Abstract
We report on the solution structure of an unprecedented intramolecular G-quadruplex formed by the guanosine-rich human chl1 intronic d(G3-N-G4-N2-G4-N-G3-N) 19-mer sequence in K+-containing solution. This G-quadruplex, composed of three stacked G-tetrads containing four syn guanines, represents a new folding topology with two unique conformational features. The first guanosine is positioned within the central G-tetrad, in contrast to all previous structures of unimolecular G-quadruplexes, where the first guanosine is part of an outermost G-tetrad. In addition, a V-shaped loop, spanning three G-tetrad planes, contains no bridging nucleotides. The G-quadruplex scaffold is stabilized by a T•G•A triple stacked over the G-tetrad at one end and an unpaired guanosine stacked over the G-tetrad at the other end. Finally, the chl1 intronic DNA G-quadruplex scaffold contains a guanosine base intercalated between an extended G-G step, a feature observed in common with the catalytic site of group I introns. This unique structural scaffold provides a highly specific platform for the future design of ligands specifically targeted to intronic G-quadruplex platforms.
Introduction
The diversity of possible G-quadruplex folds adopted by single-stranded guanosine-rich sequences containing four tracts of consecutive guanosine residues interspersed by loop sequences is currently under intensive investigation (reviewed in Davis, 2002; Burge et al., 2006; Phan et al., 2006; Patel et al., 2007; Neidle, 2009; Balasubramanian & Neidle, 2009; Lipps & Rhodes, 2009). We were interested in addressing the solution structure of a consensus motif for DNA promoter sequences found in proto-oncogenes (reviewed in Qin & Hurley, 2008), composed of four successive G3 guanosine-tracts (in bold),
G-G-G-N-G-G-G-N-N-N-N-G-G-G-N-G-G-G
with the aim of probing for new G-quadruplex folds. During sequence design within the framework of this concept, we obtained a well-resolved imino proton NMR spectrum for the 19-mer sequence with a central A-A segment, that contains successive G3, G4, G4 and G3 guanine-tracts as outlined below,
G1-G2-G3-T4-G5-G6-G7-G8-A9-A10-G11-G12-G13-G14-T15-G16-G17-G18-T19
This sequence (Figure 1A) exhibited imino proton NMR characteristics of a single conformation in K+-containing solution, amenable for structural characterization (Figure 1B). A search for the corresponding sequence pattern in the human genome revealed its presence in the 5′-intron of the human chl1 gene with the exact sequence spanning positions 214.844 to 214.863 on chromosome 3 (Kent et al., 2002) shown below
C-G-G-G-C-G-G-G-G-A-A-G-G-G-G-T-G-G-G-A
This observation is relevant considering the recent insight from bioinformatics searches of conserved guanosine-rich tracts with the potential to form polymorphic G-quadruplex folds in the first intron of human genes (Eddy & Maizels, 2008, 2009).
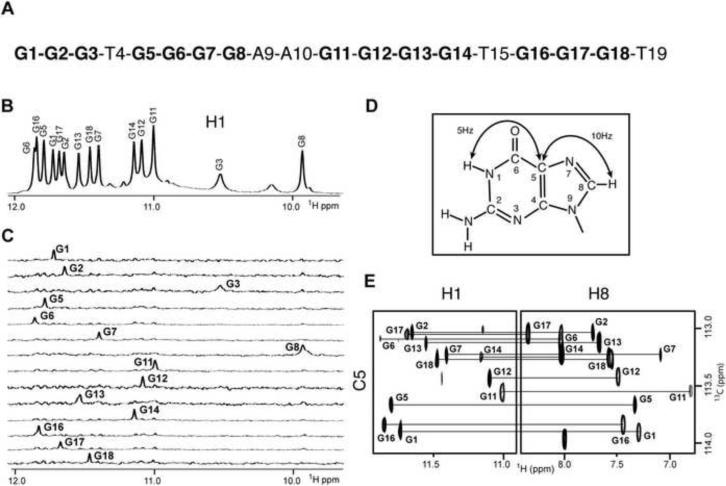
Figure 1. NMR Spectra and Assignments of human chl1 Intronic 19-mer Sequence in 50 mM K+, 5 mM Phosphate-H2O Buffer, pH 6.8, at 25 °C.
(A) Sequence of the 19-mer human chl1 intronic DNA. (B) Imino proton NMR spectrum of chl1 sequence. Unambiguous guanosine imino proton assignments are listed over the spectrum. (C) Guanosine imino protons were assigned in 15N-filtered NMR spectra of samples containing 2% uniformly-15N-labeled guanosines at the indicated positions. (D) A schematic indicating long-range J couplings used to correlate imino and H8 protons within the guanosine base. (E) H8 proton assignments of chl1 intronic sequence by through-bond correlations between guanoisne imino and H8 protons via 13C5 at natural abundance, using long-range J couplings shown in (D).
In addition, the CHL1 gene product belongs to the family of FANCJ helicases, a group of DNA dependent ATPases capable of catalytically resolving G-quadruplex-forming blocks generated at guanosine-rich tracts, that can impede DNA replication (Wu et al., 2008; 2009).
Here we report on the solution structure of the chl1 intronic G-quadruplex in K+-containing aqueous solution and demonstrate that it adopts an unprecedented G-quadruplex fold exhibiting two unanticipated conformational features, one of which has not been observed previously in the unimolecular G-quadruplex topology literature (reviewed in Burge et al., 2006; Patel et al., 2007).
Surprisingly, the G-quadruplex solution structure of this guanosine-rich consensus sequence identified for a particular intron in the human genome exhibits certain features in common with the catalytic site of group I introns.
Results
NMR Spectra and Spectral Assignments
The imino proton NMR spectrum of the chl1 intronic sequence in 50 mM K+-containing H2O buffer solution at 25 °C is characterized by twelve well-resolved resonances in the 11–12 ppm range (Figure 1B), a spectral region consistent with guanosine imino N-H protons hydrogen bonded to nitrogen acceptors (N-H••N), as observed for G-tetrads involved in G-quadruplex formation
We pursued a strategy of initially assigning the guanosine imino protons by incorporating 2% 15N-labeled-guanosines one at a time into the oligonucleotide sequence (Figure 1C) and then correlating these unambiguous assignments with guanosine H8 protons (Figure 1D), through long-range through-bond J-coupling experiments (Figure 1E) (Phan, 2000; Phan & Patel, 2002). The resulting guanosine imino proton assignments of the chl1 intronic sequence are listed above the control NMR spectrum in Figure 1B, and are analyzed in the context of the four-guanosine tracks G1-G2-G3, G5-G6-G7-G8, G11-G12-G13-G14 and G16-G17-G18 within the 19-mer sequence (Figure 1A). We anticipated that three guanosines within each track would be involved in G-quadruplex formation. Nevertheless, the imino proton of G3 at 10.55 ppm, which originates in the first G1-G2-G3 tract, is both broad and resonates upfield of the 11 to 12 ppm region (Figure 1b), suggestive of a guanosine imino proton not involved in N-H••N hydrogen bond formation. In addition the imino proton of G8, though narrow, resonates upfield at 9.95 ppm, indicative of a guanosine imino proton hydrogen-bonded to oxygen acceptors (N-H••O). These results are consistent with involvement of G1-G2, G5-G6-G7, G11-G12-G13-G14 and G16-G17-G18 guanosine-tracts in G-quadruplex formation. Thus, rather than an expected G3-G3-G3-G3 alignment of guanosine-tracts required to form a standard G-quadruplex, the chl1 intronic sequence adopts an unanticipated G2-G3-G4-G3 alignment of guanosine-tracts.
The remaining non-exchangeable base and sugar protons of the chl1 intronic sequence were assigned using standard protocols by through-space (NOESY) and through-bond (COSY, TOCSY, 13C-GHSQC) experiments in 50 mM K+-containing 2H2O buffer solution. In case of spectral overlap, resonance assignments were facilitated following 8-bromoguanosine (at positions 1, 5, 11 and 16) for guanosine substitutions, as well as deoxyuridine (at position 15) for thymine substitutions. The expanded NOESY contour plot (200 ms mixing time) in 50 mM K+-containing 2H2O buffer at 25 °C correlating base and sugar H1' protons, together with assignment connectivities (base proton to its own and 5'-flanking sugar H1' protons), is shown in Figure 2A. The complete tabulation of base and sugar proton chemical shifts are listed in Table S1 (Supplementary Materials).
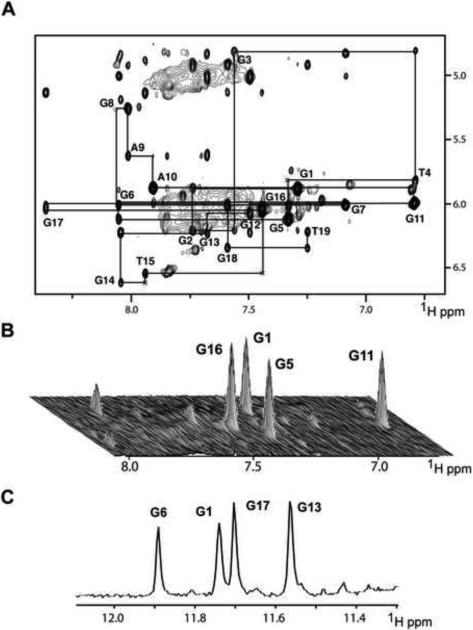
Figure 2. Non-exchangeable Proton Assignments of chl1 Intronic 19-mer Sequence in 50 mM K+, 5 mM phosphate-2H2O buffer, pH 6.8, at 25°C.
(A) Expanded NOESY contour plot (200 ms mixing time) correlating base and sugar H1' protons. The line connectivities trace NOEs between a base proton (H8 or H6) and its own and 5'-flanking sugar H1' protons. Intraresidue base to sugar H1' NOEs are labeled with residue numbers. (B) Stacked plot of short mixing time (50 ms) NOESY data set under same buffer conditions as in (A). The strong intraresidue guanosine H8-H1' cross-peaks (syn glycosidic bonds) are labeled and can be distinguished from weak cross-peaks (anti glycosidic bonds). (C) Imino proton NMR spectrum of chl1 intronic sequence recorded after 45 min following transfer after lyophilization from H2O to 2H2O solution. Assignments of slowly exchanging imino protons are listed over the spectrum.
The expanded NOESY stacked plot (50 ms mixing time) correlating base with sugar H1' NOEs of the chl1 intronic sequence in 50 mM K+-containing 2H2O buffer at 25 °C exhibits four strong cross peaks for G1, G5, G11 and G16 residues (Figure 2B), indicative of syn torsion angles for these four guanosines (short base to sugar H1' distances of 2.5 Å; Patel et al., 1982) in the folded structure of the G-quadruplex in solution. In addition, four guanosine imino protons, assigned to G1, G6, G17 and G13, exchange slowly, as recorded 45 min after transferring (following lyophilization) the chl1 intronic sequence from 50 mM K+-containing H2O solution to its 2H2O counterpart at 25 °C (Figure 2C). These results establish that four guanosines G1, G6, G17 and G13 must originate from the central G-tetrad (slowest exchanging guanosine imino protons due to shielding from exchange with solvent) of the G-quadruplex.
G-tetrad Alignments
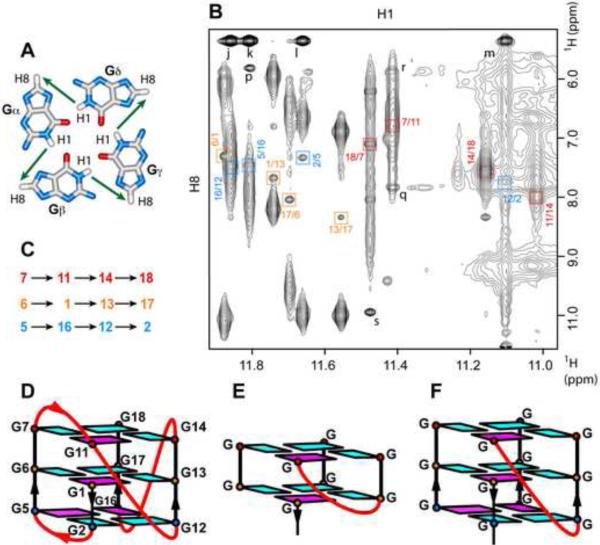
We have assigned guanosines to the three G-tetrad planes by monitoring NOEs between guanosine imino and H8 protons between adjacent guanosines around individual G-tetrad planes (Figure 3A) in the NOESY spectrum (mixing time 300 ms) of the chl1 intronic G-quadruplex in 50 mM K+-containing H2O solution at 25 °C (Figure 3B). Thus, the imino proton of G1 shows an NOE to the H8 proton of G13 (labeled G1/G13 in orange), the imino proton G13 shows an NOE to the H8 proton of G17, the imino proton of G17 shows an NOE to the H8 proton of G6, and the imino proton of G6 shows an NOE to the H8 proton of G1, thereby assigning the guanosines and their order around the G1•G13•G17•G6 G-tetrad plane (assignments in orange) (Figures 3B–D). Related NOE tracings identify the G7•G11•G14•G18 (assignments in red) and G2•G5•G16•G12 (assignments in blue) G-tetrads that form the G-quadruplex (Figures 3B–D). Since the imino protons of the G1•G13•G17•G6 G-tetrad exhibit the slowest exchange rate (Figure 2C), this central G-tetrad must be bracketed on either side by stacked G7•G11•G14•G18 and G2•G5•G16•G12 G-tetrads, resulting in the backbone tracing from G1 to G18 as shown in Figure 3D.
Figure 3. Assignment of Individual G-tetrads within the chl1 G-Quadruplex.
(A) Characteristic guanosine imino-H8 NOE connectivity patterns around a Gα•Gβ•Gγ•Gδ tetrad as indicated by arrows. (B) Expanded contour plot showing imino to H8 connectivities of chl1 intronic 19-mer sequence in 50 mM K+, 5 mM phosphate-H2O buffer, pH 6.8, at 25 °C. Cross-peaks identifying connectivities within individual G-tetrads are color-coded, with each cross-peak framed and labeled with the residue number of guanosine imino proton in the first position and that of the H8 proton in the second position. (C) Guanosine imino-H8 connectivities observed for G1•G13•G17•G6 (orange), G7•G11•G14•G18 (red) and G2•G5•G16•G12 (blue) G-tetrads. (D) Schematic representation of the chl1 intronic G-quadruplex fold. Anti bases are indicated in cyan, while syn bases are indicated in magenta. (E) Schematic of a V-shaped loop spanning two G-tetrad planes. (F) Schematic of a V-shaped loop spanning three G-tetrad planes.
G-Quadruplex Folding Topology
The chl1 intronic G-quadruplex folding topology shown in Figure 3D contains syn guanosines at G1, G5, G11 and G16, and anti guanosines at the remaining positions, consistent with the short mixing time NOESY data (Figure 2B). Unexpectedly, the first guanosine in the sequence, G1, is positioned within the central G-tetrad of the G-quadruplex. The G1-G2 segment forming the first column spans the lower two G-tetrad planes, and is followed by the G3-T4 segment which forms an edge-wise loop, leading into the G5-G6-G7 segment of the second column that spans all three G-tetrad planes. Next, the G8-A9-A10 segment forms a second edge-wise loop, leading into the G11-G12 step with its extended sugar-phosphate backbone, whereby G11 of the top G-tetrad is directly connected in a V-shaped extended alignment to G12 of the bottom G-tetrad, located on adjacent columns. The G12-G13-G14 segment that forms the third column, is connected to G16-G17-G18 segment that forms the fourth column of the G-quadruplex, by a single-residue (T15) double chain reversal loop.
Structure Calculations
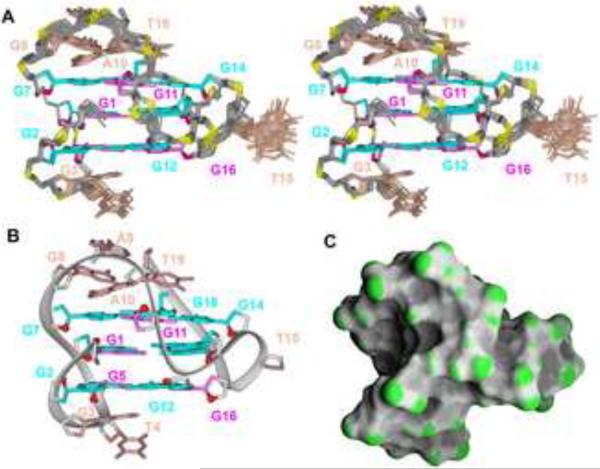
Initial distance-restrained and subsequent intensity-restrained molecular dynamics calculations (see Methods section) of the solution structure of the chl1 intronic G-quadruplex were guided by exchangeable and non-exchangeable proton restraints, with the restraints listed by category in Table 1. The ensemble of 17 refined superpositioned structures is shown in stereo in Fig 4A, with a representative refined structure shown in a ribbon representation in Figure 4B and a surface representation in Figure 4C. The ensemble of refined structures is well converged, exhibiting pair-wise root-mean-square-deviation (rmsd) values in the 0.47 range for all nucleotides except for the T15 residue (Table 1), which is confined to a cluster of energetically preferred conformations pointing outwards from the groove (Figure 3A).
Table 1.
Statistics of NMR restraints-guided computations of chl1 intronic quadruplex.
| A. NMR restraints | ||
|---|---|---|
| Distance restraintsa | Non-exchangeable | Exchangeable |
| Intra-residue distance restraints | 120 | 0 |
| Sequential (i, i+1) distance restraints | 55 | 5 |
| Long-range (i, ≥ i+2) distance restraints | 3 | 42 |
| Other restraints | ||
| Hydrogen bonding restraints (H-N, H-O, and heavy atoms) | 52 | |
| Torsion angle restraintsa | 49 | |
| Intensity restraints | ||
| Non-exchangeable protons (each of five mixing times) | 178 | |
| B. Structure statistics in 17 molecules following the intensity refinement | ||
|---|---|---|
| NOE violations | ||
| Number (> 0.2Å) | 0.0±0.0 | |
| r.m.s.d. of violations | 0.02±0.00 | |
| Deviations from the ideal covalent geometry | ||
| Bond lengths (Å) | 0.01±0.00 | |
| Bond angles (deg.) | 0.83±0.01 | |
| Impropers (deg) | 0.30±0.01 | |
| NMR R-factor (R 1/6) | 0.02±0.02 | |
| Pairwise all heavy atom r.m.s.d. values (17 refined structures) | ||
| All heavy atoms except T15 | 0.47±0.12 | |
| All heavy atoms | 0.64±0.15 | |
The residues G1, G5, G11, G16 were restrained to χ values in the 60 (±30)° range, characteristic of syn glycosidic torsion angles, identified experimentally. All other residues were restrained to χ values in the 240 (±70)° range, characteristic of anti glycosidic torsion values. The ε of the residues G1-G18 was restrained to the stereochemically allowed range 225 (±75)°. The γ torsion angle of the residues 1, 2, 4, 7, 8, 9, 17, 18, 19 was restrained to the values of 60 (±35)°, for residues 5, 10, 11 to 180 (±35)°, identified experimentally.
Figure 4. Solution Structure of the chl1 G-Quadruplex in K+-containing solution.
(A) Stereo view of 17 superpositioned refined structures. Guanosine bases in the G-tetrad core are colored cyan (anti) or magenta (syn). Bases in connecting loops are in biscuit color, with the backbone in grey and phosphorus and oxygen atoms in yellow and red, respectively. (B) Ribbon and (C) surface representations of a representative refined structure.
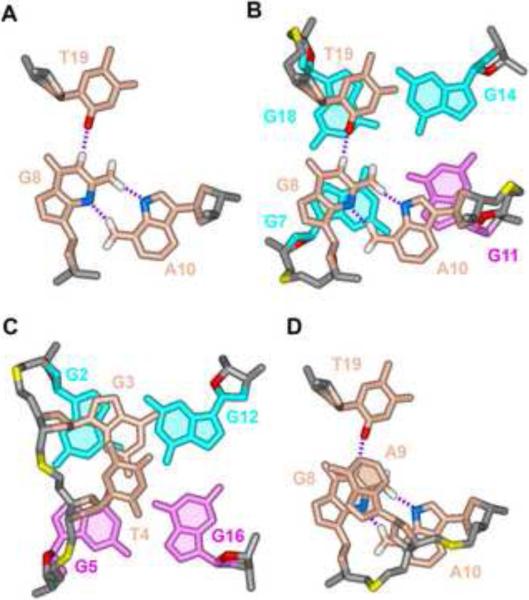
Both the top (G8-A9-A10) and bottom (G3-T4) lateral loops are structured within the chl1 intronic G-quadruplex fold. The top loop is stabilized by a T19•G8•A10 triple (Figure 5A), which stacks on the top G7•G11•G14•G18 tetrad (Figure 5B), while G3 of the bottom loop stacks on the G2•G5•G16•G12 tetrad (Figure 5C). Finally, A9 is unpaired and stacks on top of the T19•G8•A10 triple (Figure 5D).
Figure 5. Pairing and Stacking Alignments within the Loops Segments of the chl1 G-quadruplex in K+-containing Solution.
(A) Pairing alignment in the T19•G8•A10 triple. (B) Stacking between the T19•G8•A10 triple (biscuit) and the G7•G11•G14•G18 G-tetrad (cyan). (C) Stacking between the G3 base (biscuit) and the G2•G5•G16•G12 G-tetrad (cyan). (D) Stacking of A9 over the T19•G8•A10 triple.
Analysis of Modified Sequences
We have systematically probed specific structural elements within the proposed chl1 intronic G-quadruplex fold by base substitutions so as to gain insights into the range of tolerated mutations by the adopted scaffold and to derive a consensus sequence for bioinformatics analysis.
As expected from the G-quadruplex toplogy (Figure 3D), unpaired residue A9 positioned on top of the T19•G8•A10 triple (Figure 5D), can be substituted by G, T or C, with no impact on the imino proton spectrum (Figure S1, Supplementary Materials). Similarly, replacement of unpaired residue T4 by G, T or C, and unpaired residue T15 by C15, has no impact on the imino proton spectrum (Figure S2). It is noteworthy that the G-quadruplex folding topology was retained followed replacement of T4 by G4, despite formation of the resulting G8 segment, involving eight consecutive guanosines at the 5'-end of the sequence.
By contrast, replacement of unpaired residue G3 by I, T and A resulted in doubling of imino proton spectra (Figure S3), indicative of a role for the 2-amino protons of G3 in stabilizing the G3-T4 edge-wise loop of the G-quadruplex fold. Replacement of the 3'-terminal residue T19, which is involved T19•G8•A10 triple formation, by C, A and G, results in progressive deterioration of imino proton spectral quality, despite retaining the overall spectral pattern (Figure S4), consistent with T19 contributing to the stabilization of the G8-A9-A10 edge-wise loop of the G-quadruplex fold, as a consequence of hydrogen bonding to G8 of the sheared G•A non-canonical pair (Figure 5A). As a result of the mutation studies, the consensus sequence compatible with the fold reported here is G3NG4NAG3(A/C/T)G3(A/C/T) and might provide useful constraints in future bioinformatics searches for intronic G-quadruplex folds.
Discussion
The recent demonstration of guanosine-rich repeats with the potential for formation of polymorphic G-quadruplex scaffolds within the first intron of human genes (Eddy & Maizels, 2008, 2009), has highlighted the need to elucidate the diversity of G-quadruplex folding topologies adopted by human intronic guanosine-rich repeats.
To this end, we have solved the solution structure of an unique intramolecular G-quadruplex formed by the chl1 human intronic sequence in K+ solution. Inspection of the sequence
G1-G2-G3-T4-G5-G6-G7-G8-A9-A10-G11-G12-G13-G14-T15-G16-G17-G18-T19
shows that four guanosine-rich tracts are separated by a single (T4), double (A9-A10) and single (T15) connecting segments, suggesting the potential for formation of an all-parallel-stranded G-quadruplex involving three stacked G-tetrads connected by double-chain-reversal loops, as reported for the four guanosine-rich c-myc (Phan et al., 2004; Ambrus et al., 2005) and c-kit1 (Phan et al., 2007) sequences. However, this is not the case since we observe a new G-quadruplex folding topology containing two unanticipated conformational features, one of which has not been reported previously for unimolecular G-quadruplexes.
First Guanosine is Positioned within the Central G-tetrad
The first striking feature is that the first guanosine G1 is part of the central G-tetrad (Figure 3D), in contrast to the first guanosine having always been reported to be part of a terminal G-tetrad in all unimolecular G-quadruplexes reported to date (reviewed in Patel et al., 2007; Neidle, 2009) Thus G11 and the G1-G2 step that are part of the same column are not connected to each other, in contrast to the unbroken linkage for the three other G5-G6-G7, G12-G13-G14 and G16-G17-G18 columns of the chl1 intronic G-quadruplex (Figure 3D). It should be noted that an earlier report did identify a bimolecular G-quadruplex where the first guanosine was also part of a central G-tetrad for one of the strands (Crnugelj et al.,2003).
A V-shaped Loop Spanning Three G-tetrad Planes
Our group first identified the V-shaped loop topology for G-quadruplex folds (Zhang et al., 2001), where guanosines at adjacent corners of stacked G-tetrads are directly connected, resulting in a missing support column. The first example of a V-shaped loop within a G-quadruplex scaffold linked two adjacent G-tetrads as shown in Figure 3E (Zhang et al., 2001), with later examples identified for dimeric G-quadruplexes formed by d(G3T4G4) (Crnugelj et al., 2003) and d(GLGLT4GLGL), where L is a locked nucleic acid (Nielsen et al., 2009), contained V-shaped loops that spanned three G-tetrad planes as shown in Figure 3F. The V-shaped loop motif represented in Figure 3F is observed for the unimolecular chl1 intronic G-quadruplex (Figure 3D), where G11 belongs to the top G-tetrad of one column while G12 belongs to the bottom G-tetrad of an adjacent column, accompanied by an extension of the backbone at the G11-G12 step. In essence, a second striking feature is that the G1 base is partially intercalated between two guanosine bases G11 and G12 within the folding topology of the chl1 intronic G-quadruplex (Figure 4B).
Stacked T19•G8•A10 Triple Stabilizes Edge-wise G8-A9-A10 Loop
The T19•G8•A10 triple involves a sheared G8•A10 pair alignment involving pairing of the minor groove edge of G8 with the major groove edge of A10 (Figure 5A), as reflected in the NOEs within the sheared pair and with the stacked adjacent G7•G11•G14•G18 tetrad. The O4 carbonyl group of T19 is hydrogen-bonded to the imino proton of G8 in the T19•G8•A10 triple, readily explaining the upfield shifted 9.95 ppm imino proton chemical shift of G8, characteristic of a guanosine imino proton hydrogen-bonded to an oxygen acceptor (N-H••O). Replacement of T19 by A and G, but not C, results in deterioration in imino proton spectral quality (Figure S4). The O4 carbonyl is common to both T and C, but not to the purines, emphasizing its importance as an acceptor for the hydrogen bond from the imino proton of G8. In addition, the larger size of the purine ring at position 19 would be more difficult to accommodate as part of a triple on top of the adjacent G7•G11•G14•G18 tetrad.
Consistent with the structure, unpaired A9, which stacks on the T19•G8•A10 triple, can be replaced by G, T and C, without impacting on the imino proton NMR spectrum (Figure S1). Since A9 is well-defined in the structure, the conformation of the connecting sugar-phosphate is more important for the G8-A9-A10 edge-wise loop than the base at position 9.
Stacked G3 Stabilizes Edge-wise G3-T4 Loop
The two-residue bottom edge-wise loop is composed of residues G3 and T4. They form a configuration where T4 is stacked on top of the G3, which in turn is stacked on the G2•G5•G16•G12 G-tetrad (Figure 5C). We observe a single average resonance for the two amino protons of G3, in contrast to separate hydrogen-bonded and exposed amino protons for guanines involved in G-tetrad formation. The average amino protons of G3 exhibit cross-peaks to the imino protons of all four guanosines in the G2•G5•G16•G12 G-tetrad (peaks j, k, l, m in Figure 3B), consistent with the overlap geometry shown in Figure 5C, where the G3 amino protons are stacked over the center of the adjacent G-tetrad.
Replacement of G3 by I3, T3 or A3, results in doubling of the imino proton NMR spectra, including the upfield shifted imino proton of G8 at 9.95 ppm (Figure S3), indicative of an equilibrium between two G-quadruplex folds for the substituted analogs. Guanosine has an amino proton at the 2-position, which is missing in inosine and adenosine. It is conceivable that a K+ ion may be sandwiched between G3 and the G2•G5•G16•G12 G-tetrad, and that monovalent cation coordination requires the presence of an amino group at the 2-position of the purine ring, to stabilize a single G-quadruplex conformation. The proposed coordinated K+ cation could also slow down the exchange of the imino proton of G3, thereby accounting for the observation of a broadened resonance at 10.55 ppm (Figure 1B).
A Double-chain-reversal Loop
The parallel alignments of the G12-G13-G14 and G16-G17-G18 segments are bridged by a single residue (T15) double-chain-reversal loop in the structure of the chl1 intronic G-quadruplex (Figures 4A and 4B). Such double-chain-reversal loops were first reported a decade and a half ago by our group for the four-guanosine repeat Tetrahymena G-quadruplex (Wang & Patel, 1994), but came into greater prominence with their observation in the crystal structures of two- and four-guanosine repeat human telomere G-quadruplexes by the Neidle laboratory (Parkinson et al., 2002). Single residue double-chain-reversal loops spanning three G-tetrad planes, as observed for chl1 intronic G-quadruplex (Figure 3D), were first reported for the G-quadruplex formed by the four-guanosine repeat segment of the c-myc promoter (Phan et al., 2004; Ambrus et al., 2005).
Recognition properties of chl1 intronic G-quadruplex
The chl1 intronic G-quadruplex has a distinct pattern of grooves, which sets it apart from other known G-quadruplexes formed in the vicinity of transcription start sites, such as ckit (Phan et al., 2007) and c-myc (Phan et al., 2004; Ambrus et al., 2005) promoter G-quadruplexes. The start of the fold in the middle G-tetrad and zero-residue bridging of the G11-G12 step imposes restrictions on the accessibility of the groove formed by the G11-break-G1-G2 and G12-G13-G14 columns (Fig. 4A). The adjacent groove between columns G12-G13-G14 and G16-G17-G18, which is bridged by single residue T15, is accessible for hydrogen-bond recognition the guanosine edges of its G-tetrad and its outwardly-pointed T15 base. The two remaining grooves are completely accessible for hydrogen-bond recognition by small drugs or protein side chains. The two lateral loops positioned above and below the G-quadruplex also have potential for hydrogen bond recognition involving accessible base edges of the G8•A10•T21 base triple (Figure 5A), the single base A9 in the 3′-end loop (Figure 5D), and bases G3 and T4 in the 5′-end loop (Figure 5C). In contrast to c-kit and c-myc promoter G-quadruplexes, the chl1 intronic G-quadruplex has only one single-residue double-chain reversal loop and a unique pattern of guanosine bases constituting the quadruplex core. The 5′- and 3′-ends of the chl1 intronic G-quadruplex are positioned on adjacent stacked G-tetrads and positioned diagonally across the G-quadruplex (Figure 3D).
Comparison with the Active Site of Group I Introns
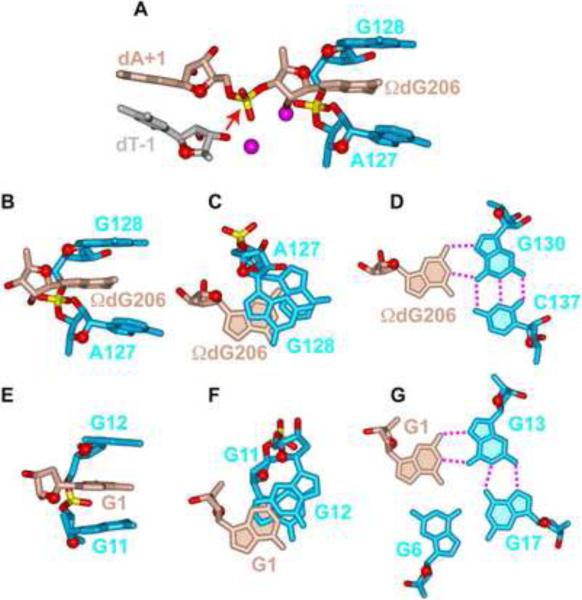
A novel feature of the solution structure of the chl1 intronic G-quadruplex is the partial intercalation of G1 between two other guanines, each of which is involved in G-tetrad formation (Figure 6E). We therefore searched data bases for related alignments amongst higher order RNA folds and found a similar arrangement within the G-binding motif in the catalytic core of group I intron ribozymes (Adams et al., 2004; Guo et al., 2004; Golden et al., 2004; reviewed in Stahley & Strobel, 2006). Indeed, in the 3.1 Å crystal structure of Azoarcus group I intron in complex with both 5'- and 3'-splice sites, corresponding to the splicing intermediate before the exon ligation step, the terminal nucleotide of the intron, ΩG206, partially intercalates between G128 and A127, as part of an anchored G-binding motif (Adams et al., 2004; Figure 6A). Partial intercalation of ΩG206 between purine bases (Figures 6B and 6C) and its pairing with the Hoogsteen edge of G130 (Figure 6D) in the Azoarcus group I intron (Adams et al., 2004) has striking parallels with partial intercalation of G1 between G11 and G12 (Figures 6E and 6F), with G1 paired with the Hoogsteen edge of G13 as part of the G1•G13•G17•G6 G-tetrad (Figure 6G) in the chl1 intronic G-quadruplex. One difference between these two alignments is that the sugar ring oxygens of ΩG206 in the intron (Figure 6B) and G1 in the G-quadruplex (Figure 6E) are oriented in opposite directions.
Figure 6. Comparison of a Guanosine Intercalated Between the G-G step in Azoarus Group I Intron and in the chl1 intronic G-Quadruplex.
(A) The G-binding motif in the crystal structure of the Azoarcus Group I intron (Adams et al., 2004). Note that partial intercalation of ΩdG206 between A127 and G128 assists in positioning the phosphate between ΩdG206 and dA+1 for in-line attack by the 2'-OH of dT-1 (designated by a red arrow). The scissible phosphate is flanked by Mg2+ cations shown as magenta balls. Phosphorus and oxygen atoms are colored in yellow and red, respectively. (B) Side and (C) look-down views of the intercalation of ΩdG206 between a G-G step and (D) hydrogen-bonding of Watson-Crick edge of ΩdG206 with Hoogsteen edge of G130 in the crystal structure of the Azoarcus Group I intron (Adams et al., 2004). (E) Side and (F) look-down views of the intercalation of G1 between the G11-G12 step and (G) hydrogen-bonding of Watson-Crick edge of G1 with Hoogsteen edge of G13 in the NMR-based solution structure of the chl1 intronic G-quadruplex reported in this study. (H) The postulated in line attack by a 3′ -OH on the scissible phosphate is shown by an arrow. Phosphorus and oxygen atoms are colored in yellow and red, respectively.
A Proposed Model for Phosphoryl Transfer Mediated by a G-Quadruplex Scaffold
A unique feature of the group I catalytic pocket is the in-line position of the activated 5'-exon 2'-OH and the conformationally constrained scissible phosphate at the intron-3'-exon junction, which is bracketed on either side by Mg2+ cations (Figure 6A; Stahley & Strobel, 2005). Such alignments, involving a constrained guanosine, define splice site selection during catalyzed phosphoryl transfer between guanosine and a substrate RNA strand (reviewed in Stahley & Strobel, 2006). We propose that the guanosine flanking the scissible phosphate can be equally well locked into position either through partial intercalation (Figures 6B and 6C) and base triple formation (Figure 6D) as observed for ΩG206 in the group I intron (Adams et al., 2004) or through partial intercalation (Figures 6E and 6F) and G-tetrad formation (Figures 6G) as observed for G1 in the chl1 intronic G-quadruplex.
We therefore propose a model of a higher order RNA structure where a constrained guanosine as part of a G-quadruplex could participate in the splicing reaction at a quadruplex-duplex junction as shown schematically in Figure S5. The RNA duplex is on the left and the RNA G-quadruplex is on the right, with the 5' and 3'-ends of the RNA labeled in Figure S5 A distinction between the experimentally determined group I intron splice site (Figure 6A) and the proposed model of the quadruplex-duplex junctional splice site (Figure S5B; expanded view of Figure S5A) is that the attacking nucleophile is a 3'-OH group in the former structure and a 5'-OH group in the latter model. The model in Figure S5A has been proposed in the spirit of stimulating further experiments at comprehensive molecular engineering of higher-order G-quadruplex folds and clearly needs experimental validation.
Our structure of the chl1 intronic G-quadruplex raises the issue as to whether a DNA segment can be accommodated 5' to the first guanosine that is part of the central G-tetrad of the chl1 intronic DNA G-quadruplex. Our modeling studies shown in Fig. S5A indicate that there is ample room to accommodate such a DNA segment without perturbing this unique G-quadruplex fold.
Summary and Future Prospects
The G-quadruplex fold adopted by the human chl1 intronic sequence in K+-containing solution, composed of three G-tetrads containing four syn guanines, exhibits conformational features not reported previously for unimolecular G-quadruplexes. The unprecedented combination of the first guanosine being part of the central G-tetrad, together with a V-shaped loop spanning three G-tetrad planes without an intervening nucleotide, results in intercalation of the G1 base between an extended G11-G12 step. Strikingly, this intercalation alignment is similar to what has been observed in the catalytic core of group I introns, an alignment that facilitates catalyzed phosphoryl transfer reactions. The novel topology of the chl1 intronic G-quadruplex makes this scaffold a unique platform for future structure-based ligand design (reviewed in Patel et al., 2007; de Cian et al., 2008; Ou et al., 2008; Monchaud & Teulade-Fichou, 2008; Neidle, 2009; Balasubramanian & Neidle, 2009).
Methods
Sample preparation
The unlabeled and the site-specific low-enrichment (2% uniformly-N15-labeled) oligonucleotides were synthesized and purified as described previously (Phan, 2000; Phan & Patel, 2002). The strand concentration of the sample was typically in the range 0.5 to 5.0 mM and was dissolved in 50 mM KCl, 5 mM K-phosphate buffer, pH 6.8.
NMR spectroscopy
Experiments were performed on 600 MHz Varian NMR spectrometers with data recorded at 25 °C. Guanosine base resonances were assigned unambiguously by using site-specific low-enrichment labeling and through-bond correlation at natural abundance (Phan, 2000; Phan & Patel, 2002). Assignment for some residues were verified and confirmed in independently synthesized samples with specific substitutions. Spectral assignments were also assisted and supported by COSY, TOCSY, 13C-HSQC and NOESY spectra. Interproton distances involving exchangeable protons were categorized as strong (2.8 to 5.2 Å), medium (3.15 to 5.85 Å) or weak (3.5 to 6.5 Å) based on cross-peak intensities recorded in a NOESY spectrum (50 and 300 ms mixing time) in H2O solution. Interproton distances involving non-exchangeable protons were measured from NOE build ups using NOESY experiments recorded at five mixing times (50, 100, 150, 200 and 250 ms) in 2H2O solution.
Structure Calculations
The structures of the chl1 intronic G-quadruplex were calculated using the X-PLOR program (Brünger, 1992) as described previously (Phan et al., 2007). The initial folds guided by NMR restraints listed in Table 1 were obtained using torsion dynamics. The structures were further refined by Cartesian dynamics and, finally, using relaxation matrix refinement.
The initial structure consisted of an extended DNA strand with randomized chain torsion angles of constituent nucleotides, whose angles and bonds were set up in accordance with most updated measurements (Gelbin et al., 1996; Clowney et al., 1996)
Torsion dynamics: In the heating stage, the regularized extended DNA chain was subjected to 60 ps of torsion-angle molecular dynamics at 40,000 K using a hybrid energy function composed of geometric and NOE terms. The van der Waals (vdW) component of the geometric term was set to 0.1, thus facilitating torsional bond rotations, while the NOE term included NOE-derived distances with the scaling factor of 150. The structures were then slowly cooled from 40,000 K to 1,000 K over period of 60 ps during which the vdW term was linearly increased from 0.1 to 1. At the third stage, the molecules were slowly cooled from 1000 K to 300 K for 6 ps of Cartesian molecular dynamics (Stein et al., 1997). The 35 best structures with no 0.5 A violations and minimal energies were selected for further refinement.
Distance restrained molecular dynamics: Cartesian molecular dynamics was initiated at 300 K and the temperature was gradually increased to 1000 K during 7 ps. The system was equilibrated for 0.5 ps, while the force constants for the distance restraints were kept at 1 Kcal mol−1 Å−2. Subsequently, the force constants were linearly scaled up to 150 during 17.5 ps. The system was then slowly cooled to 300 K in 14 ps and equilibrated at 300 K for 12 ps. The coordinates saved every 0.5 ps during the last 4.0 ps were averaged. The resulting average structure was subjected to minimization until the gradient of energy was less than 0.1 Kcal mol−1. The soft planarity restraints imposed on the G-tetrads with the weight 10 kcal mol−1 Å−2 before the heating process, were removed at the beginning of equilibration stage. The electrostatic term was excluded from the energy function to increase the weight of covalent geometry terms during minimization process. The dihedral and hydrogen-bonding restraints for G-tetrad formation were maintained throughout the computations. The 17 best structures were selected at this stage.
Relaxation matrix intensity refinement: To account for spin diffusion effects, all 17 distance refined structures were next subjected to the energy minimization with back-calculation of the NOESY spectra with X-PLOR (Nilges et al., 1991). The relaxation matrix was set up for the non-exchangeable protons, with the exchangeable imino and amino protons replaced by deuterons. NOE intensity volumes from 178 non-exchangeable cross-peaks for each of five mixing times (50, 100, 150, 200, 250) were used as restraints, with uniform upper and lower bounds of ±30%. Dynamics was started at 5 K, and the system was heated up to 300 K in 0.6 psec. During the subsequent relaxation the force constant for NOE intensities was gradually increased from 0 to 300 Kcal mol−1 Å−2 with simultaneous decrease of the distance force constant of non-exchangeable protons from 50 to 30 Kcal mol−1 Å−2. The force constant for exchangeable protons and hydrogen bonds was kept at 100 Kcal mol−1 Å−2. After equilibration at 300 K for 3.0 ps the resulting structure was subjected to minimization until the gradient of energy was less than 0.1 Kcal mol−1. The NMR R-factor (R1/6) improved from initial value of 6.0% to 2.0% with simultaneous improved structure convergency.
Proposed Model for Phosphoryl Transfer Mediated by a G-Quadruplex Scaffold
The chl1 intronic RNA G-quadruplex was modeled by rmsd fitting of individual RNA nucleotides onto the DNA G-quadruplex structure (presented here) using common base atoms as superposition targets. All torsion angles of DNA residues except the sugar pucker pseudorotation angle (P) were copied into RNA nucleotides. The junction between the G-quadruplex and duplex was modeled using the Azoarcus group I intron junction as a prototype, with inversion of duplex orientation in order to form the connection with the 5′-end G1 of the chl1 G-quadruplex. The initial model was built manually and subsequently refined by energy minimization using the Discover module of Insight II software.
Data Deposition
The coordinates for the chl1 intronic G-quadruplex have been depositied in the Protein Data Bank (accession code: 2KPR).
Supplementary Material
Acknowledgement
The research was supported by NIH Grant GM34504 to D.J.P. He is a member of the New York Structural Biology Center supported by NIH Grant GM66354.
Footnotes
Author Contributions: VK identified the potential of the chl1 intronic DNA sequence for G-quadruplex formation, as well as recorded and interpreted the NMR spectra and undertook the computations leading to structure determination under the guidance of DJP. VK noted the similarities with group I intronic sequences and formulated the proposed model for phosphoryl transfer at a quadruplex-duplex junction. DJP and VK jointly wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. A System for X-ray Crystallography and NMR. Yale University Press; New Haven: 1992. X-PLOR. [Google Scholar]
- Burge SE, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney L, Jain SC, Srinivasan AR, Westbrook J, Olson WK, Berman HM. Geometric parameters in nucleic acids: nitrogenous bases. J. Am. Chem. Soc. 1996;118:509–518. [Google Scholar]
- Crnugelj M, Sket P, Plavec J. Small change in a G-rich sequence, a dramatic change in topology: New dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- Davis GT. G-Quartets 40 years later: from 5'-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Edn. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- De Cian A, Lacroix L, Douarre C, Temeime-Smalli N, Trentesaux C, Riou J-F, Mergny J-L. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplexes in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy J, Maizels N. Selection for the G4 DNA motif at the 5' end of human genes. Mol Carcinog. 2009;48:319–325. doi: 10.1002/mc.20496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbin A, Schneider B, Clowney L, Hsieh S-H, Olson WK, Berman HM. Geometric parameters in nucleic acids: sugar and phosphate constituents. J. Am. Chem. Soc. 1996;118:519–528. [Google Scholar]
- Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2004;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: Base triple sandwich and a metal ion at the active site. Mol. Cell. 2004;16:351–32. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends in Cell Biol. 2009;19:412–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Nielsen JT, Arar K, Petersen M. Solution structure of a locked nucleic acid modified quadruplex: Introducing the V4 folding topology. Angew. Chem. Int. Edn. 2009;48:3009–3113. doi: 10.1002/anie.200806244. [DOI] [PubMed] [Google Scholar]
- Nilges M, Habazettl J, Brünger AT, Holak TA. Relaxation matrix refinement of the solution structure of squash trypsin inhibitor. J Mol Biol. 1991;21:499–510. doi: 10.1016/0022-2836(91)90189-d. [DOI] [PubMed] [Google Scholar]
- Ou T, Lu Y, Huang Z, Wong K, Gu L. G-quadruplexes: targets in anti-cancer drug design. Chem. Med. Biochem. 2008;3:690–713. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Patel DJ, Kozlowski SA, Nordheim A, Rich A. Right-handed and left-handed DNA: Studies of B-DNA and Z-DNA by using proton nuclear Overhauser effect and phosphorus NMR. Proc. Natl. Acad. Scis. USA. 1982;79:1413–1417. doi: 10.1073/pnas.79.5.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promotor and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- Phan AT, Patel DJ. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Patel DJ. DNA architecture from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promotor. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Mody YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Hurley LH. Structures, folding patterns and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of Group 1 intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- Stahley MR, Strobel SA. RNA splicing: group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis. Curr. Opin. Struct. Biol. 2006;16:319–326. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Stein EG, Rice LM, Brünger AT. Torsion angle molecular dynamics: a new, efficient tool for NMR structure calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- Wang Y, Patel DJ. Solution structure of the Tetrahymena d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM. FANCJ helicase defective in Fanconia Anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Suhasini AN, Brosh RM. Welcome to the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell. Mol. Life Sci. 2009;66:1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. V-shaped scaffold: A new architectural motif identified in an A•(G•G•G•G) pentad-containing dimeric DNA quadruplex involving stacked G(syn)•G(anti)•G(anti)•G(anti) tetrads. J. Mol. Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.