Abstract
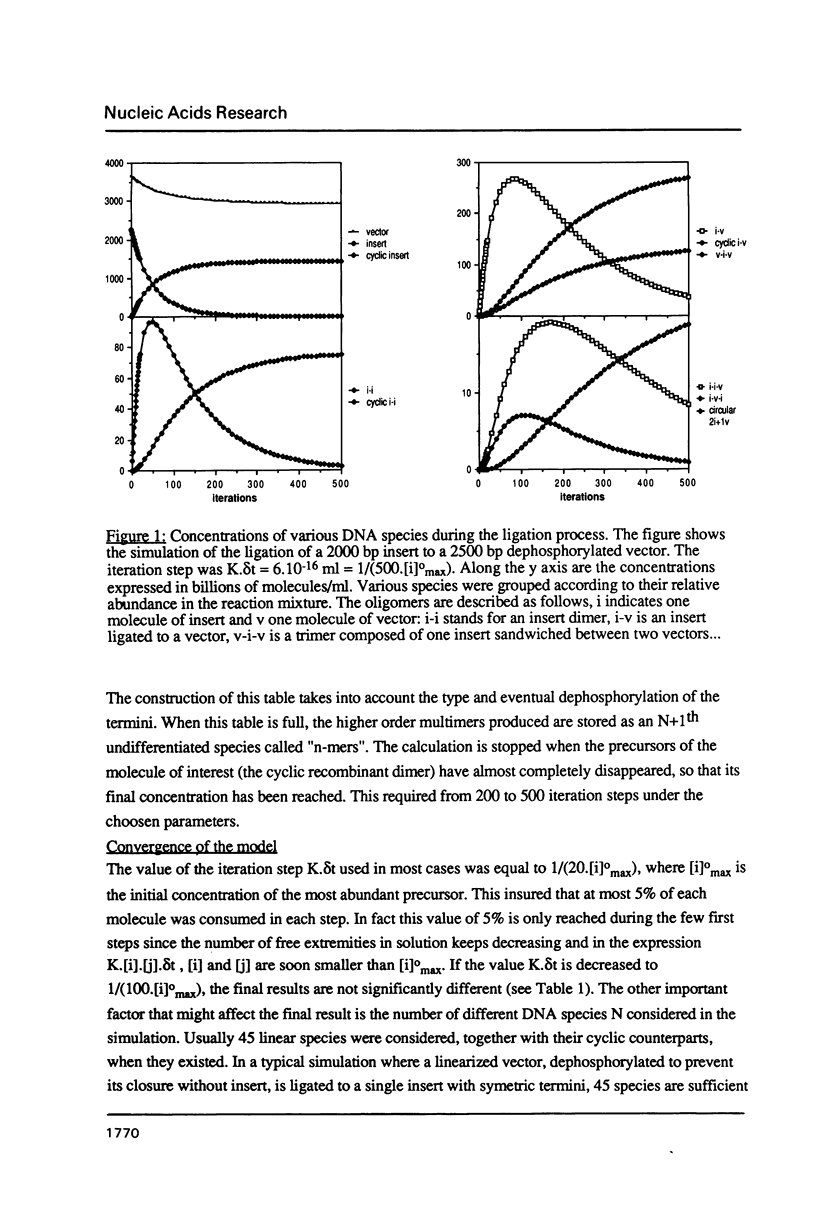
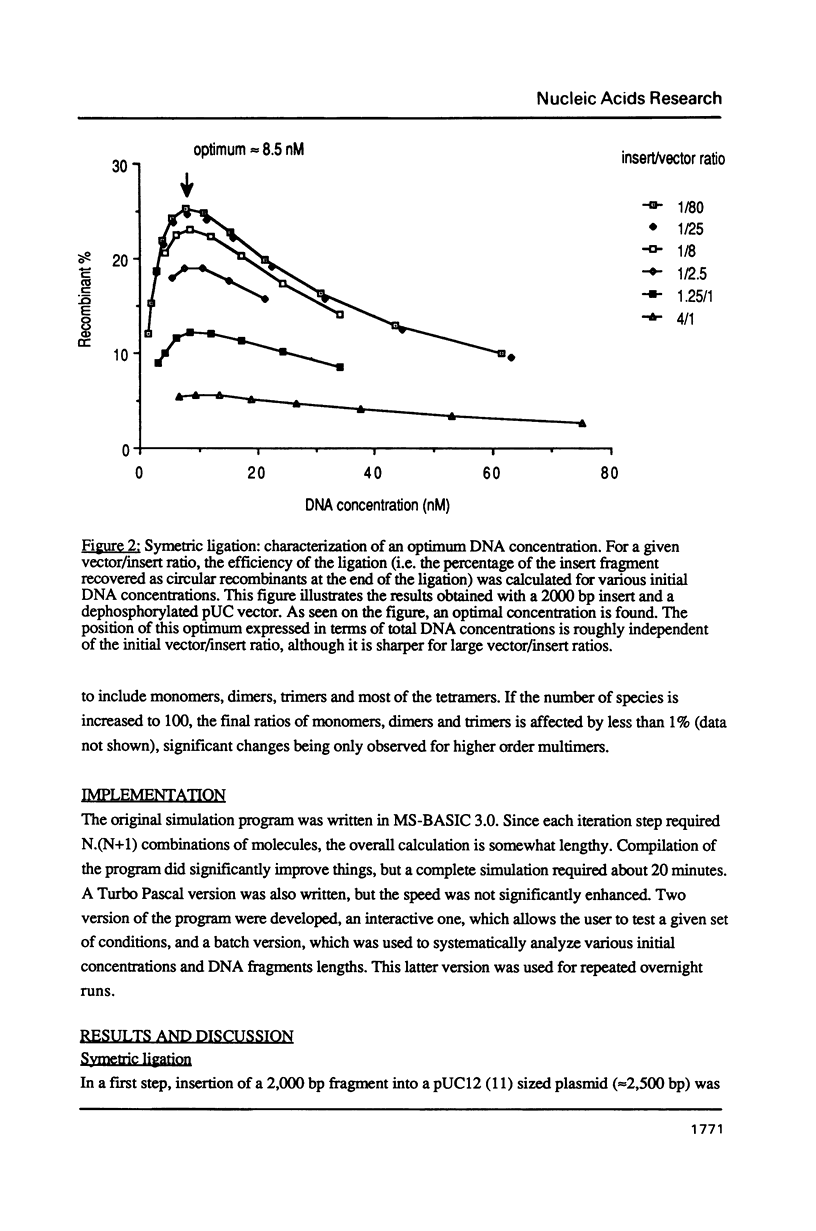
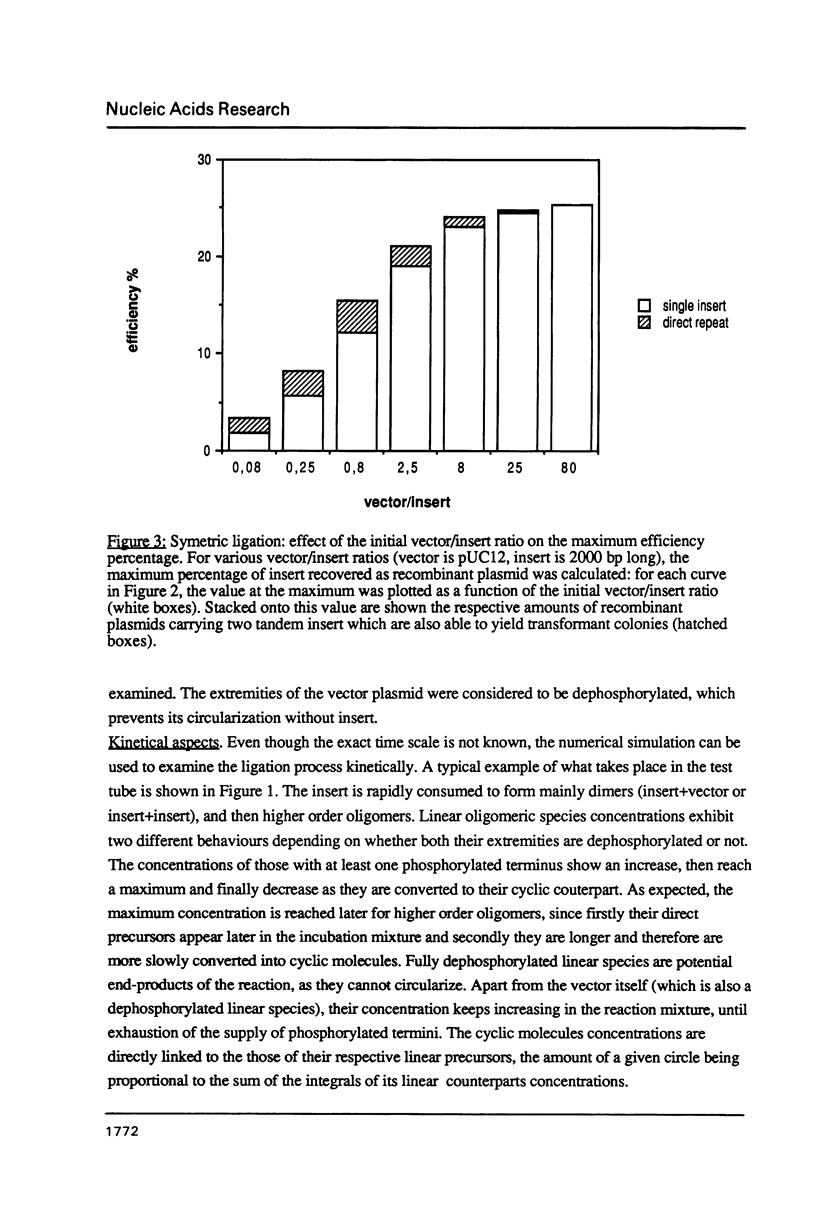
A computer program was used to simulate the dynamic process of a ligation of DNA fragments. More specifically, the influence of the initial DNA fragments lengths and concentrations on the relative abundance of the various end-products was systematically investigated. Depending on the nature of the DNA extremities (asymmetric or symmetric, dephosphorylated or not), sets of initial conditions could be found that optimized the yield of active recombinant molecules. These results can be directly used to increase the efficiency of the ligation step, in particular for the construction of cDNA or genomic libraries.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Nakazawa M., Ishizaki Y., Hiraoka N., Obayashi A. Regulation of inter- and intramolecular ligation with T4 DNA ligase in the presence of polyethylene glycol. Nucleic Acids Res. 1986 Oct 10;14(19):7617–7631. doi: 10.1093/nar/14.19.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Crothers D. M. Ring closure probabilities for DNA fragments by Monte Carlo simulation. J Mol Biol. 1986 May 5;189(1):61–72. doi: 10.1016/0022-2836(86)90381-5. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Crothers D. M. Topological distributions and the torsional rigidity of DNA. A Monte Carlo study of DNA circles. J Mol Biol. 1986 May 5;189(1):73–83. doi: 10.1016/0022-2836(86)90382-7. [DOI] [PubMed] [Google Scholar]
- Mayaux J. F., Fayat G., Panvert M., Springer M., Grunberg-Manago M., Blanquet S. Control of phenylalanyl-tRNA synthetase genetic expression. Site-directed mutagenesis of the pheS, T operon regulatory region in vitro. J Mol Biol. 1985 Jul 5;184(1):31–44. doi: 10.1016/0022-2836(85)90041-5. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Schmitter J. M., Mechulam Y., Fayat G., Anselme M. Rapid purification of DNA fragments by high-performance size-exclusion chromatography. J Chromatogr. 1986 Jun 13;378(2):462–466. doi: 10.1016/s0378-4347(00)80743-4. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weiss B., Jacquemin-Sablon A., Live T. R., Fareed G. C., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. VI. Further purification and properties of polynucleotide ligase from Escherichia coli infected with bacteriophage T4. J Biol Chem. 1968 Sep 10;243(17):4543–4555. [PubMed] [Google Scholar]


