Abstract
We have developed a computer graphics program system for the schematic representation of several protein secondary structure analysis algorithms. The programs calculate the probability of occurrence of alpha-helix, beta-sheet and beta-turns by the method of Chou and Fasman and assign unique predicted structure to each residue using a novel conflict resolution algorithm based on maximum likelihood. A detailed structure map containing secondary structure, hydrophobicity, sequence identity, sequence numbering and the location of putative N-linked glycosylation sites is then produced. In addition, helical wheel diagrams and hydrophobic moment calculations can be performed to further analyze the properties of selected regions of the sequence. As they require only structure specification as input, the graphics programs can easily be adapted for use with other secondary structure prediction schemes. The use of these programs to analyze protein structure-function relationships is described and evaluated.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Cohen G. H., Dietzschold B., Ponce de Leon M., Long D., Golub E., Varrichio A., Pereira L., Eisenberg R. J. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates the production of neutralizing antibody. J Virol. 1984 Jan;49(1):102–108. doi: 10.1128/jvi.49.1.102-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
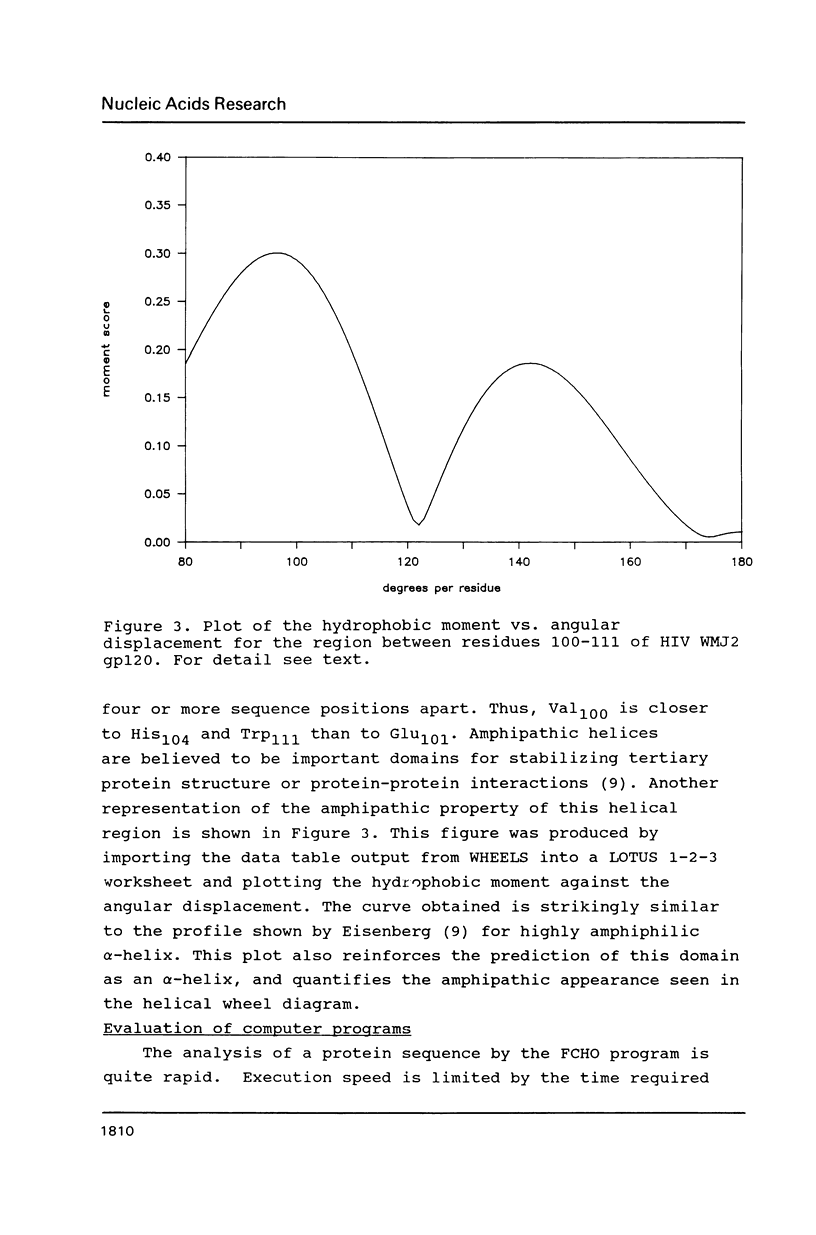
- Cornette J. L., Cease K. B., Margalit H., Spouge J. L., Berzofsky J. A., DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987 Jun 5;195(3):659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- Corrigan A. J., Huang P. C. A BASIC microcomputer program for plotting the secondary structure of proteins. Comput Programs Biomed. 1982 Dec;15(3):163–168. doi: 10.1016/0010-468x(82)90001-0. [DOI] [PubMed] [Google Scholar]
- Deleage G., Tinland B., Roux B. A computerized version of the Chou and Fasman method for predicting the secondary structure of proteins. Anal Biochem. 1987 Jun;163(2):292–297. doi: 10.1016/0003-2697(87)90226-0. [DOI] [PubMed] [Google Scholar]
- Dietzschold B., Eisenberg R. J., Ponce de Leon M., Golub E., Hudecz F., Varrichio A., Cohen G. H. Fine structure analysis of type-specific and type-common antigenic sites of herpes simplex virus glycoprotein D. J Virol. 1984 Nov;52(2):431–435. doi: 10.1128/jvi.52.2.431-435.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzschold B., Eisenberg R. J., Ponce de Leon M., Golub E., Hudecz F., Varrichio A., Cohen G. H. Fine structure analysis of type-specific and type-common antigenic sites of herpes simplex virus glycoprotein D. J Virol. 1984 Nov;52(2):431–435. doi: 10.1128/jvi.52.2.431-435.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci U S A. 1984 Jan;81(1):140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg R. J., Long D., Ponce de Leon M., Matthews J. T., Spear P. G., Gibson M. G., Lasky L. A., Berman P., Golub E., Cohen G. H. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985 Feb;53(2):634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg R. J., Long D., Ponce de Leon M., Matthews J. T., Spear P. G., Gibson M. G., Lasky L. A., Berman P., Golub E., Cohen G. H. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985 Feb;53(2):634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshita Y., Bishop D. H. The complete sequence of the M RNA of snowshoe hare bunyavirus reveals the presence of internal hydrophobic domains in the viral glycoprotein. Virology. 1984 Sep;137(2):227–240. doi: 10.1016/0042-6822(84)90215-0. [DOI] [PubMed] [Google Scholar]
- Kabat E. A., Wu T. T. The influence of nearest-neighbor amino acids on the conformation of the middle amino acid in proteins: comparison of predicted and experimental determination of -sheets in concanavalin A. Proc Natl Acad Sci U S A. 1973 May;70(5):1473–1477. doi: 10.1073/pnas.70.5.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lewis P. N., Go N., Go M., Kotelchuck D., Scheraga H. A. Helix probability profiles of denatured proteins and their correlation with native structures. Proc Natl Acad Sci U S A. 1970 Apr;65(4):810–815. doi: 10.1073/pnas.65.4.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim V. I. Structural principles of the globular organization of protein chains. A stereochemical theory of globular protein secondary structure. J Mol Biol. 1974 Oct 5;88(4):857–872. doi: 10.1016/0022-2836(74)90404-5. [DOI] [PubMed] [Google Scholar]
- Modrow S., Hahn B. H., Shaw G. M., Gallo R. C., Wong-Staal F., Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987 Feb;61(2):570–578. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson B., Pain R. H. Analysis of the code relating sequence to conformation in globular proteins. Development of a stereochemical alphabet on the basis of intra-residue information. Biochem J. 1974 Sep;141(3):869–882. doi: 10.1042/bj1410869. [DOI] [PMC free article] [PubMed] [Google Scholar]
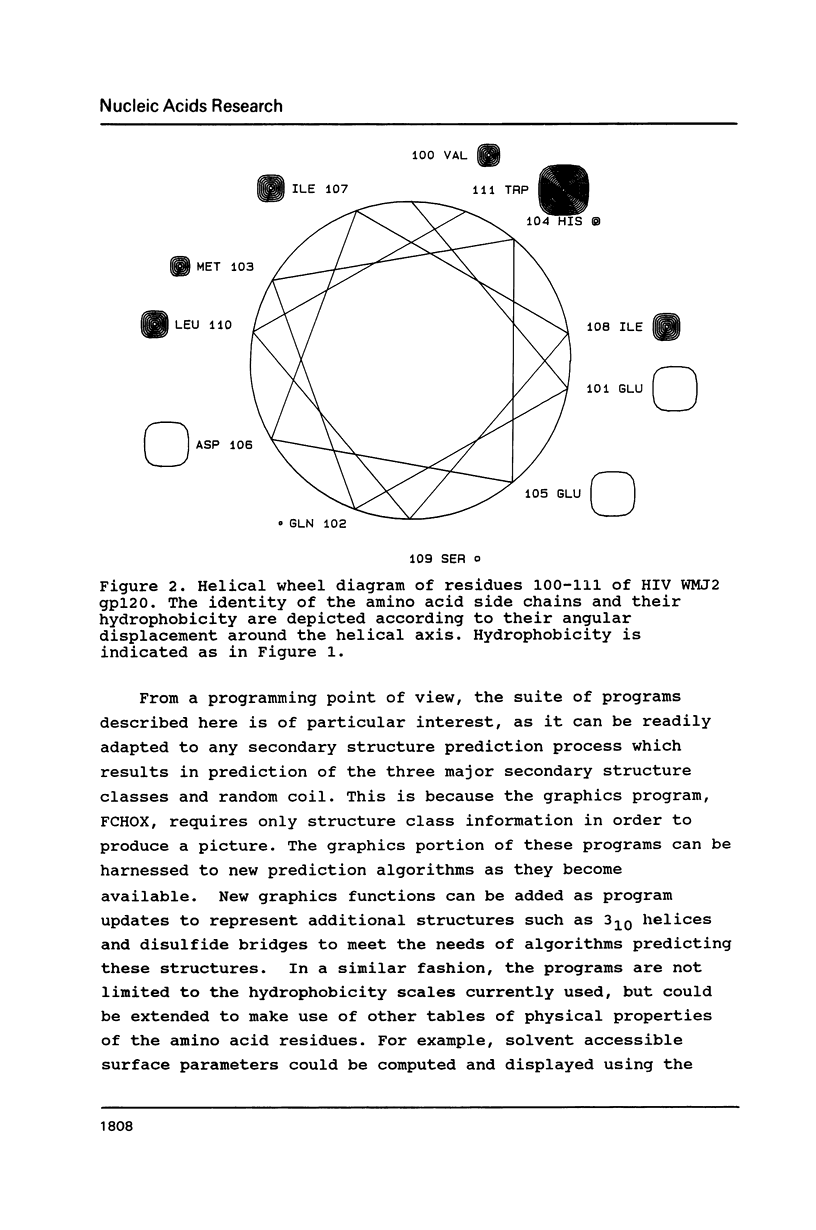
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Motz M., Kühbeck R., Seibl R., Jilg W., Bayliss G. J., Barrell B., Golub E., Zeng Y., Gu S. Y. Strategies for the economic preparation of Epstein-Barr virus proteins of diagnostic and protective value by genetic engineering: a new approach based on segments of virus-encoded gene products. IARC Sci Publ. 1984;(63):525–539. [PubMed] [Google Scholar]
- Wunner W. H., Dietzschold B., Smith C. L., Lafon M., Golub E. Antigenic variants of CVS rabies virus with altered glycosylation sites. Virology. 1985 Jan 15;140(1):1–12. doi: 10.1016/0042-6822(85)90440-4. [DOI] [PubMed] [Google Scholar]
- Yoon K., Davidson J. M., Boyd C., May M., LuValle P., Ornstein-Goldstein N., Smith J., Indik Z., Ross A., Golub E. Analysis of the 3' region of the sheep elastin gene. Arch Biochem Biophys. 1985 Sep;241(2):684–691. doi: 10.1016/0003-9861(85)90595-8. [DOI] [PubMed] [Google Scholar]


