Abstract
DNA Strider is a new integrated DNA and Protein sequence analysis program written with the C language for the Macintosh Plus, SE and II computers. It has been designed as an easy to learn and use program as well as a fast and efficient tool for the day-to-day sequence analysis work. The program consists of a multi-window sequence editor and of various DNA and Protein analysis functions. The editor may use 4 different types of sequences (DNA, degenerate DNA, RNA and one-letter coded protein) and can handle simultaneously 6 sequences of any type up to 32.5 kB each. Negative numbering of the bases is allowed for DNA sequences. All classical restriction and translation analysis functions are present and can be performed in any order on any open sequence or part of a sequence. The main feature of the program is that the same analysis function can be repeated several times on different sequences, thus generating multiple windows on the screen. Many graphic capabilities have been incorporated such as graphic restriction map, hydrophobicity profile and the CAI plot- codon adaptation index according to Sharp and Li. The restriction sites search uses a newly designed fast hexamer look-ahead algorithm. Typical runtime for the search of all sites with a library of 130 restriction endonucleases is 1 second per 10,000 bases. The circular graphic restriction map of the pBR322 plasmid can be therefore computed from its sequence and displayed on the Macintosh Plus screen within 2 seconds and its multiline restriction map obtained in a scrolling window within 5 seconds.
Full text
PDF

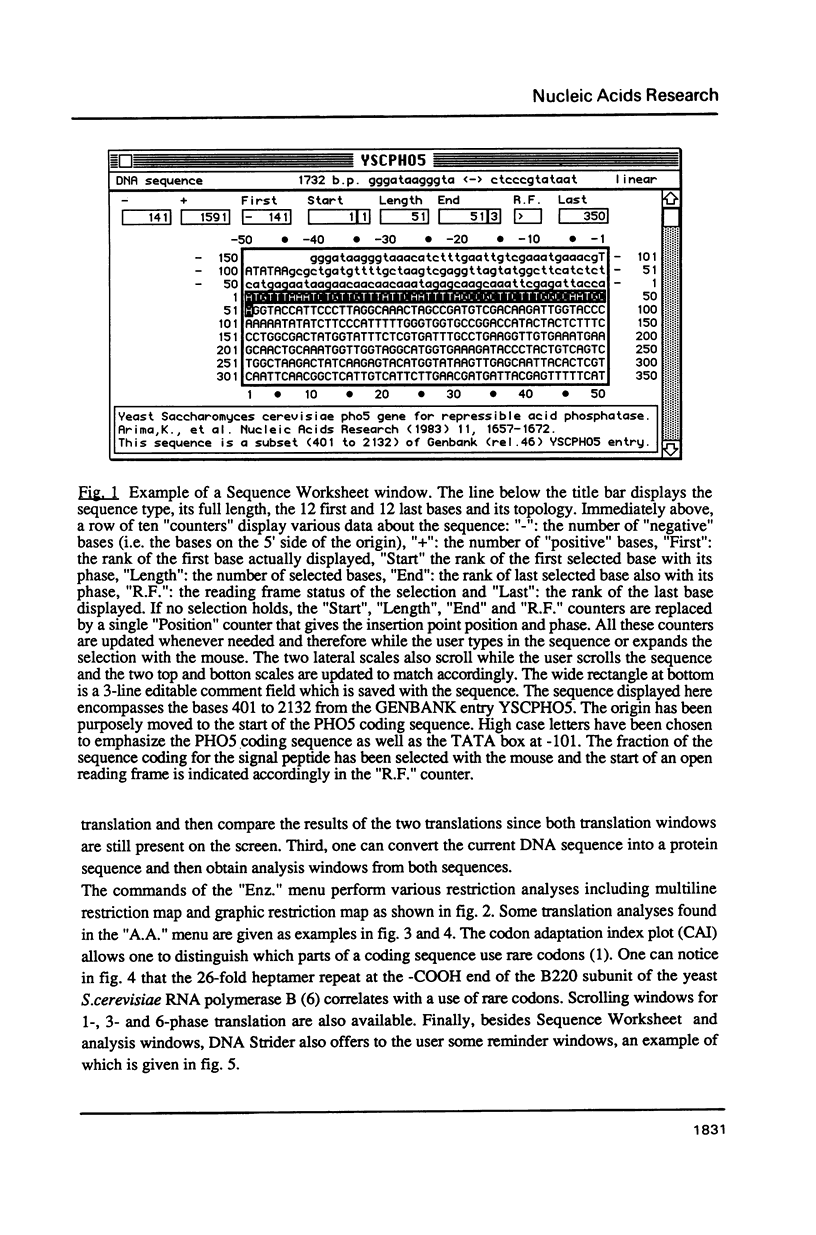
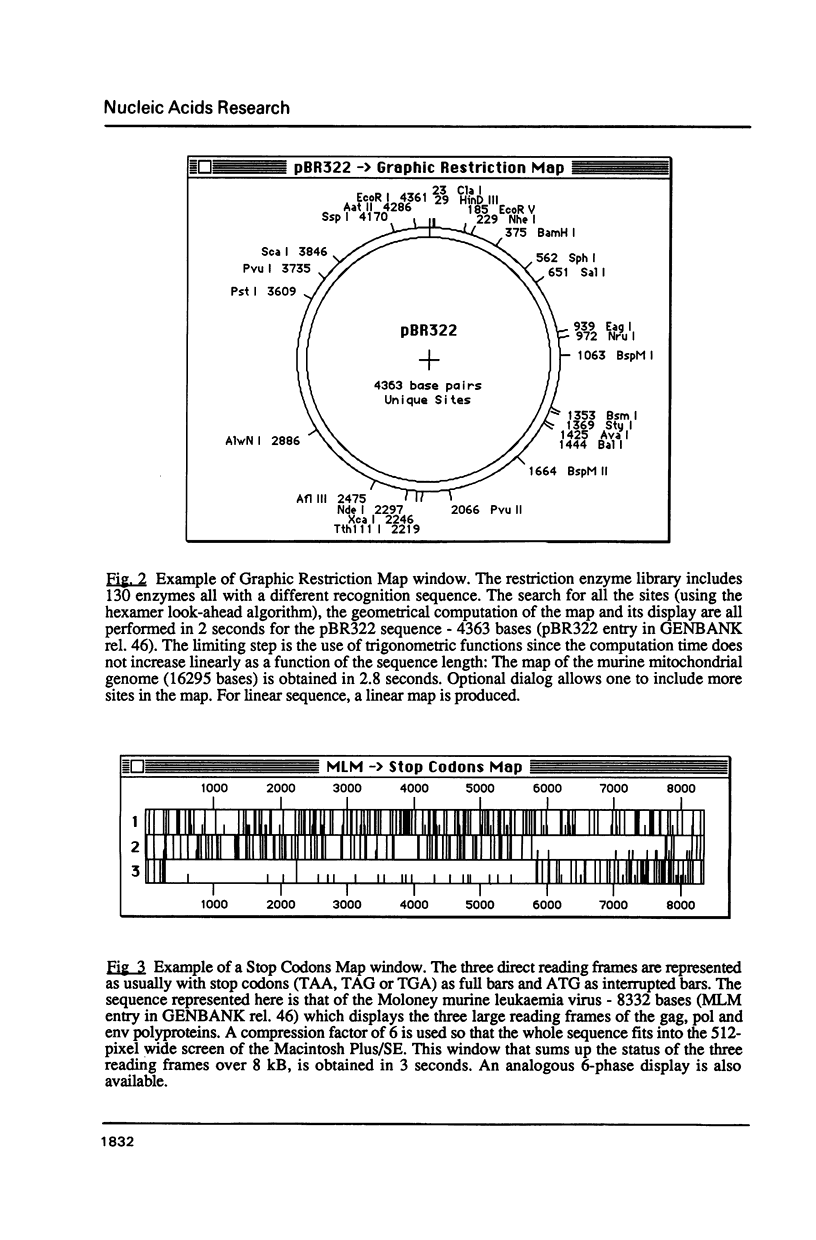
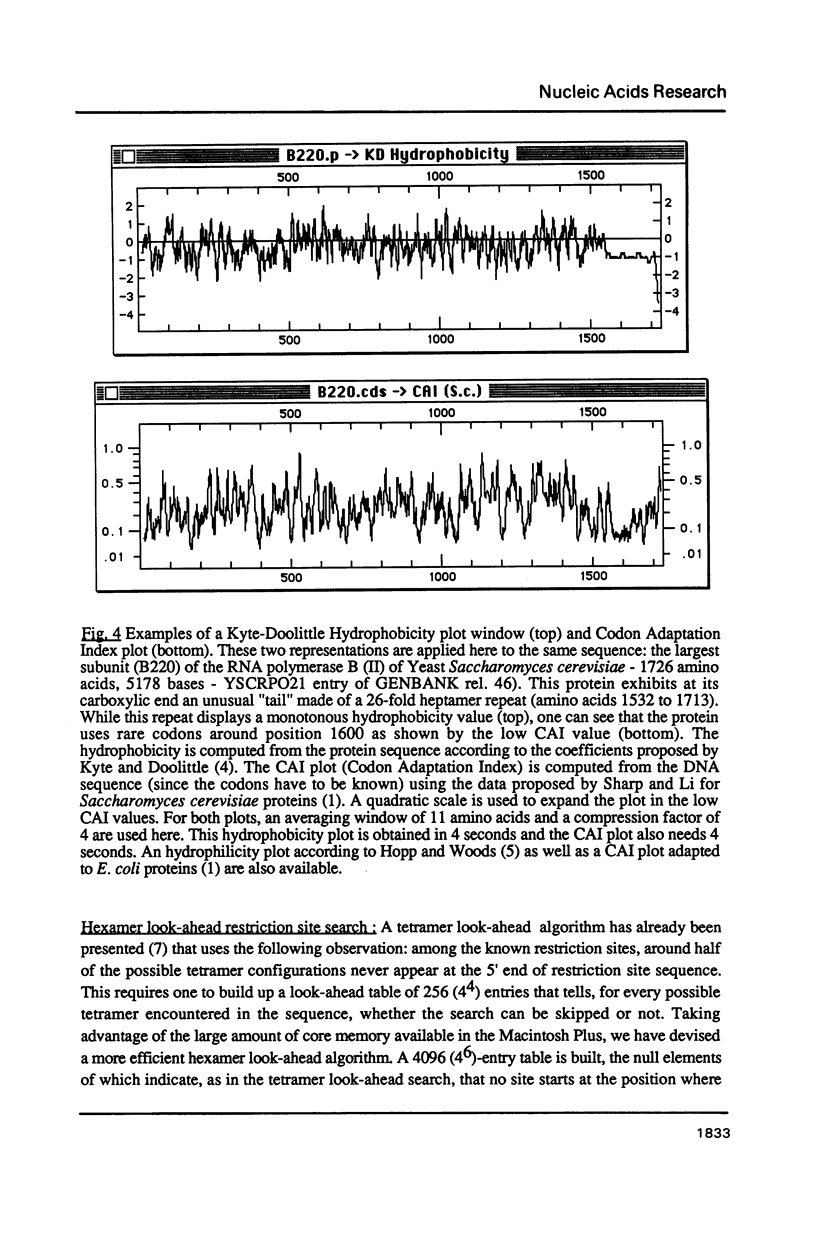



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Arima K., Oshima T., Kubota I., Nakamura N., Mizunaga T., Toh-e A. The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res. 1983 Mar 25;11(6):1657–1672. doi: 10.1093/nar/11.6.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985 May 10;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Marck C. Fast analysis of DNA and protein sequence on Apple IIe: restriction sites search, alignment of short sequence and dot matrix analysis. Nucleic Acids Res. 1986 Jan 10;14(1):583–590. doi: 10.1093/nar/14.1.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]


