denotes the position of PMe substitutions (Fig. 2a), which were made symmetrically in both DNA strands. Unless diastereomer is specified, substitution was racemic RP,SP,-PMe.
Equilibrium binding constants were measured in binding buffer plus 0.24 M K+ (Materials & Methods) at 295K.
KA(ref) is the equilibrium association constant for the unmodified DNA.
For modified complexes with equilibrium association constant KA,mod, the difference in standard binding free energy (ΔΔG°bind) relative to that for the unmodified complex is calculated as ΔΔG°bind = −RTln(KA,mod/KA,ref).
The SP-PMe-2 substitution replaces the pro-R phosphoryl oxygen with a methyl group and reduces wt binding affinity 3,000-fold relative to unmodified DNA (ΔΔG°bind = +4.7 kcal/mol; Table 1). This effect is more severe than that of the RP-PMe-2 substitution and reflects the disruption of the interactions of the pro-R phosphoryl oxygen with both T111 and R221 sidechains and a water-mediated interaction with T187, which is itself involved with networked interactions with nearby phosphates (Fig. 2c).
Considering the above analysis, it is evident that neither site-directed mutagenesis of K119 alone, (the thermodynamic cycle of Fig. 3a) nor chiral PMe-2 substitution alone (Fig. 3b), can be used to extract the contribution of the electrostatic asymmetry induced by the protein. In all cases, there are multiple offsetting contributions.
We now analyze the chiral PMe-2 substitution in the context of the K119A mutation, corresponding to the thermodynamic cycle of Fig. 3c. Comparing K119A binding to unmodified and PMe-2 modified DNA (Table 2), we observed that chiral RP-PMe substitution at position P−2 increases K119A binding affinity by 50-fold relative to unmodified DNA. This corresponds to a favorable ΔΔG°bind of −2.3 kcal/mol. The RP-PMe-2 substitution replaces only the pro-S phosphoryl oxygen with a methyl group. MD simulations suggest that the T111 contact to the non-bridging pro-R phosphoryl oxygen is conserved in the K119A-RP-PMe-2 complex. The behavior of R221 is more dynamic and complicated in the wild-type complex with unmodified DNA (see below), but remains roughly similar in the K119A-RP-PMe-2 complex. In the context of the K119A mutant protein, the K119 sidechain is not available to interact with the phosphate backbone and the RP-PMe-2 substitution does not result in the removal of any additional contacts (Fig. 2c). Thus, this comparison isolates the effect of removing the negative charge on P−2.
The question then arises: Does the charge on P−2 have an unfavorable effect on K119A binding because of repulsion during DNA bending, or because of an “unsatisfied” (unpaired) negative charge in the interface? MD simulations suggest that the latter is unlikely to be a significant factor because the phosphate remains solvated. MD simulations were performed using the AMBER suite of programs and an EcoRV-DNA-Ca2+ ternary complex as the starting coordinates (PDB ID: 1B9460) to simulate four different EcoRV-DNA complexes; wild type EcoRV-GATATC, wild type EcoRV-RP-PMe-2, K119A-GATATC, and K119A-RP-PMe-2. The desired perturbations were made to the starting coordinates in silico and 6 ns simulations were performed in the presence of explicit water molecules and ions (Materials and Methods).
When unmodified DNA is bound to K119A, the pro-S phosphoryl oxygen remains fully solvated (100% occupancy by 3 waters at 2.7–3Å) and solvent-accessible (water residence times of 100–200 ps) throughout (Supp. Fig. 1). One Na+ ion remains stably associated with the pro-S oxygen at 4.4–4.6 Å, while another Na+ (also at < 5Å) exchanges freely with the bulk solution (6 Na+ ions visit within a 6 ns simulation). This is also consistent with salt dependence data (Fig. 5, see below) showing that the K119A-unmodified DNA complex displaces fewer counterions than does the wild-type complex. Thus, this phosphate is not being inserted into a low-dielectric environment (which would be energetically unfavorable62) and there should be no penalty attributable to its desolvation when unmodified DNA binds to K119A (upper horizontal arrow, Fig. 3b). The favorable effect of the PMe substitution cannot be attributed to absence of such a penalty.
Figure 5. Salt dependence of equilibrium and kinetic parameters.
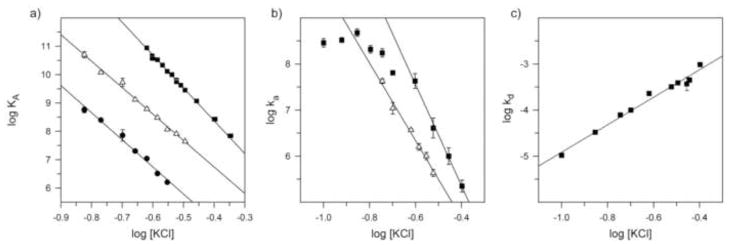
(a) Salt dependence of equilibrium association constant KA for wild-type EcoRV with unmodified DNA in binding buffer (Materials & Methods); (△) for K119A protein with RP-PMe-2 DNA; and (●) K119A protein with unmodified DNA. (b) Salt dependence of apparent association rate constant ka for (■) wild type EcoRV with unmodified DNA or (△) K119A protein with RP-PMe-2 DNA. (c) Salt dependence of apparent dissociation rate constant kd for wild-type EcoRV with unmodified DNA. Note that although KA = ka/kd (cf. Table 4), the salt-dependence of KA is not generally the sum of the separate salt-dependences of ka and kd 63. Data points show means ± std. dev. of ≥3 determinations; in some cases, error bars are too small to be seen at this scale.
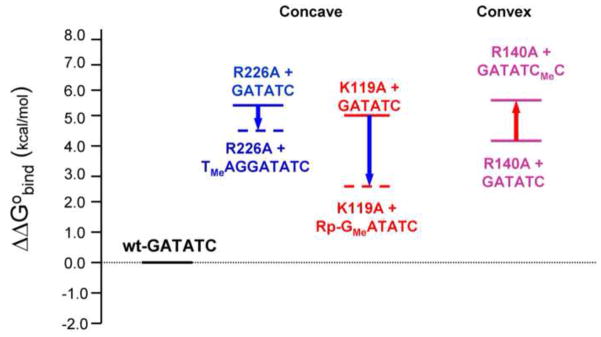
We therefore infer that relief of electrostatic repulsion at the two quasi-symmetrical P−2 positions can contribute as much as −2.3 kcal/mol to EcoRV-DNA binding. Put another way, if the protein did not ion-pair with the P−2 phosphates, repulsion among these phosphates and other phosphates on the concave DNA face would contribute an unfavorable +2.3 kcal/mol to the conversion of free protein and unbent DNA to the EcoRV complex containing bent DNA.
An important caveat for the interpretation above is that the introduction of the RP-PMe-2 does not result in a new contact and/or favorable interaction between the methyl group of the PMe and the methyl group of A119 of the mutant protein. To investigate this possibility, we used MD simulations to assess the structural impact of the RP-PMe-2 modification on the K119A mutant complex. Importantly, the RP-PMe-2 modification does not result in the formation of a new interaction between RP-PMe-2-CH3 and A119-CH3. In both half-sites (Supp. Fig. 2), the distance between these groups fluctuates from approximately 4.5 to 8.5 Å over the course of three independent 6 ns MD simulations (Supp Fig. 2), which is greater than the stable ~4 Å distance for a control methyl-methyl van der Waals interaction (Supp. Fig. 2). Thus, the simulations suggest that addition or subtraction of interactions between protein and DNA functional groups do not contribute to the observed effects of the chemical perturbation of replacing the pro-S phosphoryl oxygen with a methyl group in the context of the K119A mutant protein. The observed effects evidently arise from differences in the electrostatics at this position.
Hancock and Jen-Jacobson (manuscript in preparation) have used van’t Hoff analysis of the temperature dependence of KA to measure the contributions of changes in enthalpy (ΔH°) and entropy (ΔS°) to binding free energy for these mutant and PMe complexes, finding that the favorable effect of the RP-PMe-2 substitution on K119A binding derives from a favorable effect on ΔH°. This accords with our expectation that the RP-PMe-2 substitution results in relaxation of interphosphate repulsive forces, thereby stabilizing the bent DNA conformation in the specific EcoRV-DNA recognition complex. It is not consistent with an alternative explanation in which the PMe substitution primarily increases the nonpolar surface desolvated during protein binding.
We also analyzed the dependence of wild-type EcoRV and K119A binding on salt concentration. For wild-type EcoRV binding to unmodified DNA, KA is strongly affected by salt such that the slope of a plot of log KA vs. log [KCl] is −11.3 ± 0.2, whereas for K119A-GATATC and K119A-RP-PMe-2 binding the slopes are −9.6 ± 0.4 and −9.3 ± 0.2, respectively (Fig. 5a). Thus, formation of the K119A-DNA complex entails the release of fewer thermodynamically bound counterions 63 than does the wild-type complex. Furthermore, the nearly indistinguishable slopes for K119A-GATATC and K119A-RP-PMe-2 complex formation show that the energetic benefit of the RP-PMe-2 substitution is the same at all salt concentrations tested. This strongly implies that it is the bound EcoRV protein, rather than small cations in solution, that dominates the electrostatic environment at P−2 in the protein-DNA complex.
The P–5 position on the concave face
In 9 crystal structures of EcoRV-DNA complexes (PDB IDs: 1B94, 1RVA, 1RVB, 1AZ0, 1BGB, 1EOO, 1EOP, 1BSS, 1SUZ, 1SX8) that differ in the presence or absence of divalent metals and in crystallographic space group, we observed that the R226 sidechain usually lies approximately equidistant between the two phosphoryl oxygens of P−5 of both DNA strands, at distances (3.3Å to ~5Å) that varied somewhat among structures. P−5 also accepts a water-mediated hydrogen bond from Y219 (Fig. 2c), such that the water lies at hydrogen bonding range almost equidistant between the phosphoryl oxygens. If these circumstances also exist in solution, it would be impossible to target particular interactions selectively with a chiral PMe, but racemic RP,SP-PMe would produce about the same observed effect as either diastereomer.
In addition to its Coulombic interaction with both phosphoryl oxygens at P−5 the R226 sidechain also contacts the main chain carbonyl oxygen of E220 and T222, and the side chain hydroxyl of Y219 (Fig. 4c). The R226 side chain is disordered in the free protein11; 61, suggesting that these interactions form upon DNA binding and contribute favorable ΔG°bind. Of the four mutations of cationic sidechains we examined, the R226A mutation exacts the largest energetic penalty for binding to unmodified DNA (ΔΔG°bind = +5.5 kcal/mol; Table 1). This is likely the net of removing three favorable factors: (1) intraprotein interactions; (2) protein-phosphate interactions; (3) diminished repulsive potential near P−5.
We next observed that replacement of P−5 with racemic RP,SP-PMe had no effect on binding affinity of wild type EcoRV (Table 2). We conjecture that in this instance, unfavorable removal of contacts offsets almost exactly the favorable contributions of phosphate neutralization at this position; this exact offset is purely fortuitous.
We also analyzed the double-modification cycle (Fig. 3c) by showing that racemic RP,SP-PMe-5 substitution increased the binding affinity of R226A mutant EcoRV by about 4-fold relative to that for unmodified DNA (ΔΔG°bind = −0.9 kcal/mol; Table 1) suggesting that neutralization at P−5 contributes favorable binding free energy, although the effect is smaller than at P−2.
To illustrate the point that electrostatic effects in the EcoRV-DNA interface involve more than local point-to-point interactions, we also compared the binding of unmodified and PMe-5 DNA to both wild type and R221A mutant EcoRV. We discuss below the dynamic interactions of R221, but it is unambiguous that R221 does not interact closely with P−5. The racemic PMe-5 substitution improves R221A binding by 2-fold (ΔΔG°bind = −0.5 kcal/mol; Table 1), whereas it has no effect on binding of wild type EcoRV (Table 2). How can this be, when the same interactions to P−5 are perturbed for both wild type and R221A mutant proteins?
In the case of the R221A mutant, there should be increased repulsion on the concave face of the bent DNA due to the removal of the positively charged R221 sidechain. We conjecture that phosphate neutralization by PMe, even at a position as far removed as P−5, can relax some of this repulsion, and contribute favorably to binding. (The P−2 to P−5 distances in the complex for various combinations of phosphoryl oxygens are 18–19Å intrastrand and 17–22Å interstrand.) Wild type EcoRV provides the full set of cationic sidechains to diminish repulsion on the concave face of the bound DNA, so the concave face carries less net negative charge than it does in the mutant R221A complex. Thus, the fractional electrostatic effect of a PMe is greater in the mutant than in the wild type complex, and the effect on ΔG°bind is correspondingly more favorable.
The R221 interaction at P−2
The ability to extract the desired free-energy contribution of induced electrostatic asymmetry from a thermodynamic pseudocycle such as that in Fig. 3c depends not only on the use of this combined mutant-modification cycle, but also on the use of the correct chiral PMe-modified DNA and the correct choice of site-directed mutation. To illustrate these factors, we now compare the analysis of interaction by R221, which contacts the same P−2 phosphate as K119 (Fig. 2c), although there are several complicating factors: (1) R221 shares the pro-R oxygen with T111 (Fig. 2c, Fig. 4b); (2) A survey of 6 structures of cognate EcoRV complexes shows that the R221-P−2 interaction occurs on only one strand of the DNA; (3) MD simulations (starting coordinates PDB ID: 1B94) suggest that the R221 sidechain can transiently interact with the pro-R oxygen of either the P−2 or P−3 phosphates on one DNA strand, but does not appear to make stable intraprotein interactions. Because of these complicating factors, the desired contribution cannot be isolated and evaluated.
Of the four EcoRV mutant proteins we studied, R221A reduces EcoRV binding affinity the least relative to the wild type protein (180-fold weaker binding; ΔΔG°bind = +3.2kcal/mol; Table 1). The energetic penalty observed for R221A binding, relative to wild type EcoRV binding, likely is the net of (1) removal of R221 interactions with P−2 and/or P−3 (see below) and (2) greater interphosphate repulsion on the concave face of the R221A-bound DNA due to the absence of 2 positive charges.
In contrast to our observations for the K119A mutant protein, chemical neutralization by PMe-2 substitution did not improve R221A binding affinity. In the R221A complex, an RP-PMe-2 modification perturbs the interaction between the K119 side chain and the pro-S phosphoryl oxygen. Alternatively, the SP-PMe-2 modification reduces R221A binding affinity by 250-fold (ΔΔG°bind = +3.2 kcal/mol, Table 1) by perturbing the interactions of the pro-R phosphoryl oxygen with the T111 side chain, and a water mediated contact to T187, which also makes a water mediated contact with the adjacent phosphoryl oxygen (Fig. 2c). Thus, both PMe diastereomers perturb additional protein-DNA interactions, thereby masking any favorable effects of charge-charge interaction at P−2. As an additional complicating factor, the R221A-PMe-2 complex appears to achieve a smaller DNA bend angle than all the other complexes we have examined (see below).
The P−3 position on the concave face
Upon bending, the largest absolute decrease in interphosphate distance across the major groove occurs between the two P−3 phosphates (Fig. 1c). In crystal structures this phosphate makes no direct interactions with cationic sidechains, so we anticipated that neutralization with PMe would be favorable to binding. On the contrary, racemic PMe-3 substitution reduces wt binding affinity 12-fold relative to unmodified DNA (ΔΔG°bind = +1.5kcal/mol; Table 2).
Molecular dynamics simulations suggested a possible explanation. In our simulations, the R221 side chain transiently interacts with both P−2 and P−3 phosphates in the unmodified EcoRV-DNA complex (S. Hancock and L. Jen-Jacobson, unpublished data). We speculate that the PMe-3 substitution abolishes the transient interaction of R221 with P−3, thus constraining the R221 side chain to the P−2 phosphate, resulting in an unfavorable reduction in sidechain freedom. Such unfavorable ΔS° of two R221 sidechains could easily account for sufficient unfavorable ΔG°bind to mask any favorable ΔG° from reduced interphosphate repulsion. In support of this entropichypothesis, we find that the PMe-3 substitution has very little, if any, impact on R221A binding affinity (ΔΔG°bind = +0.2 kcal/mol; compare R221A binding with unmodified and PMe-3 DNA, Table 2). However, this observation leaves it unclear why, even with the R221A mutant protein, we observe no apparent favorable effect of neutralizing P−3.
Neutralization on the convex face opposes DNA bending
According to the hypothesis of Manning64 and Mirzabekov and Rich33, repulsive forces between phosphates on the convex face of the double helix should help drive bending toward the concave face when protein induces electrostatic imbalance between the two faces. To investigate this potentially favorable electrostatic contribution, we made electrostatic perturbations at the P+3 phosphate. At the P+3 position, a close Coulombic interaction is formed by the R140 sidechain lying approximately equidistant (3Å to 4.3Å in various structures) between the two phosphoryl oxygens, but on the other DNA strand the sidechain lies an average 6.3Å from the phosphate. The pro-S phosphoryl oxygen receives a hydrogen bond from Q69, but only in one DNA strand, while the pro-R oxygen receives a hydrogen bond from Y95-OH on both strands (Fig. 2c). There is, in addition, a van der Waals interaction between R140 and Y95 (Fig. 4d).
In experiments corresponding to the double-modification thermodynamic pseudocycle (Fig. 3c), we measured binding for R140A mutant protein to DNA with or without racemic RP, SP-PMe modification at P+3 (Tables 1 and 2). (Note that because R140 is equidistant between the phosphoryl oxygens, the PMe diastereomers would have equal effects.) The incremental effect of the PMe+3 substitution was ΔΔG°bind = +1.3 kcal/mol, a penalty that arises because the PMe+3 substitution removes a favorable repulsive element. This is consistent with a favorable role in DNA bending for phosphate repulsion on the convex face, but the observed ΔΔG° may also include perturbation of the interactions with Y95 and Q69 (Fig. 4d).
Position-dependent effects of phosphate neutralization
Our data show that the energetic benefit for phosphate neutralization increases as phosphates on the concave face are nearer to the apex of the DNA bend (Fig. 6; see positions in Fig. 1). Specifically, the energetic benefit (Fig. 6; Table 1) for phosphate neutralization by Rp-PMe-2 in the K119A context (ΔΔG°bind = −2.3 kcal/mol) is greater than that for neutralization by RP,SP-PMe-5 in the R226A context (ΔΔG°bind = −0.9 kcal/mol). (Recall that all these values are for manipulation of two symmetrically located phosphates in the two DNA strands.) Even in comparing racemic PMe-2 modifications, which to some extent convolute the effects of phosphate neutralization and perturbation of contacts, the energetic benefit is greater at P−2 than at P−5 (ΔΔG°bind = −1.5 vs. −0.9 kcal/mol).
Figure 6. Free energy diagram for the effects of phosphate neutralization.
The standard binding free energy difference (ΔΔG°bind) for each complex of a mutant protein with unmodified or PMe-DNA was calculated at 294K relative to that for binding of wild type protein to unmodified DNA as ΔΔG°bind = −RTln(KA,mod/KA,ref), where KA is the equilibrium association constant, subscript “mod” denotes the binding of a mutant protein to unmodified or PMe-DNA and subscript “ref” denotes wild type protein with unmodified DNA. PMe-5, and PMe-2 (left) on the concave face show favorable effects (blue arrows), whereasPMe+3 (right) on the convex face shows an unfavorable effect (red arrow).
We propose that this is due to greater interphosphate repulsion at P−2 relative to that at P−5, resulting in a larger energetic benefit for neutralization at P−2. Part of this results simply from the symmetrical way we made the substitutions, in that the distances (Fig. 1) between symmetrically related phosphates in the two DNA chains decrease in going from P−5 to P−3 to P−2. (This trend does not continue nearing the apex of the bend, where rotation about the helix axis dominates.) Furthermore, in the EcoRV-DNA-Ca2+ crystal structure (Fig. 1d), the major groove of the bent DNA substrate becomes more narrow on the concave face, and phosphates across the major groove are pushed closer to one another, suggesting an increase in interphosphate repulsion relative to the unbent conformation. In addition, many (not all) intrastrand distances on the concave face decrease in the bent DNA, so that, for example, particular phosphoryl oxygens of P−2 are brought as much as 1.4 Å closer to those of P−3 and 2 Å closer to those of P−4. These arguments provide a rationale for increasingly favorable effects of neutralization at phosphates closer to the center of the bend at the T-A step.
Are these free energy contributions significant? If the neutralization contributions at P−2 and P−5 were additive, then ΔΔG°bind = −3.2 kcal/mol is equivalent to about 200-fold in binding affinity at 25°C. However, it is likely that these electrostatic effects on ΔG°bind will be nonadditive, simply because neutralization affects the repulsion energy between any given phosphate and other nearby phosphates. If one is neutralized, then the benefit of neutralizing another is diminished. It should also be borne in mind that all our studies measure the contribution when P0 is bound by Ca2+, and may thus underestimate the contribution of induced electrostatic asymmetry in the absence of divalent metal.
We can nevertheless compare these values to the total bending free energy calculated according to the equation of Kahn and Crothers65, which takes into account the persistence length for random-sequence DNA, the DNA bend angle Δθ and the number of base-pairs (L) over which the bend occurs.
| (5) |
From various EcoRV-DNA crystal structures10 we take bend angles of 35° to 57°, with bends occurring over 2 to 4 bp, and thus calculate a range for ΔG°bend from +8 to +17 kcal/mol. This is probably an overestimate of bending free energy for our DNA, since the central T-A step renders the EcoRV recognition site more flexible to bending than average (see below). Computational studies of DNA bending by restrained molecular dynamics66 confirm that for bends up to 50°, ΔG°bend is quadratic with Δθ, and yield computed ΔG°bend near Δθ = 50° of ~3 kcal/mol for alternating AT DNA, in excellent agreement with experimental values from atomic force microscopy21. However, this simulation produced smooth DNA bends over 15 base-pairs, whereas the DNA bend in the EcoRV-DNA complex is sharper. Correction of the computed ΔG°bend for this difference produces a value of +11 to +22 kcal/mol. Thus, it seems likely that induced electrostatic asymmetry can significantly reduce the total free energy cost of bending by EcoRV, but cannot provide all the bending energy. Much of the balance is provided by the hydrophobic effect (see below), which supplies much larger energies67.
DNA bend angles are little affected in modified complexes
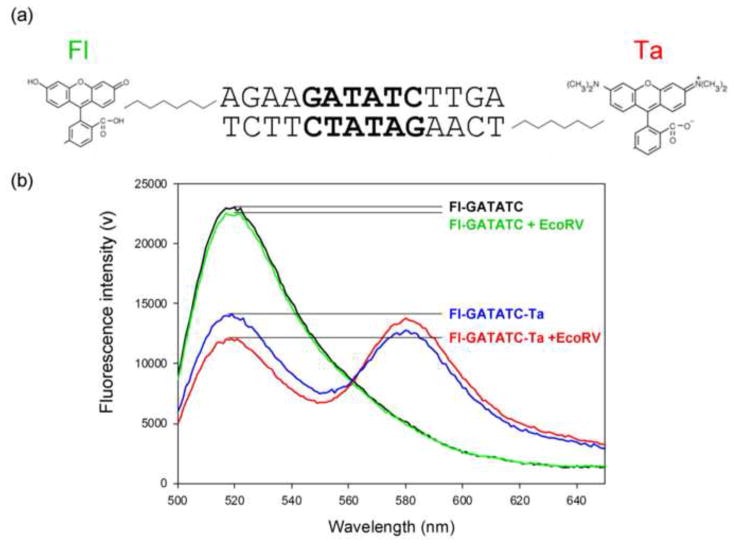
To investigate whether favorable contributions result from a modified cost of DNA bending or a modified extent of DNA bending, we have estimated the DNA bend angles in both modified and unmodified EcoRV-DNA complexes using equilibrium Förster Resonance Energy Transfer (FRET). In these assays, 14 base pair duplex DNA substrates containing the specific recognition sequence GATATC were labeled at one end with a donor fluorophore, fluorescein (Fl), and at the other end with an acceptor fluorophore, TAMRA (TA), by covalent linkage to the 5′-ends of complementary DNA strands (Fig. 7a). FRET efficiency was extracted from the donor-induced acceptor emission for both free and EcoRV-bound DNA68; 69 (Materials and Methods).
Figure 7. Equilibrium fluorescence spectra for free and EcoRV bound DNA.
(a) The fluorophore-labeled oligonucleotide substrate used in FRET experiments. Fluorophores were covalently attached to the 5′-end of each complementary DNA strand by a 6-carbon linker. Fluorescein (Fl) was linked to the top strand and TAMRA (Ta) to the bottom strand.
(b) Equilibrium fluorescence emission spectra for the 14 base-pair DNA containing the specific recognition sequence GATATC, excited at 485 nm. Equilibrium binding reactions contained 9 nM DNA and (where indicated) 90 nM EcoRV in binding buffer (Materials & Methods) with KCl added to bring the total cation concentration to 0.1 M. Spectra are shown for the DNA labeled with donor fluorophore only in the absence (black) or presence (green) of EcoRV, and for DNA labeled with both donor and acceptor fluorophores without (blue), or with (red) EcoRV.
In the absence of EcoRV, fluorescein emission (520nm) was reduced nearly 2-fold for the double labeled substrate (Fl-GATATC-Ta) relative to a single labeled substrate (Fl-GATATC) due to significant FRET in the presence of the acceptor (Fig. 7b; compare blue and black spectra). The addition of wild type EcoRV to Fl-GATATC-Ta resulted in an additional decrease in donor emission accompanied by an increase in acceptor emission, indicating a reduced donor-acceptor distance due to DNA bending (Fig. 7b; compare blue and red spectra). The interfluorophore distance, R, was calculated for either free or EcoRV-bound DNA substrates (Table 3; see Materials & Methods).
Table 3.
Effect of modifications on DNA bend angle in protein-DNA complexes
| Protein | DNAa | E (Free DNA)b,c,d | E (+EcoRV) | Rfree (Å)e | R+EcoRV (Å)e | Bend Angle (°)f |
|---|---|---|---|---|---|---|
| wild type | GATATC | 0.62 ±0.01 | 0.75 ±0.01 | 48.3 ±0.1 | 43.6 ±0.3 | 51 ±2 |
| wild type | PMe-5 | 0.63 ±0.02 | 0.76 ±0.02 | 48.1 ±0.6 | 43.4 ±0.6 | 51 ±4 |
| wild type | PMe-2 | 0.55 ±0.01 | 0.68 ±0.03 | 50.8 ±0.5 | 46.5 ±1.0 | 48 ±3 |
| wild type | PMe+3 | 0.51 ±0.03 | 0.64 ±0.04 | 52.0 ±1.0 | 47.8 ±1.3 | 47 ±2 |
| K119A | GATATC | 0.61 ±0.01 | 0.77 ±0.01 | 48.6 ±0.1 | 42.9 ±0.4 | 56 ±2 |
| K119A | PMe-2 | 0.56 ±0.01 | 0.76 ±0.05 | 50.6 ±0.6 | 43.3 ±1.9 | 56 ±8 |
| R221A | GATATC | 0.61 ±0.01 | 0.74 ±0.02 | 48.9 ±0.4 | 44.1 ±0.9 | 51 ±3 |
| R221A | PMe-2 | 0.54 ±0.01 | 0.62 ±0.03 | 51.5 ±0.5 | 48.4 ±0.5 | 39 ±3 |
| R226A | GATATC | 0.61 ±0.01 | 0.73 ±0.03 | 48.8 ±0.4 | 44.4 ±1.0 | 49 ±4 |
| R226A | PMe-5 | 0.63 ±0.02 | 0.74 ±0.04 | 48.2 ±0.6 | 44.0 ±1.3 | 48 ±5 |
| R140A | GATATC | 0.61 ±0.01 | 0.73 ±0.03 | 48.6 ±0.2 | 44.3 ±1.0 | 48 ±6 |
| R140A | PMe+3 | 0.51 ±0.02 | 0.63 ±0.03 | 52.2 ±0.6 | 48.0 ±1.0 | 46 ±3 |
The double-stranded 14 bp oligonucleotide d (AGAAGATATCTTGA) was used as the parent sequence context for all FRET experiments.
FRET efficiency (E) was measured in binding buffer plus 0.1 M K+ at 294K
Means and standard deviations are for at least 2 determinations of FRET efficiency.
FRET efficiency was calculated from the acceptor emission as described in Materials and Methods.
Interfluorophore distance (R) was calculated from the equation E = R06/(R06 + R6) where R0 is the Förster distance for the Fl/Ta pair (52.5Å).
Calculated from the equation; Bend Angle = 180-[2 (sin−1[(R+EcoRV/2)/(Rfree/2)]]; values were calculated from individual experiments before calculating mean and standard deviation.
A numerically accurate measurement of bend angle from FRET measurements must take into account the exact orientation of each fluorophore and the precise geometry of the DNA bend. However, all we require here is a reliable indicator of whether different complexes have the same or different bend angles. In our experiments, the DNA ends carrying the fluorophores lie outside the domain of the bound protein, so we assume that the fluorophore orientations are determined by their chemical interaction with the DNA ends and do not differ among various modified complexes. Although the pronounced bend at the central T-A step is common to all EcoRV-DNA crystal structures, there are minor differences in bend geometry at other base-pairs. It is impossible to know how to attribute these differences to differences in crystal packing or crystallization conditions, so we make the simplifying assumption that the DNA bend has the same general form in all modified complexes.
We therefore used a simple model, which assumes that each free DNA is unbent, that both donor and acceptor fluorophores are equidistant from the DNA end to which they are linked, and that the EcoRV-bound DNA is bent at the center of the molecule at the T-A step. From this model a bend angle of 51° was estimated for the DNA in the unmodified complex (Table 3). Although the actual uncertainty in this angle is almost certainly greater than the dispersion in the experimental measurements, it nevertheless agrees reasonably well with DNA bend angles observed in crystal structures of EcoRV-DNA complexes10; 11; 60 (~42° to 60°), suggesting that DNA bending by EcoRV in solution resembles that observed in the crystalline state. In any case, consistent use of this model allows us to compare complexes in a consistent way.
In order to probe the extent of DNA bending in EcoRV-DNA complexes with electrostatic modifications, FRET was measured for various modified and unmodified DNA substrates in the presence and absence of wild type or mutant EcoRV (Table 3). For these experiments, 9 nM DNA was used to obtain an appropriate fluorescence signal. EcoRV was then added to a final concentration that was 10-fold greater than the DNA concentration (90 nM). At these concentrations, the EcoRV in each sample was at least 20-fold greater than the experimentally determined equilibrium dissociation constant (KD) (at 0.14M KCl) so for each enzyme-DNA pair, the DNA was almost completely saturated with EcoRV.
Free DNA molecules harboring the racemic PMe-2 and PMe+3 substitutions show slightly smaller FRET values (and correspondingly larger interfluorophore distances) than those of unmodified and PMe-5 substituted DNA (Table 3). This did not derive from differences in fluorophore labeling, since absorbance measurements showed that labeling was ~95–100% efficient for all substrates. Thus, the free DNA molecules harboring the PMe-2 and PMe+3 substitutions may have a subtle conformational difference from unmodified DNA. Note that the larger Rfree values imply that these PMe DNAs cannot be more bent than unmodified DNA.
Table 3 shows FRET efficiencies, interfluorophore distances, and bend angles calculated for unmodified and modified DNA in complexes with wild type or mutant proteins. In the presence of K119A or R226A mutant proteins, FRET efficiencies for unmodified and PMe modified DNA (PMe-2 and PMe-5, respectively) were the same within experimental error, and were also within experimental error of those observed for the unmodified complex. This implies that (1) PMe substitutions did not alter the overall DNA bend in these mutant complexes and (2) the DNA bend achieved in complexes of these mutant proteins with unmodified or modified DNA is very similar to that in the unmodified complex. Thus, these electrostatic modifications do not reduce the extent of DNA bending, but instead modulate the energetic cost of achieving EcoRV-DNA complexes in which the DNA is bent to a normal extent.
Bend angles for K119A complexes were estimated to be slightly larger than those for the unmodified and R226A complexes. This follows from our modeling assumptions, in which each free DNA is assumed to be unbent, even though the various free DNAs differ somewhat from each other in FRET efficiency and consequently in the calculated distance between fluorophores (Rfree). Because of the uncertainty in the free DNA conformations for different DNAs, as well as the modeling assumptions, the experimental precisions (Table 3) probably underestimate the true uncertainty in calculating DNA bend angles in solution. Regardless, bend angles estimated for these complexes are very similar, and agree reasonably well with bend angles observed in crystal structures.
In one instance, the FRET measurements identify a complex with abnormal DNA bending. The R221A-PMe-2 complex shows the smallest degree of DNA bending (39°±3°) for any of the enzyme-DNA pairs examined. A smaller degree of DNA bending might suggest that this is an “adaptive” complex, off the path to the transition state complex required for DNA hydrolysis13. Indeed first order cleavage rate constants for the R221A-PMe-2 complex are severely compromised relative to those for the R221A-unmodified complex (see below), supporting the conjecture that the interface and active site for this complex may be malformed.
The kinetics of EcoRV-DNA association and dissociation
The favorable effects of phosphate neutralization on equilibrium EcoRV-DNA binding may derive from a slower rate of dissociation and/or a faster association rate. Several lines of evidence10; 49; 69 show that free EcoRV and DNA first associate to form a non-specific encounter complex, followed by mutually induced conformational rearrangements that include both DNA bending and repositioning of protein elements to form the specific recognition interface. The observed association (ka) and dissociation (kd) rate constants are composites of elementary rate constants associated with the binding (k1, k–1) and bending (k2, k–2 steps). Stopped-flow FRET studies69 implied that the bending step (k2), or the conversion of the non-specific encounter complex to the specific recognition complex, was either much faster than, or simultaneous with, the formation of the non-specific collision complex (k1). If this is the case, then the apparent ka is dominated by k1. In some instances, it is possible that the electrostatic perturbations made to either protein or DNA change the rate-limiting step such that the DNA bending step is no longer much faster than bimolecular association.
We used FRET stopped-flow measurements (at 0.1 M KCl) to measure the apparent association rate constants and filter-binding techniques (at 0.14 M KCl) to measure the apparent association and dissociation rate constants for modified and unmodified EcoRV-DNA complexes (Table 4). The agreement between filter binding and FRET data is generally excellent (Table 4), showing the robustness of the methodologies. (We find that apparent ka is strongly salt-dependent above 0.15 M (see below, Fig. 5b), but nearly independent of salt concentration at lower salt, as has been observed for other protein-DNA interactions70.) Some representative data are shown in Fig. 8. The association rate data (Fig. 8a) show good fits to an integrated second-order rate equation for a reversible bimolecular association reaction71; 72. As previously shown for wild-type EcoRV and unmodified DNA 69, the concentration-dependence of ka in the FRET measurements for mutant proteins and/or modified DNA is linear with enzyme concentration (data not shown), implying that the observed ka closely approximates to k173. The dissociation rate data (Fig. 8b,c) are well represented by a single first-order process for unmodified DNA or for chiral RP-PMe-DNA, but the situation for racemic RP,SP-PMe-DNA is more complicated (see below, Fig. 8b).
Table 4.
Effects of phosphate neutralization on association and dissociation kinetics
| Protein | DNA | ka (M−1s−1) FRET | ka,ref/ka FRET | ka (M−1s−1)a filter | ka,ref/ka filte | ΔΔG°ka (kcal/mol)b | kd (s−1)a filter | kd/kd,ref filter | ΔΔG°kd (kcal/mol) | KA (calc)c M−1 | KA (expt)a M−1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| wild type | GATATC | 1.1(±0.2) ×108 | 1 | 2.5(±0.8) ×108 | 1 | 0 | 3.3(±0.3) ×10−5 | 1 | 0 | 7.6(±2.5) ×1012 | 4.7(±0.8) ×1012 |
| wild type | RP-PMe-2 | NDd | ND | 2.6(±0.5) ×108 | 1 | 0 (±0.2) | 7.3(±0.9) ×10−3 | 221 | +3.2(±0.1) | 3.6(±0.8) ×1010 | 2.4(±0.1) ×1010 |
| wild type | PMe-2 | 7.0(±0.1) ×107 | 1.6 | ND | NA | +0.3 (±0.1) | 4.2(±0.8) ×10−2 | 1272 | +4.2(±0.1) | 1.7(±0.3) ×109 | ND |
| 2.8(±0.2) ×10−3 | 85 | +2.6(±0.1) | 2.3(±0.3) ×1010 | ||||||||
| wild type | PMe-5 | 1.5(±0.02) ×108 | 0.7 | 3.6(±1.0) ×108 | 0.7 | −0.2 (±0.2) | 4.1(±0.1) ×10−5 | 1 | −0.1(±0.1) | 8.8(±2.5) ×1012 | ND |
| wild type | PMe+3 | 1.7(±0.1) ×107 | 6.5 | 4.3(±0.8) ×107 | 5.8 | +1.0 (±0.2) | 5.3(±2.0) ×10−2 | 1606 | +4.3(±0.2) | 8.1(±3.4) ×108 | 1.7(±0.4) ×109 |
| K119A | GATATC | 7 (±3) ×106 | 15.7 | 2.2(±0.7) ×107 | 11.4 | +1.4 (±0.3) | 5.1(±2.0) ×10−2 | 1545 | +4.3(±0.2) | 4.3(±2.2) ×108 | 6.7(±0.6) ×108 |
| K119A | RP-PMe-2 | ND | NA | 7.0(±1.7) ×107 | 3.6 | +0.7 (±0.2) | 6.1(±0.7) ×10−4 | 19 | +1.7(±0.1) | 1.2(±0.3) ×1011 | 6.4(±0.4) ×1010 |
| K119A | PMe-2 | 1.1(±0.2) ×107 | 10 | ND | NA | +1.4 (±0.1) | NDe | NA | NA | NA | NA |
| R226A | GATATC | 6 (±2) ×106 | 18 | 1.1(±0.4) ×107 | 22.7 | +1.8 (±0.3) | 9.0(±2.0) ×10−2 | 2727 | +4.6(±0.1) | 1.2(±0.5) ×108 | 2.0(±0.3) ×108 |
| R226A | PMe-5 | 1.6(±0.3) ×107 | 6.9 | 2.0(±0.5) ×107 | 12.5 | +1.5 (±0.2) | 3.5(±0.6) ×10−2 | 1061 | +4.1(±0.1) | 5.7(±1.7) ×108 | 8.8(±0.3) ×108 |
| R140A | GATATC | 3.0(±0.7) ×107 | 3.7 | 8.5(±1.7) ×107 | 2.9 | +0.6 (±0.2) | 9.3(±0.4) ×10−3 | 282 | +3.3(±0.1 | 9.1(±1.9) ×109 | 2.4(±0.3) ×1010 |
| R140A | PMe+3 | 7.5(±0.8) ×106 | 15 | 4.2(±0.6) ×106 | 60 | +2.4 (±0.2) | ND | NA | NA | NA | 1.5(±0.2) ×109 |
Values of KA, ka and kd by filter binding (Materials & Methods) were measured at 295K in binding buffer plus 0.14M K+. Values of ka were measured at 295K by FRET (Materials & Methods) in 50 mM Tris (pH 7.5), 0.1M NaCl, 10 mM CaCl2.
Differences in standard free energy of association (ΔΔG°ka) were calculated for filter-binding data at 295K as ΔΔG°ka = −RTln(ka,mod/ka,ref) where ka,ref is the ka for wild type EcoRV with unmodified DNA. Differences in standard free energy of dissociation (ΔΔG°kd) were calculated at 294K as ΔΔG°kd = −RTln(kd,mod/kd,ref) where kd,ref is the kd for wild type EcoRV with unmodified DNA. These relate to the equilibrium values by ΔΔG°(eq) = ΔΔG°ka − ΔΔG°kd. The positive values of −ΔΔG°kd (note negative sign in column header) imply that the protein-DNA complex is destabilized by the modification.
Calculated from experimental values of ka and kd as KA = ka/kd.
ND = not determined. NA = not applicable.
The fast component of the biphasic dissociation cannot be measured at this salt concentration.
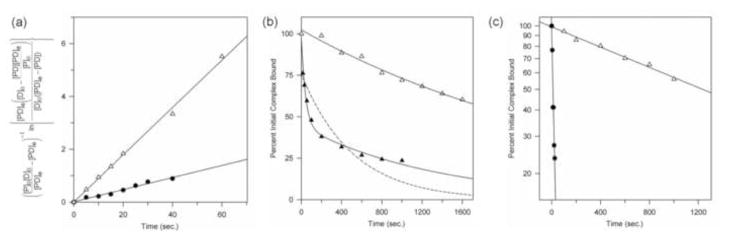
Figure 8. Effect of PMe on association and dissociation of K119A-DNA complexes.
(a) Representative kinetics of association of K119A mutant EcoRV (1 nM) with (△) unmodified DNA (0.5 nM) or (●) RP-PMe-2 DNA (0.5 nM) determined in binding buffer plus 0.14M K+. The ordinate is the integrated-rate expression for a reversible bimolecular process. The slope of the curve (△, R2 = 0.95; ●, R2 = 0.98) is the apparent association rate constant ka.
(b) Representative kinetics of dissociation of complexes of K119A mutant EcoRV with (△) RP-PMe-2 DNA or (▲) racemic RP,SP-PMe-2 DNA at 0.1M K+. (Lower salt is required to detect the faster rate component for racemic DNA.) K119A-RP-PMe-2 complexes were formed at 1.9 nM DNA, 1.5 nM enzyme (compare KD = 0.38 pM) and measurement initiated with 7.6 μM cold competitor DNA. Racemic complex data at 2.7 nM DNA, 2.2. nM enzyme (compare KD = 1.1 pM), 2.7 μM competitor DNA. The ordinate is a linear scale. For the K119A-RP-PMe-2 complex, R2 =0.98 for both single and double exponential fits. For the racemic DNA, the dashed line shows the best single-exponential fit (R2 =0.78); the solid line shows a double-exponential fit (R2 =0.99). The biphasic curve for racemic DNA implies that complexes with RP-PMe-2 DNA are much more stable than those with SP-PMe-2 DNA, because the latter substitution disrupts stabilizing interactions with R221 and T111.
c) Semilog plot of the dissociation of K119A mutant EcoRV complexes with (●) unmodified DNA and (△) RP-PMe-2 DNA at 0.14M K+. The relief of electrostatic repulsion by the PMe-2 markedly stabilizes the complex against dissociation. Unmodified complex data at 0.43 μM DNA, 0.34 μM enzyme (compare KD = 1.7 nM), 43 μM competitor. RP-PMe-2 complex data at 7.8 nM DNA, 6.3 nM enzyme (compare KD = 16 pM), 7.8 μM competitor.
KA values calculated from the ratio of ka to kd (Table 4) generally agree with those obtained by direct equilibrium measurements, although there can be up to a 2-fold difference. Furthermore, the observed equilibrium differences between wild-type and mutant proteins (ΔΔG°bind ) at 0.14M KCl are similar to those observed at 0.24M KCl (compare Tables 2 and 4).
For the association reaction, our data (Table 4) show that removal of positively charged side chains (e.g., K119A or R226A) produces relatively modest effects on the observed association rate constant (ka). The K119A mutation causes an 11-fold decrease in ka (Table 4), while the R226A mutation produces a 23-fold decrease. These data show that electrostatic complementarity is a factor in bimolecular association. Removal of the positively charged K119 or R226 sidechains (one per subunit) reduces the positive electrostatic potential associated with the DNA-interaction surface of the protein by 14 to 20 percent (calculated from the number of cationic sidechains within 6Å of the DNA backbone in the crystal structure11 of the non-specific complex PDB ID: 2RVE) thereby reducing electrostatic attraction between the protein and DNA74 and the rate of bimolecular association.
As expected, PMe had even smaller effects on association (Table 4), the largest being that RP-PMe-2 substitution increased ka for K119A by 3-fold. Electrostatic orientation effects74 on the initial encounter would be expected to decrease with this modification, but the observed effect on ka is the opposite. Even more strikingly, the introduction of PMe+3 in the context of the R140A mutant protein reduced the apparent ka by 4-fold (FRET) to 20-fold (filter binding). Although this observation in isolation is consistent with a reduced electrostatic orientation effect, a parsimonious explanation would account for the opposite effects of these two PMe modifications on opposite DNA faces.
The most likely possibility is that the DNA in the initial encounter complex is already mildly bent as previously proposed49 or samples bent conformations, albeit not bent to the degree ultimately achieved in the final recognition complex. If this is the case, chemical neutralization on the concave face would tend to lower the activation energy barrier to association and neutralization on the convex face would raise it, but these would be much smaller effects than in the equilibrium data, which primarily reflect the formation of the fully bent DNA complex. This is precisely what we observed.
A less likely alternative possibility is that RP-PMe-2 substitution affects the transition from the unbent encounter complex to the specific recognition complex in which the bound DNA is fully bent. If the K119A mutation slows the ‘bending’ rate constant k2 such that it becomes comparable to k1 or even the rate-limiting step in complex formation, then the apparent ka that we measure would no longer reflect only k1. In this case, the subtle increase in the K119A-RP-PMe-2 association rate constant relative to that for K119A-GATATC would suggest that electrostatic repulsion on the concave face must be partially overcome for the transition from an unbent encounter complex to the specific complex containing fully bent DNA. The inhibition of ka by PMe+3 in the R140A context might again suggest a change in the rate-limiting step, since bending would be slowed when a repulsive driving force is removed from the convex face. Although the linear dependence of apparent ka on enzyme concentration suggests that bimolecular association is still rate-limiting, the slope is low enough so that it might have been hard to detect curvature indicative of a change in rate-limiting step. We could not make measurements above 1 μM EcoRV because of light scattering effects and thus could not try to detect nonlinearity at higher concentration.
For the dissociation reaction, the kinetic data for PMe-DNA are represented by either one or two first-order processes, depending on position and the PMe stereochemistry. Fig. 8b, c shows that dissociation of the chiral K119A-RP-PMe-2 complex is well represented by a single exponential process. By contrast, dissociation of racemic K119A-RP,SP-PMe-2 complex is fitted poorly by a single exponential, but is fitted well by a two-phase exponential, that is, two first-order processes with markedly different rate constants (Fig. 8b). The back-extrapolation of the slower rate component to t = 0 is not far from 50% of the complex, and the slow rate closely matches that observed with the RP-PMe-2 DNA. These observations are precisely those predicted from the contacts in crystal structures (Fig. 2c): the RP-PMe-2 substitution specifically leaves intact the interactions with R221 and T111, therefore giving slower dissociation, while the presence of SP-PMe-2 in the racemic mixture disrupts these interactions and gives a faster dissociation rate.
Our data (Table 4) show that charge modifications on the concave side of the protein-DNA interface dramatically affect the dissociation rate constant (kd). The observed kd was increased 1,500-fold by the K119A mutation and 2,700-fold by the R226A mutation. Conversely, in the context of mutant proteins where the direct sidechain-phosphate interactions are already disrupted, PMe substitutions slow the dissociation, ranging from 80-fold for K119A-RP-PMe-2 (compare K119A-RP-PMe-2 to K119A-GATATC, Table 4) to 3-fold for R226A-PMe-5 (compare R226A-PMe-5 to R226A-GATATC, Table 4). Note that the effect is greater nearer the apex of the bend (PMe-2) than farther from the apex (PMe-5), as expected for relief of electrostatic repulsion.
We interpret these results as follows: In the equilibrium wild type EcoRV (bent DNA) complex, phosphates near the apex of the concave face of the bent DNA are contacted by positively-charged groups (e.g., K119, R226) that not only contribute favorable Coulombic interactions with the DNA backbone but also asymmetric reduction of repulsive interactions among phosphates. Thus, mutations that increase the repulsion have large effects in promoting dissociation, and chemical modifications that reduce repulsion have the opposite effect. These observations therefore support our hypothesis that the apposition of a positively charged protein surface to the concave DNA face reduces interphosphate repulsion and stabilizes the bent DNA conformation in the specific EcoRV-DNA recognition complex. An alternative hypothesis in which direct attractive Coulombic interactions are the principal factors in stabilizing the bent DNA would be consistent with the mutant protein effects, but inconsistent with the PMe effects.
The salt-dependence of the association and dissociation reactions is also informative. The association step is highly sensitive to salt concentration above 0.16 M (d log ka/d log[KCl] = −11.0 ± 0.1; Fig. 5b) and in fact accounts for nearly all the salt sensitivity of equilibrium binding. This implies that most of the charge-charge interactions between protein and DNA phosphates form in the initial encounter complex, and further confirms that the observed ka reflects primarily k1. This slope is decreased (−8.4 ± 0.2) for association of K119A protein with PMe-2 modified DNA (Fig. 5b), as expected.
The measured dissociation rate, on the other hand, shows relatively weak salt dependence (d log kd/d log[KCl] = +3.0 ± 0.2; Fig. 5c). This suggests that the dissociation step being measured in our assays represents k-2 instead of k-1, as k-1 has been shown to be strongly salt dependent for protein-DNA interactions75; 76. The small residual sensitivity to [KCl] in the dissociation process implies that the complex with fully bent DNA forms some additional charge-charge interactions that were not present in the initial encounter complex. This is consistent with the crystallographic observation that R221 and R226 remain disordered in the nonspecific EcoRV-DNA complex, but become ordered and interact with DNA phosphates in the specific complex. It also implies that counterion release may contribute to the formation of the encounter complex, but likely cannot provide much free energy for subsequent DNA bending.
The picture that emerges is that the diffusion-controlled formation of the initial encounter complex (ka = 1.1 to 2.5×108 M−1s−1; Table 4) is driven primarily by charge-charge interactions, and the DNA may be mildly bent. The much greater DNA bending that follows is relatively rapid and involves the formation of many stabilizing protein-DNA interactions, but only a few additional charge-charge interactions. In the reverse direction, the unbending step (k–2) is very slow because many protein-DNA interactions (interfacial hydrogen-bonds and van der Waals networks) must be broken, leaving only the charge-charge interactions to be broken much more rapidly. Cosolute studies67; 77 show that about half the water release occurs in each step (k1 and k2). However, the water release in the first step would be largely from polar surfaces and therefore contributes little to the free energy barrier between the free molecules and the encounter complex. By contrast, the second step likely releases water from nonpolar surfaces as the highly complementary interface forms, implying that the reassociation of waters with nonpolar surfaces contributes an additional factor to the large energy barrier to dissociation.
Phosphate neutralization in the transition state for DNA cleavage
It has been shown for BamHI and EcoRI endonucleases that many protein-DNA interactions are equally realized in the ground state and the transition state, such that perturbations (e.g., base analogue substitutions) that affect binding free energy do not affect first-order cleavage rate constants13; 78. To explore whether induced electrostatic asymmetry contributes equally to ground state and transition state complexes, we measured first-order cleavage rate constants kcleave (k1 and k2 for the two DNA strands) by adding Mg2+ to pre-formed EcoRV-DNA complexes. Because kcleave reflects only the energy gap between ground and transition states, a perturbation (mutation, PMe substitution) can only affect kcleave if it alters this energy gap.
For example, the R221A mutation inhibits binding 180-fold (Table 2), but kcleave is within standard deviation of that for wild type EcoRV, implying that the interaction between R221 and DNA has an essentially equal contribution to stabilizing the ground and transition states. By contrast, mutations of other basic protein sidechains that ion-pair with DNA phosphates (K119A, R226A, R140A) cause relatively mild (5 to 30-fold) inhibition of kcleave (Table 5), indicating that there is some differential effect on the transition state. These are much smaller, however, than the effects of these mutations on equilibrium binding, (Table 2), showing that most of the penalty for disrupting the charge-charge interactions with DNA phosphates is paid in the ground state, and need not be paid again in the transition state.
Table 5.
Effect of phosphate neutralization on the EcoRV-DNA transition state complex.
| Protein | DNAa | k1 (s−1)b | kref/k1 | ΔΔG1°‡ (kcal/mol)c | k2 (s−1)b | kref/k2 | ΔΔG2°‡ (kcal/mol)c |
|---|---|---|---|---|---|---|---|
| wild type | GATATC | 1.8 ± 0.1 | 1 | 0 | 1.8 ± 0.1 | 1 | 0 |
| wild type | PMe-5 | 1.1 ± 0.1 | 2 | +0.3 ± 0.1 | 1.2 ± 0.1 | 2 | +0.3 ± 0.1 |
| wild type | PMe-4 | 0.26 ± 0.04 | 7 | +1.1 ± 0.1 | 0.26 ± 0.04 | 7 | +1.1 ± 0.1 |
| wild type | PMe-3 | 1.3 ± 0.2 | 1 | +0.2 ± 0.1 | 1.3 ± 0.2 | 1 | +0.2 ± 0.1 |
| wild type | PMe-2 | 0.05 ± 0.004 | 36 | +2.1 ± 0.1 | 0.05 ± 0.008 | 36 | +2.1 ± 0.1 |
| wild type | Rp-PMe-2 | 0.08 ± 0.01 | 22 | +1.8 ± 0.1 | 0.09 ± 0.01 | 22 | +1.7 ± 0.1 |
| CONCAVE FACE | |||||||
| R226A | GATATC | 0.06 ± 0.01 | 30 | +2.0 ± 0.1 | 0.06 ± 0.01 | 30 | +2.0 ± 0.1 |
| R226A | PMe-5 | 0.28 ± 0.05 | 6 | +1.1 ± 0.2 | 0.33 ± 0.02 | 6 | +1.1 ± 0.2 |
| K119A | GATATC | 0.13 ± 0.01 | 14 | +1.6 ± 0.1 | 0.14 ± 0.01 | 14 | +1.5 ± 0.1 |
| K119A | Rp-PMe-2 | 0.56 ± 0.03 | 3 | +0.7 ± 0.1 | 0.61 ± 0.05 | 3 | +0.6 ± 0.1 |
| R221A | GATATC | 1.4 ± 0.8 | 0.8 | +0.2 ± 0.2 | 1.3 ± 0.8 | 1 | +0.2 ± 0.2 |
| R221A | Sp-PMe-2 | 0.006 ± 0.001 | 300 | +3.4 ± 0.1 | 0.006 ± 0.001 | 300 | +3.4 ± 0.1 |
| CONVEX FACE | |||||||
| R140A | GATATC | 0.37 ± 0.1 | 5 | +0.9 ± 0.2 | 0.32 ± 0.1 | 5 | +1.0 ± 0.2 |
| R140A | PMe+3 | 0.02 ± 0.001 | 90 | +2.7 ± 0.1 | 0.02 ± 0.002 | 90 | +2.6 ± 0.1 |
First order cleavage rate constants were measured for the double-stranded oligonucleotide d(TGTGTTGTAGGATATCCTACAGGT) in binding buffer + 0.1 M K+. Reactions at 295K were initiated by the addition of 10 mM MgCl2.
k1 denotes the first order cleavage rate constant for the first (top) strand and k2 that for the second (bottom) strand..
ΔΔGn°‡ = −RTln(kn/kref)
The question then arises: How much of the additional penalties in the transition state represents the role of these basic sidechains in inducing electrostatic asymmetry around the DNA, as distinct from other contributions to transition-state stabilization? We again used the strategy of measuring the effect of single-position PMe substitutions in the context of mutant proteins where the direct interaction to that phosphate had already been eliminated. As in our equilibrium measurements, the cleanest extraction of the electrostatic effect comes from the comparison of K119A mutant EcoRV cleavage of unmodified and Rp-PMe-2 DNA (Table 5). The data show that chemical neutralization of this phosphate on the concave face, in the K119A context, increases kcleave by more than 4-fold over that for unmodified DNA, although the absence of the K119-P-2 interactions still leaves kcleave about 3-fold lower than that of the wild type enzyme on unmodified DNA. Also on the concave face, cleavage of (racemic) PMe-5 DNA in the R226A context is nearly 5-fold faster than that of unmodified DNA, still leaving kcleave about 6-fold below the wild-type value. Both these comparisons indicate that protein-induced diminution of the negative electrostatic potential on the concave face contributes favorable free energy in the transition state, over and above the contribution to ground-state equilibrium binding.
We found the opposite effect on the convex face. PMe+3 substitution decreased kcleave for R140A mutant EcoRV by about 18-fold relative to that of unmodified DNA. This finding is consistent with the hypothesis that repulsion between phosphates on the convex face provides additional energy for DNA bending in the transition state, beyond that used in the ground state.
In the case of R221A-SP-PMe-2, we observed an anomalously strong inhibition of cleavage rate (300-fold; Table 5). This is the complex that showed a markedly smaller DNA bend angle in our FRET experiments (Table 3), so we infer that this complex has likely “adapted” into a structure that lies off the path to the transition state.
Our data imply that for the wild type protein interaction with unmodified DNA, the charge-charge interactions to DNA phosphates are still retained in the transition state. They also strongly support the hypothesis that induced electrostatic asymmetry contributes to the energetics of DNA bending in both ground-state and transition-state complexes, and probably contributes an additional increment in the transition state. Mg2+ soaking experiments on EcoRV-DNA co-crystals show that catalytic activity is retained only for those complexes in which the substrate DNA has achieved the greatest degree of bending10. We cannot tell from our data whether the DNA becomes somewhat more sharply bent as the complex progresses from the ground state to the transition state, or whether the electrostatics are affected by other minor structural adjustments.
Conclusions
Our data show that induction of electrostatic asymmetry by EcoRV endonuclease contributes favorable free energy to both the ground-state complex and the transition-state complex. The key observations are that the effects of reducing the negative potential on the concave DNA face are favorable for binding (Table 2) and catalytic rate (Table 5) and have larger effects on dissociation rate than on association rate (Table 4), whereas similar perturbation on the convex face has opposite effects on all parameters. Even on the concave face, the energetic contribution of these electrostatic factors is position-dependent, making a larger favorable contribution at phosphates nearer the apex of the bent DNA, and a smaller contribution at phosphates more distant from the apex. These observations are exactly those expected for electrostatic contributions to DNA bending, but would be difficult to rationalize by any other model.
We can thus relate the following molecular picture of DNA binding and cleavage by EcoRV endonuclease: The T-A step at the center of the GATATC recognition site is unusually flexible11; 23; 47; 48, by which is meant not that the free DNA is intrinsically bent, but rather that the free energy cost of axial bending is lower at a T-A step than at the other 9 possible dinucleotide steps79. MD simulations show that the harmonic force constant for roll is lowest at a T-A step79, in accord with the rank order of roll dispersions based on crystallographic survey23. A recent computational study80 shows that although the most probable bend angle at the protein-free GATATC site is close to zero (i.e., straight DNA), the fraction of the population assuming bend angles of 10° to 30° is substantially greater than for a GAATTC site lacking the T-A step.
EcoRV endonuclease approaches the (unbent or slightly bent) DNA and presents numerous cationic sidechains that contribute both an electrostatic attraction and an orientation factor between protein and DNA. It is not yet completely clear if the initial encounter complex at a GATATC recognition site is unbent like the nonspecific complex, or is already distinct from the nonspecific complex in that most complexes are mildly bent or the population of complexes samples a bent conformation more frequently. Because the GATATC site does not intrinsically have a narrowed minor groove, and EcoRV endonuclease does not present cationic sidechains to the minor groove, this is not an instance of “shape-dependent” electrostatic recognition proposed for complexes where the minor groove is narrowed 81.
Some of the cationic sidechains (e.g., R221, R226) are disordered in the free protein and do not yet become ordered upon binding nonspecific DNA11, but we do not know if they become ordered in the initial encounter complex at a GATATC site. Divalent metal ions are not yet bound at the EcoRV catalytic sites. (This is inferred from the fact that Ca2+ does not enhance equilibrium binding to nonspecific DNA57, implying that the binding sites for 4 divalent cations50; 59; 60 form only in the specific recognition complex with bent DNA.)
The asymmetric apposition of cationic sidechains to phosphates P−2, P−3 and P−5 (total of 6 in the DNA duplex) reduces repulsion on one face of the DNA, partially offset by the interaction of R140 with P+3 on the opposite face. Nevertheless, this asymmetry provides insufficient driving force for full DNA bending at sites other than GATATC, because (a) central steps other than pyrimidine-purine present too high an energy barrier to bending; (b) other DNA functional groups on the bases are not correctly positioned to form favorable interactions with the protein and/or are in position to form unfavorable interactions with protein groups11; 47. Those favorable protein-DNA interactions formed in the encounter complex are largely between charged or polar groups, so some water is released, but desolvation of polar groups is entropically favorable and enthalpically unfavorable51; 82, so there is little or no favorable free energy contribution. The strong salt-dependence of association rate suggests that cation release may make a favorable entropic contribution. This partially hydrated “nonspecific” encounter complex can diffuse linearly along the DNA74; 83; 84.
However, at a GATATC recognition site, protein-induced electrostatic asymmetry, combined with electrostatic repulsion on the opposite DNA face and the intrinsic axial flexibility of the T-A step, greatly lowers the energy of activation for axial DNA bending, such that conversion of the encounter complex (whether with straight or mildly bent DNA) to the specific recognition complex is faster than the initial bimolecular collision69. As the DNA becomes increasingly bent and protein sidechains adjust concomitantly to form the complementary protein-base contacts of the recognition interface, water is squeezed from newly apposed nonpolar surfaces, contributing a large free energy drive for formation of the specific recognition complex67; 77. This additional energy generation is crucial because the free energy cost of bending rises approximately quadratically with bend angle65; 66. At the same time, D90, D74 and D45 from both subunits are brought into proximity with phosphates P0, forming the cation binding sites50; 59; 60. Four Mg2+ ions bind (coordinating to P0/D74/D90 and to P0/D45/D74 in each site) and neutralize P0, not only providing the last elements required to initiate catalysis, but also reducing repulsion between P0 and other nearby phosphates on the concave face (e.g., P−2 at 11.5Å distance), thereby further stabilizing the bent DNA conformation. Our cleavage-rate data (Table 5) and crystallographic studies10; 85 suggest that the transition state may require further minor structural adjustments with electrostatic consequences. Thus, because EcoRV endonuclease discriminates among closely related sites in both binding and cleavage steps57, the favorable energetic contribution of protein-induced electrostatic asymmetry is an important element of specificity determination.
Materials and Methods
Mutagenesis and protein purification
Plasmids containing EcoRV mutations were generated using the QuikChange strategy (Stratagene) as previously described61. Oligonucleotides for PCR were purchased from Integrated DNA Technologies and enzymes were purchased from New England Biolabs. The entire coding region of the DNA was sequenced (Iowa State University DNA Facility) for all mutant plasmids to verify that only the desired mutation was introduced.
Wild type EcoRV and mutant enzymes K119A and R226A were purified by chromatography essentially as described86, except that a third column (Heparin Sepharose, GE Healthcare) was added to the two-column procedure. The R221A and R140A mutant enzymes were purified by the original two-column procedure86. Purified enzyme preparations (2 mg/ml in 20mM phosphate, 0.4 M NaCl, 1mM DTT, 10% glycerol; pH 7.3) were flash frozen and stored at −80 °C.
Oligodeoxynucleotide substrates
Methylphosphonate (PMe) substitutions were made in a parent 24 nt sequence (abbreviated GATATC) d(TGTGTTGTAGGATATCCTACAGGT), in which the EcoRV recognition site (underlined) is positioned off-center so that cleavage in each of the DNA strands produces distinguishable products. Unmodified and racemic RP,SP-PMe-substituted DNA oligonucleotides were synthesized at the University of Pittsburgh DNA synthesis facility on an Expedite synthesizer (Model 8909). In all cases, PMe modifications were made in symmetry-related positions in both strands. Oligodeoxynucleotides were synthesized on Q supports (Glen Research) using standard phosphoramidite chemistry with the following modifications: (1) PMe linkages were incorporated using an increased coupling time of 6 minutes. (2) Acetyl-protected dC monomer instead of the commonly used benzoylated derivative was used to prevent transamination of dC during deprotection with ethylenediamine. (3) Anhydrous tetrahydrofuran instead of anhydrous acetonitrile was used as a diluent for dG-methylphosphonamidite. Cleavage of PMe substituted deoxyoligomers from the Q-supports and deprotection was carried out according to the “one-pot” procedure developed by Hogrefe et al.87. PMe-oligodeoxynucleotides were desalted over C18 columns (Alltech) and purified by gel electrophoresis as described57; 88. The chiral RP-PMe-2 and SP-PMe-2 oligomers were gifts of Dr. Richard Hogrefe (TriLink Biotechnologies) and Dr. Wojciech J. Stec (Polish Academy of Sciences, Centre of Molecular and Macromolecular Studies) respectively.
Concentrations of purified single-stranded oligodeoxynucleotides were determined spectrophotometrically from extinction coefficients calculated by the nearest neighbor method89; 90. Stoichiometric amounts of complementary single strands were annealed, and the products were confirmed to be > 98% in duplex form by non-denaturing gel electrophoresis (12% polyacrylamide in TBE (89 mM Tris-borate,2 mM Na2EDTA, pH 8.3). DNA constructs were 5′-end labeled with 32P-ATP using T4 polynucleotide kinase as described88.
For FRET experiments, PMe substitutions were made in the parent 14 nt sequence d(AGAAGATATCTTGA). Fluorescein was coupled directly at the 5′-end using 5′-fluorescein phosphoramidite with a 6-carbon spacer (6-(3′,6′-dipivaloylfluoresceinyl-6-carboxamido)-hexyl-1-O-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite; Glen Research 10-5901). 6-carboxy-tetramethylrhodamine (TAMRA; Molecular Probes C-6123) as the succinimidyl ester was coupled post-synthetically to an amino-modified 6-carbon linker (Glen Research) incorporated at the 5′-end of the oligonucleotide. PMe incorporation, cleavage, and deprotection of these substrates were carried out as above. Concentrations of labeled oligonucleotides were determined as described above accounting for the contribution of fluorescein and TAMRA to A260. The extent of fluorophore labeling was 95–100% (molar absorptivity coefficients used were ε494 = 61,000M−1 cm−1 for fluorescein and ε555 = 91,000 M−1cm−1 for TAMRA).
Equilibrium binding studies
Equilibrium association constants KA were determined on nitrocellulose filters by either direct-binding or competition methods as described88; 91–93 with minor adaptations. All equilibrium binding reactions were carried out at 295K in binding buffer (20 mM cacodylic acid, 10 mM CaCl2, 100 mM dithiothreitol, 100 μg/ml bovine serum albumin, pH 7.25) plus KCl added to achieve the desired total K+ concentration. The dependence of KA on [K+] was essentially identical in bis-tris-propane buffer. Those cases (some mutant proteins, modified DNA) that gave unusually weak direct binding were checked by equilibrium competition, which gave KA values essentially indistinguishable from those by direct binding. Engler et al. had previously shown close concordance with values from gel retardation57. In addition, values of KA from equilibrium measurements agreed very well with KA calculated from independently measured ka and kd (Table 4), and values of ka by filter binding agreed closely with those from solution FRET (Table 4). Finally, in measuring the salt-dependence of KA, values at low salt were obtained by direct filter binding while the weaker KA values at higher salt were measured by equilibrium competition, yet the overall plots remained linear (Fig. 5). Because competition measurements use the weaker-binding DNA as competitor against labeled, unmodified DNA that forms a maximally stable complex, we can exclude the possibility that kinetically unstable complexes gave inaccurately low values of KA in direct filter-binding measurements.
Kinetics of complex association and dissociation
Association rate constants for the formation of modified and unmodified EcoRV-DNA complexes were determined in both stopped flow FRET69 and nitrocellulose filter binding94 experiments. Association rates determined by stopped flow FRET were measured on an Applied Photophysics SX.18MV stopped flow reaction analyzer. The buffer for these analyses contained 50 mM Tris (pH 7.5), 100 mM NaCl, and 10 mM CaCl2. Acceptor emission was monitored through a 570 nm cutoff filter. In all cases, at least 5 runs were averaged and fit to a single exponential function to obtain the observed rate constant at each concentration. Plots of observed rate versus enzyme concentration were fit to the following equation:
| (7) |
This equation applies to second-order events with enzyme in excess of substrate73. For each enzyme/substrate pair, the full range of kobs versus concentration was collected in triplicate. Each set was fit separately to eqn. 7. The mean and standard error of the three trials are reported. For the filter binding analyses, sufficiently dilute solutions of EcoRV protein (0.25 nM to 6 nM) and 24-bp 32P-labeled oligonucleotide (0.12 nM to 4 nM) in binding buffer plus 0.14M K+ were rapidly mixed at 295K and filtered over a nitrocellulose filter at desired time intervals. Details are given in the legends to Table 4 and Fig. 8. EcoRV-32P-DNA complex retained on the filter was measured by scintillation counting and data were fit to the integrated rate expression for a reversible bimolecular interaction71; 72 as follows
| (6) |
where [P]0 and [D]0 are the initial concentrations of EcoRV and DNA respectively, [PD] is the concentration of complex formed, [PD]e is the amount of complex at equilibrium, ka is the observed association rate constant, and t is time. The slope of a plot (cf. Fig. 8) of the left hand side of equation vs. t yields the apparent ka. We obtained very similar values of apparent ka by a nonlinear fit 95.
Dissociation rate constants (kd) for EcoRV endonuclease-DNA complexes were determined by filter binding at 295K as described91; 94. Experimental details are in the legends to Table 4 and Fig. 8.
Kinetics of DNA cleavage
Single-turnover cleavage rates were determined as described88; 92. Enzyme (1 μM) and DNA (0.5 μM) were pre-equilibrated in binding buffer plus 0.1M KCl for 90 minutes at 295K. Reactions were initiated by the addition of MgCl2 (final concentration 10 mM) and quenched at appropriate time intervals with stop solution (9M urea, 50 mM EDTA). A rapid quench-flow instrument (KinTek RQF-3, KinTek Corp., Austin TX) was used to verify the very fast first-order cleavage kinetics (kcleave > 1 s−1) of wild type EcoRV-GATATC complexes. Pre-formed wild type EcoRV-GATATC complex (2 μM EcoRV, 1.6 μM GATATC) was rapidly mixed with an equal volume of 20mM MgCl2 to initiate the reaction. At time intervals ranging from 100 ms to 10 s, reactions were stopped by the addition of an equal volume of stop solution. For both the “manual” and rapid quench-flow experiments, changing the order of addition (e.g. initiating reactions by adding DNA-Mg2+ to enzyme-Mg2+ solutions) had no effect on the results. Substrate and products were separated by denaturing (8.3 M urea) polyacrylamide gel electrophoresis and quantified by microdensitometric scans of X-ray film or phosphoimaging plates (Fujifilm FLA-5100 imager). Both methods gave similar results.
Molecular Dynamics Simulations
Atomic coordinates from PDB ID 1B9460 were used as the starting coordinates for simulations of EcoRV-DNA complexes. Modifications (sidechain mutations and PMe substitutions) to this coordinate set were made in silico with the MIDAS molecular graphics package96. Hydrogen atoms were added to the protein, DNA, and crystallographic water molecules using the AMBER 8 package97 with the all atom force field described by Cornell et al.98 as modified by Wang et al.99. Force field parameters for methylphosphonate groups were from Hamelberg et al.54. TIP3P water molecules were added to extend 10Å beyond any atoms of the EcoRV-DNA complex, creating a 514,000 Å3 octahedral water box (about 12,000 water molecules in total). 74 Na+ and Cl− ions were added to give a salt concentration of 0.24 M, supplemented by additional Na+ ions to achieve electroneutrality (4 to 8 Na+ depending on the complex). SHAKE100 was used to restrain bonds involving hydrogens (tolerance 0.0001Å ). Equations of motion were integrated using a 0.0015 ps time step in an isothermal-isobaric ensemble. Electrostatic interactions were treated with an Ewald sum101 and the temperature was regulated using the bath coupling method102. Positions of the added salt and water atoms were refined through energy minimization followed by successive rounds of molecular dynamics to equilibrate T and V. In order to escape any local minima, we heated the system to 373K and annealed back to 300K and allowed P, V, and ρ to equilibrate. Production dynamics were then initiated on the Cray XT3 MPP system at the Pittsburgh Supercomputing Center. Similar trajectories were obtained by either continuous 6 ns runs or by re-minimization at 0.5ns intervals during 6 ns of production dynamics.
Steady-state Förster resonance energy transfer
Steady state FRET experiments was carried out as described68; 69 with slight modifications. Steady-state fluorescence measurements were performed on a Photon Technologies International fluorometer (QuantaMaster) at 298K. Excitation and emission slit widths were 1.5 mm and 3.0 mm, respectively, corresponding to spectral bandwidths of 2.7 nm and 5.4 nm. The donor fluorophore (fluorescein, Fl) was excited at 488 nm and emission spectra were collected from 500–625 nm. The acceptor fluorophore (TAMRA,TA) was directly excited at 555 nm and emission spectra were collected from 570–650nm. The efficiency of energy transfer was extracted from the increase in acceptor emission68; 69. A normalized Fl-GATATC spectrum was subtracted from a spectrum of the Fl-GATATC-Ta excited at 485 nm to isolate the TAMRA emission. The area under this corrected spectrum from 575 to 585 nm was divided by area under the directly excited emission spectrum, also integrated from 575–585nm. This gave the value Ratio A. Ratio A was then used to calculate the efficiency of energy transfer (E) using
| (8) |
where εA(λ) and εD(λ) are the extinction coefficients at wavelength λ for the acceptor and donor fluorophores respectively.
The interfluorophore distance (R) was calculated from E by
| (9) |
where R0 is the Förster distance (52.5Å) for the donor-acceptor pair 69. A simple model was employed to calculate the DNA bend angle from interfluorophore distance R such that the bend angle θ is described by the expression
| (10) |
where Rfree and Rbent are the interfluorophore distances measured for the free DNA and bound/bent DNA, respectively. This model assumes that the free DNA molecule is completely B-form, and that the conjugated fluorophores are extended from the DNA such that the DNA bend occurs at the very center of the fluorophore labeled DNA molecule. It is likely that TAMRA in fact interacts with the DNA end 103; 104 and is not fully extended, but our simplified model permits consistent comparison of relative bend angles. The bend angle calculation is insensitive to the numerical value of the Förster distance in the range 49–61Å.
Supplementary Material
Snapshot (4.5 ns) from a 6ns MD simulation, showing water molecules (red) and sodium ions phosphate. See text for (blue) surrounding the pro-S phosphoryl oxygen (O1P) of the P−2 discussion of the occupancies of these sites and dwell times of individual waters and ions. The pro-R oxygen (O2P) is hydrogen bonded to T111. At this point in the MD trajectories, the R221 (see text for discussion of R221 dynamics). sidechain has migrated to interact with P−3
Snapshots from a 6ns MD simulation, showing the K119A-GATATC (panel a) and K119A-Rp-PMe-2 (panel b) complexes. The protein backbone is grey, the DNA is magenta and waters are red spheres. The A119 sidechain and PMe methyl group at P−2 (panel b) are in green. (c) Distances between functional groups during a 6ns MD simulation at 300K for the K119A-Rp-PMe-3 modified complex. Distances between A119:CH3 and PMe-2:CH3 are shown for half site A (black) and half-site B (red). The stable van der Waals interaction between T186:CH3 and the C7:CH3 of Thymine at GATATC is shown in green.
Acknowledgments
Supported by NIH grant GM29207 (to L.J-.J.) and NIH grant GM53763 (to J.J.P.)
This work was supported by NIH MERIT award 5R37-GM029207 to L.J.-J., NIH grant R01-GM53763 to J. J.P. and allocations of advanced computing resources to L.J.-J. from the Pittsburgh Supercomputing Center (PSC), supported by NSF grants MCB050027P, MCB060029P, MCB030007P, and MCB090189. The PSC is supported by several federal agencies, the Commonwealth of Pennsylvania and private industry. We thank Dr. Richard Hogrefe (TriLink Biotechnologies) for synthesis of the chiral RP-PMe-2 oligomer and Dr. Wojciech J. Stec (Polish Academy of Sciences, Centre of Molecular and Macromolecular Studies) for synthesis of the chiral SP-PMe-2 oligomer. We are grateful to Dr. Lauren Ernst (Molecular and Biosensor Imaging Center, Carnegie Mellon University) for help with fluorescence spectroscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: The DNA is bent by 90°. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 3.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 4.Goosen N, Van de Putte P. The regulation of transcription initiation by integration host factor. Molec Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 5.Kalodimos CG, Biris N, Bonvin AMJJ, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 6.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 7.Sam MD, Cascio D, Johnson RC, Clubb RT. Crystal structure of the excisionase-DNA complex from bacteriophage lambda. J Mol Biol. 2004;338:229–240. doi: 10.1016/j.jmb.2004.02.053. [DOI] [PubMed] [Google Scholar]
- 8.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J Struct Biol. 2006;156:262–72. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Horton NC, Perona JJ. Crystallographic snapshots along a protein-induced DNA-bending pathway. Proc Nat Acad Sci USA. 2000;97:5729–5734. doi: 10.1073/pnas.090370797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albright RA, Matthews BW. How Cro and lambda-repressor distinguish between operators: The structural basis underlying a genetic switch. Proc Nat Acad Sci USA. 1998;95:3431–3436. doi: 10.1073/pnas.95.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jen-Jacobson L. Protein-DNA recognition complexes: Conservation of structure and binding energy in the transition state. Biopolymers. 1997;44:153–180. doi: 10.1002/(SICI)1097-0282(1997)44:2<153::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 15.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc Nat Acad Sci USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairney KL, Harrington RE. Flow birefringence of T7 phage DNA: dependence on salt concentration. Biopolymers. 1982;21:923–934. doi: 10.1002/bip.360210506. [DOI] [PubMed] [Google Scholar]
- 17.Porschke D. Persistence length and bending dynamics of DNA from electrooptical measurements at high salt concentrations. Biophys Chem. 1991;40:169–179. doi: 10.1016/0301-4622(91)87006-q. [DOI] [PubMed] [Google Scholar]
- 18.Sobel ES, Harpst JA. Effects of Na+ on the persistence length and excluded volume of T7 bacteriophage DNA. Biopolymers. 1991;31:1559–1564. doi: 10.1002/bip.360311311. [DOI] [PubMed] [Google Scholar]
- 19.Hagerman PJ. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 20.Manning GS, Ebralidse KK, Mirzabekov AD, Rich A. An estimate of the extent of folding of nucleosomal DNA by laterally asymmetric neutralization of phosphate groups. J Biomol Struct Dyn. 1989;6:877–89. doi: 10.1080/07391102.1989.10506519. [DOI] [PubMed] [Google Scholar]
- 21.Wiggins PA, van der Heijden T, Moreno-Herrero F, Spakowitz A, Phillips R, Widom J, Dekker C, Nelson PC. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat Nanotechnol. 2006;1:137–41. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 22.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Molec Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 23.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Nat Acad Sci, USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorin AA, Zhurkin VB, Olson WK. B-DNA twisting correlates with base-pair morphology. J Mol Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 25.SantaLucia J., Jr A unified view of polymer, dumbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Searle MS, Williams DH. On the stability of nucleic acid structures in solution: Enthalpy - entropy compensations, internal rotations and reversibility. Nucl Acids Res. 1993;21:2051–2056. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tchernaenko V, Radlinska M, Drabik C, Bujnicki J, Halvorson HR, Lutter LC. Topological measurement of an A-tract bend angle: comparison of the bent and straightened states. J Mol Biol. 2003;326:737–749. doi: 10.1016/s0022-2836(02)01468-7. [DOI] [PubMed] [Google Scholar]
- 28.Zinkel SS, Crothers DM. Comparative gel electrophoresis measurement of the DNA bend angle induced by the catabolite activator protein. Biopolymers. 1990;29:29–38. doi: 10.1002/bip.360290106. [DOI] [PubMed] [Google Scholar]
- 29.Griffith JD, Makhov A, Zawel L, Reinberg D. Visualization of TBP oligomers binding and bending the HIV-1 and adeno promoters. J Mol Biol. 1995;246:576–584. doi: 10.1016/s0022-2836(05)80107-x. [DOI] [PubMed] [Google Scholar]
- 30.Rice PA, Yang SW, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 31.Maher LJ., 3rd Mechanisms of DNA bending. Curr Opin Chem Biol. 1998;2:688–94. doi: 10.1016/s1367-5931(98)80104-x. [DOI] [PubMed] [Google Scholar]
- 32.Elcock AH, McCammon JA. The low dielectric interior of proteins is sufficient to cause major structural changes in DNA on association. J Am Chem Soc. 1996;118:3787–3788. [Google Scholar]
- 33.Mirzabekov AD, Rich A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc Nat Acad Sci USA. 1979;76:1118–1121. doi: 10.1073/pnas.76.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paolella DN, Liu Y, Fabian MA, Schepartz A. Electrostatic mechanism for DNA bending by bZIP proteins. Biochem. 1997;36:10033–10038. doi: 10.1021/bi970515b. [DOI] [PubMed] [Google Scholar]
- 35.Metallo SJ, Paolella DN, Schepartz A. The role of a basic amino acid cluster in target site selection and non-specific binding of bZIP peptides to DNA. Nucleic Acids Res. 1997;25:2967–72. doi: 10.1093/nar/25.15.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams LD, Maher LJ., 3rd Electrostatic mechanisms of DNA deformation. Annu Rev Biophys Biomol Struct. 2000;29:497–521. doi: 10.1146/annurev.biophys.29.1.497. [DOI] [PubMed] [Google Scholar]
- 37.Manning GS. Is a small number of charge neutralizations sufficient to bend nucleosome core DNA onto its superhelical ramp? J Am Chem Soc. 2003;125:15087–92. doi: 10.1021/ja030320t. [DOI] [PubMed] [Google Scholar]
- 38.Strauss JK, Maher LJ., 3rd DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–34. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 39.Hardwidge PR, Zimmerman JM, Maher LJ., 3rd Charge neutralization and DNA bending by the Escherichia coli catabolite activator protein. Nucleic Acids Res. 2002;30:1879–85. doi: 10.1093/nar/30.9.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurlie R, Duong TH, Zakrzewska K. The role of DNA-protein salt bridges in molecular recognition: a model study. Biopolymers. 1999;49:313–27. doi: 10.1002/(SICI)1097-0282(19990405)49:4<313::AID-BIP6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 41.Zakrzewska K, Gurlie R. Protein-induced DNA bending: the role of phosphate neutralization. Theoret Chem Accts. 2001;106:8. [Google Scholar]
- 42.Strauss JK, Prakash TP, Roberts C, Switzer C, Maher LJ., 3rd DNA bending by a phantom protein. Chem Biol. 1996;3:571–678. doi: 10.1016/s1074-5521(96)90135-0. [DOI] [PubMed] [Google Scholar]
- 43.Strauss JK, Roberts C, Nelson MG, Switzer C, Maher LJ., 3rd DNA bending by hexamethylene-tethered ammonium ions. Proc Nat Acad Sci USA. 1996;93:9515–9520. doi: 10.1073/pnas.93.18.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardwidge PR, Wu J, Williams SL, Parkhurst KM, Parkhurst LJ, Maher LJ., 3rd DNA bending by bZIP charge variants: A unified study using electrophoretic phasing and fluorescence resonance energy transfer. Biochem. 2002;41:7732–7742. doi: 10.1021/bi020213w. [DOI] [PubMed] [Google Scholar]
- 45.Strauss-Soukup JK, Maher LJ., 3rd DNA bending by GCN4 mutants bearing cationic residues. Biochem. 1997;36:7. doi: 10.1021/bi970215u. [DOI] [PubMed] [Google Scholar]
- 46.Hardwidge PR, Parkhurst KM, Parkhurst LJ, Maher LJ., 3rd Reflections on apparent DNA bending by charge variants of bZIP proteins. Biopolymers. 2003;69:110–117. doi: 10.1002/bip.10321. [DOI] [PubMed] [Google Scholar]
- 47.Martin AM, Sam MD, Reich NO, Perona JJ. Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nat Struct Biol. 1999;6:269–77. doi: 10.1038/6707. [DOI] [PubMed] [Google Scholar]
- 48.Mack DR, Chiu TK, Dickerson RE. Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: a comparative analysis of T-A and A-T steps within A-tracts. J Mol Biol. 2001;312:1037–49. doi: 10.1006/jmbi.2001.4994. [DOI] [PubMed] [Google Scholar]
- 49.Hiller DA, Perona JJ. Positively charged C-terminal subdomains of EcoRV endonuclease: Contributions to DNA binding, bending, and cleavage. Biochem. 2006;45:11453–11463. doi: 10.1021/bi0606400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kostrewa D, Winkler FK. Mg2+ binding to the active site of EcoRV endonuclease: A crystallographic study of complexes with substrate and product DNA at 2 A? resolution. Biochem. 1995;34:683–696. doi: 10.1021/bi00002a036. [DOI] [PubMed] [Google Scholar]
- 51.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 52.Kumar S, Nussinov R. Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys J. 2002;83:1595–1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong H, Freed KF. Electrostatic solvation energy for two oppositely charged ions. Biophys J. 2010;98:470–477. doi: 10.1016/j.bpj.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamelberg D, Williams LD, Wilson WD. Effect of a neutralized phosphate backbone on the minor groove of B-DNA: Molecular dynamics simulation studies. Nucl Acids Res. 2002;30:3615–3623. doi: 10.1093/nar/gkf472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chacko KK, Lindner K, Saenger W, Miller PS. Molecular structure of deoxyadenylyl-3′-methylphosphonate-5′-thymidine dihydrate, (d-ApT x 2H2O), a dinucleoside monophosphate with neutral phosphodiester backbone. An X-ray crystal study. Nucleic Acids Res. 1983;11:2801–14. doi: 10.1093/nar/11.9.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vipond IB, Halford SE. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochem. 1995;34:1113–9. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 57.Engler LE, Welch KK, Jen-Jacobson L. Specific binding by EcoRV endonuclease to its DNA recognition site GATATC. J Mol Biol. 1997;269:82–101. doi: 10.1006/jmbi.1997.1027. [DOI] [PubMed] [Google Scholar]
- 58.Martin AM, Horton NC, Lusetti S, Reich NO, Perona JJ. Divalent metal dependence of site-specific DNA binding by EcoRV endonuclease. Biochem. 1999;38:8430–9. doi: 10.1021/bi9905359. [DOI] [PubMed] [Google Scholar]
- 59.Horton NC, Newberry KJ, Perona JJ. Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases. Proc Natl Acad Sci U S A. 1998;95:13489–94. doi: 10.1073/pnas.95.23.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas MP, Brady RL, Halford SE, Sessions RB, Baldwin GS. Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: A catalytic role for a main chain carbonyl group. Nucl Acids Res. 1999;27:3438–3445. doi: 10.1093/nar/27.17.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perona JJ, Martin AM. Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis. J Mol Biol. 1997;273:207–225. doi: 10.1006/jmbi.1997.1315. [DOI] [PubMed] [Google Scholar]
- 62.Hendsch ZS, Jonsson T, Sauer RT, Tidor B. Protein stabilization by removal of unsatisfied polar groups: computational approaches and experimental tests. Biochem. 1996;35:7621–5. doi: 10.1021/bi9605191. [DOI] [PubMed] [Google Scholar]
- 63.Record MT, Jr, Ha JH, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between protein and helical DNA. Meth Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 64.Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 65.Kahn JD, Crothers DM. DNA bending in transcription initiation. Cold Spring Harb Symp Quant Biol. 1993;58:115–22. doi: 10.1101/sqb.1993.058.01.015. [DOI] [PubMed] [Google Scholar]
- 66.Curuksu J, Zacharias M, Lavery R, Zakrzewska K. Local and global effects of strong DNA bending induced during molecular dynamics simulations. Nucl Acids Res. 2009;37:3766–3773. doi: 10.1093/nar/gkp234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jen-Jacobson L, Jacobson LA. Role of Water and Effects of Small Ions in Site-specific Protein-DNA Interactions. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions: Structural Biology. Royal Society of Chemistry Press; 2008. pp. 14–37. [Google Scholar]
- 68.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–88. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 69.Hiller DA, Fogg JM, Martin AM, Beechem JM, Reich NO, Perona JJ. Simultaneous DNA Binding and Bending by EcoRV Endonuclease Observed by Real-Time Fluorescence. Biochem. 2003;42:14375–14385. doi: 10.1021/bi035520w. [DOI] [PubMed] [Google Scholar]
- 70.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3 The Escherichia coli lac repressor operator interaction: Kinetic measurements and conclusions. Biochem. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 71.Weiland GA, Molinoff PB. Quantitative analysis of drug-receptor interactions: I. Determination of kinetic and equilibrium properties. Life Sci. 1981;29:313–330. doi: 10.1016/0024-3205(81)90324-6. [DOI] [PubMed] [Google Scholar]
- 72.Cheung AT, Johnson DA, Taylor P. Kinetics of interaction of N epsilon-fluorescein isothiocyanate-lysine-23-cobra alpha-toxin with the acetylcholine receptor. Biophys J. 1984;45:447–54. doi: 10.1016/S0006-3495(84)84167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochem. 2002;41:6032–44. doi: 10.1021/bi020122z. [DOI] [PubMed] [Google Scholar]
- 74.Halford SE. An end to 40 years of mistakes in DNA–protein association kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 75.Lohman TM, DeHaseth PL, Record MT., Jr Analysis of ion concentration effects of the kinetics of protein-nucleic acid interactions. Application to lac repressor-operator interactions. Biophys Chem. 1978;8:281–94. doi: 10.1016/0301-4622(78)80011-8. [DOI] [PubMed] [Google Scholar]
- 76.Lohman TM. Kinetics of protein-nucleic acid interactions: Use of salt effects to probe mechanisms of interaction. CRC Crit Rev Biochem. 1985;19:191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- 77.Cao D. PhD Thesis. University of Pittsburgh; Pittsburgh, PA: 2002. Cosolute effects as probes of the role of water in DNA bindins and catalysis by three restriction endonucleases. [Google Scholar]
- 78.Engler LE. PhD Thesis. University of Pittsburgh; Pittsburgh, PA: 1998. Specificity determinants in the BamHI-DNA interaction. [Google Scholar]
- 79.Lankas F, Sponer J, Langowski J, Cheatham TE., 3rd DNA basepair step deformability inferred from molecular dynamics simulations. Biophys J. 2003;85:2872–83. doi: 10.1016/S0006-3495(03)74710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zahran M, Daidone I, Smith JC, Imhof P. Mechanism of DNA recognition by the restriction enzyme EcoRV. J Mol Biol. 2010;401:415–32. doi: 10.1016/j.jmb.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 81.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1254. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Makhatadze GI, Privalov PL. Energetics of protein structure. Adv Prot Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 83.von Hippel PH, Berg OG. Facilitated target location in biological system. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 84.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–52. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Horton NC, Perona JJ. DNA cleavage by EcoRV endonuclease: Two metal ions in three metal ion binding sites. Biochem. 2004;43:6841–6857. doi: 10.1021/bi0499056. [DOI] [PubMed] [Google Scholar]
- 86.Luke PA, McCallum SA, Halford SE. The EcoR V restriction endonuclease. Gene Amplif Anal. 1987;5:185–207. [PubMed] [Google Scholar]
- 87.Hogrefe RI, Vaghefi MM, Reynolds MA, Young KM, Arnold LJ., Jr Deprotection of methylphosphonate oligonucleotides using a novel one-pot procedure. Nucleic Acids Res. 1993;21:2031–8. doi: 10.1093/nar/21.9.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lesser DR, Grajkowski A, Kurpiewski MR, Koziolkiewicz M, Stec WJ, Jen-Jacobson L. Stereoselective interaction with chiral phosphorothioates at the central DNA kink of the EcoRI endonuclease-GAATTC complex. J Biol Chem. 1992;267:24810–24818. [PubMed] [Google Scholar]
- 89.Warshaw MM, Cantor CR. Oligonucleotide interactions. IV Conformational differences between deoxy- and ribodinucleoside phosphates. Biopolymers. 1970;9:1079–103. doi: 10.1002/bip.1970.360090910. [DOI] [PubMed] [Google Scholar]
- 90.Senior M, Jones RA, Breslauer KJ. Influence of dangling thymidine residues on the stability and structure of two DNA duplexes. Biochem. 1988;27:3879–85. doi: 10.1021/bi00410a053. [DOI] [PubMed] [Google Scholar]
- 91.Jen-Jacobson L, Lesser D, Kurpiewski M. The enfolding arms of EcoRI endonuclease: Role in DNA binding and cleavage. Cell. 1986;45:619–629. doi: 10.1016/0092-8674(86)90294-1. [DOI] [PubMed] [Google Scholar]
- 92.Lesser DR, Kurpiewski MR, Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease-DNA interaction. Science. 1990;250:776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- 93.Connolly BA, Liu HH, Parry D, Engler LE, Kurpiewski MR, Jen-Jacobson L. Assay of restriction endonucleases using oligonucleotides. Meth Molec Biol. 2001;148:465–490. doi: 10.1385/1-59259-208-2:465. [DOI] [PubMed] [Google Scholar]
- 94.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3 Kinetic studies. J Mol Biol. 1970;53:401–17. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 95.Morton TA, Myszka DG, Chaiken IM. Interpreting complex binding kinetics from optical biosensors: A comparison of analysis by linearization, the integrated rate equation and numerical integration. Anal Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 96.Ferrin TE, Huang CC, Jarvis LE, Langridge R. The MIDAS display system. J Molec Graph. 1988;6:13–27. [Google Scholar]
- 97.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. Amber. Vol. 8. University of California; San Francisco: 2004. [Google Scholar]
- 98.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, DMFJ, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 99.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comp Chem. 2000;21:1049–1074. [Google Scholar]
- 100.Rykeart JP, Ciccotti G, Brenderson HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 101.de Leeuw SW, Perram JW, Smith ER. Ewald summations and dielectric constants. Proc Roy Soc Lond Ser A. 1980;373:27–56. [Google Scholar]
- 102.Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 103.Vamosi G, Gohlke C, Clegg RM. Fluorescence characteristics of 5-carboxytetramethylrhodamine linked covalently to the 5′ end of oligonucleotides: multiple conformers of single-stranded and double-stranded dye-DNA complexes. Biophys J. 1996;71:972–94. doi: 10.1016/S0006-3495(96)79300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lorenz M, Hillisch A, Payet D, Buttinelli M, Travers A, Diekmann S. DNA bending induced by high mobility group proteins studied by fluorescence resonance energy transfer. Biochem. 1999;38:12150–8. doi: 10.1021/bi990459+. [DOI] [PubMed] [Google Scholar]
- 105.Hays FA, Teegarden A, Jones ZJR, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. How sequence defines structure: A crystallographic map of DNA structure and conformation. Proc Nat Acad Sci USA. 2005;102:7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Snapshot (4.5 ns) from a 6ns MD simulation, showing water molecules (red) and sodium ions phosphate. See text for (blue) surrounding the pro-S phosphoryl oxygen (O1P) of the P−2 discussion of the occupancies of these sites and dwell times of individual waters and ions. The pro-R oxygen (O2P) is hydrogen bonded to T111. At this point in the MD trajectories, the R221 (see text for discussion of R221 dynamics). sidechain has migrated to interact with P−3
Snapshots from a 6ns MD simulation, showing the K119A-GATATC (panel a) and K119A-Rp-PMe-2 (panel b) complexes. The protein backbone is grey, the DNA is magenta and waters are red spheres. The A119 sidechain and PMe methyl group at P−2 (panel b) are in green. (c) Distances between functional groups during a 6ns MD simulation at 300K for the K119A-Rp-PMe-3 modified complex. Distances between A119:CH3 and PMe-2:CH3 are shown for half site A (black) and half-site B (red). The stable van der Waals interaction between T186:CH3 and the C7:CH3 of Thymine at GATATC is shown in green.